Abstract
MAL2 (myelin and lymphocyte protein 2) is thought to regulate at least two steps in the hepatic apical transcytotic pathway. As vesicle budding and delivery at each step are driven by complex machineries, we predicted that MAL2 participates in several large protein complexes with multiple binding partners. To identify novel MAL2 interactors, we performed split-ubiquitin yeast two-hybrid assays and identified STK16 (serine/threonine kinase 16) as a putative interactor which we verified morphologically and biochemically. As STK16 is a Golgi-associated constitutively active kinase implicated in regulating secretion and because of the massive constitutive secretory capacity of hepatic cells, we tested whether MAL2 and STK16 function in secretion. Expression of a dominant-negative kinase-dead STK16 mutant (E202A) or knockdown of MAL2 impaired secretion that correlated with decreased expression of albumin and haptoglobin. By using 19 °C temperature blocks and lysosome deacidification, we determined that E202A expression or MAL2 knockdown did not interfere with albumin synthesis or processing, but led to albumin lysosomal degradation. We conclude that MAL2 and the constitutively active STK16 function to sort secretory soluble cargo into the constitutive secretory pathway at the TGN (trans-Golgi network) in polarized hepatocytes.
Keywords: albumin, constitutive secretion, hepatocyte, myelin and lymphocyte protein 2 (MAL2), serine/threonine kinase 16 (STK16)
INTRODUCTION
The plasma membrane of a polarized hepatocyte is physically continuous, but functionally and compositionally divided into two distinct domains: the apical and basolateral surfaces. Each domain is characterized by distinct proteins and lipids which are required for maintaining the domain-specific activities at each surface. Our research addresses the fundamental question of how hepatic plasma membrane polarity is established and maintained. Our focus is to understand the mechanisms regulating delivery of newly synthesized domain-specific secretory and membrane proteins in polarized hepatocytes.
Recently, we have been examining the role of MAL2 (myelin and lymphocyte protein 2) in regulating indirect delivery of newly synthesized apical proteins in polarized hepatocytes. MAL2 has been implicated in regulating the transcytotic delivery of both pIgA-R (polymeric IgA receptor) and the glycosylphosphatidylinositol-anchored protein CD59 from the early endosome to the subapical compartment in HepG2 cells [1]. Using AS (antisense) MAL2 recombinant viruses, we have confirmed these results in polarized hepatic WIF-B cells [2]. In the course of those studies, we determined further that MAL2 also selectively regulates delivery of newly synthesized pIgA-R (but not dipeptidyl peptidase IV or haemagglutinin) from the TGN (trans-Golgi network) to the basolateral membrane, indicating that MAL2 may regulate protein sorting at multiple transport steps.
Protein sorting, vesicle formation and targeting are driven by complex protein machineries [3–8]. As MAL2 regulates at least two trafficking steps, we predict that it participates in multiple large protein complexes. Consistent with this hypothesis is the identification of several MAL2-binding partners, including members of the tumour protein D52 family, mucin1 and the inverted formin INF2 [3,9–11]. To identify other MAL2-binding partners, we used the split-ubiquitin Y2H (yeast two-hybrid) system. Using human MAL2 as bait, 19 putative novel interactors were identified from human liver cDNA libraries. Of particular interest is STK16 (serine/threonine kinase 16), an atypical lipid-modified kinase originally identified in embryonic murine livers and originally named PKL12 [12–15].
Although STK16 is ubiquitous, it is highly expressed in liver, testis and kidney [14]. Interestingly, STK16 overexpression in the epithelial cells of the developing murine mammary gland led to enhanced ductal end-bud formation [16]. Because end-bud proliferation is promoted by a host of secreted cytokines and morphogens, the authors postulated that STK16 may regulate secretion of these factors [16], a hypothesis that has not yet been tested. As hepatocytes constitutively synthesize and secrete copious amounts of serum proteins, and because of the high expression levels of STK16 in the liver, we chose to characterize further STK16 and MAL2 interactions and function in secretion in polarized hepatic WIF-B cells.
EXPERIMENTAL
Reagents and antibodies
F12 (Coon’s modification) medium and BFA (brefeldin A) were purchased from Sigma–Aldrich. BFA was stored at −20°C as a 10 mg/ml stock in DMSO. Lactacystin was stored as a 5 mM stock at −20°C. MG-132 (Cell Signaling Technology) was stored at −20°C as a 10 mM stock in DMSO. FBS and newborn calf serum were purchased from Gemini Bio-Products. TaqSelect DNA polymerase was purchased from Lucigen. High-speed plasmid mini kits were from MidSci. HRP (horseradish peroxidase)-conjugated secondary antibodies and Western Lightning ECL reagent were purchased from Sigma–Aldrich and PerkinElmer respectively. Alexa Fluor®-conjugated secondary antibodies were purchased from Invitrogen. Monoclonal antibodies against the V5 epitope tag were from AbD Serotec or against mannosidase II were purchased from Covance. ProteoStat® was purchased from Enzo Life Sciences and cells were stained according to the manufacturer’s instructions. LC3A/B antibodies were from Cell Signaling Technology. Anti-MAL2 polyclonal antibodies were generated and affinity-purified as described previously [2]. Polyclonal antibodies against haptoglobin, HA321, CE9, rat serum albumin, APN (aminopeptidase N) and lysosomal glycoprotein 120 were provided by Dr A. Hubbard (Johns Hopkins University School of Medicine, Baltimore, MD, U.S.A.).
Construction of bait plasmid and Y2H screening
Split-ubiquitin Y2H screens were performed with the Y2H Membrane Protein System (MoBiTec) according to the manufacturer’s instructions. Full-length human MAL2 with an SfiI linker at both the 5′ and 3′ ends was generated by PCR primers 5′-GGAACAGGCCATTACGGCCGGCAGCATGTCG-3′ and 5′-GCTTCAGGCCGAGGCGGCCACGGACGGTCGCCATCT-3′. The resulting PCR product was cloned in-frame into the pBT3-STE vector in the x-Cub orientation and the pBT3-N vector in the Cub-x orientation. The bait vectors contained the selectable marker LEU2 and the LexA-VP16 DNA-binding and transactivation domains. An adult human liver cDNA library in the x-NubG orientation (MoBiTec) or NubG-x orientation (Dualsystems Biotech) was transformed into the bait yeast strain (Saccharomyces cerevisiae NMY51 transformed with the bait-expressing vector). Clones were selected on leucine/histidine/tryptophan selection plates supplemented with 7.5 mM 3-aminotriazole (Sigma–Aldrich). Plasmids were isolated from positive colonies using Zymoprep Yeast Plasmid Miniprep II (Zymo Research) and transformed into XL10-Gold Escherichia coli (Agilent). Plasmids were re-isolated from E. coli colonies with the QIAprep Spin Miniprep Kit (Qiagen) and sequenced (Retrogen). Sequences were identified using the BLAST database.
Cell culture
WIF-B cells were grown in a humidified 7% CO2 incubator at 37°C as described previously [17]. Briefly, the cells were grown in F12 medium (Coon’s modification), pH 7.0, supplemented with 5% (v/v) FBS, 10 μM hypoxanthine, 40 nM aminoterpin and 1.6 μM thymidine. Clone 9 cells were grown at 37°C in a 5% CO2 incubator in F12 medium supplemented with 10% (v/v) newborn calf serum. WIF-B cells were seeded on to glass coverslips at 1.3×104 cells/cm2 , whereas Clone 9 cells were seeded on to coverslips in six-well dishes at 0.5–1.0×106 cells/well. Clone 9 cells were cultured for 1–2 days and WIF-B cells for 8–12 days until they reached maximal density and polarity.
