Abstract
Ralstonia solanacearum is one of the most lethal phytopathogens in the world. Due to its broad host range, it can cause wilting disease in many plant species of economic interest. In this work, we identified the O-oligosaccharyltransferase (O-OTase) responsible for protein O-glycosylation in R. solanacearum. An analysis of the glycoproteome revealed that 20 proteins, including type IV pilins are substrates of this general glycosylation system. Although multiple glycan forms were identified, the majority of the glycopeptides were modified with a pentasaccharide composed of HexNAc-(Pen)-dHex3, similar to the O antigen subunit present in the lipopolysaccharide of multiple R. solanacearum strains. Disruption of the O-OTase led to the total loss of protein glycosylation, together with a defect in biofilm formation and reduced pathogenicity towards tomato plants. Comparative proteomic analysis revealed that the loss of glycosylation is not associated with widespread proteome changes. Only the levels of a single glycoprotein, the type IV pilin, were diminished in the absence of glycosylation. In parallel, disruption of glycosylation triggered an increase in the levels of a surface lectin homologous to Pseudomonas PA-IIL. These results reveal the important role of glycosylation in the pathogenesis of R. solanacearum.
Keywords: biofilm, protein O-glycosylation, Type IV pili
Introduction
Ralstonia solanacearum is a soil-borne β-proteobacterium known to cause lethal wilts in >200 plant species in all continents leading to enormous economic losses (Genin and Denny 2012; Peeters et al. 2013). In soil, the pathogen relies on flagellar-driven swimming to access the plant vascular system via the roots (Tans-Kersten et al. 2001; Yao and Allen 2006). Once inside the host, R. solanacearum rapidly colonize the xylem tissues and produce large amounts of extracellular polysaccharides (Orgambide et al. 1991; Saile et al. 1997). Accumulation of the released polysaccharides is required for the establishment of colonization as well as vascular occlusion of the infected plant (Husain and Kelman 1958; Denny and Baek 1991; Kao et al. 1992; Araud-Razou et al. 1998). Additionally, R. solanacearum uses its type III secretion system (T3SS) to hijack host cellular pathways in order to avoid detection by the plant immune system (Erhardt et al. 2010; Coll and Valls 2013; Peeters et al. 2013). Ralstonia solanacearum is known to secrete a large number of T3SS effectors, many of which were acquired by horizontal gene transfer (Poueymiro and Genin 2009). Other virulence factors in R. solanacearum include a type II secretion system, extracellular cellulases and pectinases, and type IV pili (Peeters et al. 2013). Type IV pili are involved in many biological processes including adhesion, twitching motility, biofilm formation and horizontal gene transfer (Strom and Lory 1993; Fussenegger et al. 1997; Merz et al. 2000). These filamentous appendages are formed by the polymerization of pilin monomers that can reach a few micrometers in length (Strom and Lory 1993; Fernandez and Berenguer 2000). Type IV pili were found to be dispensable for the virulence of some phytopathogens like Xanthomonas campestris (Ojanen-Reuhs et al. 1997). However, the type IV pilus is essential for pathogenesis in R. solanacearum. Strains lacking the type IV pili display impaired twitching motility and biofilm formation, resulting in virulence attenuation in tomato plant models (Liu et al. 2001; Kang et al. 2002).
Pilin, the monomer units of the type IV pili, are often glycosylated within Gram negative pathogens, such as Neisseria meningitidis, N. gonorrhoeae, Pseudomonas aeruginosa, Francisella tularensis, Acinetobacter nosocomialis, A. baylyi and Burkholderia cenocepacia (Castric 1995; Marceau et al. 1998; Egge-Jacobsen et al. 2011; Jennings et al. 2011; Lithgow et al. 2014; Harding et al. 2015). The identification and characterization of protein glycosylation in these pathogens have increasingly demonstrated that glycosylation, once thought to be a rare event, is widespread in bacteria. One of the most common mechanisms for bacterial O-linked protein glycosylation is mediated by a newly discovered family of enzymes named O-OTase (Iwashkiw et al. 2013). O-OTases catalyze the transfer of glycans previously assembled onto the undecaprenyl-phophate lipid carrier in the inner membrane to acceptor protein/s in the periplasm space. This class of OTase-dependent glycosylation systems have been described in many bacteria including Neisseria spp., Bacteroides fragilis, F. tularensis, Acinetobacter baumannii and B. cenocepacia (Marceau et al. 1998; Fletcher et al. 2009; Balonova et al. 2012; Iwashkiw et al. 2012; Lithgow et al. 2014). Moreover, functional OTases were found in Vibrio cholerae and B. thailandensis (Gebhart et al. 2012). Although protein O-glycosylation is required for the virulence of many bacteria, its exact role remains unclear (Iwashkiw et al. 2013).
O-OTases and the closely related WaaL enzymes, which catalyze the attachment of the O antigen to the lipid A core during the last step of lipopolysaccharide (LPS) synthesis, share the Wzy_C domain. For this reason, genome annotation projects have typically misidentified O-OTases as WaaL enzymes. Recently, bioinformatics and biochemical approaches have been employed to differentiate between these two enzymatic families (Gebhart et al. 2012; Schulz et al. 2013). In silico analysis of R. solanacearum GMI1000 genome revealed the presence of a protein carrying the Wzy_C domain (Power et al. 2006). The putative O-OTase (Rsc0559) is located downstream of PilA, the type IV pilin subunit protein. This suggested the presence of a functional O-glycosylation system in R. solanacearum, with pilin as one of the glycoproteins. In this work, we demonstrate that O-glycosylation in R. solanacearum actually extends to 20 proteins. Our assays show that O-glycosylation is important for biofilm formation and is required for virulence of R. solanacearum towards tomato plants. Furthermore, we analyzed the changes in the proteome of R. solanacearum in response to disruption of glycosylation.
Results
The Ralstonia O-OTase (Rsc0559) is functional in Escherichia coli
Power et al. (2006) and Schulz et al. (2013) suggested the presence of an O-OTase (Rsc0559) in the genome of R. solanacearum GMI1000 (Power et al. 2006; Schulz et al. 2013). Although some O-OTases are specific to pilin, others are classified as general O-OTases with a broad specificity towards their protein targets (Iwashkiw et al. 2013). Our phylogenetic analysis suggested that Rsc0559 is a general O-OTase similar to Neisseria PglL (Supplementary data, Figure S1). To test the activity of the putative O-OTase in R. solanacearum, we first employed an in vivo enzymatic assay (Gebhart et al. 2012). Rsc0559 was recombinantly expressed in E. coli CLM24, a W3110 strain that lacks the WaaL O antigen ligase (Feldman et al. 2005). Deletion of the waaL gene eliminates the competition between the O antigen ligase and the O-OTase for the lipid-linked glycans. Concurrently with Rsc0559 expression, both an acceptor protein and the genes coding for the biosynthesis of undecaprenyl-linked 2,4-di-N-acetylbacillosamine (diNAcBac) were co-expressed in trans. This sugar is a good substrate for O-OTases and a specific antibody for diNAcBac is available. We employed a C-terminal hexahistidine-tagged version of the disulfide oxidoreductase protein DsbA (Ng_1706) from N. meningitidis as the acceptor protein in this assay (Vik et al. 2009) as this protein has been demonstrated to be a compatible glycosylation substrate for general O-OTases (Lithgow et al. 2014; Scott et al. 2014; Harding et al. 2015). Cell lysates of E. coli CLM24 strain expressing Rsc0559, Neisseria PglL, or containing an empty vector, were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and analyzed by western blot to detect glycosylated DsbA. We used a monoclonal anti-hexa histidine antibody (green channel) to detect DsbA expression while diNAcBac was visualized using a polyclonal antibody (red channel). Overlapping the two signals will yield a yellow color that is indicative of DsbA glycosylation with bacillosamine. DsbA was glycosylated only when Rsc0559 or PglL from N. meningitidis (PglLNm) were expressed (Figure 1). These results suggest that Rsc0559 is a functional O-OTase. We therefore named Rsc0559 as PglLRs.
Fig. 1.
