Abstract
Both chemokine oligomerization and binding to glycosaminoglycans (GAGs) are required for their function in cell recruitment. Interactions with GAGs facilitate the formation of chemokine gradients, which provide directional cues for migrating cells. In contrast, chemokine oligomerization is thought to contribute to the affinity of GAG interactions by providing a more extensive binding surface than single subunits alone. However, the importance of chemokine oligomerization to GAG binding has not been extensively quantified. Additionally, the ability of chemokines to form different oligomers has been suggested to impart specificity to GAG interactions, but most studies have been limited to heparin. In this study, several differentially oligomerizing chemokines (CCL2, CCL3, CCL5, CCL7, CXCL4, CXCL8, CXCL11 and CXCL12) and select oligomerization-deficient mutants were systematically characterized by surface plasmon resonance to determine their relative affinities for heparin, heparan sulfate (HS) and chondroitin sulfate-A (CS-A). Wild-type chemokines demonstrated a hierarchy of binding affinities for heparin and HS that was markedly dependent on oligomerization. These results were corroborated by their relative propensity to accumulate on cells and the critical role of oligomerization in cell presentation. CS-A was found to exhibit greater chemokine selectivity than heparin or HS, as it only bound a subset of chemokines; moreover, binding to CS-A was ablated with oligomerization-deficient mutants. Overall, this study definitively demonstrates the importance of oligomerization for chemokine–GAG interactions, and demonstrates diversity in the affinity and specificity of different chemokines for GAGs. These data support the idea that GAG interactions provide a mechanism for fine-tuning chemokine function.
Keywords: chemokine, heparan sulfate, heparin, oligomerization, surface plasmon resonance
Introduction
Chemokines belong to an ∼45-member family of small (8–12 kDa) proteins that are involved in regulating the migration of cells in a wide range of developmental, homeostatic and inflammatory processes (Salanga and Handel 2011). They are produced by virtually all cell types, either constitutively or inducibly depending on context, and serve as directional signals for migrating cells. Chemokine-mediated cell recruitment is triggered upon interaction with G protein-coupled chemokine receptors on leukocytes and many other cell types (Baggiolini 1998), which promotes migration of receptor-bearing cells along increasing chemokine gradients, as shown in vitro (Rot 1993; Haessler et al. 2011) and more recently in tissues (Weber et al. 2013). Formation of chemokine gradients on cell surfaces is considered to be critical for haptotactic cell migration (Middleton et al. 1997; Patel et al. 2001) and the absence of such gradients leads to impaired migration either because of the lack of directional signals (Weber et al. 2013) or due to bulk receptor desensitization (Ali et al. 2007).
Glycosaminoglycans (GAGs) on cell surfaces and in the extracellular matrix (ECM) provide one of the main mechanisms for establishing chemokine gradients. Recent studies show that blood vessels create steep gradients of heparan sulfate (HS) between their lumenal and basolateral surfaces and that further inflammation significantly increases HS deposition in the ECM, thereby providing a mechanism for patterning chemokine gradients (Stoler-Barak et al. 2014). Chemokine–GAG interactions are also involved in the transport of chemokines across endothelial cells from their site of production at inflammatory foci (Wang et al. 2005), and they play a role in the secretion of chemokines from tumor cells (Soria et al. 2012) and in the storage and release of chemokines from T cells (Wagner et al. 1998). Moreover, several studies have shown that the ability of some chemokines to bind to GAGs is important for their function using in vivo models of inflammation (Proudfoot et al. 2003; Peterson et al. 2004; Ali et al. 2005). While the importance of these interactions has motivated numerous studies to determine binding affinities of chemokines with GAGs, there are few quantitative comparisons of the affinities of different chemokines for GAGs apart from some early studies (Hoogewerf et al. 1997; Kuschert et al. 1999). Compiling comparisons based on studies of individual chemokines is, however, challenging due to the use of a wide range of techniques, solution conditions and types/sources of GAGs in these studies (Hamel et al. 2009). Nevertheless, such comparisons are important as they may reveal differences in the specificity of chemokines for GAGs and/or be relevant to the role of such interactions in chemokine function.
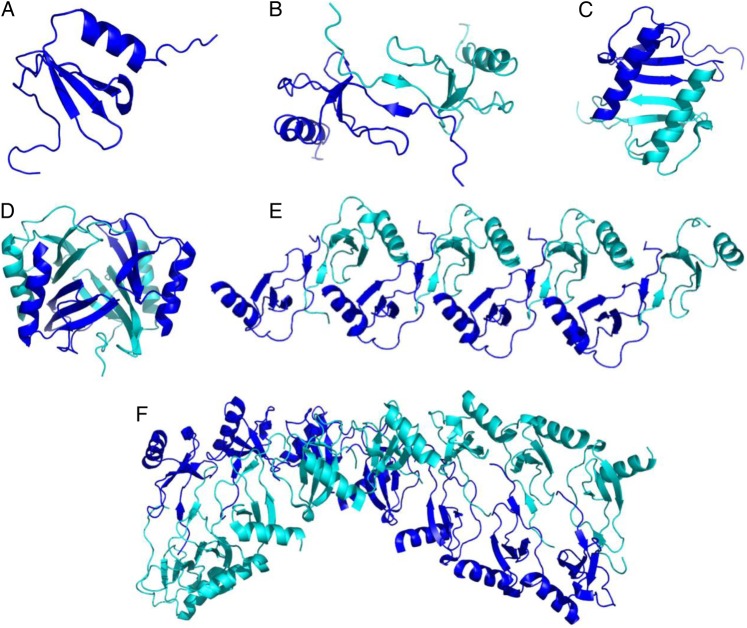
The molecular details of how chemokines bind to GAGs are also poorly understood. Numerous chemokine structures have been determined, all showing a highly conserved motif consisting of a disordered N-terminus that is the key signaling domain, an irregular “N-loop”, a three-stranded β-sheet and a C-terminal helix (Figure 1A). GAG-binding epitopes have been defined primarily by targeting BBXB and related motifs (where B is a basic and X is any amino acid) on the surface of these conserved tertiary structures (Cardin and Weintraub 1989). However, while some chemokines function as monomers, many chemokines oligomerize and form diverse quaternary structures including dimers, tetramers and polymers (Figure 1; Salanga and Handel 2011). Additionally, similar to their requirement for binding to GAGs, the ability of some chemokines to oligomerize has been shown to be important for their in vivo function (Proudfoot et al. 2003; Campanella et al. 2006; Gangavarapu et al. 2012). Using oligomerization-deficient variants of chemokines, it has been shown that chemokine monomers bind and activate chemokine receptors to induce cell migration (Rajarathnam et al. 1994; Paavola et al. 1998), ruling out chemokine oligomers in receptor activation. In contrast, several studies have shown that chemokines oligomerize upon binding to GAGs and/or that their oligomeric state is stabilized by GAG binding (Salanga and Handel 2011). Thus, it has been hypothesized that oligomerization and GAG binding are coupled and that oligomerization might provide a mechanism for enhancing the affinity of chemokines for GAGs (Hoogewerf et al. 1997; Salanga and Handel 2011) and possibly influencing GAG specificity (Lortat-Jacob et al. 2002; Handel et al. 2005). However, only a few studies have been undertaken to broadly determine the extent to which oligomerization contributes to GAG-binding affinity (Ziarek et al. 2013; Salanga et al. 2014).
Fig. 1.
Chemokines exhibit a wide range of oligomeric structures. While the tertiary structure of chemokines are highly conserved and generally resemble the CCL7 monomer (PDB ID 1BO0) (A), chemokines can also form diverse oligomeric states (B–F). For example, CC-type chemokines dimers are often extended structures involving the N-terminus, like the CCL2 dimer shown (PDB ID 1DOL) (B) in contrast to the more compact dimer structure of CXCL8 (PDB ID 1IL8) (C), formed through interactions involving the first beta-strand, and common to the CXC-type chemokines. On the other hand, CXCL4 forms a stable tetramer (PDB ID 1RHP) (D) and CCL5 (model provided by Dr. Xu Wang) and CCL3 (PDB ID 3KKH) have been shown to form higher order oligomers (E and F). Structures were generated with PyMol. This figure is available in black and white in print and in color at Glycobiology online.
This study was initiated to systematically investigate the ability of a set of differentially oligomerizing chemokines to bind to GAGs including the monomeric chemokine CCL7, dimeric chemokines CXCL8, CXCL12 and CCL2, the tetrameric chemokine CXCL4, polymeric chemokines CCL3 and CCL5 as well as CXCL11 whose oligomerization state has not been well characterized (Allen et al. 2007; Severin et al. 2010; Salanga and Handel 2011). Several oligomerization-deficient or oligomerization-stabilizing variants of a subset of these chemokines were also examined to define the contribution of oligomerization to GAG binding. Surface plasmon resonance (SPR) was used to obtain kinetic and thermodynamic information on the interactions, as it is one of the more quantitative methods for determining chemokine–GAG-binding affinities (Hamel et al. 2009). SPR also allows one to use surface immobilized rather than soluble GAGs, which alleviates problems with precipitation of chemokines when investigated with solution techniques (Hamel et al. 2009). In contrast to most studies that use heparin as a model GAG, we also compare interactions with HS and chondroitin sulfate-A (CS-A). Studies with heparin have provided valuable information; however, this GAG is only secreted by mast cells, whereas the predominant GAG involved in chemokine biology is HS, with some contribution from CS and dermatan sulfate (Xu and Esko 2014). Finally, following prior studies that suggest oligomeric chemokines cross-link GAG chains (Migliorini et al. 2014; Salanga et al. 2014), we also investigate the role of HS density on the affinity of WT and oligomerization-deficient chemokines. Overall, the results demonstrate that chemokine oligomerization is critical for GAG-binding affinity and in regulating the overall accumulation of chemokines on GAGs, and in some cases it contributes to GAG-binding specificity.
Results
Chemokines have a wide range of binding affinities for GAGs
The aim of this study was to systematically analyze a variety of chemokines with varying oligomeric propensities using SPR to determine their relative affinities for different GAGs and the contribution of oligomerization to these affinities. In addition to providing kinetic association/dissociation rates and affinities, an advantage of SPR over other techniques is that the setup involves solid-phase interactions with GAGs immobilized on SPR chips, which alleviates issues with precipitation; this problem is frequently observed with solution-based techniques (e.g., isothermal fluorescence titration, Krieger et al. 2004, and isothermal titration calorimetry, Kuschert et al. 1999), particularly when analyzing interactions with longer GAGs (degree of polymerization >10), which have higher affinity for chemokines than short GAGs (Kuschert et al. 1999). In our experience with solution-based methods, chemokines frequently oligomerize upon addition of GAG, and then dissociate as the ratio of GAG:chemokine exceeds 1:1 (Jansma et al. 2010). This behavior complicates analysis of affinities and GAG-induced oligomerization, but is not observed in SPR studies where the GAG is constrained on a two-dimensional surface.
Implementation of the SPR assay involved passing different concentrations of a given chemokine over the surface of a neutravidin-coated C1 chip onto which biotinylated GAG was immobilized (see Materials and methods). One caveat of the immobilization method that we used, however, is that each GAG chain may have more than one biotinylation group due to the nonspecific nature of the chemistry where any carboxyl group can be modified. Nevertheless, as described later, the results are qualitatively consistent with prior affinity studies and with binding studies of select chemokines/mutants on cell surfaces.