Virus production and infection
Recombinant adenoviruses encoding V5 epitope-tagged full-length and AS MAL2, and full-length and kinase-dead STK16 were generated using the ViraPower Adenoviral Expression System (Invitrogen) according to the manufacturer’s instructions. A V5 epitope-tag was added to the C-terminus of MAL2 or the STK16 constructs. The point mutation was verified by plasmid sequencing. The full-length and kinase-dead STK16 plasmids have been described and characterized previously [18]. For the AS constructs, MAL2 was PCR-amplified with primers against the full-length sequences that have additional sequences that encode the recombination sites in reverse order from how they appear in the Invitrogen Gateway® vector. Thus, when recombined, the fragment is inserted in the opposite AS orientation, which was verified by plasmid sequencing. WIF-B or Clone 9 cells were infected with recombinant adenovirus particles for 60 min at 37°C as described previously [19]. Complete medium was added to the cells and they were incubated an additional 16– 20 h (for overexpression) or 48 h (for AS adenovirus-mediated knockdown).
Immunoblotting
Samples were mixed with Laemmli sample buffer [20] and boiled for 3 min. Proteins were electrophoretically separated using SDS/PAGE and transferred on to nitrocellulose before immunoblotting with the indicated antibodies. HRP-conjugated secondary antibodies were used at 5 ng/ml, and immunoreactivity was detected with ECL. Relative protein levels were determined by densitometric analysis of immunoreactive bands using the ImageJ software (NIH).
Immunofluorescence microscopy and imaging
Control or infected cells were fixed on ice with ice-cold PBS containing 4% paraformaldehyde for 1 min and permeabilized with ice-cold methanol for 10 min. Cells were processed for indirect immunofluorescence as described previously [21]. Alexa Fluor® 488- or 568-conjugated secondary antibodies were used at 3–5 μg/ml. Labelled cells were visualized at room temperature by epifluorescence with an Olympus BX60 Fluorescence Microscope (OPELCO) using an UPlanFl ×60/NA (numerical aperture) 1.3 phase 1 oil-immersion objective. Images were taken with an HQ2 CoolSnap digital camera (Roper Scientific) and IP Labs software (BD Biosciences). Adobe Photoshop (Adobe Systems) was used to process images and to compile Figures.
Immunoprecipitations
WIF-B cells were lysed in 0.5 ml of RIPA buffer (150 mM NaCl, 50 mM Tris/HCl, 1% Nonidet P40, 0.5% deoxycholic acid and 1% SDS, pH 8.0) with protease inhibitors (1 μg/ml each of leupeptin, antipain, PMSF and benzamidine) and incubated on ice for 30 min. Lysates were cleared by centrifugation at 120 000 g for 30 min at 4°C. Supernatants were incubated with affinity-purified anti-MAL2 (0.8 μg/ml) or anti-V5 (1.0 μg/ml) antibodies overnight at 4°C on a fixed-speed tube rotator. Protein A– or G–Sepharose [50 μl of a 50% (v/v) slurry] was added for 2h at 4°C and samples were processed as described previously [22]. Immunoprecipitates were separated by SDS/PAGE and transferred on to nitrocellulose before immunoblotting for the indicated proteins. HRP-conjugated secondary antibodies were used at 5 ng/ml and immunoreactivity was detected by ECL.
Secretion assays
Cells were infected with recombinant AS MAL2 for 48 h, or wild-type STK16 or kinase-dead STK16 (E202A) for 20 h. Cells were rinsed five times with prewarmed serum-free medium and then re-incubated in serum-free medium. At 0, 15, 30 and 60 min after re-incubation, aliquots of medium were collected and analysed for albumin secretion by immunoblotting. The cell lysates were collected by solubilization directly into SDS/PAGE sample buffer. Samples were processed for Western blotting and densitometric analysis of immunoreactive bands.
Antibody trafficking in live cells
Cells were continuously labelled with anti-APN antibodies (1:50 dilution) for 60 min at 37°C in complete medium as described previously [23]. As tight junctions restrict access of the antibodies to the apical membrane, only antigens at the basolateral surface were labelled. Cells were washed three times for 2 min with prewarmed medium and fixed as described above. Cells were labelled with Alexa Fluor®-conjugated secondary antibodies to detect the trafficked antigen–antibody complexes.
RESULTS
Split-ubiquitin Y2H screens identify STK16 and 18 other putative MAL2-binding partners
Since both the MAL2 C- and N-termini are cytosolic, we constructed two bait constructs: Cub–MAL2 (Cub fused to the MAL2 N-terminus) and MAL2–Cub (Cub fused to the MAL2 C-terminus). We screened two human cDNA liver libraries: X-NubG (NubG fused to C-termini of Type I membrane proteins) and NubG-X (NubG fused to N-termini of Type II membrane proteins). Screening of all colonies resulted in 54 clones from which 19 putative binding partners were identified (Supplementary Table S1 at http://www.biochemj.org/ bj/463/bj4630201add.htm). The first screen yielded a particularly interesting candidate, the atypical serine/threonine kinase STK16, which reproducibly bound MAL2 in the screens. As expression of this lipid-modified kinase is enriched in liver and it distributes to the Golgi [14,16,18], we further characterized its relationship with MAL2 in regulating basolateral secretion in polarized hepatic WIF-B cells.
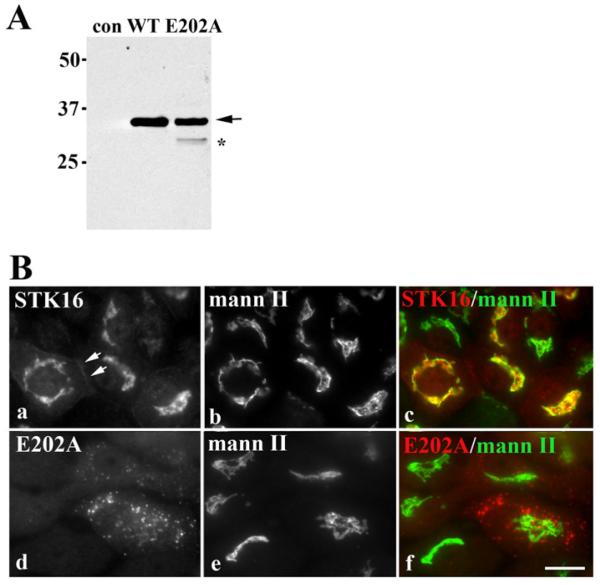
For our analysis, we generated recombinant adenoviruses expressing full-length V5-tagged STK16 or a previously characterized kinase-dead STK16 mutant (E202A) [18]. In lysates from cells expressing either wild-type STK16 or E202A, a 35 kDa immunoreactive species was detected corresponding to the predicted STK16 molecular mass (Figure 1A). In general, Ad-STK16 infection efficiencies were high (>80% of cells), whereas Ad-E202A efficiencies were typically and reproducibly lower (25–50% of cells) which is reflected in the relative intensities of the immunoreactive bands. In E202A lysates, a 30 kDa band is also observed (marked with an asterisk) that is probably a partially degraded E202A species (see Figure 5).
Figure 1. Wild-type STK16 distributes to the Golgi while E202A is present on peripheral puncta.
(A) Cell lysates from control (uninfected), STK16- or E202A-expressing cells were immunoblotted with anti-V5 antibodies. Molecular mass markers are indicated on the left-hand side in kDa. The arrow is pointing to the 35 kDa wild-type and E202A STK16 species. The band marked with an asterisk is probably a partially degraded E202A. (B) WIF-B cells expressing STK16 (a–c) or E202A (d–f) were double-labelled for mannosidase II (mann II) and the V5 tag. Merged images are shown in c and f. Scale bar, 10 μm.