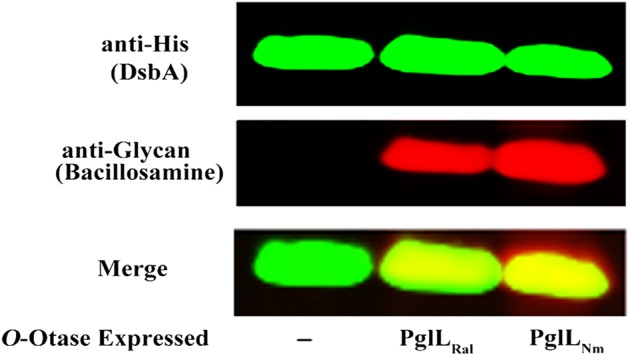
Glycosylation of Neisseria protein (DsbA) by Ralstonia O-OTase (PglLRs) in E. coli. Whole-cell lysates of E. coli CLM24 cells expressing DsbA as an acceptor protein, together with bacillosamine and different O-OTases, were separated by SDS–PAGE and analyzed by western blotting. His-tagged DsbA was detected using the monoclonal anti-His antibody (green). The bacillosamine sugar was detected using specific antibody (red). The overlapping signals are shown in yellow (merge). Both O-OTases from Ralstonia (PglLRs) and Neisseria (PglLNm) were able to glycosylate DsbA (Lanes 2 and 3, respectively).
PglLRs is essential for O-glycosylation in R. solanacearum
To investigate the O-OTase activity of PglLRs, we constructed an unmarked deletion mutant of the gene. In addition, we complemented the mutation by expressing PglLRs in cis in the mutant strain. PglLRs, like all the O-OTases, shares the Wzy_C domain with WaaL ligases. To analyze if PglLRs plays a role as a WaaL ligase in LPS synthesis, we analyzed the LPS from the wild-type and mutant strains using SDS–PAGE followed by specific staining. LPS appeared identical in the two strains, excluding a role of PglLRs in O antigen biosynthesis (Supplementary data, Figure S2).
We sequenced the genomes of both, the ΔpglLRs and the wild-type R. solanacearum GMI1000 strains using PacBio sequencing, to search for any secondary mutations or genomic rearrangements that might have occurred as a result of the mutagenesis process. The alignment of the two genomes revealed that the absence of pglLRs in the oligosaccharyltransferase deficient strain is the only difference with respect to the wild-type strain (data not shown).
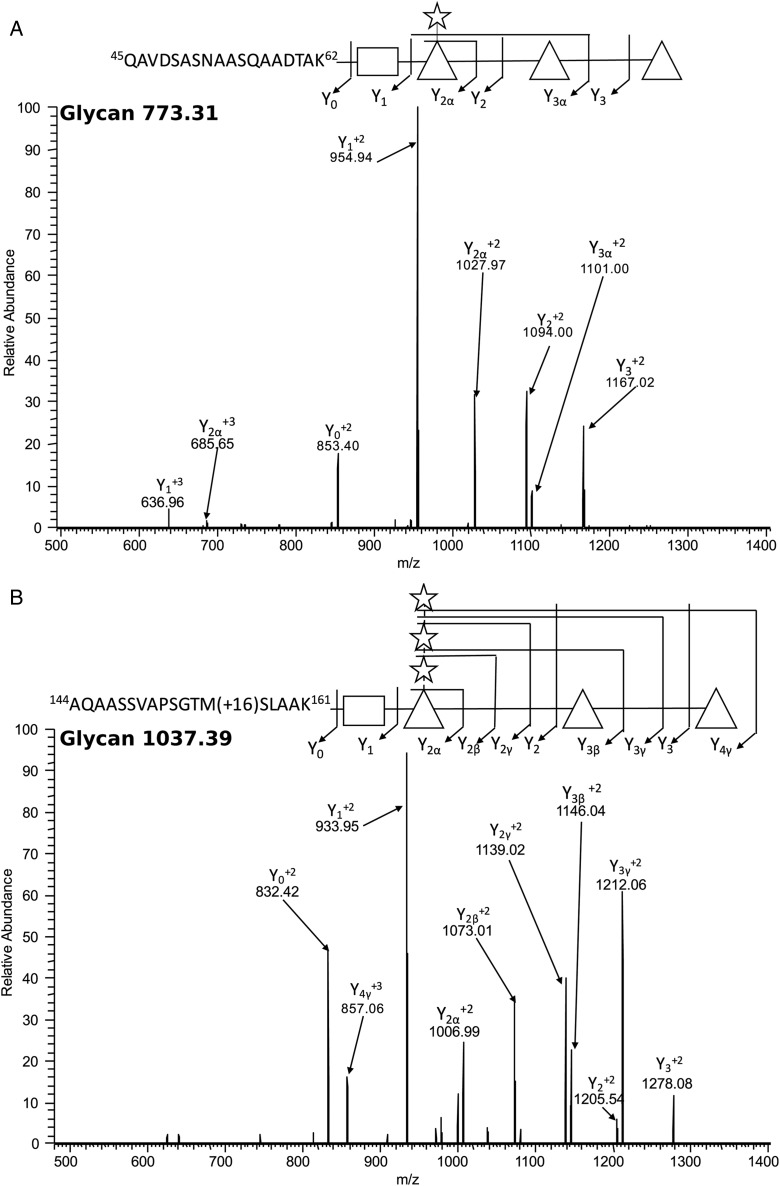
To identify the glycoproteins in R. solanacearum and their decorating glycan structures, we employed glycopeptide enrichment coupled to MS/MS. We used ZIC-HILIC enrichment, which exploits the hydrophilic properties of glycans to enable their enrichment from complex lysates (Iwashkiw et al. 2012; Nothaft et al. 2012; Lithgowet al. 2014), and multiple MS/MS fragmentation approaches to provide glycan and peptide information (Scott et al. 2011; Nothaft et al. 2012). A total of 53 unique glycopeptides were identified corresponding to 20 glycoproteins (Table I), supporting the presence of a functional general protein glycosylation system in wild-type R. solanacearum GMI1000. All of the detected glycopeptides were always found to be glycosylated in the wild-type strain during our analysis. Similar to previously identified O-OTase modified substrates (Anonsen et al. 2012; Lithgow et al. 2014; Scott et al. 2014), O-glycosylation appeared to occur on serine residues within peptides located in disordered regions of proteins (Table I, Figure 2, Supplementary data, Figure S3). Interestingly, within the detected glycoproteome a diverse array of glycopeptides were identified from the type IV pilin protein (PilA) confirming that this protein is subjected to glycosylation at least two sites within the peptides: 72LSVGSSVFTPTK83 and 144AQAASSVAPSGTMSLAAK161.
Table I.
Summary of the glycoproteins detected in R. solanacearum GMI1000 showing the detected glycans masses and the glycosylation sites
| Uniprot number | Protein annotation | Gene locus or name | Mass of attached glycan/s | Glycosylated sites/peptides |
|---|---|---|---|---|
| HBOH_RALSO | d-(−)-3-hydroxybutyr-ate oligomer hydrolase | RSc1334 | 773.32 | 63HDGANDDLLTAGLGAAGLASASAPSVATPTAPTAAELR100 |
| Q8XQM5_RALSO | Putative membrane fusion protein | RSp1197 | 773.31 | S418 |
| 815.32 | S418 | |||
| 815.31 | S442 | |||
| Q8XR30_RALSO | Probable lipoprotein transmembrane | RSp1038 | 773.31 | S120 |
| Q8XRR4_RALSO | Probable transmembrane protein | RSp0767 | 773.30 | S160 |
| Q8XRR8_RALSO | Probable transmembrane protein | RSp0763 | 773.34 | 1264LPTSIADATASQNTATPPAPAGSRPASAAAATTQATAR1301 |
| Q8XS78_RALSO | Probable serine protease protein | RSp0603 | 773.34 | 339FADQPIDPNGTGTPGRPLNFDPSGASQVYALPVR372 |
| Q8XSI7_RALSO | Probable m20-related peptidase | RSp0487 | 773.34 | 378LLPGDSASSVIAHVEQAVR396 |
| 905.36 | ||||
| Q8XV57_RALSO | Probable fimbrial type-4 assembly membrane transmembrane protein PilN | RSc2974 | 773.30 | 192AEPATPAKPGSAASAVAGK210 |
| Q8XVC9_RALSO | Probable lipoprotein | RSc2902 | 773.31 |
45QAVDSASNAASQAADTAK62 63SGVAEVASGAQAAVNAASGAMADAK87 |
| 815.32 | ||||
| 905.34 | ||||
| 947.36 | ||||
| Q8XWI3_RALSO | Hypothetical signal peptide protein | RSc2491 | 641.26 | S61 |
| 815.31 | ||||
| 773.30 | ||||
| Q8XX43_RALSO | Probable polysaccharide transport system component | ragB | 773.31 | S346 |
| Q8XXY5_RALSO | Probable transmembrane protein | RSc1978 | 815.33 | 129ADGAAPQQAQALDQGEEVVSSAGGTSAASTPAAAKPSPK167 |
| Q8XZ41_RALSO | Peptidyl-prolyl cis-trans isomerase | RSc1565 | 773.31 | 23ASAVSAAPAESLPSGVTIQHVAK45 |
| Q8Y030_RALSO | Probable transmembrane protein | RSc1214 | 773.31 | 243SDAGAMAAPATAVDATRPAVVSVDASSVPAVPAAEVASK281 |
| Q8Y078_RALSO | Probable tpr domain signal peptide protein | RSc1166 | 203.08 | S210 |
| Q8Y1X9_RALSO | Type 4 fimbrial pilin signal peptide protein | PilA | 1037.40 |
59ALVSENAANAQSDLSVGSSVFTPTK83 144AQAASSVAPSGTMSLAAK161 |
| 203.09 | ||||
| 1169.43 | ||||
| 905.35 | ||||
| Q8Y2P9_RALSO | Probable cell division FtsN transmembrane protein | ftsN | 947.37 | 50NGAQPKPSEPGSVVNPLPAPVQPAPQASAPPADPNAPLWSR90 |
| 773.32 | S78 | |||
| 905.35 | ||||
| 815.33 | ||||
| Q8Y3G9_RALSO | Probable acriflavin resistance lipoprotein | acrA | 773.31 | S393 |
| 815.32 | ||||
| Q8Y2I4_RALSO | Probable peptidase transmembrane protein | RSc0352 | 773.31 | 523AAPASEPAAPSGPASGVIPAPEPTGAR549 |
| 815.32 | ||||
| Q8XVI0_RALSO | Cell division protein FtsL | ftsL | 773.32 | 81TQYLQGFADLPAAASAAASAPAASGVQP108 |
| 905.35 |
Fig. 2.
