Preface
Ageing is affected by both genetic and non-genetic factors. Here, we review the chromatin-based epigenetic changes that occur during ageing, the role of chromatin modifiers in modulating lifespan and the importance of epigenetic signatures as biomarkers of ageing. We also discuss how epigenome remodeling by environmental stimuli impacts several aspects of transcription and genomic stability, with important consequences on longevity, and outline epigenetic differences between the ‘mortal soma’ and the ‘immortal germline’. Finally, we discuss the inheritance of ageing characteristics and potential chromatin-based strategies to delay or reverse hallmarks of ageing or age-related diseases.
Introduction
The lifespan of an organism encompasses a period of growth that culminates in sexual maturity, a period of maximal fitness and fertility, and a period of ageing that is characterized by functional decline and an increased probability of death. Ageing is associated with loss of functionality at the cellular, tissue and organismal level and a wide range of diseases, including cardiovascular and neurodegenerative diseases, metabolic disorders and many cancers. Highly relevant to human ageing, ‘healthspan’ is the duration of disease-free physiological health (for example high cognition and mobility). Understanding the changes that occur during ageing, and identifying regulators of lifespan and healthspan, should pave the way for interventions to promote a longer youthful period, increase vigor and, potentially, reverse some hallmarks of ageing.
The discovery of long-lived mutants in invertebrate model systems supports the idea that the ageing process can be genetically modulated1. In addition to genetic inputs, evidence implicates non-genetic factors in ageing. Indeed, studies in humans have estimated the non-heritable portion of lifespan regulation to be ~70%2. Environmental stimuli, such as dietary manipulations or stress, can potently influence the lifespan and healthspan of animals across various species3. The importance of non-genetic factors is further underscored in eusocial insects that have a caste system with queens and workers (for example, honey bees), where individuals with similar genomes exhibit large differences in lifespan (~10 fold difference between queen and workers)4. Such differences can be triggered by early exposure to environmental stimuli and are relatively stable throughout life, although some remodeling is still possible later on4. Even in relatively controlled environments, lifespan among isogenic individuals is highly variable with large differences between the first and last death5. Although these differences could be purely stochastic, this result suggests that even minute environmental variations may cumulatively influence lifespan. While a portion of this non-genetic variation may stem from somatic mutations, the majority likely results from other non-genetic, regulated factors that stably impact healthspan and longevity.
In this Review, we examine evidence for an epigenetic component in the regulation of ageing. We will use the term epigenetic broadly to refer to changes in genomic regulation, although we note that in its strictest definition, this term encompasses only heritable phenotypic changes without changes to the underlying gene sequence6. Among the different modes of genome regulation, we will particularly focus on chromatin (Box 1) for several reasons. First, changes to chromatin states influence transcription and could underlie at least in part the transcriptional changes that are observed with ageing7, 8. Second, as chromatin can be influenced by the environment9, it could act as an interface through which environmental signals interact with genetic components throughout lifespan. Finally, stable changes to the chromatin landscape could preserve memory of past environmental exposures, leading to long-lasting phenotypic effects10, 11 that may be particularly relevant for ageing.
Box 1. Chromatin structure and regulation.
Chromatin is the nucleo-proteic complex that allows the genome to be packaged inside the nucleus. It can be broadly categorized into two states: euchromatin, a loose and transcription-permissive compartment, and heterochromatin, a dense and compact compartment that harbors repressed DNA. The basic repeating units of chromatin, ‘nucleosomes’, consist of ~150bp of DNA wrapped around histone protein octamers (octamers contain two each of the H2A, H2B, H3 and H4 histone proteins)176. Post-translational modifications of histone protein N-terminal tails are thought to regulate the accessibility and expression potential of underlying genes177, a hypothesis referred to as the ‘histone code’ hypothesis177. Interestingly, in addition to canonical histone proteins, the transcription of which is usually restricted to S-phase, some histone variants can be expressed at any point during the cell cycle. These specific histone variants have been involved in regulatory aspects of gene expression (for example, H2A.Z or H3.3) or chromatin structure (for example, CENP-A)178. The best-studied core histone modifications include Lys and Arg methylation, Lys acetylation, Ser/Thr phosphorylation and Lys poly- or mono-ubiquitinylation (reviewed in179). These modifications can be added and removed by ‘chromatin modifiers’, including histone acetyltransferases (HAT), histone deacetylases (HDAC), histone methyltransferases (HMT) and histone demethylases (KDM).