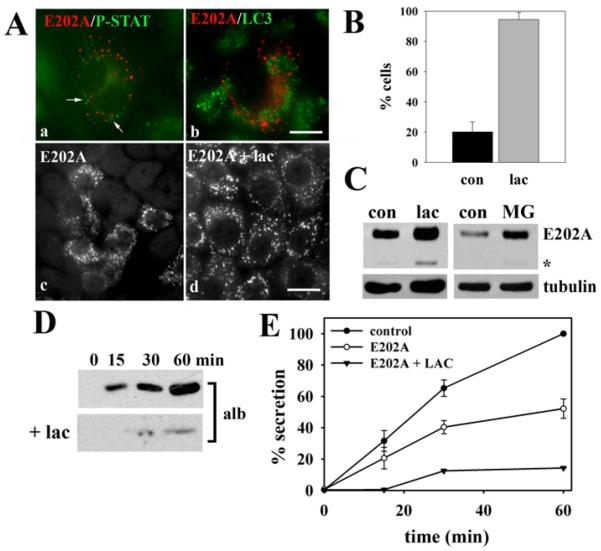
Figure 5. E202A associates with proteasomes.
(A) Cells expressing E202A were double-labelled for E202A and ProteoStat (a) or LC3 (b). E202A-expressing cells were incubated in the absence (c) or presence (d) of 5 μM lactacystin (lac) for 4 h and stained for E202A. Scale bar, 10 μm. (B) WIF-B cells were scored for E202A labelling after incubation in the absence (con) or presence (Lac) of 5 μm lactacystin for 4 h. The percentage of total cells positive for E202A labelling is plotted. Values are expressed as the means±S.E.M. Measurements were performed on at least three independent experiments. (C) E202A-expressing cells were incubated with 5 μM lactacystin for 4 h or 5 μM MG132 (MG) for 3 h. Cells wer−e lysed and immunoblotted for E202A and tubulin (as a loading control) as indicated. (D) E202A-expressing cells were treated in the absence or presence of 5 μM lactacystin for 4 h and the secretion assay was performed as described in Figure 3. (E) Secretion levels of control, E202A-infected, and E202A-infected plus lactacystin (LAC) were calculated from the densitometric analysis of immunoreactive bands shown in (D). Values for control and E202A are expressed as the means±S.E.M. measured from three independent experiments. Values for E202A plus lactacystin are expressed as the mean from two independent experiments.
Consistent with previous reports [18], STK16 localized primarily to the Golgi with near perfect overlap with mannosidase II in WIF-B cells (Figure 1B). However, there was also a small, but reproducible, STK16 subpopulation detected at the basolateral membrane (Figure 1B, panel a, arrows), consistent with a role in basolateral secretion. STK16 was also detected at the Golgi in Clone 9 cells (non-polarized hepatic cells lacking MAL2 expression) (Supplementary Figure S1 at http://www.biochemj.org/bj/463/bj4630201add.htm), indicating that STK16 Golgi association is independent of MAL2 expression and cell polarity. Also consistent with previous reports [14,18], E202A localized to bright peripheral puncta in both WIF-B (Figure 1B) and Clone 9 cells (Supplementary Figure S1).
MAL2 and wild-type STK16 interact
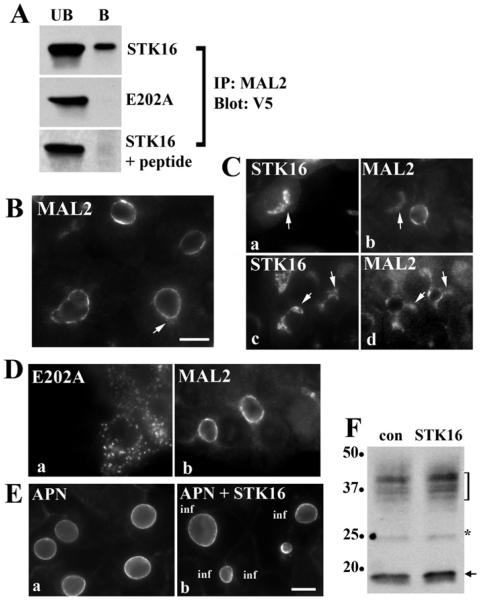
To confirm the Y2H results, we performed co-immunoprecipitations using anti-MAL2 or -V5 antibodies in wild-type or mutant expressing cells. As shown in Figure 2(A), only wild-type STK16 co-precipitated with anti-MAL2 (ranging from 0.8 to 1.7% of total STK16). Addition of the peptide against which the anti-MAL2 antibodies were made prevented STK16 co-precipitation, indicating that the interactions are specific. Although less robust, reciprocal precipitations with anti-V5 antibodies confirmed these results (results not shown). MAL2 distributions in overexpressing cells are remarkably consistent with these results. In uninfected cells, MAL2 is present predominantly at the apical membrane and in small sub-apical puncta (Figure 2B). However, in cells overexpressing the wild-type kinase, a dim MAL2 population was observed at the Golgi overlapping with STK16. Two such examples are shown in Figure 2(C) where the MAL2 images were intentionally overexposed to better visualize the intracellular labelling. This was a highly reproducible effect with almost all STK16-overexpressing cells positive for intracellular MAL2. In contrast and consistent with the co-precipitations, no overlapping MAL2 staining was observed with E202A (Figure 2D) and MAL2 expression levels did not change (results not shown). Despite the interactions of MAL2 with the wild-type kinase, there is no evidence that MAL2 is an STK16 substrate (A.C. Striz and P.L. Tuma, unpublished work and see the Discussion section).
Figure 2. MAL2 and STK16 interact.
(A) Lysates from cells overexpressing STK16 or E202A were immunoprecipitated with affinity-purified MAL2 antibodies (top two panels). Lysates from STK16-expressing cells were immunoprecipitated in the presence of a 10-fold molar excess of a MAL2 N-terminal peptide (bottom panel). Unbound (UB) and bound (B) fractions were immunoblotted for V5-tagged STK16 or E202A as indicated. The MAL2 peptide specifically abolished STK16 co-precipitation. (B) Uninfected WIF-B cells were immunolabelled for endogenous MAL2. The arrow marks sub-apical MAL2 labelling. (C) Cells exogenously expressing STK16 were immunolabelled for MAL2. Two examples are shown. Images were intentionally overexposed to highlight the intracellular MAL2 labelling. Arrows mark overlapping MAL2 and STK16 labelling at the Golgi. (D) WIF-B cells expressing E202A were immunolabelled for MAL2. No changes in MAL2 distributions were observed. (E) Cells overexpressing STK16 were immunolabelled for the apical resident protein APN. In b, APN apical distributions do not change in infected (inf) cells. (F) Uninfected (con) and STK16-expressing cells were immunoblotted for MAL2. Molecular mass markers are indicated on the left-hand side in kDa. The arrow marks the species with the predicted MAL2 molecular mass. The bracket highlights a 30–38 kDa diffuse set of bands and the asterisk indicates a 25 kDa species also detected by others (see text). Scale bar, 10 μm.