Major O-glycan structures identified in R. solanacearum glycoproteins. O-Glycan structures were identified using ITMS-CID fragmentation of Ralstonia glycopeptides. (A) A pentamer glycan of HexNAc-(Pen)-dHex3 attached to 45QAVDSASNAASQAADTAK62 of Q8XVC9_RALSO. (B) A heptamer glycan of HexNAc-(Pen3)-dHex3 attached to 144AQAASSVAPSGTM(+16)SLAAK161 of Q8Y1X9_RALSO. Sugar monomers were represented graphically as follows: rectangles for N-acetyl hexosamine (HexNAc) units, triangles for deoxyhexose (dHex) units and stars for pentose (Pen) units.
The majority of the glycopeptides were found modified with a pentasaccharide composed of HexNAc-(Pen)-dHex3, which is similar to the O antigen subunit characterized in the LPS structures of multiple R. solanacearum strains (Figure 2A) (Varbanets et al. 2003). However, the whole glycoproteome exhibited eight unique glycoforms (Figure 2, Supplementary data, Figure S3) ranging in size from a single HexNAc (N-acetylhexosamine) (Supplementary data, Figure S3A) to an octasaccharide composed of HexNAc-(Pen)-dHex3-Pen3 (dHex; deoxyhexose, pen; pentose) (Supplementary data, Figure S3F). Multiple glycoforms displayed an atypical carbohydrate of 188 Da potentially corresponding to the mass of an acetylated deoxyhexose sugar (Supplementary data, Figure S3C and E). Examination of the oxonium ion of this 188 Da atypical carbohydrate (189.07438 MH+) suggested that its structure might be C8H12O5 (data not shown). Interestingly, different glycans were found decorating the same glycosylation sites within some proteins (Table I). These results support the presence of glycan heterogeneity at the glycosylation sites of some R. solanacearum proteins. As expected, no glycopeptides were detected in the ΔpglLRs strain proteome while the non-glycosylated forms of the same peptides were observed confirming that protein O-glycosylation in Ralstonia is O-OTase dependent. Conversely, the in-genome expression of PglLRs in the ΔpglLRs strain led to the restoration of all the glycoproteins.
Quantitative proteomics reveals that disruption of glycosylation is not pleiotropic in R. solanacearum
The role of bacterial O-linked glycosylation is still largely unknown. The detection of multiple glycoproteins in R. solanacearum suggested an important role of O-glycosylation in this bacterium. Therefore, we hypothesized that determining what pathways are affected by disruption of glycosylation could help to understand the role of glycosylation in the physiology or the pathogenesis of this bacterium this microorganism. To accomplish this goal, the total proteomes of wild type, ΔpglLRs and the complemented strains were analyzed by the peptide stable isotope dimethyl labelling technique (Boersema et al. 2009).
Surprisingly, in response to the loss of glycosylation only a few proteins displayed altered protein levels compared with wild type, as determined by a 2-fold change cut-off (Table II, Supplementary data, Table SI). PilA, the major pilin subunit, displayed about 4-fold reduction in ΔpglLRs compared with wild-type strain. Similarly, the putative type VI secretion protein (Q8XRT8_RALSO) showed a 2-fold reduction in the ΔpglLRs strain. Conversely, the RS-IIL lectin levels in ΔpglLRs were about 3.5-fold more than in the wild-type strain. RS-IIL binds sugars containing mannose and fucose, which are widely distributed among the plant polysaccharides (Sudakevitz et al. 2004; Kostlanova et al. 2005). Expressing pglLRs in cis from its native promoter restored the wild-type levels of the three proteins in ΔpglLRs strain as shown by our proteomic analysis. Additionally, we monitored the levels of PilA using immunoblotting in the three R. solanacearum strains. As shown in Figure 3, the pilin subunit displayed a weak signal in the ΔpglLRs strain compared with the wild-type and complemented strains, confirming the mass spectrometry (MS) data. Moreover, PilA band appeared at a lower size in the strain lacking pglL confirming that PilA is glycosylated in R. solanacearum as shown by our MS analysis. RNA polymerase detection by immunoblotting was included as a loading control.
Table II.
List of proteins that displayed altered levels in OTase− compared with wild-type strain
| Uniprot number | Protein annotation | Gene locus | Relative protein abundance in O-Otase− vs WT straina | Relative protein abundance in cis complemented vs. WT straina |
|---|---|---|---|---|
| Q8Y1X9_RALSO | PilA | RSc0558 | 0.23 | 1 |
| Q8XRT8_RALSO | Putative type VI secretion protein | RSp0743 | 0.46 | 0.96 |
| Q8XUA5_RALSO | RS-II lectin | RSc3288 | 3.45 | 0.9 |
aPresented is the average of two readings.
Fig. 3.
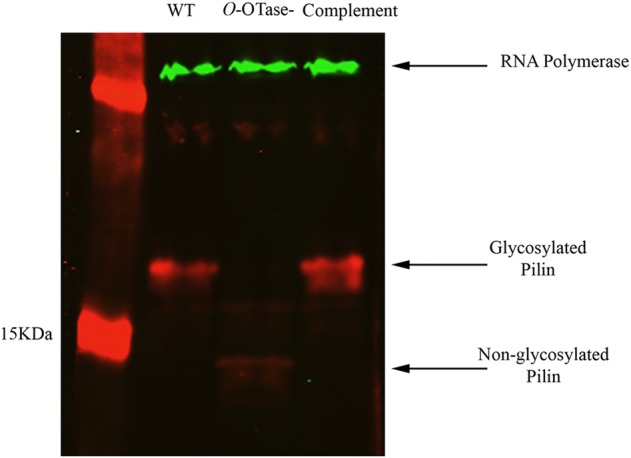
Loss of O-glycosylation altered the levels of pilin in R. solanacearum. Whole-cell lysates of different R. solanacearum strains were run on SDS–PAGE gel followed by immunoblotting using rabbit polyclonal anti-pilin and mouse monoclonal anti-RNA polymerase (1:2500, RNAP ɑ-subunit; Neoclone). Membranes were then probed with IRDye conjugated anti-mouse and anti-rabbit antibodies and visualized on an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE).