To make the most robust comparative analyses of different chemokines and mutants, special attention was made to collect the data under similar experimental conditions with the same source of GAG, similar or identical GAG densities immobilized on the chips and the same chip. “Apparent affinities” were then calculated from association (ka) and dissociation (kd) rates as KD = kd/ka using a 1:1 Langmuir model. This model is unlikely to be an accurate description of all chemokine–GAG interactions given the diverse propensities of chemokines to oligomerize both in solution and on GAGs. Furthermore, heterogeneity in the GAG interactions of a given chemokine, both in terms of the actual oligomerization state of the free and bound chemokine and the heterogeneity of GAG-binding sites, is likely. However, as these interaction details are unknown, inclusion of specific models to improve fits to the data cannot be justified; thus, we chose the 1:1 Langmuir model and designate the results as “apparent affinities”.
For datasets that reached or approached saturation at high chemokine concentration, steady-state analysis was used as an alternative or additional method for determining apparent affinities. To compare the overall accumulation of different chemokines and mutants to one another, the maximum signal (RU) at each chemokine concentration was also evaluated as it reflects a combination of affinity and oligomerization propensity. Thus, the apparent affinities derived from kinetic data, steady-state affinity analysis, the overall accumulation of chemokine on the GAG surfaces as well as visual analysis of the associated sensorgrams, were used together to provide a robust comparison between different chemokines and their associated mutants. Additionally, the relative binding of WT chemokines and chemokine variants on cell surfaces was used to validate the SPR results.
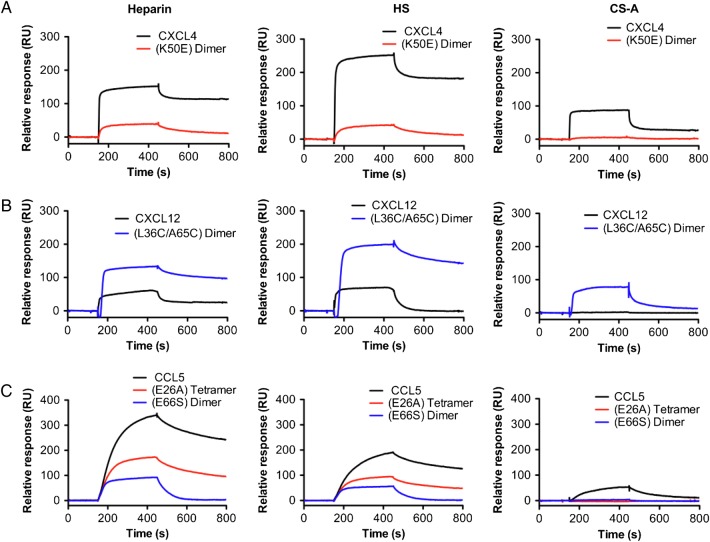
Figure 2 shows representative sensorgrams for select chemokines and mutants on heparin, HS and CS-A, and Table I summarizes the KD values determined from the kinetic rates and/or steady-state analyses; notably, all values presented in Table I were derived from the datasets shown. More complete sensorgram datasets along with fits and/or steady-state affinity analyses are provided in Supplementary data, Figures S1–S3. Additionally, the mean affinity of replicate experiments from two independently generated surfaces is reported in Supplementary data, Table SI. As demonstrated by these results, the apparent affinities of chemokines for heparin and HS span a wide range and cluster into three affinity classes: (i) CXCL4, CCL5 and CXCL11 have the highest apparent affinity (<10 nM); (ii) CXCL12 and CCL2 (as previously reported in (Salanga et al. 2014)) have intermediate apparent affinities (<100 nM); (iii) CXCL8 has the weakest apparent affinity (<500 nM); and CCL3 shows no detectable interaction. In contrast to heparin and HS, which bind most of the chemokines investigated, CS-A shows more selectivity and only binds CXCL4, CCL5 and CXCL11, although with reduced affinity compared with heparin and HS (Table I).
Fig. 2.
Differential chemokine and oligomeric mutant interactions with Heparin, HS or CS-A. SPR sensorgrams of chemokine and associated oligomeric mutant(s): WT CXCL4 tetramer and CXCL4(K50E) dimer (40 nM) (A), WT CXCL12 and CXCL12(L36C/A65C) disulfide-locked dimer (200 nM) (B) or WT CCL5 polymer, CCL5(E26A) tetramer and CCL5(E66S) dimer (50 nM) (C) passed over immobilized heparin, HS or CS-A on a BIAcore C1 chip. The curves represent the maximum response signal (RU) minus the reference cell signal (no GAG). In most cases, the oligomeric mutants exhibited decreased affinity compared with WT for the GAGs tested (see Table I). Experiments were performed in duplicate with a representative sensorgram shown. This figure is available in black and white in print and in color at Glycobiology online.
Table I.
Surface plasmon resonance analysis of chemokine and mutant affinities for surfaces of heparin, heparan sulfate and CS-A
| Chemokine | Heparin |
High-density HS |
CS-A |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ka (M−1 s−1) | kd (s−1) | KD (nM) | χ2 | Steady-state affinity (nM) | ka (M−1 s−1) | kd (s−1) | KD (nM) | χ2 | Steady-state affinity (nM) | ka (M−1 s−1) | kd (s−1) | KD (nM) | χ2 | Steady-state affinity (nM) | |
| CXCL4 | 4.7 × 106 | 1.3 × 10−4 | 0.03a | 13.5 | 2.4 | 3.3 × 106 | 2.5 × 10−4 | 0.08a | 31.8 | 3.8 | PFb | PF | PF | PF | 9.3 |
| (K50E) Dimer | 1.3 × 106 | 3.0 × 10−3 | 2.3 | 1.0 | 51 | 1.7 × 105 | 3.3 × 10−3 | 19 | 3.5 | 20 | NPAc | NPA | NPA | NPA | NPA |
| CXCL8 | PF | PF | PF | PF | 480 | PF | PF | PF | PF | 450 | NPA | NPA | NPA | NPA | NPA |
| CXCL11 | 8.0 × 105 | 1.8 × 10−3 | 2.3 | 4.9 | NEd | 5.3 × 105 | 3.2 × 10−3 | 6.0 | 3.1 | NE | 6.4 × 104 | 2.9 × 10−3 | 45 | 2.9 | NS |
| CXCL12 | 7.8 × 105 | 2.3 × 10−2 | 29 | 4.5 | 39 | 4.9 × 105 | 3.0 × 10−2 | 61 | 2.3 | 60 | NOIe | NOI | NOI | NOI | NOI |
| (L36C/A65C) dimer | PF | PF | PF | PF | 6.4 | PF | PF | PF | PF | 12 | PF | PF | PF | PF | 59 |
| CCL3 | NOI | NOI | NOI | NOI | NOI | NOI | NOI | NOI | NOI | NOI | NOI | NOI | NOI | NOI | NOI |
| CCL5 | 2.0 × 105 | 7.3 × 10−4 | 3.7 | 32 | NE | 1.9 × 105 | 8.6 × 10−4 | 4.5 | 7.4 | NE | 8.8 × 104 | 4.5 × 10−3 | 51 | 1.7 | NE |
| (E26A) tetramer | 2.9 × 105 | 1.2 × 10−3 | 4.1 | 13.1 | NE | 3.2 × 105 | 1.4 × 10−3 | 4.4 | 4.5 | NE | NPA | NPA | NPA | NPA | NPA |
| (E66S) dimer | 1.6 × 105 | 1.9 × 10−2 | 120 | 2.7 | NE | 2.1 × 105 | 1.6 × 10−2 | 76 | 0.7 | NE | NOI | NOI | NOI | NOI | NOI |
Affinities were calculated from rates of association (ka) and dissociation (kd) where KD = kd/ka and/or by steady-state affinity analysis. χ2 values are a measurement of the quality of the data fitting as described under Material and methods. Shown here are data from one experiment, representative of two independent experiments undertaken on different GAG surfaces. For the averaged affinities, see Supplementary data, Table SI.
aThese values are unlikely to be accurate, and are possibly due to rebinding effects, as discussed in the main text.
bPF: values are excluded due to poor fit of the kinetic data.
cNPA: no possible analysis due to insufficient signal.
dNE: equilibrium was not reached so steady-state affinity analysis was not possible.
eNOI: no observable interaction.
The data also illustrate the complexities of chemokine–GAG interactions, which involve biologically relevant chemokine oligomerization in solution and on GAGs, where the oligomers are not necessarily homogeneous species. This is because chemokine oligomers need to dissociate into monomers to bind receptor, and thus are in dynamic equilibrium. Furthermore, some chemokines form different types of oligomers depending on the nature of the GAG (e.g., dimers and tetramers, Salanga et al. 2014). As a consequence of the inherent heterogeneity, fitting the sensorgram data is often challenging because both association and dissociation rates often show non-single exponential behavior and concentration dependencies. The complex nature of the sensorgrams for CXCL4 binding to heparin and HS surfaces provide good examples of the commonly observed deviation from “ideal” behavior for SPR analysis. At the lowest concentrations, the sensorgrams are characterized by a slower on-rate than at higher concentrations, which may be due to differences in the oligomerization state of the chemokine (e.g., with monomer or dimer dominating at low concentrations and dimer or tetramer at higher concentrations). The dissociation phase also shows non-ideal behavior; at the lowest concentrations, the sensorgrams are characterized by a very slow dissociation rate, which may reflect direct interactions of CXCL4 with GAG, while at higher concentrations (>5 nM), an initial phase of rapid dissociation is observed before a secondary slower rate dominates, which may be due to dissociation of chemokine subunits from CXCL4 oligomers (e.g., tetramers) (Supplementary data, Figure S1A). Alternatively, the slow dissociation rates could be a consequence of rebinding effects, where at lower analyte concentrations, dissociated chemokine may rebind to unoccupied binding sites on the GAG chain. This effect may partly explain the extremely tight affinity (0.03 nM) calculated for binding of CXCL4 to heparin, which must be an overestimate, compared with the more reasonable affinity estimate from steady-state analysis (2.4 nM). Nevertheless, despite these complexities, the rank order of “apparent affinities” provide results that are consistent with available data in the literature (summarized in Discussion).
Despite the challenges with accurately fitting the data, the data reveal that differences in the affinities of the chemokines for heparin and HS are primarily due to differences in the rates of dissociation. For example, CCL5 and CXCL4 have much slower dissociation rates from heparin and HS, compared with CXCL12 (Table I), which is also supported by visual analysis of the sensorgrams (Figure 2, Supplementary data, Figures S1 and S2). These slower dissociation rates are likely due to the ability of CCL5 and CXCL4 to form polymers and tetramers, respectively, whereas CXCL12 forms dimers. An intermediate dissociation rate was observed for CXCL11 (Table I), as supported by visual sensorgram analysis (Supplementary data, Figure S1D); however, due to its relatively fast rate of association, an overall high affinity was calculated. CXCL8, the chemokine with the weakest affinity, shows a very rapid dissociation rate (Supplementary data, Figure S1C).