Another layer of regulation resides in the methylation status of Cytosines in CG dinucleotides (referred to as ‘CpG’), and the most frequent type of methylation occurs on carbon 5 of cytosine (5-mC)180. In mammals, DNA methylation is generally associated with gene repression, though whether methylation plays an instructive role in repression is still unclear180. Approximately 60–90% of all CpGs are methylated in mammalian genomes, with the notable exception of hypomethylated ‘CpG islands’ (large regions with concentrated CpGs in gene-rich regions)180. DNA-methyltransferases (DNMTs) methylate DNA, and the removal process is initiated by the Ten-Eleven Translocation (TET) proteins180.
Finally, chromatin can be regulated through the positioning of nucleosomes with respect to regulatory sequences and the compaction of chromatin in a 3D structure. ATP-dependent chromatin remodelers (for example, SWI/SNF or ISWI) can impact the chromatin and transcriptional landscape by modifying nucleosome positioning, chromatin higher-order structure and overall nuclear organization77.
We will bring ageing and epigenetic research together to provide an integrated picture of the non-genetic regulation of lifespan and healthspan. We describe chromatin changes that occur during ageing and present emerging evidence for a role of chromatin modifiers in modulating lifespan. We also highlight the use of epigenetic signatures as ‘ageing clocks’ (Box 2). We then explore the influence of environmental inputs that affect ageing on epigenetic remodeling, and the potential consequences of this remodeling on genome stability and transcription. We evaluate the contributions of epigenetics in long-lasting, even heritable longevity phenotypes. Finally, we discuss remaining outstanding questions and challenges, including how to develop epigenetic strategies to counter or reverse age-related diseases. Understanding the importance of chromatin-based epigenetic mechanisms in ageing is timely because of recent advances in ultra-high throughput technology (Supplementary information S1 (Box)), which have revolutionized our knowledge of epigenetic factors and their relationships to gene regulation. Reference epigenomes provided by ENCODE and Roadmap epigenomics12, 13 should also aid our understanding of the remodeling and persistence of epigenomic patterns during ageing. These questions are all the more pressing given the increasing life expectancy in all parts of the world, and the socioeconomic and medical challenges to meet growing health demands.
Box 2. Epigenetic modifications as biomarkers of age.