Lack of O-glycosylation affects biofilm formation and virulence in a tomato plant infection of R. solanacearum
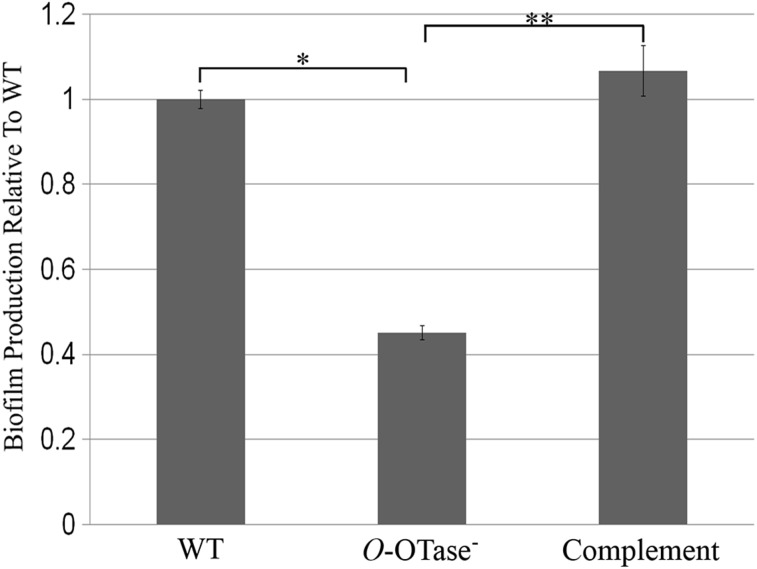
Type IV pilins are important for biofilm formation in many bacteria, including R. solanacearum (Kang et al. 2002; van Schaik et al. 2005; Flemming and Wingender 2010). Biofilm formation aids R. solanacearum in evading the host defenses and the entrapment of nutrients from the xylem flow (Yao and Allen 2007; Álvarez et al. 2010). To test the role of O-glycosylation in biofilm formation, we compared both ΔpglLRs and complemented strains to the wild-type strain in a standard microtiter plate-based biofilm assay. The three strains were grown in non-shaking conditions to allow attachment and matrix formation. The resulting biofilms were stained with crystal violet and quantified spectrophotometrically. Less biofilm was produced in the ΔpglLRs strain compared with the wild type (Figure 4). Expressing PglLRs from its native promoter in the genome restored biofilm formation in the mutant strain. Since lacking O-glycosylation affected the biofilm forming ability of R. solanacearum, we hypothesized that the ΔpglLRs strain would be defective in virulence towards the plant host. To test this hypothesis, we employed a tomato plant infection model to compare the virulence of the R. solanacearum wild-type and ΔpglLRs strains. The two strains were inoculated into wounded petioles and the infected plants were monitored for wilting symptoms (Figure 5). The WT caused the typical disease symptoms: initial chlorosis of the leaves and then loss of firmness and wilting. On the other hand, the ΔpglLRs mutant did not display wilting symptoms under the conditions tested. Expressing PglLRs in trans restored virulence in tomato plants suggesting a role of O-glycosylation in R. solanacearum pathogenesis.
Fig. 4.
Ralstonia O-OTase mutant is defective in biofilm formation. Different R. solanacearum strains were grown for 48 h in 96-well plates at 30°C. The formed biofilms were washed and stained with 0.1% crystal violet, then dissolved in 95% ethanol. Biofilm formation was measured spectrophotometrically by measuring absorbance at λ = 590 nm. Readings were normalized by the OD600 values of the corresponding strains after 48 h. Biofilm production in O-OTase− and complemented strains was presented relative to WT. The data shown were obtained from three independent experiments (n = 3). *P < 0.01, **P < 0.001.
Fig. 5.

O-Glycosylation may be involved in the virulence of Ralstonia towards tomato plants. Different Ralstonia strains were grown overnight at 30°C, then diluted to 106 CFU/mL using sterile distilled water. Inocula concentrations were checked using serial dilution plating on BG agar. Leaf petioles were inoculated with 3 µL of the normalized inocula from the three Ralstonia strains. A set of six tomato plants were inoculated with each strain and placed in separated trays in order to prevent cross contamination. After 16 h photoperiod incubation at 28°C, only plants infected with WT and the complemented strains caused wilting symptoms. The experiment was repeated twice under the same conditions to a total of three experiments.
Discussion
Protein glycosylation is a well-known post-translational modification that is widespread in bacteria. Bacteria can glycosylate their proteins by sequentially adding sugars to their target proteins via cytoplasmic glycosyltransferases. Alternatively, the lipid-linked glycans can be transferred en bloc to the target proteins by an OTase. Although initially thought to be limited to N-glycosylation, it was shown that many bacteria exploit OTases to O-glycosylate their proteins (Nothaft and Szymanski 2010; Iwashkiw et al. 2013). Some O-OTases are pilin-specific, like PilO/TfpO in Pseudomonas, while other O-OTases evolved to be more promiscuous towards their protein substrates (Castric 1995; Kus et al. 2008). Examples of OTase-dependant general O-glycosylation systems include A. baumannii, Neisseria spp., B. cepacia complex and F. tularensis (Marceau et al. 1998; Power et al. 2003; Borud et al. 2011; Jennings et al. 2011; Balonova et al. 2012; Iwashkiw et al. 2012; Lithgow et al. 2014). Some Acinetobacter strains evolved to keep both a pilin-specific and a general O-OTase (Harding et al. 2015). Protein O-glycosylation was shown to be important for the virulence of many pathogens, albeit, the exact role of O-glycosylation in pathogenesis remains unknown (Iwashkiw et al. 2013). In this work, we identified a general O-glycosylation system in the important phytopathogen R. solanacearum, one of the leading causes of plant diseases worldwide (Genin and Denny 2012). Using MS-based proteomic tools, we demonstrated that R. solanacearum GMI1000 produces at least twenty glycoproteins including type IV pilin and the type IV fimbrial membrane-assembly proteins (Table I), in a process that is PglL-dependent. We demonstrated that PglLRs was able to transfer a foreign glycan (diNAcBac) to a Neisseria protein when all the components of the reaction were co-expressed in E. coli (Figure 1). This indicates that, like previous characterized O-OTases, PglLRs has relaxed glycan specificity and reinforces the concept that, despite their divergent sequences, O-OTases from different bacteria recognize similar sequences in their target proteins. Within the detected glycoproteins, we observed heterogenous glycans decorating the proteins. The most abundant oligosaccharides detected were similar to the O antigen characterized in a number of R. solanacearum strains (Varbanets et al. 2003). Varbanets et al. (2003) detected N-acetyl glucosamine, rhamnose (deoxyhexose) and xylose (pentose) in the O-antigens characterized. Our in silico analysis of R. solanacearum GMI1000 revealed a homologue of a rhamnosyltransferase (RSc0687) within a typical glycan biosynthetic cluster that also contain enzymes usually involved in the synthesis of the O antigen. The same gene cluster included a homologue of undecaprenyl phosphate N-acetylglucosaminyltransferase transmembrane protein (RSc0689). This protein belongs to the initiating glycosyl transferases family, which is responsible for linking the first sugar in the O-antigen glycan to the lipid carrier in the inner membrane, a step essential for O-antigen synthesis. Interestingly, Li et al. demonstrated that the deletion of RSc0689 resulted in the loss of O-antigen formation in R. solanacearum GMI1000 (Li et al. 2014). Additionally, a UDP-4-keto-pentose/UDP-xylose synthase (Uxs) was previously identified in R. solanacearum GMI1000 (Gu et al. 2010), suggesting that xylose is the pentose contained in the O-glycans attached to proteins. However, this gene is not encoded in the cluster containing RSc0687. A differential regulation of these genes could explain the detection of heterogeneous glycan structures in R. solanacearum glycoproteins. Our results suggest that R. solanacearum shares its O-antigen between two different pathways; LPS synthesis and protein O-glycosylation. This is in agreement with what was observed in a number of bacteria and was proposed to be a strategy by which bacteria can save energy and resources (Cuccui and Wren 2013;Lees-Miller et al. 2013). The role of the putative initiating glycosyl transferase (RSc0689) in protein glycosylation remains to be demonstrated.
Protein O-glycosylation was completely abolished in the ΔpglLRs strain, which produced less biofilm. Our virulence test on tomato plants suggested a possible role for O-glycosylation in the pathogenesis of R. solanacearum. Nevertheless, a more comprehensive infection model will be required to confirm this finding. MS data showed that the expression of PglLRs in cis restored O-glycosylation. Type IV pilin was among the restored glycoproteins in the complemented strain. The work by Kang et al. showed that type IV pili are important for autoaggregation, biofilm development and pathogenesis of R. solanacearum (Kang et al. 2002). Our work showed that pilin is one of the few proteins affected by lack of glycosylation, and therefore, it is possible that the ΔpglLRs is defective in biofilm formation and virulence due to the impaired function of the non-glycosylated pilin.