To determine whether STK16 induced selective MAL2 redistribution to the Golgi, we labelled cells for another apical resident, APN. As shown in Figure 2(E), APN apical labelling was not altered by STK16 expression, indicating that the redistribution is at least partially selective. To rule out that STK16 expression induced MAL2 protein synthesis thereby overloading the biosynthetic pipeline allowing Golgi detection, we immunoblotted lysates for MAL2. Our antibodies detect a 19 kDa species (the predicted MAL2 molecular mass, arrow), a 25 kDa band (marked by an asterisk) and a diffuse set of bands from 30 to 38 kDa (marked by a bracket) (Figure 2F). These additional bands have been observed by others using their custom antibodies and MS analysis has confirmed that the 25 kDa species is MAL2 (O.F. Omotade and J.Q. Zheng, personal communication). At present the higher molecular mass species are thought to represent as yet unidentified post-translationally modified forms of MAL2 [1,24]. Nonetheless, no changes in expression were observed for any of the species in STK16-expressing cells, indicating that Golgi-associated MAL2 reflects STK16-induced redistribution rather than enhanced MAL2 synthesis. Together these results confirm the Y2H results and further indicate that MAL2 interacts with STK16.
STK16 and MAL2 regulate constitutive basolateral secretion
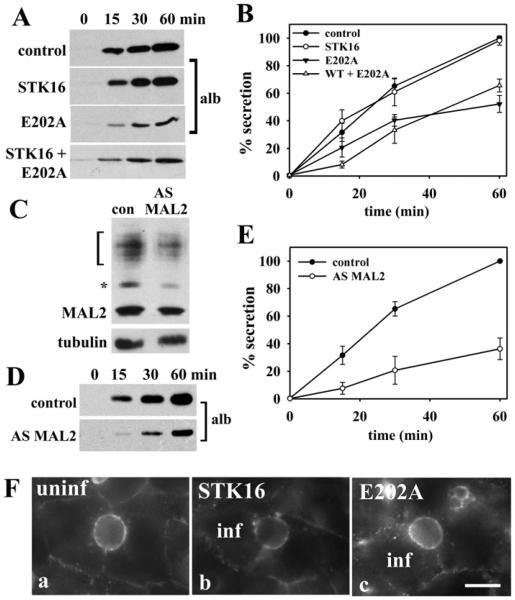
Although STK16 has been implicated in regulating secretion, this hypothesis has not been tested. To examine directly the role of STK16 in secretion, we monitored albumin release into the culture medium from cells expressing wild-type or kinase-dead STK16. As albumin is synthesized by hepatocytes to such high levels and its constitutive basolateral secretion is so robust, it is easily detected on immunoblots of the collected aliquots of medium. As shown in Figure 3(A), both uninfected and STK16-overexpressing cells had comparably high levels of secreted albumin, and when quantified, nearly overlapping plots for secretion were observed (Figure 3B). In contrast, in E202A-expressing cells, secreted albumin levels were significantly reduced to 30% of control cells (Figures 3A and 3B). Since the rate of secretion did not significantly change when normalized for total albumin for each condition (Supplementary Table S2 at http://www.biochemj.org/bj/463/bj4630201add.htm), the decreased albumin secretion probably reflects E202A infection levels. That is, the uninfected cells continue to secrete normally, whereas secretion in the E202A-expressing cells is significantly impaired. Interestingly, overexpression of wild-type STK16 in E202A-expressing cells failed to rescue the albumin secretion defect (Figures 3A and 3B), suggesting that the mutant is dominant-negative.
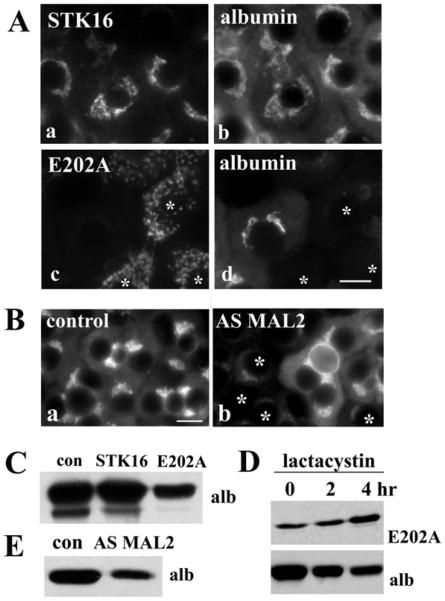
Figure 3. Figure 3 E202A expression and MAL2 knockdown impair basolateral secretion.
(A) Cells expressing STK16, E202A or both were placed in serum-free medium and aliquots of medium were collected at 0, 15, 30 and 60 min after re-incubation and immunoblotted for albumin. (B) The percentage of albumin secreted was calculated from densitometric analysis of immunoreactive bands on blots as shown in (A). Values are expressed as the means±S.E.M. Measurements were done on at least three independent experiments. (C) WIF-B cells were infected with recombinant adenovirus expressing AS MAL2 for 48 h. Lysates from control or AS−MAL2-expressing cells were immunoblotted for MAL2 and for α-tubulin (as the loading control). Decreased expression for all immunoreactive species was observed. (D) Secretion assays were performed as described in (A) in cells knocked down for MAL2 expression and aliquots immunoblotted for albumin. (E) The percentage of albumin secreted was calculated from densitometric analysis of immunoreactive bands on blots as shown in (D). Values are expressed as the means±S.E.M. Measurements were done on at least three independent experiments. (F) Uninfected cells (uninf) or cells overexpressing STK16 or E202A were continuously labelled with anti-APN antibodies for 60 min at 37°C. Cells were fixed and labelled with Alexa Fluor®-conjugated secondary antibodies to detect the trafficked antigen–antibody complexes. Cells were double-labelled with anti-V5 antibodies to monitor infection. Infected (inf) cells are indicated in panels b and c. Scale bar, 10 μm.
If MAL2 is a bona fide STK16-binding partner, the simple prediction is that its knockdown should also lead to decreased albumin secretion. As shown in Figure 3(C), in cells expressing AS MAL2 adenoviruses, MAL2 expression was routinely decreased to 40–50% of control. All of the immunoreactive species were decreased to similar extents, confirming they are indeed MAL2 species. The extent of MAL2 knockdown correlated remarkably with the decrease in albumin secretion to 50% of control (Figures 3D and 3E). Thus we conclude that both MAL2 and STK16 are both important regulators of constitutive basolateral secretion.
On the basis of its Golgi distribution, we further predicted that STK16 is not a regulator of basolateral-to-apical transcytosis, a process known to require MAL2 [1,2]. To test this hypothesis, we assayed transcytosis in control or overexpressing cells by monitoring the trafficking of antibody-labelled APN from the basolateral membrane. As shown in Figure 3(F), APN was successfully delivered to the apical membrane in uninfected cells and in cells overexpressing either wild-type STK16 or E202A. In all cases, robust apical labelling was observed after 60 min of chase with a subpopulation present in small sub-apical puncta indicating that STK16 does not regulate transcytosis. Thus STK16 selectively regulates secretion in WIF-B cells, whereas MAL2 participates in several trafficking steps.