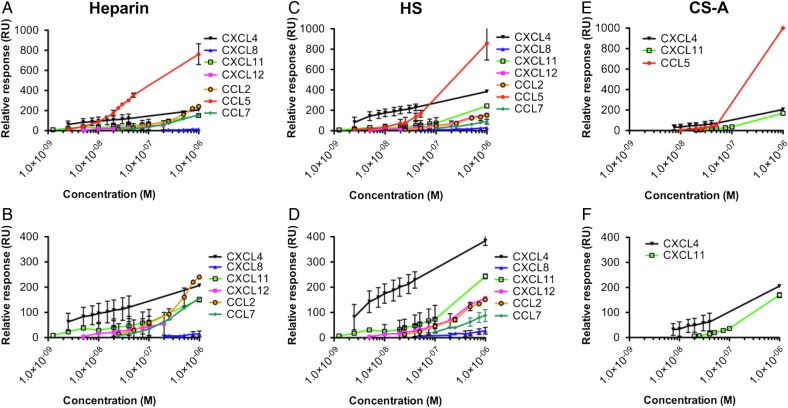
The maximum signal produced for each chemokine at varying concentrations (low nanomolar to micromolar) was also compared to determine the effect of their GAG-binding affinity and propensity to oligomerize on their overall accumulation on GAGs (Figure 3). Since most chemokines have similar molecular weights, SPR response units (RUs) serve as a good quantitative metric of surface accumulation. At low concentration (<10 nM), CXCL4 exhibited the highest signal, likely due to its exceptionally high affinity for GAGs relative to the other chemokines examined. However, >10 nM chemokine on heparin and at 1000 nM on HS and CS-A, CCL5 showed the highest level of accumulation (Figure 3A, C and E, respectively), presumably due to its high GAG-binding affinity coupled with its propensity to form polymers (Wang et al. 2011). To distinguish between the maximum signals produced by the other chemokines, similar data are plotted in the absence of CCL5 (Figure 3B, D and F). For chemokine concentrations in the range of 10–100 nM, CXCL4 demonstrates the next highest level of accumulation followed by CXCL11, whereas CXCL12 required higher concentrations (100–1000 nM) to reach comparable RU levels on heparin and HS, as did CCL2 and CCL7 in our previous study (Salanga et al. 2014). Interestingly, at 1000 nM, the highest concentration tested, CCL2 produced a greater signal than CXCL11 and was comparable with CXCL4 at the same concentration on heparin; this may be because CCL2 forms tetramers on heparin and dimers on HS (Salanga et al. 2014). In contrast, CXCL8 consistently produced the lowest signal on heparin and HS, consistent with its lower affinity interaction with GAGs.
Fig. 3.
Chemokine accumulation on Heparin, HS or CS-A. Heparin (A and B), HS (C and D) or CS-A (E and F) was immobilized onto a BIAcore C1 chip and CXCL4, CXCL8, CXCL11, CXCL12, CCL2, CCL5 and CCL7 were passed over at a range of concentrations and the interaction monitored. The maximum signal (RU) reached during injection of each chemokine was plotted against concentration for direct comparison between different chemokines (A–F). Given the high signal produced by CCL5 the other chemokine signal responses are plotted with (A, C and E) and without (B, D and F) CCL5 data included. Data plotted are the mean (±SE) of two experiments undertaken on the same chip surface. This figure is available in black and white in print and in color at Glycobiology online.
Chemokine oligomerization enhances the affinity of some chemokines for GAGs
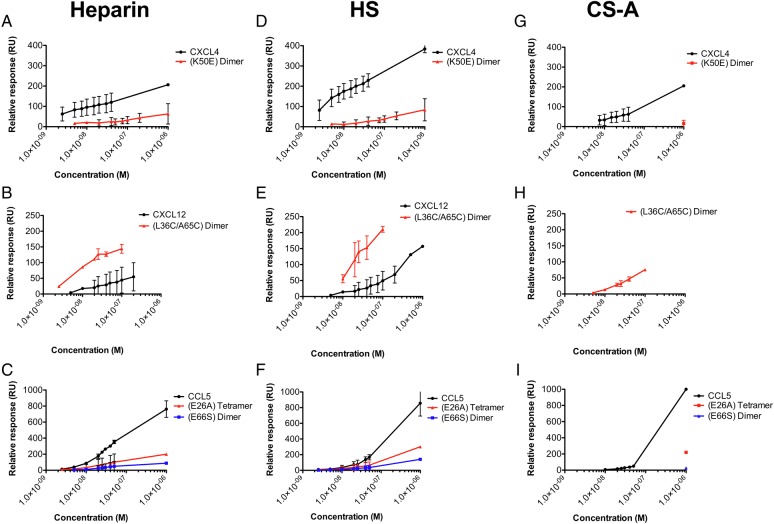
Having established that chemokines exhibit a wide range of affinities for the GAGs under study, we next set out to determine the role of chemokine oligomerization on their binding affinities and ability to accumulate on GAGs using three oligomerization-deficient mutants: the CCL5(E66S) dimer, the CCL5(E26A) tetramer (Czaplewski et al. 1999) and the CXCL4(K50E) dimer (Rauova et al. 2005). As shown in Figure 2 and Table I, the mutants showed a significant reduction in affinity for heparin, HS and CS-A compared with their WT counterparts (see Supplementary data, Figure S2, for complete datasets). Compared with WT CXCL4, the affinity of the CXCL4(K50E) dimer for heparin was reduced by 21- to 77-fold (determined from steady-state and kinetic analysis, respectively), 5- to 240-fold for HS, and for CS-A the affinity dropped from 9.3 nM to nearly undetectable binding. The CCL5(E66S) dimer also showed a significantly reduced affinity compared with WT CCL5: 32-fold for heparin, 17-fold for HS, and for CS-A there was almost no detectable interaction whereas the affinity of WT CCL5 was 51 nM. For both the CXCL4 and CCL5 mutants, the reduced affinities for heparin and HS are largely due to significantly increased rates of dissociation compared with WT (Supplementary data, Figure S2D), in line with the slow dissociation of the WT chemokines being responsible for the exceptionally high affinities for HS and heparin. Interestingly, tetrameric CCL5(E26A) showed a reduction in overall chemokine accumulation for heparin and HS similar to dimeric CCL5(E66S) (Figure 4C and F, respectively), but the apparent affinity of CCL5(E26A) for both HS and heparin was similar to WT (∼4 nM) (Table I). Nevertheless, it produced insufficient signal to enable analysis of its interaction with CS-A (Supplementary data, Figure S2B). All oligomerization mutants also displayed a reduction in the maximum signal compared with their WT counterparts at all concentrations tested (Figure 4), confirming that oligomerization contributes to both the affinity and level of accumulation of chemokines on GAGs.
Fig. 4.
Chemokine and mutant accumulation on Heparin, HS or CS-A. Heparin (A–C), HS (D–F) or CS-A (G–I) were immobilized onto a BIAcore C1 chip. Chemokines and associated oligomeric mutants, WT CXCL4 tetramer and CXCL4(K50E) dimer (40 nM) (A, D and G), WT CXCL12 and CXCL12(L36C/A65C) disulfide-locked dimer (200 nM) (B, E and H) or WT CCL5 polymer, CCL5(E26A) tetramer and CCL5(E66S) dimer (50 nM) (C, F and I) were characterized by SPR analysis. The maximum signal (RU) reached during injection of each chemokine or mutant was plotted against chemokine concentration for comparison of accumulation. Data plotted are the mean (±SE) of two experiments undertaken on the same chip surface. This figure is available in black and white in print and in color at Glycobiology online.
In a reverse approach to analyze the role of oligomerization, we investigated a disulfide-locked dimer of CXCL12, CXCL12(L36C/A65C) (Veldkamp et al. 2008); this mutant enables one to quantify the contribution of the dimer alone to GAG affinity in contrast to WT CXCL12, which exists as an equilibrium mixture of monomer and dimer (Veldkamp et al. 2005). When compared with WT CXCL12 by steady-state affinity analysis, this mutant resulted in a 6- and 5-fold increase in affinity for heparin and HS, respectively (Figure 2B, Table I), in line with previously reported results (Ziarek et al. 2013), as well as an increase in overall accumulation compared with WT CXCL12 (Figure 4B and E). Visual inspection of the data clearly shows slow dissociation of the locked dimer from heparin and HS compared with WT CXCL12, consistent with the high-affinity GAG interaction. This result is due to the non-dissociating nature of the CXCL12(L36C/A65C) locked dimer relative to WT CXCL12. Notably, while WT CXCL12 did not show any observable interaction with CS-A, the CXCL12 locked dimer exhibited an appreciable interaction (KD = 59 nM) (Table I, Figures 2B and 4H, Supplementary data, Figure S1), again due to the covalent nature of the dimer.
Overall, the reduced GAG-binding affinity and ability to accumulate on GAGs by oligomerization-deficient mutants, and the apparent enhancement of affinity and accumulation with the locked CXCL12 dimer, demonstrate the importance of chemokine oligomerization for GAG interactions. Additionally, the only chemokines capable of binding to CS-A are the oligomerizing chemokines with high affinity for HS and heparin.
Surface GAG density may play a role in regulating chemokine–HS interactions
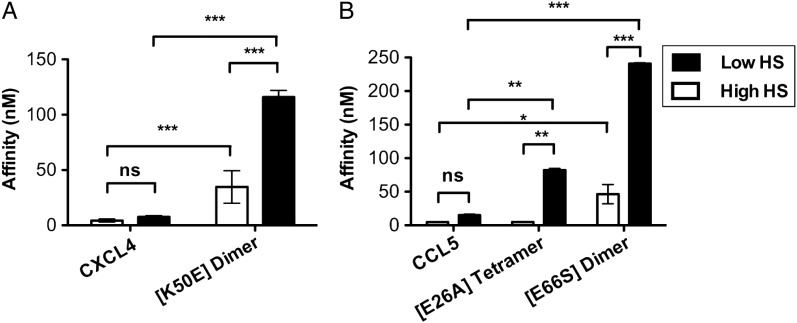
In a previous study, we demonstrated that the density of HS immobilized on SPR chips can markedly affect the GAG-binding affinity of the monomeric chemokine, CCL7, but that oligomerization renders the homologous chemokine, CCL2, less sensitive to HS density (Salanga et al. 2014). To investigate the generality of this phenomenon, we examined our set of WT and oligomerization-deficient chemokines on low- and high-density HS surfaces (Figure 5 and Supplementary data, Figure S3). Similar to CCL2, the chemokines CXCL4 and CCL5 showed no significant difference in affinity on SPR chips coated with low-density HS compared with high-density HS (Figure 5). In contrast, the GAG-binding affinities of the CCL5 and CXCL4 oligomerization-deficient mutants were significantly lower on the low-density HS surface compared with the high-density surface (Figure 5). The CXCL4(K50E) mutant has significantly reduced affinity compared with WT on both surface densities of HS, but the difference is even more exaggerated on the low HS surface (Figure 5A). As described above, the CCL5(E26A) tetramer shows no difference in affinity compared with WT CCL5 on the high-density HS surface; however, its affinity was significantly reduced on the low-density HS surface (Figure 5B and Supplementary data, Figure S3). As expected, the CCL5(E66S) dimer exhibited the weakest binding to the low HS surface (KD = 240 nM). The greater sensitivity of the oligomerization-deficient chemokines to HS density compared with the WT chemokines suggests that oligomerization facilitates cross-linking of HS chains, as proposed previously (Salanga et al. 2014).
Fig. 5.
Chemokine oligomerization mediates interaction with low-density HS. Chemokines and associated oligomeric mutants (CXCL4 and mutant (A) and CCL5 and mutants (B)) were passed over immobilized HS (high or low density) and GAG interactions were compared by SPR analysis. Affinities (nM) for chemokines and mutants are plotted as mean values (±SE) (n = 2, experiments from two independently generated surfaces). Statistical significance was determined using repeated measures ANOVA analysis with a Bonferroni post hoc test where ns denotes not significant, *P < 0.05, **P < 0.01, ***P < 0.001.