The glycoproteome of R. solanacearum proved to be relatively large compared with that of other bacteria (Iwashkiw et al. 2013). Therefore, we hypothesized that the lack of O-glycosylation might impact other physiological functions in Ralstonia. To investigate additional effects of glycosylation loss, we performed a comparative proteomic analysis between wild-type and ΔpglLRs strains, using peptide stable isotope dimethyl labelling (Boersema et al. 2009). Surprisingly, our analysis revealed a change in the expression levels of only three proteins as response to the lack of glycosylation. These proteins are PilA, RS-IILlectin and a putative type VI secretion protein. Using immunoblotting, we were able to validate our proteomic analysis by monitoring the levels of pilin proteins in different R. solanacearum strains. In addition to being unglycosylated, R.solanacearum pilin was detected at lower levels in the ΔpglLRs strain relative to wild type as suggested by our MS analysis. Native pilin protein levels were restored upon cis complementation (Figure 3). One possible explanation is that in R. solanacearum, when O-glycosylation is absent, pilin is more susceptible to degradation by proteases. The role of protein glycosylation in protection against degradation is well-established (Iwashkiw et al. 2013). Nonetheless, levels of pilin protein do not appear to be reduced in glycosylation deficient strains of other bacterial species such as Neisseria and Acinetobacter (Marceau et al. 1998; Harding et al. 2015). Due to its role in biofilm formation, the reduced pilin levels might contribute to the lower levels of biofilm displayed by ΔpglLRs strain in vitro (Figure 3). However, our experiments do not show which of the two, loss of pilin glycosylation and lower pilin levels, has more impact on biofilm formation in ΔpglLRs strain. We observed higher levels of RS-IIL lectin in the OTase mutant via quantitative proteomics. RS-II is fucose/mannose binding lectin in R. solanacearum that is homologous to PA-IIL in the phylogenetically related P. aeruginosa. PA-IIL was shown to contribute to host cell adhesion and biofilm formation (Sudakevitz et al. 2004). A similar role was suggested for RS-IIL in Ralstonia pathogenesis towards plants (Valls et al. 2006). It is tempting to speculate that the lectin upregulation might act as a compensatory attempt of the cells to restore interbacterial or host–bacterial interactions mediated by the O-glycans and the pilin in wild-type bacteria. We also detected a 2-fold decrease in the levels of a putative type VI secretion protein (Q8XRT8_RALSO) in ΔpglLRs strain (Table II). Type VI secretion system is a newly discovered system in Gram negative bacteria that is capable of delivering effectors to prokaryotic and eukaryotic cells (Jani and Cotter 2010). Recently, a type VI secretion system was identified in R. solanacearum and was shown to be involved in biofilm formation and virulence (Zhang et al. 2014). It remains to be demonstrated if the type VI machinery in the ΔpglLRs strain is affected by the reduction in Q8XRT8_RALSO levels. To our knowledge, this is the first report studying global protein changes related to a deficiency in protein glycosylation in a prokaryote. Given the extensive glycoproteome present in R. solanacearum, we expected significant changes in the levels of multiple proteins in the absence of glycosylation. However, only a few proteins were altered in our experimental conditions. It is possible that in other condition such as stress, biofilms, or during plant infection, protein glycosylation plays a more important role and therefore additional changes it the proteome of R. solanacearum occur in these conditions. Further work will be necessary to understand the role of protein glycosylation in the biology of this important phytopathogen.
Materials and methods
Bacterial strains and growth conditions
Ralstonia solanacearum GMI1000 strains were grown on BG medium at 30°C as previously described (Kang et al. 2002). When needed, tetracycline (Tc) was added at concentration 3 µg/mL. Escherichia coli CLM24 was grown on LB medium at 37°C.
In vivo protein glycosylation assay
Rsc0559 was amplified from the genome of R. solanacearum GMI1000 via PCR using primers; GMOtaSmaIFw (5′-AATTCCCGGGATGTTGTGGCCGGTCTGG) and GMOtaHindIIIHisRv (5′-TGGT AAGCTTTTAGTGGTGGTGGTGGTGGTGATCTACACCGACAACCAAGT), then cloned into SmaI/HindIII sites of pEXT20. The in vivo glycosylation experiment was carried out as described by Gebhart et al. (2012) andGebhart et al. (2012). Briefly, E. coli CLM24 strains (lacking WaaL ligase), expressing bacillosamine biosynthesis genes from Neisseria, were grown at 37°C. At mid-log phase, 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and 0.2% arabinose were added to induce the expression of different O-OTases and DsbA, respectively. Cells were incubated until stationary phase and glycosylated proteins were detected via immunoblotting using the monoclonal anti-Histidine and the polyclonal anti-bacillosamine antibodies (Gebhart et al. 2012).
Construction of O-OTase mutant in R. solanacearum GMI1000 and complementation
The suicide vector pTOK3 was used to make clean deletions in R. solanacearum. pTOK3 was obtained after cloning sacB from pFLP2 into pTOK2 (Kang et al. 2002; Iwashkiw et al. 2012). Briefly, pTOK2 was cut using BamHI, then both the vector and sacB were blunt-ended using klenow fragment (ThermoScientific) followed by ligation. In order to make a clean deletion of Rsc0559 in R. solanacearum GMI1000, ∼500 bp of the flanking regions around Rsc0559 were PCR amplified using primers; GMOtaUpFwSmaI (5′-AATTCCCGGGTATTCTGGCTGCGATTGCC) and GMOtaUpRv (5′-AATTTTCTTTTTGAGACGCAATCCCAGGGAACGATGAACTGGAA) for the upstream region, while primers GMOtaDwnFw (5′-AATTTTCCAGTTCATCGTTCCCTGGGATTGCGTCTCAAAAAGAA) and GMOtaDwnRvHindIII (5′-TATTAAGCTTAGCGGACGTCGGATTTGATC) were used to amplify the downstream region. Both regions were cloned into pTOK3 using SmaI and HindIII sites, and the resulting plasmid was used to transform R. solanacearum using electroporation. Tc resistant colonies were grown on BG broth overnight then subcultured on sucrose-supplemented minimal medium (Liu et al. 2005). Sucrose resistant colonies were screened by PCR to confirm the second recombination event followed by sequencing to confirm the deletion of Rsc0559. For in cis complementation, Rsc0559 plus the upstream and downstream regions were amplified using primers: GMOtaUpFwSmaI and GMOtaDwnRvHindIII, and cloned into pTOK3. A silent mutation was introduced into Rsc0559 by changing the codon for the serine residue at position 380 from “TCG” to “TCA” as a scar to discriminate the in cis complemented and the wild-type strains. To complement the mutation in trans, Rsc0559 was amplified via PCR using primers: GMOtaSacIFw (5′-AATTGAGCTCATGTTGTGGCCGGTCTGG) and GMOtaBamHIHisRv (5′-TGGTGGATCCTTAGTGGTGGTGGTGGTGGTGATCTACACCGACAACCAAGT), then cloned into pHC60, a vector that is stable in planta, using SacI and BamHI sites (Cheng and Walker 1998).
LPS analysis by silver-stained SDS–PAGE gel
LPS was prepared from R. solanacearum strains using 10 mg of dried cells using the method of Yi and Hackett (Yi and Hackett 2000). LPS was run on a 15% SDS–PAGE and visualized by the silver staining method described by Tsai and Frasch (1982).
Digestion of membrane enriched samples of R. solanacearum
Peptide lysates for glycopeptide enrichment and quantitative analysis were prepared according to Lithgow et al. with minor modifications (Lithgow et al. 2014). Briefly, 2 mg of dried membrane enriched protein samples were solubilized in 6 M urea, 2 M thiourea, 40 mM NH4HCO3 and reduced with 10 mM dithiothreitol (DTT) followed by alkylated with 25 mM iodoacetamide for 1 h in the absence of light. The resulting alkylated protein mixture was then digested with Lys-C (1/100 w/w) for 4 h, diluted 1:5 in 40 mM NH4HCO3 and digested with trypsin (1/50 w/w) overnight at 25°C. Digestion was terminated with the addition of 1% trifluoroacetic acid (TFA) and peptide digests were purified using C18 empore (Sigma-Aldrich, St. Louis, MO). STop And Go Extraction (STAGE) tips were used to remove primary amide and salts which can interfere with dimethyl labeling and ZIC-HILIC glycopeptide enrichment protocols described below.
Enrichment of R. solanacearum glycopeptides using ZIC-HILIC purification
ZIC-HILIC enrichment was performed according to Scott et al. (2011) with minor modifications. Micro-columns composed of 10 µm ZIC-HILIC resin (Sequant, Umeå, Sweden) packed into P10 tips containing a 1 mm2 excised C8 Empore™ disc (Sigma). Prior to use, the columns were washed with ultra-pure water, followed by 95% acetonitrile (ACN) and then equilibrated with 80% ACN and 5% formic acid (FA). Samples were resuspended in 80% ACN and 5% FA, and insoluble material was removed by centrifugation at 16,100 × g for 5 min at 4°C. Samples adjusted to a concentration of 3 µg/µL and 150 µg of peptide material were loaded onto a column and washed with 10 loading volumes of 80% ACN, 5% FA. Unbound fractions were collected, pooled and dried by vacuum centrifugation. ZIC-HILIC bound peptides were eluted with 3 loading volumes of ultra-pure water and concentrated using vacuum centrifugation. Biological replicates of R. solanacearum strains were subjected to ZIC-HILIC independently using freshly prepared reagents.