E202A is present in post-Golgi structures and associates with proteasomes
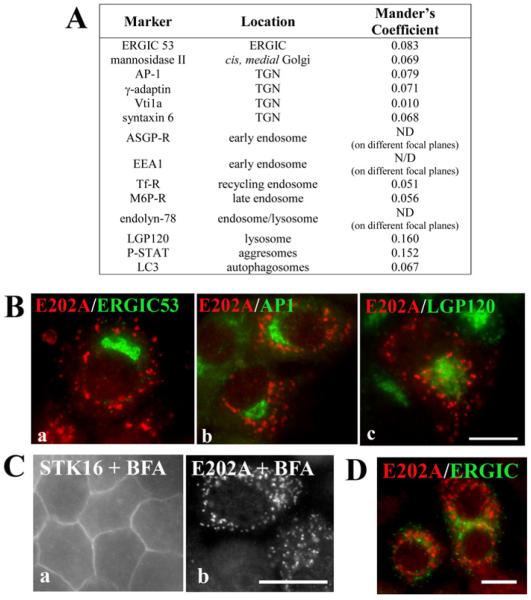
To better identify the molecular basis for impaired secretion, we hoped to identify the E202A-positive peripheral puncta. However, Mander’s coefficients of colocalization revealed virtually no overlapping staining with E202A and a host of organelle markers (Figure 4A). Examples of this lack of E202A colocalization with ERGIC53 (endoplasmic reticulum–Golgi intermediate compartment 53), AP-I and LGP-120 are shown in Figure 4(B). Importantly, the distributions of all of the tested Golgi/TGN markers did not change in E202A-expressing cells or MAL2-knockdown cells [2], ruling out that impaired secretion is simply due to Golgi/TGN disruption. To determine at minimum whether the puncta are post-Golgi structures, we treated cells with BFA. As shown in Figure 4(C), the puncta persisted in treated cells (95% of total cells contained puncta), indicating that the compartment is a post-Golgi structure. Because ERGIC is also BFA-resistant in WIF-B cells, we double-labelled cells with ERGIC53. However, no overlapping labelling was detected (Figure 4D). In comparison and in contrast, wild-type STK16 Golgi staining was lost in BFA-treated cells with reciprocal increases in diffuse cytosolic and basolateral labelling (Figure 4C), consistent with its steady-state distribution to the Golgi/TGN [25].
Figure 4. E202A is present on post-Golgi structures.
(A) Double labelling of indirect immunofluorescence with anti-V5 antibodies (to detect E202A) and antibodies specific for each of the compartment markers listed was performed. The Mander’s coefficients for colocalization of E202A with the indicated markers were determined. AP-1, adaptin-1; ASGP-R, asialoglycoprotein receptor; EEA1, early endosome antigen-1; LGP120, lysosomal glycoprotein-120; M6P-R, mannose 6-phosphate receptor; ND, not determined; Tf-R, transferrin receptor. (B) Cells expressing E202A were double-labelled for E202A and ERGIC53 (a), AP-1 (b) or LGP120 (c). Merged images are shown. (C and D) STK16-or E202A-overexpressing cells were incubated with 10 μg/ml BFA for 1 h at 37°C and labelled with anti-V5 antibodies, and in (D), double-labelled for E202A and ERGIC. Scale bar, 10 μm
The presence of the likely degradative species on E202A immunoblots (see Figure 1A) prompted us to further explore whether the E202A-positive puncta were other degradative compartments. Although the puncta minimally colocalized with markers for aggresomes (ProteoStat; P-Stat) and autophagosomes (LC3 A/B) (Figures 4A and 5A), we noticed a dramatic increase in E202A-positive cells in the presence of the proteasome inhibitor lactacystin (Figure 5A). We determined that 20.2±6.7% of control cells expressed E202A, whereas 94.4±4.7% of lactacystin-treated cells were E202A-positive (Figure 5B). This enhanced E202A labelling is reflected in increased E202A detection on immunoblots in cells treated with lactacystin or another proteasome inhibitor, MG-132 (Figure 5C). The increased E202A levels are also reflected in a further decrease in albumin secretion in lactacystin-treated cells to only 10% of control (Figures 5D and 5E). From these results, we cannot unambiguously identify the E202A-positive structures, but we can conclude that E202A is associated with and is degraded by proteasomes, and its enhanced expression by proteasome inhibition leads to a greater impairment of secretion.
E202A expression or MAL2 knockdown leads to decreased soluble secretory protein levels
To determine whether impaired secretion was due to decreased secretory protein expression or enhanced intracellular retention, we examined albumin levels and distributions in control or infected WIF-B cells. In uninfected (results not shown) and in STK16-expressing cells (Figure 6A), the robustly and constitutively synthesized albumin was readily observed in the Golgi and in the ER (endoplasmic reticulum) (as a diffuse cytoplasmic stain). In contrast and somewhat surprisingly, the overall staining of albumin was dramatically decreased in E202A-expressing cells (Figure 6A) or cells where MAL2 was knocked down (Figure 6B). Immunoblotting lysates from control or overexpressing cells confirmed these observations and decreased albumin levels were detected (Figure 6C). In cells additionally treated with lactacystin, intracellular albumin levels decreased in a time-dependent manner correlating with increased E202A expression (Figure 6D). Similarly, albumin expression was decreased to 47.7±14.8% of control in cells knocked down for MAL2 expression (Figure 6E).
Figure 6. Albumin levels are decreased in cells overexpressing E202A or knocked down for MAL2.
(A) Cells overexpressing STK16 or E202A were immunolabelled for albumin. Asterisks mark E202A-expressing cells that lack albumin expression. (B) Control cells (a) or cells knocked down for MAL2 expression (b) were immunolabelled for albumin. Asterisks mark cells knocked down for MAL2 expression that have decreased albumin labelling. Scale bar, 10 μm. (C) WIF-B lysates from uninfected (control; con), or STK16- or E2020A-expressing cells were immunoblotted for albumin. (D) Cells overexpressing E202A were incubated with 5 μM lactacystin for the indicated times, and lysates were immunoblotted for E202A and albumin. Note the increase in E202A expression and reciprocal decrease in albumin expression with prolonged treatment. (E) Lysates from uninfected or MAL2-knockdown cells were immunoblotted for albumin.
To determine whether the effect was specific for albumin or a general secretory defect, we stained STK16- and E202A-expressing WIF-B cells for haptoglobin. As shown in Supplementary Figure S2(A) (http://www.biochemj.org/bj/463/bj4630201add.htm), expression of this secretory protein was also greatly diminished in E202A-expressing cells. To determine whether the defect was specific to soluble secretory cargo, we examined the distributions of the single-spanning basolateral resident HA321. As shown in Supplementary Figure S2(B), E202A expression did not change HA321 distributions or expression levels. Similar results were observed for another apical resident, CE9 (results not shown), indicating that only soluble secretory protein levels were decreased by E202A expression.
E202A expression or MAL2 knockdown leads to lysosomal degradation of secretory proteins
Decreased albumin and haptoglobin expression can be explained by two straightforward ways: (i) decreased secretory protein synthesis and processing; or (ii) redirection of secreted cargo for degradation. To discriminate between these possibilities, we first examined whether albumin was synthesized and processed in E202A-expressing cells. For these experiments, we used a 19°C temperature shift to block post-Golgi trafficking and allow albumin processing and accumulation. After only 2 h at 19°C, intracellular albumin fluorescence was observed in E202A-expressing cells; 54.7±9.9% cells were positive for ER and/or Golgi albumin labelling (albeit dimly) (Figures 7A and 7C). By 4 h, further enhanced levels of albumin were observed with 96.7±2.4% of E202A-expressing cells positive for ER- and Golgi-associated albumin staining (Figure 7C), indicating that robust albumin synthesis and processing was not altered. Similarly, albumin staining was partially recovered in MAL2-knockdown cells after 4 h at 19°C (Figure 7B). Interestingly, E202A labelling shifted from peripheral puncta to the Golgi during the temperature block, correlating with increased albumin detection. After 2 h at 19°C, E202A Golgi labelling was observed, and, by 4 h, E202A-positive puncta were virtually absent with only Golgi and diffuse cytosolic staining detected (Figure 7A).