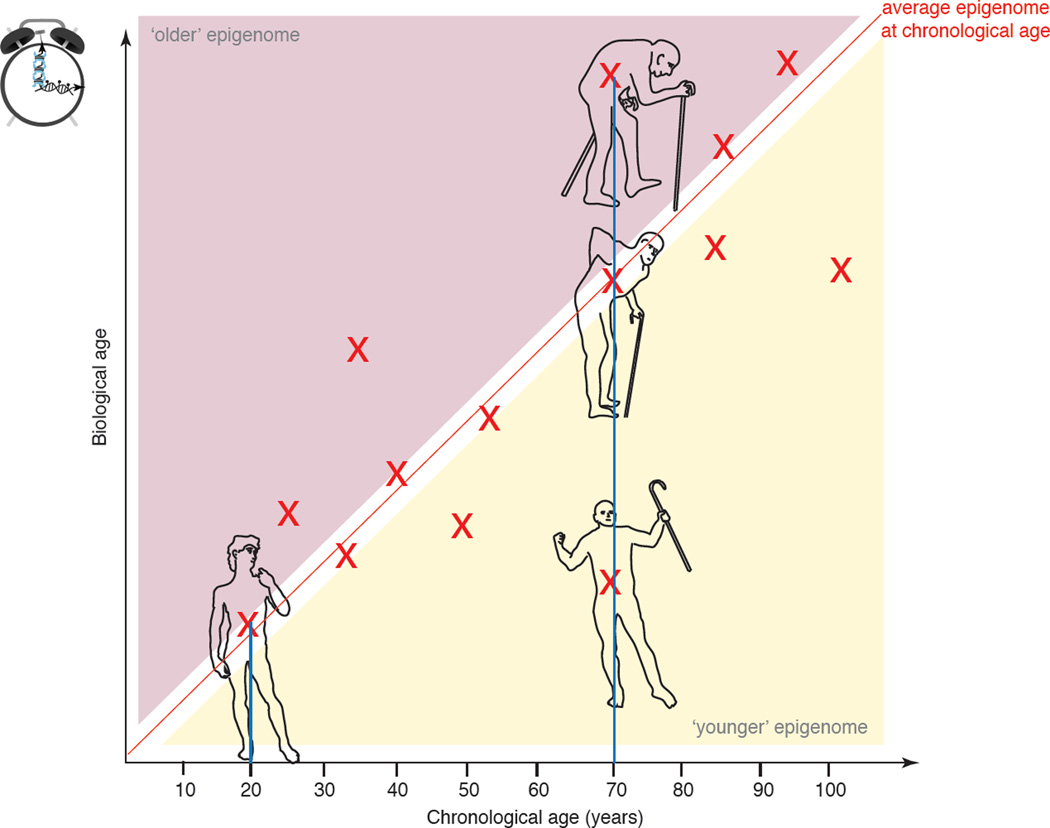
Epigenetic modifications can persist in cells even between cell divisions, and an emerging hypothesis is that epigenetic landscapes are ‘biological sensors’ that reflect cellular identity, cell health and youthfulness. But do chromatin landscapes track with chronological age (that is, the age of the individual in time units) or biological age (that is, the age in years of the population average most similar to the individual) (see model in the figure)?
Studies have explored the use of changes in DNA methylation (DNA methylation age) as a sensor for both chronological age and biological age across species and cell types. Based on the methylation status of a defined number of CpGs, highly accurate predictors of chronological age have been constructed in a given tissue, such as saliva181 or peripheral blood cells26–28. Surprisingly, these types of analyses have resulted in accurate estimators of age that seems to apply to human tissues and cell types regardless of their developmental potential28, 86. For example, embryonic stem cells and induced pluripotent stem cells are estimated to be ‘ageless’ by such models, and the DNA methylation age of sperm cells (which are part of the ‘immortal’ germline) is estimated to be far younger than that of the somatic cells of the same donor26, 28. While DNA methylation allows accurate modeling of chronological age with high correlation coefficients27, 28, there is significant deviation from the linear fit, suggesting that methylation patterns may also reflect youthfulness or health (biological age) (see model in figure). Indeed, the use of DNA methylation age may predict the health of various tissues. For example, the liver tissue of obese subjects displays increased DNA methylation age compared to muscle or blood tissue from the same individual86. Furthermore, an increased DNA methylation age of blood cells can be observed in subjects with some progeroid diseases, including aplastic anemia and dyskeratosis congenita26, or Down syndrome182. Moreover, DNA methylation-based age of blood cells can predict mortality by all causes in later life, even after adjusting for traditional risk factors and blood cell counts183.