Identification of glycopeptides using reversed-phase LC–MS, CID MS/MS and HCD MS/MS
Purified glycopeptides/peptides were resuspended in Buffer A (0.5% acetic acid) and separated using reversed-phase chromatography on either an Agilent 1290 Series HPLC (Agilent Technologies, Mississauga, ON) coupled to LTQ-Orbitrap Velos (Thermo Scientific, San Jose, CA) for qualitative analysis of glycopeptides or on EASY-nLC1000 system coupled to a Q-exactive for quantitative studies. For qualitative analysis of R. solanacearum glycopeptides, an in-house packed 20 cm, 75 µm inner diameter, 360 µm outer diameter, ReproSil—Pur C18 AQ 1.9 µm (Dr. Maisch, Ammerbuch-Entringen, Germany) column was used, while for quantitative studies a in house packed 45 cm, 50 µm inner diameter, 360 µm outer diameter, ReproSil—Pur C18 AQ 1.9 µm column was used. In both systems, samples were loaded onto a trap column, an in-house packed 2 cm, 100 µm inner diameter, 360 µm outer diameter column containing Aqua 5 µm C18 (Phenomenex, Torrance, CA), at 5 µL/min prior to gradient separation and infused for MS. A gradient was run from 0% Buffer B (80% ACN, 0.5% acetic acid) to 32% B over 140 min. Concentration of Buffer B was increased from 32 to 40% in the next 5 min. Next Buffer B concentration was increased to 100% over 2.5 min period, held at 100% for 2.5 min, and then dropped to 0% for another 20 min. Unbound fractions from ZIC-HILIC glycopeptide enrichment were subjected to analysis using the same instrumental set up as qualitative analysis of glycopeptides. Both instruments were operated using Xcalibur v2.2 (Thermo Scientific) with a capillary temperature of 275°C in a data-dependent mode automatically switching between MS, CID MS/MS and HCD MS/MS for qualitative analysis as previously described and using a top 10 data-dependent approach switching between MS (resolution 70 k, AGC target of 1 × 106), and HCD MS/MS events (resolution 17.5 k, AGC target of 1 × 106 with a maximum injection time of 60 ms, NCE 28 with 20% stepping) for quantitative studies. High-resolution CID analysis was performed on an LTQ-Orbitrap Velos with CID fragmentation (NCE 35, 10 ms activation) analyzed within the orbital trap (resolution 7.5K, AGC 5.0 × 104), to enable the detection of high m/z ion, the high mass range setting was used (Scott et al. 2011). Raw files were processed as previously described by Scott et al. (2011). Briefly, Proteome Discoverer v. 1.2 (Thermo Scientific) was used to search the resulting glycopeptide data using MASCOT v2.4 against the R. solanacearum GMI1000 database (obtained from UNIPROT, http://www.uniprot.org/, 2013-04-4, Taxon identifier: 26, 7608 containing 5014 protein sequences). Mascot searches were performed using the following parameters: peptide mass accuracy 20 ppm; fragment mass accuracy 0.02 Da; no enzyme specificity, fixed modifications—carbamidomethyl, variable modifications—methionine oxidation and deamidated N, Q. The instrument setting of MALDI-QUAD-TOF was chosen as previous studies show quadrupole-like fragmentation within HCD spectra (Olsen et al. 2007). Scan events that did not result in peptide identifications from MASCOT searches were exported to Microsoft Excel (Microsoft, Redmond, WA). To identify possible glycopeptides within exported non-match scans, the MS/MS module of GPMAW 8.2 (http://www.gpmaw.com/) called “mgf graph” was used to identify HCD scan events that contained the 204.08 m/z oxonium of HexNAc. All scan events containing the oxonium 204.08 m/z ion were manually inspected to identify possible glycopeptides. To facilitate glycopeptide assignments HCD scan events containing the 204.08 oxonium were manual inspected to identify potential deglycosylated peptides ions. Within these HCD scans the MS features (m/z, charge and intensity), which corresponded to masses below that of the deglycosylated peptide were extracted using the Spectrum list function of Xcalibur v2.2. The resulting numerical values of the detected MS features were scripted into mgf files and the peptide mass set to that of the deglycosylated peptide mass. The resulting mgf files were then searched using MASCOT. All spectra were searched with the decoy option enabled and no matches to this database were detected; the false discovery rate (FDR) was 0%. To further validate all glycopeptide matches, all HCD spectra were annotated using the Expert Annotation tool (http://www.biochem.mpg.de/mann/tools/). Glycan annotation is based on the nomenclature of Domon and Costello (Domon and Costello 1988).
Quantitative dimethylation of R. solanacearum membrane extracts
Quantitative dimethylation of WT, O-OTase− and the complemented strains were undertaken using dimethylation as outlined by Boersema et al. (2009). Two biological replicates of each strain were used in the analysis. Briefly, 1 mg of peptide lysate from each strain was resuspended in 30 µL of 100 mM Tetraethylammonium bromide and mixed with the following combinations of 200 mM formaldehyde (30 µL) and 1 M sodium cyanoborohyride (3 µL) isotopologues. For diplex experiments, wild-type samples were labeled with light formaldehyde (CH2O) and light sodium cyanoborohyride (NaBH3CN) and O-OTase− samples with medium formaldehyde (CD2O) and light sodium cyanoborohyride. For triplex experiments wild-type samples were labeled with light formaldehyde (CH2O) and light sodium cyanoborohyride (NaBH3CN), O-OTase− samples with medium formaldehyde (CD2O) and light sodium cyanoborohyride and the complemented strain samples with heavy formaldehyde (13CD2O) and heavy sodium cyanoborodeuteride (NaBD3CN). Reagents were mixed and samples incubated at room temperature for 1 h. Dimethylation reactions were repeated twice to ensure complete labeling of all amine groups. Dimethylation reactions were terminated by the addition of 30 µL of 1 M NH4Cl for 20 min at room temperature. Samples were acidified by addition of 5% (v/v) acetic acid and allowed to equilibrate in the dark for 1 h before pooling of the three samples in at 1:1:1 ratio. Pooled samples were then cleaned up by STAGE tip-based C18 clean up, lyophilized, stored at −20°C and used for ZIC-HILIC enrichment or directly for total proteome analysis.
Quantitative proteomic comparison of R. solanacearum strains
MaxQuant (v1.4.1.2; http://www.maxquant.org/) was used for identification and quantification of the resulting experiments (Cox and Mann 2008). Database searching was carried out against the UniProt R. solanacearum GMI1000 database (Taxon identifier: 267608 containing 5014 protein sequences) with the following search parameters: carbamidomethylation of cysteine as a fixed modification; oxidation of methionine, acetylation of protein N-terminal trypsin/P cleavage with a maximum of two missed cleavages. A multiplicity of two and three was used for diplex and triplex experiment, respectively, with each multiplicity denoting one of the dimethylation channels (light, medium and heavy, respectively). The precursor mass tolerance was set to 6 ppm and MS-MS tolerance 20 ppm in accordance with previously reports with a maximum false discovery rate of 1.0% set for protein identifications. The resulting protein group output was processed within the Perseus (v1.4.0.6; http://www.maxquant.org/) analysis environment to remove reverse matches and common proteins contaminants prior to analysis with Matlab R2012a (http://www.mathworks.com).
Preparation of cell lysates for SDS–PAGE and immunoblotting
Whole cells pellets were obtained from overnight cultures of different R. soalnacearum strains after normalization based on OD600 values. The harvested cell pellets were then solubilized in urea buffer at 37°C for 30 min then loaded on 15% SDS–PAGE gel. Following separation, proteins were transferred to a nitrocellulose membrane and pilin proteins were probed using rabbit polyclonal antibody. As a loading control, cytoplasmic RNA polymerase levels were monitored using mouse monoclonal anti-RNA polymerase (1:2500, RNAP ɑ-subunit; Neoclone). The membrane was incubated with IRDye conjugated anti-mouse and anti-rabbit antibodies to visualize the bands using Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE).