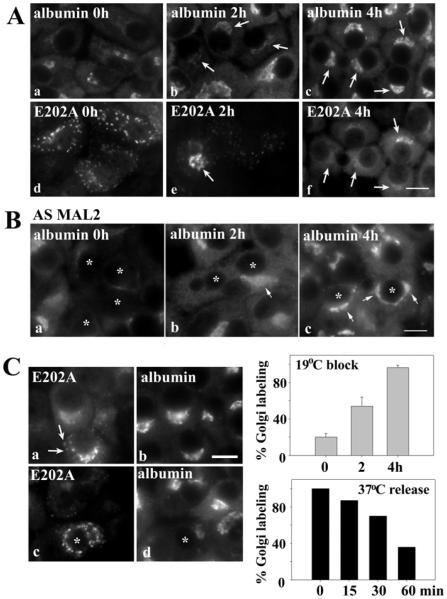
Figure 7. E202A expression or MAL2 knockdown does not impair albumin synthesis or Golgi delivery.
(A) E202A-expressing cells were incubated at 19°C for 0, 2 or 4 h as indicated. Cells were immunolabelled for albumin (a–c) or E202A (d–f). Arrows indicate recovered Golgi labelling of albumin in cells expressing E202A after the temperature block. (B) MAL2-knockdown cells were incubated at 19°C for 0, 2 or 4 h as indicated. Cells were immunolabelled for albumin. Asterisks indicate cells with knocked down MAL2 expression. Arrows mark recovered albumin labelling in knockdown cells after the temperature block. (C) Albumin and E202A distributions were monitored after incubation at 19°C for 4 h, then released back to 37°C for 15 or 30 min as indicated. Arrows mark E202A at the Golgi prior to redistribution back to the peripheral puncta. Scale bar, 10 μm. E202A-expressing cells were scored for the presence of albumin labelling in the Golgi after release from 19°C−for the indicated times. The percentage of infected cells positive for albumin labelling is plotted and shown in the top histogram. Values are expressed as the means ± S.E.M. Measurements were performed on at least three independent experiments. E202A-expressing cells were scored for the percentage of infected cells positive for albumin labelling is plotted in the bottom histogram. Values are expressed as the mean of two independent experiments.
To determine the fate of the Golgi-accumulated albumin and E202A, we monitored their distributions after release of the block to 37°C. After only 15 min, E202A labelling in puncta was observed with a reciprocal decrease in Golgi staining, and by 30 min, only E202A-positive puncta were observed (Figure 7C). This E202A redistribution was mirrored by decreased albumin detection. When quantified, we determined that, after 30 min at 37°C, 30% of the E202A-expressing cells had lost any visible albumin staining, and by 60 min, staining was absent in over 60% of cells (Figure 7C). There were no discernible albumin-positive structures/vesicles after the release, only an overall loss of albumin labelling. From these results we conclude that albumin is synthesized and processed in E202A-expressing cells and MAL2-knockdown cells and it is delivered from the ER to Golgi. Thus STK16 and MAL2 probably function in albumin sorting at the TGN.
To determine whether E202A expression led to albumin degradation, we monitored albumin expression after lysosomal deacidification with NH4Cl. After 1 h of deacidification, low albumin levels were observed in the Golgi, and by 2 h, even higher levels of Golgi labelling were detected (Figure 8A). Similarly, albumin detection was recovered in MAL2-knockdown cells (AS MAL2) after deacidification for 2 h (Figure 8B). Immunoblots confirmed these results where increased albumin expression correlated with increased deacidification (Figure 8C). Unlike in lactacystin-treated cells, no change in E202A levels was observed (Figure 8C). For comparison, lysosome deacidification in STK16-expressing cells did not change expression of STK16, albumin or MAL2 (Figure 8C). Taken together, these results indicate that decreased albumin levels in E202A- or AS MAL2-expressing cells is due to its degradation in lysosomes. As albumin was not observed in lysosomes or late endosomes after deacidification, we conclude further that STK16 and MAL2 probably regulate sorting at the TGN.
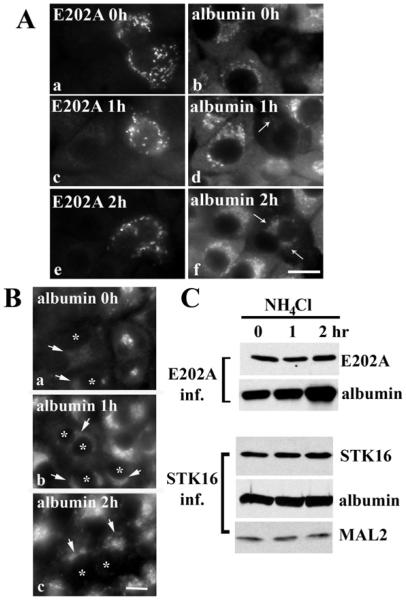
Figure 8. Albumin is degraded by lysosomes in E202A-expressing cells or in cells knocked down for MAL2 expression.
(A) E202A-expressing cells were treated with 50 mM NH4Cl for the indicated times and double-labelled for E202A and albumin as indicated. Arrows point to the emerging population of albumin after NH4Cl treatment. Scale bar, 10 μm. (B) Cells knocked down for MAL2 expression were treated with 50 mM NH4Cl for the indicated times and immunolabelled for albumin. Asterisks indicate cells with knocked down MAL2 expression. Arrows mark recovered albumin labelling in deacidified cells. (C) Whole-cell lysates from cells expressing E202A (top panels) or STK16 (bottom panels) after incubation with NH4Cl were immunoblotted for E202A, STK16, albumin and MAL2 as indicated. Albumin expression levels in E202A-expressing cells increased with prolonged NH4Cl treatment.
DISCUSSION
Using split-ubiquitin Y2H, we identified STK16 as a potential MAL2-binding partner. Overexpression of wild-type, but not a kinase-dead version of STK16 (E202A), led to the partial redistribution of MAL2 to the Golgi, suggesting that MAL2 and wild-type STK16 interact, which was confirmed by co-immunoprecipitations. Overexpression of the dominant-negative E202A led to a significant impairment in albumin expression and thus secretion, indicating that kinase activity is required for proper STK16 function, which further implies that constitutively phosphorylated substrates (probably not including MAL2, see below) must also participate in regulating albumin trafficking. Knockdown of MAL2 also impaired albumin expression and secretion, further confirming its association with STK16. Incubation of E202A-expressing or MAL2-knockdown cells at 19°C led to the reappearance of albumin expression, indicating that albumin synthesis or processing was not impaired. As lysosome deacidification also led to albumin reappearance in E202A-expressing or MAL2-knockdown cells, we conclude that albumin was being rerouted to lysosomes for degradation. As albumin was not observed in lysosomes or late endosomes after deacidification, we conclude further that STK16 and MAL2 regulate sorting at the TGN. Thus MAL2 and the constitutively active STK16 function to sort secretory soluble cargo into the constitutive secretory pathway at the TGN in polarized hepatocytes. To our knowledge, this is the first report that provides direct evidence for a role for STK16 and MAL2 in basolateral secretion regulation.
MAL2 and wild-type STK16 interact
The Y2H results and co-immunoprecipitations identified MAL2 and STK16 as binding partners. Although the binding domains have not yet been mapped, we have clues to their identities. We noticed that the Y2H screens that used MAL2 with Cub fused to its N-terminus were generally less successful than the screens where MAL2-Cub fusion proteins were used. One possible explanation for this observation is that the MAL2 N-terminus may mediate interactions with other proteins. This domain is the most divergent among MAL family members, and it is longer and more proline-rich (ten out of 34 amino acids are proline residues). Not only do proline-rich regions bind to SH3 (Src homology 3) domain proteins, they also confer structural rigidity, making the domain more available for protein–protein interactions. Sequence gazing also revealed that only MAL2 encodes VPPPP and FPPP sequences that resemble the F/L/W/YPPPP recognition sites for EVH1 [enabled, VASP (vasodilator-stimulated phosphoprotein), homology 1] motifs present in Ena/VASP proteins [26,27], suggesting MAL2 may bind members of this protein family. The N-terminal domain has also been shown to mediate interactions of MAL2 with members of the tumour protein D52 family [9]. The STK16 domain that mediates binding to MAL2 or its other binding partners (see below) has also not been identified. However, the clone identified in our Y2H screen encoded the last 118 amino acids of STK16 (178–305 amino acids) corresponding to 42% coverage. All but the last ten amino acids of this fragment encode part of the catalytic site. Although these ten amino acids are perfectly conserved among all STK16 species cloned to date, sequence gazing reveals no particularly obvious feature or sequence motif. We are currently doing mutational analysis to define important features of the MAL2 N-terminus and STK16 C-terminus that mediate protein–protein interactions.