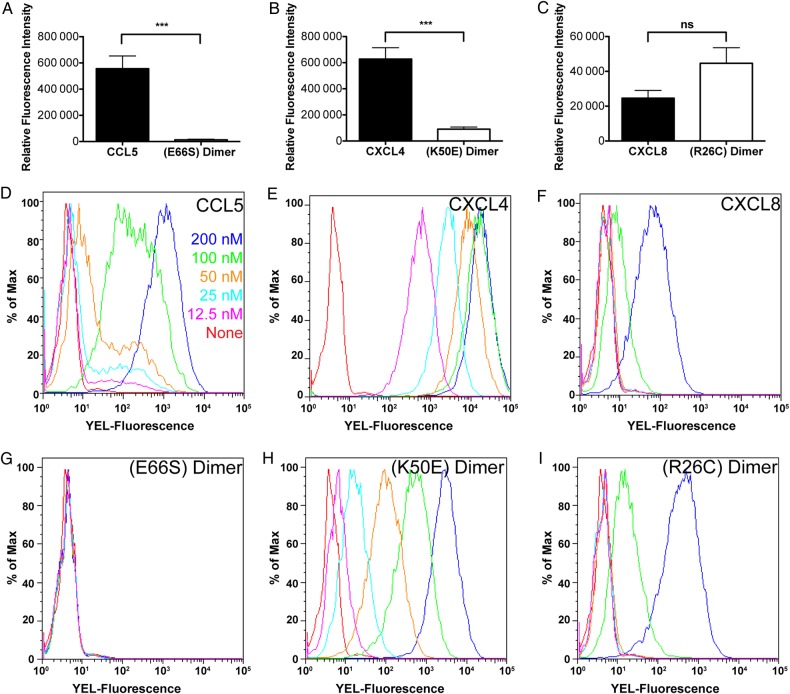
Accumulation of CCL5 and CXCL4 on cells is dependent on their ability to oligomerize
Consistent with the effects of the oligomerization mutants on GAG-binding as observed by SPR, the dimeric variants, CCL5(E66S) and CXCL4(K50E), showed significantly reduced accumulation on human umbilical vein endothelial cells (HUVECs) compared with the WT chemokines (Figure 6A and B, respectively). Chemokine accumulation was also investigated with CHO-K1 cells by flow cytometry (Figure 6D–I), and demonstrated a total and a 6- to 168-fold loss of signal across the concentrations tested for the CCL5 and CXCL4 dimer variants compared with their WT counterparts, respectively (Figure 6D, E, G and H). The disulfide-locked CXCL8 dimer, CXCL8(R26C) (Clark-Lewis et al. 1995) showed slightly elevated levels of accumulation on HUVECs relative to WT (Figure 6C), but the difference was not statistically significant. However, accumulation of the locked CXCL8 dimer on CHO-K1 cells showed enhanced accumulation at the highest concentration tested (63-fold) compared with WT CXCL8 (Figure 6F and I). By comparison, CCL3, which exhibited no binding to GAGs by SPR, also showed significantly reduced accumulation on HUVECs compared with the other chemokines tested, and no binding to CHO-K1 cells (data not shown). Notably, the levels of accumulation of CXCL8, CCL5, CXCL4 and CCL3 correlate with the affinity hierarchy observed by SPR where CXCL4 > CCL5 > CXCL8, and no detectable interaction is observed with CCL3. Additionally, PGS-745 cells, which lack xylosyl-transferase and therefore do not add GAG chains to the proteoglycan core protein, showed a significant reduction or total loss in accumulation for the WT chemokines and no binding of the oligomerization-deficient mutants (data not shown), confirming the GAG-dependence of the interactions.
Fig. 6.
Accumulation of chemokines on cell surfaces is mediated by oligomerization and GAG binding. Biotinylated WT and mutant chemokine (CCL5 and CCL5(E66S) dimer (A, 500 nM), CXCL4 and CXCL4(K50E) dimer (B, 500 nM) and CXCL8 and CXCL8(R26C) dimer (C, 125 nM)) were incubated on HUVEC monolayers and detected by streptavidin-800CW using the LI-COR Odyssey imaging system. (D–I) Varying concentrations of biotinylated chemokine (CCL5 (D) and CCL5(E66S) dimer (G); CXCL4 (E) and CXCL4 (K50E) dimer (H); CXCL8 (F) and CXCL8(R26C) dimer (I)) were incubated on CHO-K1 cells and detected by streptavidin-PE using flow cytometry. Endothelial cell-binding data are plotted as the mean value (±SE) of three independent experiments where ns denotes not significant, *P < 0.05, **P < 0.01, ***P < 0.001, as determined using Student's t-test. CHO-K1 binding experiments were performed in triplicate with a representative plot shown. This figure is available in black and white in print and in color at Glycobiology online.
2-O-Sulfation contributes to the chemokine–heparin interaction
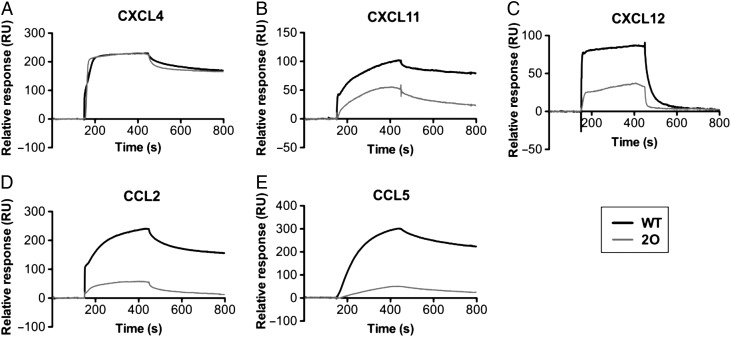
Previous studies have demonstrated the importance of O-sulfation of heparin for mediating chemokine interactions (Stringer and Gallagher 1997; Kuschert et al. 1999; de Paz et al. 2007); however, the contributions of specific sulfation motifs are not well-understood. Therefore, to better understand the role of heparin sulfation in mediating interactions with the current set of chemokines and oligomerization mutants, we conducted SPR studies with 2-O-desulfated heparin in comparison with WT heparin (Figure 7 and 8 and Supplementary data, Figure S4). The use of partially sulfated heparin significantly reduced the affinity of most of the WT chemokines tested (CXCL11, CXCL12 and CCL5) with the exception of CXCL4 (Figure 7A). While one must be cautious in comparing the SPR curves produced on different surfaces due to inherent differences in immobilization and source of GAG, the lack of an effect for CXCL4 contrasts with the other chemokines. Overall, these results demonstrate that the positions of sulfate groups on heparin differentially contribute to chemokine–GAG affinity and accumulation, suggesting specificity in these interactions.
Fig. 7.
2-O-desulfation of heparin differentially affects chemokine binding. Heparin and 2-O-desulfated heparin were immobilized onto separate flow cells on a BIAcore C1 chip and various chemokines were passed over to compare the effects of heparin 2-O-sulfation on GAG affinity. Shown are SPR sensorgrams of CXCL4 (40 nM) (A), CXCL11 (100 nM) (B), CXCL12 (200 nM) (C), CCL2 (1000 nM) (D) and CCL5 (50 nM) (E). The resulting signal reveals a reduction in chemokine accumulation and affinity of most chemokines (see also Supplementary data, Figure S4), with the exception of CXCL4 for 2-O-desulfated heparin compared with WT heparin.
Fig. 8.
Inhibition of chemokine oligomerization affects interactions with 2-O-desulfated heparin. 2-O-desulfated heparin was immobilized onto a BIAcore C1 chip and chemokines passed over. Shown are sensorgrams of WT CXCL4 tetramer and CXCL4(K50E) dimer (40 nM) (A), WT CCL5 polymer and CCL5(E66S) dimer (50 nM) (B) and WT CCL2 dimer and CCL2(P8A) monomer (1000 nM) (C).
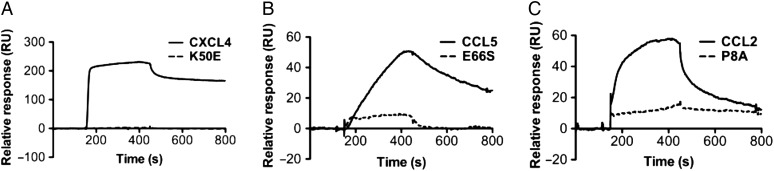
Given the striking observation that 2-O-desulfation of heparin had little obvious effect on its interaction with WT CXCL4, we investigated whether this was also the case with the CXCL4(K50E) dimer. The results were dramatically different as CXCL4(K50E) showed no detectable binding to 2-O-desulfated heparin (Figure 8A). Similarly, the oligomerization-deficient mutants CCL5(E66S) and CCL2(P8A) showed a significant loss of affinity and ability to accumulate on 2-O-desulfated heparin compared with the WT chemokines (Figure 8B and C, respectively). These results illustrate the contributions of heparin fine structure and chemokine oligomerization in promoting chemokine–GAG interactions.
Discussion
Many studies have been conducted to investigate the interaction of chemokines and GAGs, generally focusing on individual chemokines (Hamel et al. 2009). In this study, we sought to broadly characterize the interactions of a large group of chemokines and oligomerization mutants with GAGs, using SPR under similar or identical experimental conditions. The apparent affinities of CXCL12 and CXCL4 for heparin determined in the present study by SPR are in close agreement with previously reported results (Amara et al. 1999; Dubrac et al. 2010; Ziarek et al. 2013). In the case of CCL5, we determined the affinity for heparin to be 3.7 nM, which is lower than a previous SPR study (32 nM) (Martin et al. 2001) and may be due to differences in assay conditions such as the source of GAG, SPR chip preparations, and/or the challenges in accurately fitting the data. CXCL8 has been reported to have a micromolar affinity for heparin as determined by cell and immobilized heparin binding competition experiments (Kuschert et al. 1999), in line with the relatively weak affinities reported herein for heparin and HS. In the case of CXCL11, heparin-affinity chromatography suggests a tight interaction as 1 M NaCl was required to elute CXCL11 from heparin beads (Severin et al. 2010), consistent with the tight (2–6 nM) affinity reported in this study. By comparison, ∼0.6 M NaCl was required to elute CCL2, CCL7 and CXCL8 from heparin beads (Ali et al. 2005; Handel et al. 2005), suggesting that they all have weaker affinities than CXCL11, which is also consistent with the present study. Our data differ from previous observations in the low signal detected for CCL3 binding to SPR and cell surfaces in this study, compared with previous reports where binding to cells or heparin beads was observed (Hoogewerf et al. 1997; Kuschert et al. 1999). However, in the present and all of these previous studies, compared with other chemokines, CCL3 has one of the lowest affinities for heparin, likely due to its acidic nature (Proudfoot et al. 2003). Being on the low end of the affinity scale, assay conditions could easily shift results between observable binding and no detectable binding, accounting for such discrepancies.
The vast majority of previously reported studies have utilized heparin for studying GAG interactions, and a surprising result of the present study is the fact that binding affinities of chemokines for heparin and HS are generally similar (with some exceptions noted in the text). This similarity is despite the fact that the overall sulfation content differs by a factor of ∼3.3 (see below), which suggests that the presence of clustered domains of sulfate groups in HS are as efficient in binding chemokines as the uniformly high sulfation structure of heparin.