Biofilm assays
Biofilms were measured as described before (Siri et al. 2014). Briefly, different R. solanacearum strains were grown overnight then diluted to OD600 0.1 mL−1 (108 CFU/mL) and 20 µL were used to inoculate 200 µL of BG broth in 96-wells plate. Cells were grown for ∼48 h at 30°C. The formed biofilms were washed gently with sterile water and stained with 0.1% crystal violet for 30 min. Excess crystal violet was removed and the plate was washed with water. Stained biofilms were solubilized in 100 µL 95% ethanol, then quantified spectrophotometrically by measuring absorbance at λ = 590 nm. Readings were normalized by the OD600 values of the corresponding strains after 48 h. The comparative statistical analysis of results was performed using SigmaPlot® for Windows (Systat Software, CA). A one-way analysis of variance (ANOVA) followed by Holm-Sidak post hoc test to assess the significance of differences between groups were performed. The differences were considered significant when P < 0.05.
Virulence assays in tomato plants
To test the virulence of R. solanacearum strains on tomato plants, seeds (Solanum lycopersicum Mill., cultivar Platense) were germinated in commercial soil mix and 2-week old seedlings were transplanted into 10 cm-plastic pots and incubated in a growth chamber at 25°C for 16 h photoperiod for 4–5 weeks before inoculation. To prepare the inocula, strains were grown overnight at 28°C in liquid BG supplemented with appropriate antibiotics. Cells were pelleted by centrifugation, suspended in sterile distilled water and adjusted to 106 CFU/mL. Inocula concentrations were checked by plating on BG-agar supplemented with glucose (5 g/L) and triphenyltetrazolium chloride (50 mg/L) to observe typical smooth colonies after 2-day incubation at 28°C. Leaf petioles were inoculated by making a cut in the first leaves above the cotyledon (0.5–1 cm from their base), then immediately applying a 3 µL droplet of the bacterial suspensions to the wounded surfaces. A set of six tomato plants were inoculated with each strain and placed in separated trays in order to prevent cross contamination. After inoculation, plants were incubated in a growth chamber at 28°C and 16 h photoperiod. The experiment was repeated twice under the same conditions to a total of three independent experiments.
Supplementary Data
Supplementary data for this article are available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC). N.E.S. was supported by a National Health and Medical Research Council of Australia (NHMRC) Overseas (Biomedical) Fellow (APP1037373) and is currently a Michael Smith Foundation for Health Research Trainee Post-Doctoral Fellow (award no. 5363). Operational funding support for N.E.S. and L.J.F. was provided by CIHR Operating grant no. MOP-77688.
Conflict of interest statement
None declared.
Abbreviations
ACN, acetonitrile; diNAcBac, undecaprenyl-linked 2,4-di-N-acetylbacillosamine; FA, formic acid; LPS, lipopolysaccharides; OD, optical density; O-OTase, O-oligosaccharyltransferase; PglLNm, PglL from N. meningitidis; PglLRs, PglL from R. solanacearum; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; STAGE, STop And Go Extraction; TFA, trifluoroacetic acid.
Supplementary Material
Acknowledgements
We thank Tim Denny for providing us with the R. solanacearum strains and plasmids, as well as Brent Weber and Christian Harding for critical reading of the manuscript. pHC60 was a generous gift from Nicolas Vozza. We are grateful for the assistance with genomes analysis by Joshua Irwin.
References
- Álvarez B, Biosca EG, López MM. 2010. On the life of Ralstonia solanacearum, a destructive bacterial plant pathogen. Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology. 1:267–279. [Google Scholar]
- Anonsen JH, Vik A, Egge-Jacobsen W, Koomey M. 2012. An extended spectrum of target proteins and modification sites in the general O-linked protein glycosylation system in Neisseria gonorrhoeae. J Proteome Res. 11:5781–5793. [DOI] [PubMed] [Google Scholar]
- Araud-Razou I, Vasse J, Montrozier H, Etchebar C, Trigalet A. 1998. Detection and visualization of the major acidic exopolysaccharide of Ralstonia solanacearum and its role in tomato root infection and vascular colonization. Eur J Plant Pathol. 104:795–809. [Google Scholar]
- Balonova L, Mann BF, Cerveny L, Alley WR Jr, Chovancova E, Forslund AL, Salomonsson EN, Forsberg A, Damborsky J, Novotny MV et al. . 2012. Characterization of protein glycosylation in Francisella tularensis subsp. holarctica: identification of a novel glycosylated lipoprotein required for virulence. Mol Cell Proteomics. 11:M111 015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. 2009. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 4:484–494. [DOI] [PubMed] [Google Scholar]
- Borud B, Viburiene R, Hartley MD, Paulsen BS, Egge-Jacobsen W, Imperiali B, Koomey M. 2011. Genetic and molecular analyses reveal an evolutionary trajectory for glycan synthesis in a bacterial protein glycosylation system. Proc Natl Acad Sci USA. 108:9643–9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric P. 1995. pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology. 141(Pt 5):1247–1254. [DOI] [PubMed] [Google Scholar]
- Cheng HP, Walker GC. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol. 180:5183–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Valls M. 2013. Current knowledge on the Ralstonia solanacearum type III secretion system. Microbial Biotechnol. 6:614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- Cuccui J, Wren BW. 2013. Bacteria like sharing their sweets. Mol Microbiol. 89:811–815. [DOI] [PubMed] [Google Scholar]
- Denny TP, Baek S-R. 1991. Genetic evidence that extracellular polysaccharide is a virulence factor of Pseudomonas solanacearum. Mol Plant-Microbe Interact. 4:198–206. [Google Scholar]
- Domon B, Costello CE. 1988. A systematic nomenclature for carbohydrate fragmentations in Fab-Ms Ms spectra of glycoconjugates. Glycoconjugate J. 5:397–409. [Google Scholar]
- Egge-Jacobsen W, Salomonsson EN, Aas FE, Forslund AL, Winther-Larsen HC, Maier J, Macellaro A, Kuoppa K, Oyston PC, Titball RW et al. . 2011. O-linked glycosylation of the PilA pilin protein of Francisella tularensis: identification of the endogenous protein-targeting oligosaccharyltransferase and characterization of the native oligosaccharide. J Bacteriol. 193:5487–5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Namba K, Hughes KT. 2010. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol. 2:a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman MF, Wacker M, Hernandez M, Hitchen PG, Marolda CL, Kowarik M, Morris HR, Dell A, Valvano MA, Aebi M. 2005. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc Natl Acad Sci USA. 102:3016–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez LA, Berenguer J. 2000. Secretion and assembly of regular surface structures in Gram-negative bacteria. FEMS Microbiol Rev. 24:21–44. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol. 8:623–633. [DOI] [PubMed] [Google Scholar]
- Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. 2009. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell. 137:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussenegger M, Rudel T, Barten R, Ryll R, Meyer TF. 1997. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae – A review. Gene. 192:125–134. [DOI] [PubMed] [Google Scholar]
- Gebhart C, Ielmini MV, Reiz B, Price NL, Aas FE, Koomey M, Feldman MF. 2012. Characterization of exogenous bacterial oligosaccharyltransferases in Escherichia coli reveals the potential for O-linked protein glycosylation in Vibrio cholerae and Burkholderia thailandensis. Glycobiology. 22:962–974. [DOI] [PubMed] [Google Scholar]
- Genin S, Denny TP. 2012. Pathogenomics of the Ralstonia solanacearum species complex. Annu Rev Phytopathol. 50:67–89. [DOI] [PubMed] [Google Scholar]
- Gu X, Glushka J, Yin Y, Xu Y, Denny T, Smith J, Jiang Y, Bar-Peled M. 2010. Identification of a bifunctional UDP-4-keto-pentose/UDP-xylose synthase in the plant pathogenic bacterium Ralstonia solanacearum strain GMI1000, a distinct member of the 4,6-dehydratase and decarboxylase family. J Biol Chem. 285:9030–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding CM, Nasr MA, Kinsella RL, Scott NE, Foster LJ, Weber BS, Fiester SE, Actis LA, Tracy EN, Munson RS Jr et al. . 2015. Acinetobacter strains carry two functional oligosaccharyltransferases, one devoted exclusively to type IV pilin, and the other one dedicated to O-glycosylation of multiple proteins. Mol Microbiol. 96:1023–1041. [DOI] [PubMed] [Google Scholar]
- Husain A, Kelman A. 1958. Relation of slime production to mechanism of wilting and pathogenicity of Pseudomonas solanacearum. Phytopathology. 48:155–165. [Google Scholar]