STK16 is an atypical serine/threonine kinase with unknown substrates
STK16 belongs to the unique small family of numb-associated kinases. These divergent kinases share less than 25% identity with their nearest neighbour and are not well characterized. STK16 is a particularly interesting and enigmatic member of this family. Its structure reveals an atypical activation loop, and out of its 305 amino acids, only 29 are outside the catalytic site: the first 19 and the last ten amino acids [28]. Within the first 19 amino acids are three acylation sites: Gly2 is myristoylated and Cys6 and Cys8 are palmitoylated [12]. STK16 encodes no regulatory domains and is constitutively active [28]. From peptide library screens, it was determined that STK16 has a strong preference for threonine over serine as a phosphoacceptor, and an optimal threonine phosphorylation sequence of X-X-P/V/I-φ-H/Y-T*-N/G-X-X-X was identified [28]. DRG1 (developmentally regulated GTP-binding protein 1) is another known STK16-binding partner that is the only known substrate for STK16 [28]. MS analysis revealed that DRG1 is phosphorylated at Thr100 present in the proposed consensus sequence [28]. However, the physiological relevance or functional consequences of these findings are not yet understood.
To date, there is no evidence that MAL2 is a substrate for STK16. MAL2 does not encode the threonine-based consensus site described above nor does it encode any predicted threonine phosphorylation sites. Only one potential cytoplasmically orientated serine phosphorylation site (Ser17 ) has been predicted using various algorithms, but with relatively mediocre confidence scores. Furthermore, preliminary MS analysis of MAL2 N-terminal fragments (containing Ser17 ) isolated from either the 19 or 25 kDa immunoreactive species show no evidence of phosphorylation Omotade and J.Q. Zheng, personal communication). Similarly, N-acetylglucosamine kinase has been shown to be a STK16-binding partner although in vitro kinase assays indicate that it is not an STK16 substrate [29]. We have recently initiated a proteomics approach to identify potential STK16 substrates and possible regulators of constitutive secretion.
Possible mechanism for MAL2-mediated sorting
So far, MAL2 has been implicated in regulating hepatic transcytosis from basolateral endosomes to the sub-apical compartment and in pIgA-R (but not dipeptidyl peptidase IV or haemagglutinin) sorting from the TGN to the basolateral membrane [1,2,30]. These studies have now placed MAL2 as an important regulator of hepatic secretory cargo sorting. How does MAL2 recognize apical and/or secretory cargo? How are the different vesicles delivered to the correct cellular destination? From our earlier studies with pIgA-R, we predict MAL2 and the receptor interact via MAL2 cytoplasmic domains [2]. In contrast, mucin1 (a single spanning apical resident) and MAL2 probably interact via transmembrane domains [10]. It is not yet known whether the short luminal loops of MAL2 mediate interactions with secretory proteins. Alternatively, because MAL2 has been shown to be detergent-insoluble in WIF-B cells and its distribution dependent on cholesterol [31,32], MAL2 may promote lipid domain formation and/or stabilization that in turn recruits apical and/or secretory proteins to distinct vesicle populations. This is consistent with results from cholesterol-depleted cells where secretion was found to be impaired ([33] and J.G. In and P.L. Tuma, unpublished work) and with the identification of sorting signals on pancreatic constitutive secretory cargo in β-cells [34]. Once the apical cargo is packaged into discrete vesicles, we propose that sorting specificity may be conferred, in part, by interactions of distinct MAL2-binding partners with the divergent MAL2 N-terminal domain. However, a major question this model poses is what signals the binding of the different regulators to MAL2 to specify targeting. These studies suggest that, for at least soluble cargo, STK16-mediated phosphorylation may be an important signal.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Ann Hubbard (Johns Hopkins University School of Medicine, Baltimore, MD, U.S.A.) for providing many of the antibodies and reagents used in this study. We are also grateful to Dr John Golin (The Catholic University of America, Washington, DC, U.S.A.) for his assistance in performing the yeast two-hybrid screens and analysis.
FUNDING
This work was supported by the National Institutes of Health [grant numbers GM070801 and DK082890 (to P.L.T.)].
Abbreviations
- APN
aminopeptidase N
- AS
antisense
- BFA
brefeldin A
- DRG1
developmentally regulated GTP-binding protein 1
- ER
endoplasmic reticulum
- ERGIC
endoplasmic reticulum–Golgi intermediate compartment
- HRP
horseradish peroxidase
- MAL2
myelin and lymphocyte protein 2
- pIgA-R
polymeric IgA receptor
- STK16
serine/threonine kinase 16
- TGN
trans-Golgi network
- VASP
vasodilator-stimulated phosphoprotein
- Y2H
yeast two-hybrid
Footnotes
AUTHOR CONTRIBUTION
Pamela Tuma and Antonio Bernad were responsible for the experimental design. Julie In and Anneliese Striz performed all of the experiments. All of the authors analysed and interpreted the data. Figures were compiled by Pamela Tuma, Julie In and Anneliese Striz. Pamela Tuma wrote and edited the paper before submission.