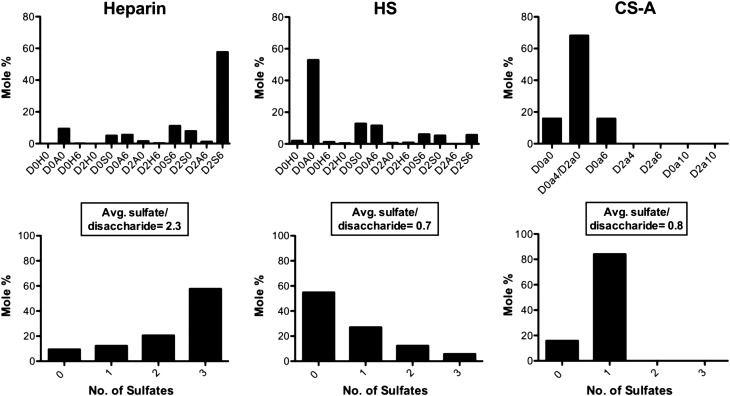
Significant differences were, however, observed between heparin/HS and CS-A. Of all of the chemokines tested, only CXCL4, CXCL11 and CCL5 bind to CS-A, and interestingly, CS has been shown to be an integral mediator of CCL5 and CXCL4 mediated monocyte adhesion on the endothelium (Baltus et al. 2005). The exclusive interaction of these chemokines with CS-A could be due to the differing disaccharide composition of CS GAGs compared with heparin and HS, and/or the fact that CS-A is reported to have on average only one sulfation site per disaccharide unit (Mizumoto et al. 2013), compared with four potential sites in heparin and HS (Xu and Esko 2014). To gain insight into this issue, we conducted quantitative glycan reductive isotope labeling with liquid chromatography and mass spectrometry (GRIL-LC/MS) (Lawrence, Olson, et al. 2008) disaccharide analysis of the heparin, HS and CS-A samples used for SPR. This analysis confirmed the overall marked differences in the GAG fine structure and sulfate distribution/composition among these GAGs. Heparin exhibited the greatest degree of sulfation (average 2.3 sulfates/disaccharide) compared with HS and CS-A, which showed average sulfate levels of 0.7 and 0.8, respectively (Figure 9). While the average sulfation per disaccharide is similar between HS and CS-A, there is a notable difference in the distribution and type of sulfation present between the GAGs (Figure 9). CS-A has a fairly uniform distribution of monosulfated disaccharides (∼84 mol%), whereas HS has a large proportion of unsulfated regions (∼55 mol%) intermixed with concentrated regions of sulfation varying from one to three sulfates per disaccharide. Again, this suggests that differences in sulfate distribution may be more important than the actual average sulfate content, in determining the selectivity and specificity observed for chemokine interactions with these GAGs (although in the case of CS-A, we cannot rule out the contribution of differences in disaccharide composition). Unlike the other chemokines studied, CXCL4 and CCL5 are also known to form stable oligomeric structures larger than dimers (tetramers and polymers, respectively) even in the absence of GAGs. As shown below, the oligomeric properties of CXCL4 and CCL5 are critical for binding to CS-A. Oligomerization may enable these chemokines to bridge multiple sulfation sites along a GAG chain, contributing to their relatively unique ability to bind CS-A as well as their rank as the highest affinity chemokines for HS and heparin. CS is most prevalent in cartilage, but can also be found with HS on syndecan-1 and syndecan-4 proteoglycans (PGs) on epithelial cells (Deepa et al. 2004). As our data suggest that chemokines cross-link GAG chains, it is possible that CXCL4 and CCL5 could simultaneously interact with CS and HS such that these GAGs cooperate in mediating chemokine presentation on PGs.
Fig. 9.
GRIL-LC/MS analysis of heparin, HS and CS-A disaccharides. (A) The disaccharides identified from digests of Heparin (left), HS (center) and CS-A (right) are presented as mole percent of the total digest. Disaccharide abbreviations are presented in accordance with the disaccharide structure code nomenclature as described previously (Lawrence, Lu, et al. 2008). (B) From the disaccharides identified for Heparin (left), HS (center) and CS-A (right), the number of sulfates was quantified as a mole percent of the total digest with the average number of sulfates per disaccharide indicated for each.
A key aim of this study was to broadly explore the role of chemokine oligomerization in binding to GAGs. The SPR data suggest that the ability of most chemokines to oligomerize contributes significantly to GAG-binding affinity, and in some cases has a dramatic effect. Mutants with impaired ability to oligomerize (CXCL4(K50E), CCL5(E66S) and as previously shown, CCL2(P8A) (Salanga et al. 2014)) show reduced binding affinity for both heparin and HS, with the effects being more pronounced for chemokines that form larger oligomers (CXCL4, CCL5) and for interactions with HS. In the case of CS-A, oligomerization-deficient mutants of CCL5 and CXCL4 showed little to no binding, while a covalently locked CXCL12 dimer, enabled this otherwise weakly dimerizing chemokine not only to bind heparin and HS with higher affinity than WT CXCL12 (Ziarek et al. 2013) but also to bind CS-A. Results of the SPR studies strongly correlated with the presentation of chemokines on cell surfaces where oligomerization-deficient mutants of CXCL4 and CCL5 showed significantly diminished or undetectable binding compared with their WT counterparts. In contrast, the disulfide-locked dimeric variant of CXCL8 exhibited enhanced binding compared with WT CXCL8. Importantly, the levels of chemokine immobilized on the cell surface reflect their affinity ranking determined by SPR with CXCL4 > CCL5 > CXCL8, and no detectable interaction by CCL3. The SPR data showing that the CXCL4(K50E) dimer has a greatly diminished affinity for heparin is also consistent with a natural variant of CXCL4 (CXCL4L1) that has a significantly reduced affinity for heparin and HS, and no ability to bind CS (Dubrac et al. 2010). CXCL4L1 has three mutations that render it unable to form tetramers (Sarabi et al. 2011), and according to our data would contribute to the reduced GAG-binding affinity. As a consequence, it has much greater diffusibility in vivo and functions as a paracrine regulator over greater distances than WT CXCL4. Additionally, a recent study of CCL5 showed that WT CCL5 forms filamentous structures on vascular endothelial cells while mutant dimers and tetramers could not form such structures or were not detectable on the cell surface at all (Oynebraten et al. 2015), supporting the biological relevance of oligomerization.
In this study, as well as a previous study from our lab (Salanga et al. 2014), the effect of HS density on the binding affinity of chemokines and oligomerization-deficient variants was investigated. Little difference was observed in the affinity of CXCL4 and CCL5 for low- or high-density HS surfaces. However, the CXCL4(K50E) dimer, the CCL5(E66S) dimer and the CCL5(E26A) tetramer all had significantly reduced affinities when measured on the low- versus high-density HS SPR surface. These observations suggest that oligomerization of CXCL4 and CCL5 may enable these chemokines to tolerate some level of variable GAG chain density and still be presented on cell surfaces. On the other hand, the phenomena of density dependence could play a regulatory role in controlling presentation of chemokines and recruitment of leukocytes as PGs are known to be shed in different physiological and pathological situations such as cancer (Afratis et al. 2012). The fact that oligomerization-deficient chemokines are sensitive to GAG density also leads to the hypothesis that chemokines can cross-link GAG chains, potentially providing a mechanism to modulate the thickness and permeability of the glycocalyx (Reitsma et al. 2007), for example, in platelet interactions with vessel walls (Reitsma et al. 2011). This cross-linking effect of chemokines may also play a role in the ability of chemokines (e.g., CXCL12 and CCL5) to signal through cell surface PGs following interactions with GAG side chains (Charnaux, Brule, Hamon, et al. 2005).
The spectrum of GAG-binding affinities observed with the subset of chemokines involved in the present study strongly suggests that GAG-binding properties of individual chemokines may be important in fine-tuning their function in different biological processes. The fact that many chemokines bind the same receptor and that many receptors respond to multiple chemokines has given rise to the concept of redundancy as a mechanism to make the immune system robust (Baggiolini 1998). While this is likely true to some extent, the results of this study support the notion that chemokine ligands of the same receptor may not always be redundant, and that their GAG-binding properties may confer specificity even if they have similar effects on receptor signaling. CCL21 and CCL19 represent a classic case of a non-redundant chemokine pair that are both ligands of CCR7 (Sullivan et al. 1999). They not only show differences in signaling (Kohout et al. 2004; Zidar et al. 2009), but CCL19 has also been described as a soluble chemokine whereas CCL21 binds tightly to GAGs on the cell surface until proteolytically cleaved from its C-terminal domain (Hiroseet al. 2002; de Paz et al. 2007; Schumann et al. 2010). Thus soluble CCL19 and immobilized CCL21 may collaborate to promote haptokinesis and chemotaxis and enable directional migration in lymphatic organs (Schumann et al. 2010). Similarly, CXCL4 and CXCL4L1 are ligands of CXCR3 (Dubrac et al. 2010; Struyf et al. 2011); however, CXCL4L1 displays a much weaker interaction with GAGs than CXCL4, resulting in greater bioavailability and ability to inhibit angiogenesis and tumor growth (Struyf et al. 2004, 2007). The gamma isoform of CXCL12 (CXCL12γ) shows significantly higher GAG-binding affinity than CXCL12α, with the consequence of reduced binding and signaling through CXCR4, presumably because of competition between binding to GAGs versus receptor (Laguri et al. 2007). Finally, CCL3 has very low affinity for GAGs, while CCL5 belongs to the subset of highest affinity GAG binders, and both are ligands of CCR1 and CCR5 (Czaplewski et al. 1999). While challenging, an important goal will be to understand the physiological context in which different chemokine ligands of the same receptor are operative. Moreover, although a hierarchy of GAG-binding properties presented in this study correlate well with presentation of chemokines on cells, the experimental conditions used in these studies may not reflect physiological contexts where some apparently weak binders (e.g., CXCL8 and CCL3), bind with high affinity. Thus another challenge will be to identify the circumstances in which chemokines bind to GAGs with high affinity versus when they act as soluble, non-GAG-binding chemoattractants (Weber et al. 1999).
The present study also demonstrates with multiple examples that oligomerization is critical for the GAG-binding affinity of many chemokines, and in some cases, confers GAG specificity. While high-affinity GAG binding is clearly important for chemokine presentation on cells, it is also possible that oligomerization contributes to cell signaling through chemokine receptors (Drury et al. 2011) and through PGs. Although full agonist signaling through receptors involves monomeric chemokines (Allen et al. 2007), Veldkamp et al. have shown how the dimeric form of the CXC chemokine CXCL12 acts as a biased agonist of its receptor CXCR4 by activating G proteins but not β-arrestin-2 recruitment with a concomitant loss of CXCR4-mediated cell migration (Veldkamp et al. 2008; Drury et al. 2011). Similarly, dimeric CXCL8 differentially activates CXCR1 and CXCR2 compared with the monomeric form (Nasser et al. 2009). On the other hand, CC chemokine dimers do not even bind their receptors (Jin et al. 2007; Tan et al. 2012). Moreover, GAGs such as heparin have been shown to inhibit chemokine binding to receptors (Kuschert et al. 1999) and while part of the mechanism seems to be due to the partial overlap of receptor- and GAG-binding sites on some chemokines (Proudfoot et al. 2001; Lau et al. 2004), another component may be stabilization of non-interacting chemokine oligomers.