- Iwashkiw JA, Seper A, Weber BS, Scott NE, Vinogradov E, Stratilo C, Reiz B, Cordwell SJ, Whittal R, Schild S et al. . 2012. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog. 8:e1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashkiw JA, Vozza NF, Kinsella RL, Feldman MF. 2013. Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol Microbiol. 89:14–28. [DOI] [PubMed] [Google Scholar]
- Jani AJ, Cotter PA. 2010. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe. 8:2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MP, Jen FE, Roddam LF, Apicella MA, Edwards JL. 2011. Neisseria gonorrhoeae pilin glycan contributes to CR3 activation during challenge of primary cervical epithelial cells. Cell Microbiol. 13:885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Liu H, Genin S, Schell MA, Denny TP. 2002. Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol Microbiol. 46:427–437. [DOI] [PubMed] [Google Scholar]
- Kao CC, Barlow E, Sequeira L. 1992. Extracellular polysaccharide is required for wild-type virulence of Pseudomonas solanacearum. J Bacteriol. 174:1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostlanova N, Mitchell EP, Lortat-Jacob H, Oscarson S, Lahmann M, Gilboa-Garber N, Chambat G, Wimmerova M, Imberty A. 2005. The fucose-binding lectin from Ralstonia solanacearum. A new type of beta-propeller architecture formed by oligomerization and interacting with fucoside, fucosyllactose, and plant xyloglucan. J Biol Chem. 280:27839–27849. [DOI] [PubMed] [Google Scholar]
- Kus JV, Kelly J, Tessier L, Harvey H, Cvitkovitch DG, Burrows LL. 2008. Modification of Pseudomonas aeruginosa Pa5196 type IV Pilins at multiple sites with D-Araf by a novel GT-C family Arabinosyltransferase, TfpW. J Bacteriol. 190:7464–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller RG, Iwashkiw JA, Scott NE, Seper A, Vinogradov E, Schild S, Feldman MF. 2013. A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol Microbiol. 89:816–830. [DOI] [PubMed] [Google Scholar]
- Li CH, Wang KC, Hong YH, Chu TH, Chu YJ, Chou IC, Lu DK, Chen CY, Yang WC, Lin YM et al. . 2014. Roles of different forms of lipopolysaccharides in Ralstonia solanacearum pathogenesis. Mol Plant Microbe Interact. 27:471–478. [DOI] [PubMed] [Google Scholar]
- Lithgow KV, Scott NE, Iwashkiw JA, Thomson EL, Foster LJ, Feldman MF, Dennis JJ. 2014. A general protein O-glycosylation system within the Burkholderia cepacia complex is involved in motility and virulence. Mol Microbiol. 92:116–137. [DOI] [PubMed] [Google Scholar]
- Liu H, Kang Y, Genin S, Schell MA, Denny TP. 2001. Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology. 147:3215–3229. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang S, Schell MA, Denny TP. 2005. Pyramiding unmarked deletions in Ralstonia solanacearum shows that secreted proteins in addition to plant cell-wall-degrading enzymes contribute to virulence. Mol Plant Microbe Interact. 18:1296–1305. [DOI] [PubMed] [Google Scholar]
- Marceau M, Forest K, Beretti JL, Tainer J, Nassif X. 1998. Consequences of the loss of O-linked glycosylation of meningococcal type IV pilin on piliation and pilus-mediated adhesion. Mol Microbiol. 27:705–715. [DOI] [PubMed] [Google Scholar]
- Merz AJ, So M, Sheetz MP. 2000. Pilus retraction powers bacterial twitching motility. Nature. 407:98–102. [DOI] [PubMed] [Google Scholar]
- Nothaft H, Scott NE, Vinogradov E, Liu X, Hu R, Beadle B, Fodor C, Miller WG, Li J, Cordwell SJ et al. . 2012. Diversity in the protein N-glycosylation pathways within the Campylobacter genus. Mol Cell Proteomics. 11:1203–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H, Szymanski CM. 2010. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol. 8:765–778. [DOI] [PubMed] [Google Scholar]
- Ojanen-Reuhs T, Kalkkinen N, Westerlund-Wikstrom B, van Doorn J, Haahtela K, Nurmiaho-Lassila EL, Wengelnik K, Bonas U, Korhonen TK. 1997. Characterization of the fimA gene encoding bundle-forming fimbriae of the plant pathogen Xanthomonas campestris pv. vesicatoria. J Bacteriol. 179:1280–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. 2007. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 4:709–712. [DOI] [PubMed] [Google Scholar]
- Orgambide G, Montrozier H, Servin P, Roussel J, Trigalet-Demery D, Trigalet A. 1991. High heterogeneity of the exopolysaccharides of Pseudomonas solanacearum strain GMI 1000 and the complete structure of the major polysaccharide. J Biol Chem. 266:8312–8321. [PubMed] [Google Scholar]
- Peeters N, Guidot A, Vailleau F, Valls M. 2013. Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol Plant Pathol. 14:651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymiro M, Genin S. 2009. Secreted proteins from Ralstonia solanacearum: a hundred tricks to kill a plant. Curr Opin Microbiol. 12:44–52. [DOI] [PubMed] [Google Scholar]
- Power PM, Roddam LF, Rutter K, Fitzpatrick SZ, Srikhanta YN, Jennings MP. 2003. Genetic characterization of pilin glycosylation and phase variation in Neisseria meningitidis. Mol Microbiol. 49:833–847. [DOI] [PubMed] [Google Scholar]
- Power PM, Seib KL, Jennings MP. 2006. Pilin glycosylation in Neisseria meningitidis occurs by a similar pathway to wzy-dependent O-antigen biosynthesis in Escherichia coli. Biochem Biophys Res Commun. 347:904–908. [DOI] [PubMed] [Google Scholar]
- Saile E, McGarvey JA, Schell MA, Denny TP. 1997. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology. 87:1264–1271. [DOI] [PubMed] [Google Scholar]
- Schulz BL, Jen FE, Power PM, Jones CE, Fox KL, Ku SC, Blanchfield JT, Jennings MP. 2013. Identification of bacterial protein O-oligosaccharyltransferases and their glycoprotein substrates. PLoS ONE. 8:e62768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott NE, Kinsella RL, Edwards AV, Larsen MR, Dutta S, Saba J, Foster LJ, Feldman MF. 2014. Diversity within the O-linked protein glycosylation systems of acinetobacter species. Mol Cell Proteomics. 13:2354–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott NE, Parker BL, Connolly AM, Paulech J, Edwards AV, Crossett B, Falconer L, Kolarich D, Djordjevic SP, Hojrup P et al. . 2011. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol Cell Proteomics. 10:M000031–MCP000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siri MI, Sanabria A, Boucher C, Pianzzola MJ. 2014. New type IV pili-related genes involved in early stages of Ralstonia solanacearum potato infection. Mol Plant Microbe Interact. 27:712–724. [DOI] [PubMed] [Google Scholar]
- Strom MS, Lory S. 1993. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 47:565–596. [DOI] [PubMed] [Google Scholar]
- Sudakevitz D, Kostlanova N, Blatman-Jan G, Mitchell EP, Lerrer B, Wimmerova M, Katcoff DJ, Imberty A, Gilboa-Garber N. 2004. A new Ralstonia solanacearum high-affinity mannose-binding lectin RS-IIL structurally resembling the Pseudomonas aeruginosa fucose-specific lectin PA-IIL. Mol Microbiol. 52:691–700. [DOI] [PubMed] [Google Scholar]
- Tans-Kersten J, Huang H, Allen C. 2001. Ralstonia solanacearum needs motility for invasive virulence on tomato. J Bacteriol. 183:3597–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CM, Frasch CE. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 119:115–119. [DOI] [PubMed] [Google Scholar]
- Valls M, Genin S, Boucher C. 2006. Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum. PLoS Pathog. 2:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik EJ, Giltner CL, Audette GF, Keizer DW, Bautista DL, Slupsky CM, Sykes BD, Irvin RT. 2005. DNA binding: A novel function of Pseudomonas aeruginosa type IV pili. J Bacteriol. 187:1455–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varbanets LD, Vasil'ev VN, Brovarskaia OS. 2003. [Characterization of lipopolysaccharides from Ralstonia solanacearum]. Mikrobiologiia. 72:19–25. [PubMed] [Google Scholar]
- Vik A, Aas FE, Anonsen JH, Bilsborough S, Schneider A, Egge-Jacobsen W, Koomey M. 2009. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 106:4447–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Allen C. 2006. Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J Bacteriol. 188:3697–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Allen C. 2007. The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. J Bacteriol. 189:6415–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi EC, Hackett M. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. The Analyst. 125:651–656. [DOI] [PubMed] [Google Scholar]
- Zhang L, Xu J, Zhang H, He L, Feng J. 2014. TssB is essential for virulence and required for type VI secretion system in Ralstonia solanacearum. Microb Pathogenesis. 74:1–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.