REFERENCES
- 1.De Marco MC, Martin-Belmonte F, Kremer L, Albar JP, Correas I, Vaerman JP, Marazuela M, Byrne JA, Alonso MA. MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells. J. Cell Biol. 2002;159:37–44. doi: 10.1083/jcb.200206033. CrossRefPubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.In JG, Tuma PL. MAL2 selectively regulates polymeric IgA receptor delivery from the Golgi to the plasma membrane in WIF-B cells. Traffic. 2010;11:1056–1066. doi: 10.1111/j.1600-0854.2010.01074.x. CrossRefPubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madrid R, Aranda JF, Rodriguez-Fraticelli AE, Ventimiglia L, Andres-Delgado L, Shehata M, Fanayan S, Shahheydari H, Gomez S, Jimenez A, et al. The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev. Cell. 2010;18:814–827. doi: 10.1016/j.devcel.2010.04.001. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 4.De Matteis MA, Luini A. Exiting the Golgi complex. Nat. Rev. Mol. Cell Biol. 2008;9:273–284. doi: 10.1038/nrm2378. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 5.Jahn R, Scheller RH. SNAREs – engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 6.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 7.Weisz OA, Rodriguez-Boulan E. Apical trafficking in epithelial cells: signals, clusters and motors. J. Cell Sci. 2009;122:4253–4266. doi: 10.1242/jcs.032615. CrossRefPubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Boulan E, Kreitzer G, Musch A. Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell Biol. 2005;6:233–247. doi: 10.1038/nrm1593. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 9.Byrne JA, Nourse CR, Basset P, Gunning P. Identification of homo- and heteromeric interactions between members of the breast carcinoma-associated D52 protein family using the yeast two-hybrid system. Oncogene. 1998;16:873–881. doi: 10.1038/sj.onc.1201604. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 10.Fanayan S, Shehata M, Agterof AP, McGuckin MA, Alonso MA, Byrne JA. Mucin 1 (MUC1) is a novel partner for MAL2 in breast carcinoma cells. BMC Cell Biol. 2009;10:7. doi: 10.1186/1471-2121-10-7. CrossRefPubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madrid R, Aranda JF, Rodriguez-Fraticelli AE, Ventimiglia L, Andres-Delgado L, Shehata M, Fanayan S, Shahheydari H, Gomez S, Jimenez A, et al. The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev. Cell. 2010;18:814–827. doi: 10.1016/j.devcel.2010.04.001. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 12.Berson AE, Young C, Morrison SL, Fujii GH, Sheung J, Wu B, Bolen JB, Burkhardt AL. Identification and characterization of a myristylated and palmitylated serine/threonine protein kinase. Biochem. Biophys. Res. Commun. 1999;259:533–538. doi: 10.1006/bbrc.1999.0811. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 13.Kurioka K, Nakagawa K, Denda K, Miyazawa K, Kitamura N. Molecular cloning and characterization of a novel protein serine/threonine kinase highly expressed in mouse embryo. Biochim. Biophys. Acta. 1998;1443:275–284. doi: 10.1016/s0167-4781(98)00224-3. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 14.Ligos JM, Gerwin N, Fernandez P, Gutierrez-Ramos JC, Bernad A. Cloning, expression analysis, and functional characterization of PKL12, a member of a new subfamily of Ser/Thr kinases. Biochem. Biophys. Res. Commun. 1998;249:380–384. doi: 10.1006/bbrc.1998.9163. [DOI] [PubMed] [Google Scholar]
- 15.Stairs DB, Perry Gardner H, Ha SI, Copeland NG, Gilbert DJ, Jenkins NA, Chodosh LA. Cloning and characterization of Krct, a member of a novel subfamily of serine/threonine kinases. Hum. Mol. Genet. 1998;7:2157–2166. doi: 10.1093/hmg/7.13.2157. [DOI] [PubMed] [Google Scholar]
- 16.Stairs DB, Notarfrancesco KL, Chodosh LA. The serine/threonine kinase, Krct, affects endbud morphogenesis during murine mammary gland development. Transgenic Res. 2005;14:919–940. doi: 10.1007/s11248-005-1806-6. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 17.Shanks MS, Cassio D, Lecoq O, Hubbard AH. An improved rat hepatoma hybrid cell line. Generation and comparison with its hepatoma relatives and hepatocytes in vivo. J. Cell Sci. 1994;107:813–825. doi: 10.1242/jcs.107.4.813. PubMed. [DOI] [PubMed] [Google Scholar]
- 18.Guinea B, Ligos JM, Lain de Lera T, Martin-Caballero J, Flores J, Gonzalez de la Pena M, Garcia-Castro J, Bernad A. Nucleocytoplasmic shuttling of STK16 (PKL12), a Golgi-resident serine/threonine kinase involved in VEGF expression regulation. Exp. Cell Res. 2006;312:135–144. doi: 10.1016/j.yexcr.2005.10.010. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 19.Bastaki M, Braiterman LT, Johns DC, Chen YH, Hubbard AL. Absence of direct delivery for single transmembrane apical proteins or their “secretory” forms in polarized hepatic cells. Mol. Biol. Cell. 2002;13:225–237. doi: 10.1091/mbc.01-07-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Ihrke G, Neufeld EB, Meads T, Shanks MR, Cassio D, Laurent M, Schroer TA, Pagano RE, Hubbard AL. WIF-B cells: an in vitro model for studies of hepatocyte polarity. J. Cell Biol. 1993;123:1761–1775. doi: 10.1083/jcb.123.6.1761. CrossRefPubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartles JR, Feracci HM, Stieger B, Hubbard AL. Biogenesis of the rat hepatocyte plasma membrane in vivo: comparison of the pathways taken by apical and basolateral proteins using subcellular fractionation. J. Cell Biol. 1987;105:1241–1251. doi: 10.1083/jcb.105.3.1241. CrossRefPubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.In JG, Ihrke G, Tuma PL. Analysis of polarized membrane traffic in hepatocytes and hepatic cell lines. Curr. Protoc. Cell Biol. 2012;15 doi: 10.1002/0471143030.cb1517s54. Unit 15.17 PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marazuela M, Martin-Belmonte F, Garcia-Lopez MA, Aranda JF, de Marco MC, Alonso MA. Expression and distribution of MAL2, an essential element of the machinery for basolateral-to-apical transcytosis, in human thyroid epithelial cells. Endocrinology. 2004;145:1011–1016. doi: 10.1210/en.2003-0652. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 25.Dahan S, Dominguez M, McNiven MA. Molecular mechanisms of hepatocellular vesicle formation. In: Arias I, Boyer J, Chisari F, Fausto N, Schachter D, Shafritz D, editors. The Liver: Biology and Pathology. Lippincott Williams and Wilkins; Philadelphia: 2001. pp. 97–117. [Google Scholar]
- 26.Renfranz PJ, Beckerle MC. Doing (F/L)PPPPs: EVH1 domains and their proline-rich partners in cell polarity and migration. Curr. Opin. Cell Biol. 2002;14:88–103. doi: 10.1016/s0955-0674(01)00299-x. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Pulido L, Martin-Belmonte F, Valencia A, Alonso MA. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem. Sci. 2002;27:599–601. doi: 10.1016/s0968-0004(02)02229-6. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 28.Eswaran J, Bernad A, Ligos JM, Guinea B, Debreczeni JE, Sobott F, Parker SA, Najmanovich R, Turk BE, Knapp S. Structure of the human protein kinase MPSK1 reveals an atypical activation loop architecture. Structure. 2008;16:115–124. doi: 10.1016/j.str.2007.10.026. CrossRefPubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ligos JM, de Lera TL, Hinderlich S, Guinea B, Sanchez L, Roca R, Valencia A, Bernad A. Functional interaction between the Ser/Thr kinase PKL12 and N-acetylglucosamine kinase, a prominent enzyme implicated in the salvage pathway for GlcNAc recycling. J. Biol. Chem. 2002;277:6333–6343. doi: 10.1074/jbc.M105766200. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 30.de Marco MC, Puertollano R, Martinez-Menarguez JA, Alonso MA. Dynamics of MAL2 during glycosylphosphatidylinositol-anchored protein transcytotic transport to the apical surface of hepatoma HepG2 cells. Traffic. 2006;7:61–73. doi: 10.1111/j.1600-0854.2005.00361.x. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 31.Ramnarayanan SP, Cheng CA, Bastaki M, Tuma PL. Exogenous MAL reroutes selected hepatic apical proteins into the direct pathway in WIF-B cells. Mol. Biol. Cell. 2007;18:2707–2715. doi: 10.1091/mbc.E07-02-0096. CrossRefPubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramnarayanan SP, Tuma PL. MAL, but not MAL2, expression promotes the formation of cholesterol-dependent membrane domains that recruit apical proteins. Biochem. J. 2011;439:497–504. doi: 10.1042/BJ20110803. CrossRefPubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Thiele C, Huttner WB. Cholesterol is required for the formation of regulated and constitutive secretory vesicles from the trans-Golgi network. Traffic. 2000;1:952–962. doi: 10.1034/j.1600-0854.2000.011205.x. CrossRefPubMed. [DOI] [PubMed] [Google Scholar]
- 34.Lara-Lemus R, Liu M, Turner MD, Scherer P, Stenbeck G, Iyengar P, Arvan P. Lumenal protein sorting to the constitutive secretory pathway of a regulated secretory cell. J. Cell Sci. 2006;119:1833–1842. doi: 10.1242/JCS.02905. CrossRefPubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.