Signaling of CXCL12 and CCL5 directly through PGs has also been reported (Chang et al. 2002; Roscic-Mrkic et al. 2003; Charnaux, Brule, Hamon, et al. 2005; Maillard et al. 2014). For example, activation of mitogen-activated protein kinase pathways (MAPKs) by CCL5 is dependent on GAGs and independent of receptor, and leads to enhanced infectivity of HIV (Roscic-Mrkic et al. 2003). Given the results presented herein, it is not surprising that the effect was shown to be dependent on the ability of CCL5 to oligomerize (Trkola et al. 1999). CCL5 and CXCL12 signaling through PGs has also been shown to accelerate shedding of PGs from the surface of cells (Charnaux, Brule, Chaigneau, et al. 2005; Brule et al. 2006). In the case of CXCL12, the process is CXCR4 independent and PG dependent (Brule et al. 2006). Thus our expectation is that these processes require chemokine oligomerization. Our current and prior data also suggest that chemokine oligomers cross-link GAG chains, and thus it is possible that such cross-linking is relevant to PG signaling. Since shedding of the glycocalyx can have profound effects on many processes such as leukocyte adhesion (Kolarova et al. 2014), it stands to reason that in addition to localization for directional cues, chemokine–GAG interactions and thus oligomerization, may have other effects such as preparing endothelial cells for transmigration. A final key question is whether oligomerization enables chemokines to simultaneously bridge GAGs and receptors or whether chemokine oligomerization on GAGs simply provides a concentrated source of chemoattractant to establish gradients. Further investigation into these questions is currently underway.
Materials and methods
Chemokine expression and purification
CXCL11 was kindly provided by Dr. Amanda Proudfoot. WT and biotinylated chemokines CCL2, CCL3, CCL7, CXCL8, CXCL12 and associated mutants were expressed and purified as described previously (Veldkamp et al. 2008; O'Hayre et al. 2009; Allen et al. 2011; Takekoshi et al. 2012; Salanga et al. 2014). All other chemokines were prepared as follows. CXCL4 was subcloned into a His-Ub fusion construct (pHUE 3D3) (Allen et al. 2011) and CCL5 into a (His)8 tag construct, with an enterokinase recognition site (O'Hayre et al. 2009); chemokine mutants were generated using QuikChange site-directed mutagenesis (Stratagene). CXCL4, CCL5 and associated mutants were insolubly expressed in BL21(DE3)pLysS Escherichia coli cells. Cultures were grown at 37°C in Luria Broth (with 30 µg/mL kanamycin for pHUE 3D3 constructs or 100 µg/mL carbenicillin for (His)8 constructs) and induced with 0.5 mM isopropyl-d-1-thiogalactopyranoside once OD600nm reached 0.6. Cell pellets were harvested and solubilized in denaturing buffer (6 M guanidine–HCl, 50 mM NaCl, 50 mM Tris, pH 8.0) and tagged protein was extracted using Ni-nitrilotriacetic acid (NTA) affinity chromatography. Protein was then refolded in 0.5 M arginine, 1 mM GSSG, 1 mM EDTA, 50 mM Tris, pH 8.0 (for CXCL4 and mutant) or at pH 7.5 (for CCL5 and mutants) and refolding confirmed by reversed-phase high-pressure liquid chromatography (HPLC). Chemokine fusion was then cleaved with ubiquitinase (for pHUE 3D3 constructs) or enterokinase (New England Biolabs; for (His)8 constructs) at a 1:100 molar ratio (chemokine:enzyme) for ∼18–36 h, as monitored by HPLC. The resulting products were then passed over Ni-NTA beads to remove unwanted cleavage products followed by a final purification with HPLC equipped with a C18 semi-prep column. Protein purity and size was confirmed by electrospray-ionization mass spectrometry and SDS-PAGE analysis.
Glycosaminoglycan biotinylation
HS from bovine kidney (Sigma), CS-A (Sigma), 2-O-desulfated heparin and 6-O-desulfated heparin (Neoparin Inc.) were biotinylated as described previously (Salanga et al. 2014) and biotinylated porcine intestinal heparin (average molecular mass = 15 kDa) was purchased (Calbiochem). Briefly, GAG was resuspended at 5 mg/mL in 100 mM MES, pH 5.0, followed by addition of 6.5 mM 1-ethyl-3-((3-dimethylaminopropyl) carbodiimide Hydrochloride) solution (EDC)) (GE Healthcare) and 1.25 mM EZ-Link Hydrazide-LC-Biotin (Pierce); the solution was then incubated with rotation for 18 h at room temperature. Excess biotin was removed with extensive dialysis into water.
Sulfation analysis of glycosaminoglycans
Glycan reductive isotope labeling with liquid chromatography and mass spectrometry analysis (GRIL-LC/MS) of lyase-derived disaccharides was used to determine the sulfate content and composition of heparin, HS and CS-A used in the SPR studies. All analyses were carried out by the UCSD Glycotechnology Core as described previously (Lawrence, Olson, et al. 2008).
Surface plasmon resonance
SPR was performed on a BIAcore 3000 instrument (GE Healthcare) using a C1 (no dextran) chip as described previously (Salanga et al. 2014). Briefly, the chip was equilibrated in running buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.05% Tween-20, pH 7.4), activated with 200 µL of a 1:1 mixture of NHS (0.2 M) and EDC (0.1 M) and immobilized with neutravidin (Invitrogen) (0.2 mg/mL in 20 mM sodium acetate, pH 6.0) until saturation was reached, followed by extensive washing with regeneration buffer (0.1 M glycine, 1 M NaCl, 0.1% Tween-20, pH 9.5). Biotinylated GAG was then passed over an individual flow cell (0.2 mg/mL) until saturation was reached (40 response units (RU) for heparin and 97 RU for CS-A). In the case of HS, a high-density (250 RU) and low-density (106 RU) surface was generated. Apparent affinity determinations of chemokine–GAG interactions were performed by passing varying concentrations of chemokines over the chip. Data were acquired with a flow rate of 40 µL/min to limit the effect of mass transfer (Amara et al. 1999; Dubrac et al. 2010; Tanino et al. 2010; Ziarek et al. 2013); increasing the flow rate had little effect on pilot chemokine–GAG interactions and associated residual plots (which can reveal mass transfer effects), suggesting that mass transfer was limited at this flow rate (Karlsson and Falt 1997). Washing of the surface between each cycle was performed using regeneration buffer. Signal specific to chemokine–GAG interactions was determined by subtracting the response signal from a blank flow cell (no immobilized GAG). Data were analyzed with the BIAevaluation software (GE Healthcare) using the 1:1 Langmuir interaction model and χ2 values and visual inspection were used to assess the fitting of the data, where a value of <10 was accepted as a good fit. Fitting with alternative models such as the bivalent model did not improve the fit of any of these data sets. In some cases, a “bulk correction” was included to account for the rapid phase of non-single exponential dissociation of some chemokines (related to their known ability to oligomerize on GAGs as described in the text). A few datasets have less than desirable fits as observed visually and reflected by high χ2 values. In these instances, additional qualitative interpretation of the SPR curves was used to validate the relative apparent affinities reported. For example, the fits for CXCL4-HS and CCL5-heparin are poor (χ2 = 31.8 and 32.0, respectively); however, the associated sensorgrams (Figures 1A and 2A, respectively) clearly show that these chemokines have slow dissociation rates, supporting the calculated higher apparent affinity, compared with the other chemokines examined. In datasets where a sufficient number of sensorgrams reached steady state, the maximal response signal was plotted against concentration and analyzed using the 1:1 steady-state affinity model in the BIAevaluation software (GE Healthcare), to provide alternative equilibrium estimates of the affinities. Full datasets showing sensorgrams with associated fits and equilibrium analyses (when appropriate) are provided in Supplementary data, Figures S1–S4.
Endothelial chemokine presentation assay
A clear-bottomed, black-walled 96-well polystyrene plate (Corning) was prepared as described previously (Salanga et al. 2014). Briefly, wells were coated with 100 µg/mL collagen (PureCol, Advanced Biomatrix) for 1 h at 37°C after which HUVECs (Lonza) were added to each well (20,000 cells/well) in EBM2 basal medium supplemented with the EGM2 bullet kit (Lonza; endothelial medium) and incubated for 18 h to confluence at 37°C, 5% CO2. Non-adherent cells were removed by washing with endothelial medium followed by two washes with PBS supplemented with 1 mM CaCl2 and 0.5 mM MgCl2 [complete PBS (cPBS)]. Biotinylated chemokine (500 nM) was then added to the endothelial cells and incubated at 37°C, 5% CO2 for 1 h. Unbound chemokine was removed by washing three times with cPBS, followed by fixation of cells with ice-cold 4% (w/v) paraformaldehyde in PBS for 20 min at room temperature. Cells were washed four times with PBS + 0.05% Tween-20, blocked with Odyssey blocking buffer for 90 min (LI-COR Biosciences), stained with streptavidin conjugated IRDye 800CW (LI-COR Biosciences) diluted 1:1000 in Odyssey blocking buffer (LI-COR Biosciences) and then incubated for 90 min at room temperature with gentle rocking. Finally, cells were washed four times with PBS + 0.05% Tween-20, followed by detection of biotinylated chemokine using an Odyssey imaging system (LI-COR Biosciences). Data are plotted as the mean of three independent experiments performed in at least duplicate. Statistical analysis was performed using a Student's t-test, where n.s. denotes not significant, *P < 0.05, **P < 0.01 and ***P < 0.001.
Chemokine CHO cell binding
CHO-K1 (with GAG) or PGS-745 (without GAG) cells (kind gifts of Dr. Jeffrey Esko, UCSD) were maintained in a Dulbecco's modified Eagle medium/F12 mixture (Gibco) containing 10% fetal bovine serum. For assays, cells were lifted with 1 mM EDTA in PBS, washed with PBS and then resuspended at 2.0 × 106 cells/mL in FACs buffer (PBS + 0.1% BSA) and placed on ice. Subsequently, 50 µL of cells was transferred to a v-bottom 96-well plate (100,000 cells/well) and incubated with varying concentrations of biotinylated chemokine or associated mutants (12.5–200 nM) prepared in FACs buffer. Chemokine was detected by incubation with phycoerythrin-conjugated streptavidin (1:200) (Invitrogen) and incubated for 45 min on ice in darkness. Cells were then washed three times with FACs buffer and analyzed by a Guava EasyCyte 8HT flow cytometer (EMD Millipore). Postacquisition analysis was performed with FlowJo (Tree Star, Inc.); shown are representative data of experiments performed in triplicate.
Supplementary data
Supplementary data are available at http://glycob.oxfordjournals.org/ online.
Conflict of interest statement
None declared.
Funding
This work was supported by National Institute of Health grants R01 AI37113 to T.M.H. and R01 AI058072 to B.V.
Abbreviations
GAG, glycosaminoglycan; CCR, CC-type chemokine receptor; CCL, CC-type chemokine ligand; CXCL, CXC-type chemokine ligand; HUVECs, human umbilical vein endothelial cells; HS, heparan sulfate; CS, chondroitin sulfate; SPR, surface plasmon resonance; PG, proteoglycan; ECM, extracellular matrix; GRIL-LC/MS, glycan reductive isotope labeling with liquid chromatography/mass spectrometry.
Supplementary Material
Acknowledgements
We thank Dr. Xu Wang for the coordinates of the CCL5 model and the UCSD Glycotechnology Core and Biswa Choudhury for the GRIL-LC/MS analysis.
References
- Afratis N, Gialeli C, Nikitovic D, Tsegenidis T, Karousou E, Theocharis AD, Pavao MS, Tzanakakis GN, Karamanos NK. 2012. Glycosaminoglycans: Key players in cancer cell biology and treatment. FEBS J. 279:1177–1197. [DOI] [PubMed] [Google Scholar]
- Ali S, O'Boyle G, Mellor P, Kirby JA. 2007. An apparent paradox: Chemokine receptor agonists can be used for anti-inflammatory therapy. Mol Immunol. 44:1477–1482. [DOI] [PubMed] [Google Scholar]
- Ali S, Robertson H, Wain JH, Isaacs JD, Malik G, Kirby JA. 2005. A non-glycosaminoglycan-binding variant of CC chemokine ligand 7 (monocyte chemoattractant protein-3) antagonizes chemokine-mediated inflammation. J Immunol. 175:1257–1266. [DOI] [PubMed] [Google Scholar]
- Allen SJ, Crown SE, Handel TM. 2007. Chemokine: Receptor structure, interactions, and antagonism. Annu Rev Immunol. 25:787–820. [DOI] [PubMed] [Google Scholar]
- Allen SJ, Hamel DJ, Handel TM. 2011. A rapid and efficient way to obtain modified chemokines for functional and biophysical studies. Cytokine. 55:168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara A, Lorthioir O, Valenzuela A, Magerus A, Thelen M, Montes M, Virelizier JL, Delepierre M, Baleux F, Lortat-Jacob H et al. 1999. Stromal cell-derived factor-1alpha associates with heparan sulfates through the first beta-strand of the chemokine. J Biol Chem. 274:23916–23925. [DOI] [PubMed] [Google Scholar]
- Baggiolini M. 1998. Chemokines and leukocyte traffic. Nature. 392:565–568. [DOI] [PubMed] [Google Scholar]
- Baltus T, von Hundelshausen P, Mause SF, Buhre W, Rossaint R, Weber C. 2005. Differential and additive effects of platelet-derived chemokines on monocyte arrest on inflamed endothelium under flow conditions. J Leukoc Biol. 78:435–441. [DOI] [PubMed] [Google Scholar]
- Brule S, Charnaux N, Sutton A, Ledoux D, Chaigneau T, Saffar L, Gattegno L. 2006. The shedding of syndecan-4 and syndecan-1 from HeLa cells and human primary macrophages is accelerated by SDF-1/CXCL12 and mediated by the matrix metalloproteinase-9. Glycobiology. 16:488–501. [DOI] [PubMed] [Google Scholar]
- Campanella GSV, Grimm J, Manice LA, Colvin RA, Medoff BD, Wojtkiewicz GR, Weissleder R, Luster AD. 2006. Oligomerization of CXCL10 is necessary for endothelial cell presentation and in vivo activity. J Immunol. 177:6991–6998. [DOI] [PubMed] [Google Scholar]
- Cardin AD, Weintraub HJ. 1989. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 9:21–32. [DOI] [PubMed] [Google Scholar]
- Chang TLY, Gordon CJ, Roscic-Mrkic B, Power C, Proudfoot AEI, Moore JP, Trkola A. 2002. Interaction of the CC-chemokine RANTES with glycosaminoglycans activates a p44/p42 mitogen-activated protein kinase-dependent signaling pathway and enhances human immunodeficiency virus type 1 infectivity. J Virol. 76:2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnaux N, Brule S, Chaigneau T, Saffar L, Sutton A, Hamon M, Prost C, Lievre N, Vita C, Gattegno L. 2005. RANTES (CCL5) induces a CCR5-dependent accelerated shedding of syndecan-1 (CD138) and syndecan-4 from HeLa cells and forms complexes with the shed ectodomains of these proteoglycans as well as with those of CD44. Glycobiology. 15:119–130. [DOI] [PubMed] [Google Scholar]
- Charnaux N, Brule S, Hamon M, Chaigneau T, Saffar L, Prost C, Lievre N, Gattegno L. 2005. Syndecan-4 is a signaling molecule for stromal cell-derived factor-1 (SDF-1)/CXCL12. FEBS J. 272:1937–1951. [DOI] [PubMed] [Google Scholar]
- Clark-Lewis I, Kim KS, Rajarathnam K, Gong JH, Dewald B, Moser B, Baggiolini M, Sykes BD. 1995. Structure-activity relationships of chemokines. J Leukoc Biol. 57:703–711. [DOI] [PubMed] [Google Scholar]
- Czaplewski LG, McKeating J, Craven CJ, Higgins LD, Appay V, Brown A, Dudgeon T, Howard LA, Meyers T, Owen J et al. 1999. Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants. J Biol Chem. 274:16077–16084. [DOI] [PubMed] [Google Scholar]
- de Paz JL, Moseman EA, Noti C, Polito L, von Andrian UH, Seeberger PH. 2007. Profiling heparin-chemokine interactions using synthetic tools. ACS Chem Biol. 2:735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepa SS, Yamada S, Zako M, Goldberger O, Sugahara K. 2004. Chondroitin sulfate chains on syndecan-1 and syndecan-4 from normal murine mammary gland epithelial cells are structurally and functionally distinct and cooperate with heparan sulfate chains to bind growth factors. A novel function to control binding of midkine, pleiotrophin, and basic fibroblast growth factor. J Biol Chem. 279:37368–37376. [DOI] [PubMed] [Google Scholar]
- Drury LJ, Ziarek JJ, Gravel S, Veldkamp CT, Takekoshi T, Hwang ST, Heveker N, Volkman BF, Dwinell MB. 2011. Monomeric and dimeric CXCL12 inhibit metastasis through distinct CXCR4 interactions and signaling pathways. Proc Natl Acad Sci USA. 108:17655–17660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrac A, Quemener C, Lacazette E, Lopez F, Zanibellato C, Wu WG, Bikfalvi A, Prats H. 2010. Functional divergence between 2 chemokines is conferred by single amino acid change. Blood. 116:4703–4711. [DOI] [PubMed] [Google Scholar]
- Gangavarapu P, Rajagopalan L, Kolli D, Guerrero-Plata A, Garofalo RP, Rajarathnam K. 2012. The monomer-dimer equilibrium and glycosaminoglycan interactions of chemokine CXCL8 regulate tissue-specific neutrophil recruitment. J Leukocyte Biol. 91:259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haessler U, Pisano M, Wu M, Swartz MA. 2011. Dendritic cell chemotaxis in 3D under defined chemokine gradients reveals differential response to ligands CCL21 and CCL19. Proc Natl Acad Sci USA. 108:5614–5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel DJ, Sielaff I, Proudfoot AEI, Handel TM. 2009. Chapter-4 interactions of chemokines with glycosaminoglycans. Methods Enzymol. 2009;461:71–102. [DOI] [PubMed] [Google Scholar]
- Handel TM, Johnson Z, Crown SE, Lau EK, Proudfoot AE. 2005. Regulation of protein function by glycosaminoglycans – as exemplified by chemokines. Annu Rev Biochem. 74:385–410. [DOI] [PubMed] [Google Scholar]
- Hirose J, Kawashima H, Swope Willis M, Springer TA, Hasegawa H, Yoshie O, Miyasaka M. 2002. Chondroitin sulfate B exerts its inhibitory effect on secondary lymphoid tissue chemokine (SLC) by binding to the C-terminus of SLC. Biochim Biophys Acta. 1571:219–224. [DOI] [PubMed] [Google Scholar]
- Hoogewerf AJ, Kuschert GS, Proudfoot AE, Borlat F, Clark-Lewis I, Power CA, Wells TN. 1997. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry. 36:13570–13578. [DOI] [PubMed] [Google Scholar]
- Jansma AL, Kirkpatrick JP, Hsu AR, Handel TM, Nietlispach D. 2010. NMR analysis of the structure, dynamics, and unique oligomerization properties of the chemokine CCL27. J Biol Chem. 285:14424–14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Shen X, Baggett BR, Kong X, LiWang PJ. 2007. The human CC chemokine MIP-1beta dimer is not competent to bind to the CCR5 receptor. J Biol Chem. 282:27976–27983. [DOI] [PubMed] [Google Scholar]
- Karlsson R, Falt A. 1997. Experimental design for kinetic analysis of protein-protein interactions with surface plasmon resonance biosensors. J Immunol Methods. 200:121–133. [DOI] [PubMed] [Google Scholar]
- Kohout TA, Nicholas SL, Perry SJ, Reinhart G, Junger S, Struthers RS. 2004. Differential desensitization, receptor phosphorylation, beta-arrestin recruitment, and ERK1/2 activation by the two endogenous ligands for the CC chemokine receptor 7. J Biol Chem. 279:23214–23222. [DOI] [PubMed] [Google Scholar]
- Kolarova H, Ambruzova B, Svihalkova Sindlerova L, Klinke A, Kubala L. 2014. Modulation of endothelial glycocalyx structure under inflammatory conditions. Mediators Inflamm. 2014:694312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, Geretti E, Brandner B, Goger B, Wells TN, Kungl AJ. 2004. A structural and dynamic model for the interaction of interleukin-8 and glycosaminoglycans: Support from isothermal fluorescence titrations. Proteins. 54:768–775. [DOI] [PubMed] [Google Scholar]
- Kuschert GS, Coulin F, Power CA, Proudfoot AE, Hubbard RE, Hoogewerf AJ, Wells TN. 1999. Glycosaminoglycans interact selectively with chemokines and modulate receptor binding and cellular responses. Biochemistry. 38:12959–12968. [DOI] [PubMed] [Google Scholar]
- Laguri C, Sadir R, Rueda P, Baleux F, Gans P, Arenzana-Seisdedos F, Lortat-Jacob H. 2007. The novel CXCL12gamma isoform encodes an unstructured cationic domain which regulates bioactivity and interaction with both glycosaminoglycans and CXCR4. PLoS ONE. 2:e1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau EK, Paavola CD, Johnson Z, Gaudry J-P, Geretti E, Borlat F, Kungl AJ, Proudfoot AE, Handel TM. 2004. Identification of the glycosaminoglycan binding site of the CC chemokine, MCP-1: Implications for structure and function in vivo. J Biol Chem. 279:22294–22305. [DOI] [PubMed] [Google Scholar]
- Lawrence R, Lu H, Rosenberg RD, Esko JD, Zhang L. 2008. Disaccharide structure code for the easy representation of constituent oligosaccharides from glycosaminoglycans. Nat Methods. 5:291–292. [DOI] [PubMed] [Google Scholar]
- Lawrence R, Olson SK, Steele RE, Wang L, Warrior R, Cummings RD, Esko JD. 2008. Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J Biol Chem. 283:33674–33684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lortat-Jacob H, Grosdidier A, Imberty A. 2002. Structural diversity of heparan sulfate binding domains in chemokines. Proc Natl Acad Sci USA. 99:1229–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard L, Saito N, Hlawaty H, Friand V, Suffee N, Chmilewsky F, Haddad O, Laguillier C, Guyot E, Ueyama T et al. 2014. RANTES/CCL5 mediated-biological effects depend on the syndecan-4/PKCalpha signaling pathway. Biol Open. 3:995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L, Blanpain C, Garnier P, Wittamer V, Parmentier M, Vita C. 2001. Structural and functional analysis of the RANTES-glycosaminoglycans interactions. Biochemistry. 40:6303–6318. [DOI] [PubMed] [Google Scholar]
- Middleton J, Neil S, Wintle J, Clark-Lewis I, Moore H, Lam C, Auer M, Hub E, Rot A. 1997. Transcytosis and surface presentation of IL-8 by venular endothelial cells. Cell. 91:385–395. [DOI] [PubMed] [Google Scholar]
- Migliorini E, Thakar D, Sadir R, Pleiner T, Baleux F, Lortat-Jacob H, Coche-Guerente L, Richter RP. 2014. Well-defined biomimetic surfaces to characterize glycosaminoglycan-mediated interactions on the molecular, supramolecular and cellular levels. Biomaterials. 35:8903–8915. [DOI] [PubMed] [Google Scholar]
- Mizumoto S, Fongmoon D, Sugahara K. 2013. Interaction of chondroitin sulfate and dermatan sulfate from various biological sources with heparin-binding growth factors and cytokines. Glycoconj J. 30:619–632. [DOI] [PubMed] [Google Scholar]
- Nasser MW, Raghuwanshi SK, Grant DJ, Jala VR, Rajarathnam K, Richardson RM. 2009. Differential activation and regulation of CXCR1 and CXCR2 by CXCL8 monomer and dimer. J Immunol. 183:3425–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hayre M, Salanga CL, Dorrestein PC, Handel TM. 2009. Phosphoproteomic analysis of chemokine signaling networks. Methods Enzymol. 460:331–346. [DOI] [PubMed] [Google Scholar]
- Oynebraten I, Barois N, Bergeland T, Kuchler AM, Bakke O, Haraldsen G. 2015. Oligomerized, filamentous surface presentation of RANTES/CCL5 on vascular endothelial cells. Sci Rep. 5:9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavola CD, Hemmerich S, Grunberger D, Polsky I, Bloom A, Freedman R, Mulkins M, Bhakta S, McCarley D, Wiesent L et al. 1998. Monomeric monocyte chemoattractant protein-1 (MCP-1) binds and activates the MCP-1 receptor CCR2B. J Biol Chem. 273:33157–33165. [DOI] [PubMed] [Google Scholar]
- Patel DD, Koopmann W, Imai T, Whichard LP, Yoshie O, Krangel MS. 2001. Chemokines have diverse abilities to form solid phase gradients. Clin Immunol. 99:43–52. [DOI] [PubMed] [Google Scholar]
- Peterson FC, Elgin ES, Nelson TJ, Zhang F, Hoeger TJ, Linhardt RJ, Volkman BF. 2004. Identification and characterization of a glycosaminoglycan recognition element of the C chemokine lymphotactin. J Biol Chem. 279:12598–12604. [DOI] [PubMed] [Google Scholar]
- Proudfoot AE, Fritchley S, Borlat F, Shaw JP, Vilbois F, Zwahlen C, Trkola A, Marchant D, Clapham PR, Wells TN. 2001. The BBXB motif of RANTES is the principal site for heparin binding and controls receptor selectivity. J Biol Chem. 276:10620–10626. [DOI] [PubMed] [Google Scholar]
- Proudfoot AEI, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, Borlat F, Wells TNC, Kosco-Vilbois MH. 2003. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci USA. 100:1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarathnam K, Sykes BD, Kay CM, Dewald B, Geiser T, Baggiolini M, Clark-Lewis I. 1994. Neutrophil activation by monomeric interleukin-8. Science. 264:90–92. [DOI] [PubMed] [Google Scholar]
- Rauova L, Poncz M, McKenzie SE, Reilly MP, Arepally G, Weisel JW, Nagaswami C, Cines DB, Sachais BS. 2005. Ultralarge complexes of PF4 and heparin are central to the pathogenesis of heparin-induced thrombocytopenia. Blood. 105:131–138. [DOI] [PubMed] [Google Scholar]
- Reitsma S, Oude Egbrink MG, Heijnen VV, Megens RT, Engels W, Vink H, Slaaf DW, van Zandvoort MA. 2011. Endothelial glycocalyx thickness and platelet-vessel wall interactions during atherogenesis. Thromb Haemost. 106:939–946. [DOI] [PubMed] [Google Scholar]
- Reitsma S, Slaaf DW, Vink H, van Zandvoort MA, oude Egbrink MG. 2007. The endothelial glycocalyx: Composition, functions, and visualization. Pfluegers Arch. 454:345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscic-Mrkic B, Fischer M, Leemann C, Manrique A, Gordon CJ, Moore JP, Proudfoot AEI, Trkola A. 2003. RANTES (CCL5) uses the proteoglycan CD44 as an auxiliary receptor to mediate cellular activation signals and HIV-1 enhancement. Blood. 102:1169–1177. [DOI] [PubMed] [Google Scholar]
- Rot A. 1993. Neutrophil attractant/activation protein-1 (interleukin-8) induces in vitro neutrophil migration by haptotactic mechanism. Eur J Immunol. 23:303–306. [DOI] [PubMed] [Google Scholar]
- Salanga CL, Dyer DP, Kiselar JG, Gupta S, Chance MR, Handel TM. 2014. Multiple glycosaminoglycan-binding epitopes of monocyte chemoattractant protein-3/CCL7 enable it to function as a non-oligomerizing chemokine. J Biol Chem. 289:14896–14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanga CL, Handel TM. 2011. Chemokine oligomerization and interactions with receptors and glycosaminoglycans: The role of structural dynamics in function. Exp Cell Res. 317:590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarabi A, Kramp BK, Drechsler M, Hackeng TM, Soehnlein O, Weber C, Koenen RR, Von Hundelshausen P. 2011. CXCL4L1 inhibits angiogenesis and induces undirected endothelial cell migration without affecting endothelial cell proliferation and monocyte recruitment. J Thromb Haemost. 9:209–219. [DOI] [PubMed] [Google Scholar]
- Schumann K, Lammermann T, Bruckner M, Legler DF, Polleux J, Spatz JP, Schuler G, Forster R, Lutz MB, Sorokin L et al. 2010. Immobilized chemokine fields and soluble chemokine gradients cooperatively shape migration patterns of dendritic cells. Immunity. 32:703–713. [DOI] [PubMed] [Google Scholar]
- Severin IC, Gaudry J-P, Johnson Z, Kungl A, Jansma A, Gesslbauer B, Mulloy B, Power C, Proudfoot AEI, Handel T. 2010. Characterization of the chemokine CXCL11-heparin interaction suggests two different affinities for glycosaminoglycans. J Biol Chem. 285:17713–17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Lebel-Haziv Y, Ehrlich M, Meshel T, Suez A, Avezov E, Rozenberg P, Ben-Baruch A. 2012. Mechanisms regulating the secretion of the promalignancy chemokine CCL5 by breast tumor cells: CCL5's 40s loop and intracellular glycosaminoglycans. Neoplasia. 14:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler-Barak L, Moussion C, Shezen E, Hatzav M, Sixt M, Alon R. 2014. Blood vessels pattern heparan sulfate gradients between their apical and basolateral aspects. PLoS ONE. 9:e85699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer SE, Gallagher JT. 1997. Specific binding of the chemokine platelet factor 4 to heparan sulfate. J Biol Chem. 272:20508–20514. [DOI] [PubMed] [Google Scholar]
- Struyf S, Burdick MD, Peeters E, Van den Broeck K, Dillen C, Proost P, Van Damme J, Strieter RM. 2007. Platelet factor-4 variant chemokine CXCL4L1 inhibits melanoma and lung carcinoma growth and metastasis by preventing angiogenesis. Cancer Res. 67:5940–5948. [DOI] [PubMed] [Google Scholar]
- Struyf S, Burdick MD, Proost P, Van Damme J, Strieter RM. 2004. Platelets release CXCL4L1, a nonallelic variant of the chemokine platelet factor-4/CXCL4 and potent inhibitor of angiogenesis. Circ Res. 95:855–857. [DOI] [PubMed] [Google Scholar]
- Struyf S, Salogni L, Burdick MD, Vandercappellen J, Gouwy M, Noppen S, Proost P, Opdenakker G, Parmentier M, Gerard C et al. 2011. Angiostatic and chemotactic activities of the CXC chemokine CXCL4L1 (platelet factor-4 variant) are mediated by CXCR3. Blood. 117:480–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SK, McGrath DA, Grigoriadis D, Bacon KB. 1999. Pharmacological and signaling analysis of human chemokine receptor CCR-7 stably expressed in HEK-293 cells: High-affinity binding of recombinant ligands MIP-3beta and SLC stimulates multiple signaling cascades. Biochem Biophys Res Commun. 263:685–690. [DOI] [PubMed] [Google Scholar]
- Takekoshi T, Ziarek JJ, Volkman BF, Hwang ST. 2012. A locked, dimeric CXCL12 variant effectively inhibits pulmonary metastasis of CXCR4-expressing melanoma cells due to enhanced serum stability. Mol Cancer Ther. 11:2516–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JH, Canals M, Ludeman JP, Wedderburn J, Boston C, Butler SJ, Carrick AM, Parody TR, Taleski D, Christopoulos A et al. 2012. Design and receptor interactions of obligate dimeric mutant of chemokine monocyte chemoattractant protein-1 (MCP-1). J Biol Chem. 287:14692–14702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanino Y, Coombe DR, Gill SE, Kett WC, Kajikawa O, Proudfoot AEI, Wells TNC, Parks WC, Wight TN, Martin TR et al. 2010. Kinetics of chemokine-glycosaminoglycan interactions control neutrophil migration into the airspaces of the lungs. J Immunol. 184:2677–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Gordon C, Matthews J, Maxwell E, Ketas T, Czaplewski L, Proudfoot AE, Moore JP. 1999. The CC-chemokine RANTES increases the attachment of human immunodeficiency virus type 1 to target cells via glycosaminoglycans and also activates a signal transduction pathway that enhances viral infectivity. J Virol. 73:6370–6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldkamp CT, Peterson FC, Pelzek AJ, Volkman BF. 2005. The monomer-dimer equilibrium of stromal cell-derived factor-1 (CXCL 12) is altered by pH, phosphate, sulfate, and heparin. Protein Sci. 14:1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldkamp CT, Seibert C, Peterson FC, De la Cruz NB, Haugner JC, Basnet H, Sakmar TP, Volkman BF. 2008. Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12. Sci Signal. 1:ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner L, Yang OO, Garcia-Zepeda EA, Ge Y, Kalams SA, Walker BD, Pasternack MS, Luster AD. 1998. Beta-chemokines are released from HIV-1-specific cytolytic T-cell granules complexed to proteoglycans. Nature. 391:908–911. [DOI] [PubMed] [Google Scholar]
- Wang L, Fuster M, Sriramarao P, Esko JD. 2005. Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat Immunol. 6:902–910. [DOI] [PubMed] [Google Scholar]
- Wang X, Watson C, Sharp JS, Handel TM, Prestegard JH. 2011. Oligomeric structure of the chemokine CCL5/RANTES from NMR, MS, and SAXS data. Structure. 19:1138–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hauschild R, Schwarz J, Moussion C, de Vries I, Legler DF, Luther SA, Bollenbach T, Sixt M. 2013. Interstitial dendritic cell guidance by haptotactic chemokine gradients. Science. 339:328–332. [DOI] [PubMed] [Google Scholar]
- Weber KS, von Hundelshausen P, Clark-Lewis I, Weber PC, Weber C. 1999. Differential immobilization and hierarchical involvement of chemokines in monocyte arrest and transmigration on inflamed endothelium in shear flow. Eur J Immunol. 29:700–712. [DOI] [PubMed] [Google Scholar]
- Xu D, Esko JD. 2014. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem. 83:129–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziarek JJ, Veldkamp CT, Zhang F, Murray NJ, Kartz GA, Liang X, Su J, Baker JE, Linhardt RJ, Volkman BF. 2013. Heparin oligosaccharides inhibit chemokine (CXC motif) ligand 12 (CXCL12) cardioprotection by binding orthogonal to the dimerization interface, promoting oligomerization, and competing with the chemokine (CXC motif) receptor 4 (CXCR4) N terminus. J Biol Chem. 288:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidar DA, Violin JD, Whalen EJ, Lefkowitz RJ. 2009. Selective engagement of G protein coupled receptor kinases (GRKs) encodes distinct functions of biased ligands. Proc Natl Acad Sci USA. 106:9649–9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.