Previously proposed biomarkers of ageing include expression levels of the cell cycle inhibitor p16-INK4A184 and telomere length attrition rate185. However, CpG DNA-methylation outperforms these molecular markers as predictors of chronological age for unknown reasons28, 86, 183. Whether greater deviations between chronological (‘real’) age and biological (predicted) age reflect actual disparities in ageing rates or represent potential oversimplifications (overfitting) of specific models remains unclear. Other DNA modifications, such as 5-hmC and other new oxidation derivatives186 or 6-mA23–25, could also be integrated into models of ‘epigenetic’ ageing clocks. Finally, although histone modification patterns are still unexplored as epigenetic biomarkers of age, they may increase the accuracy and scope of current biological clock models. As ‘clock measurements’ are mostly taken on entire tissues, it will be crucial to determine the role of age-related changes in cell composition within tissues in the predictive nature of the epigenetic clocks19.
Chromatin modifications and ageing
A role for epigenetics in ageing is supported by evidence that chromatin is altered during the ageing process and that interfering with chromatin regulatory complexes impacts lifespan. We will present findings from model organisms (yeast, worms, flies, and mice) and from cellular models of replicative senescence. We will also discuss results from organismal and cellular models of diseases with accelerated signs of ageing (progeroid syndromes), such as Hutchinson-Gilford progeria syndrome (HGPS) or Werner syndrome (WS). In presenting this evidence, we note that the precise role of chromatin in lifespan is difficult to establish. Indeed, chromatin-modifying enzymes can often also affect non-chromatin substrates, or target multiple chromatin sites that could influence ageing. Furthermore, swapping modified DNA or histone residues by ones that cannot be modified, and to do so in a locus-specific manner, is technically difficult to achieve. Nevertheless, the evidence accumulated so far highlights the importance of many chromatin features during ageing.
DNA methylation in ageing
Methylation of the 5-carbon of cytosines in CpG dinucleotide sites is a conserved epigenetic modification that is classically linked to transcriptional silencing in vertebrates (Box 1). DNA methylation undergoes remodeling during ageing in various tissues in mice and humans (Table 1 and supplementary information S2 (table)). In human cells, the global levels of 5-methylcytosine (5-mC) are reduced in senescent compared to actively cycling cells14, 15 (Table 1). Studies probing the methylation status of specific CpG sites across the genome suggest that the methylation of such sites outside of promoter CpG ‘islands’ tends to decrease in a variety of tissues in humans16. In contrast, in both mice and humans, CpG ‘islands’ near promoters are typically hypermethylated with age, most notably on genes involved in differentiation or development16–18. Although changes in DNA methylation in heterogeneous tissues may partly reflect alterations in cell composition19, it is interesting to note that age-dependent changes in DNA methylation are detected in more homogeneous cell populations, such as hematopoietic stem cells (HSCs)20, 21. The consequences of age-dependent changes in DNA methylation for cellular function are not yet completely clear, but interestingly, in HSCs, regions with altered DNA methylation overlap binding sites of specific sets of transcription factors21. This observation raises the possibility that these methylation changes could influence the targeting of transcription factors, and thereby contribute to misregulation of gene expression during ageing21. While many studies have focused on 5-mC, the discovery of other forms of DNA methylation, including 5-hydroxymethylcytosine (5-hmC), cytosine methylation at non-CpG dinucleotides22, and N6-methylation of adenines (6-mA)23–25, deserve exploration in the context of ageing. Interestingly, recent studies in humans suggest a link between 5-mC DNA methylation, chronological age, and even ‘biological’ age26–28 (Box 2). Thus, DNA methylation could represent an ‘ageing clock’.
Table 1.
Epigenetic changes observed during ageing¥
| Epigenetic mark |
Functional role |
Ageing paradigm |
Change in epigenetic mark with ageing |
Change observed in tissue/cells |
Detection method |
Species |
|---|---|---|---|---|---|---|
| 5-mC | Transcriptional repression (?) | Organismal (WS) | No global change | MSCs | RRBS | H. sapiens |
| Cellular (senescence) | Global decrease, local increases | Fibroblasts | Immunostaining, WGBS, chromatography | H. sapiens, M. musculus, M. auratus | ||
| Organismal | Minor global increase, local increasesand some local decreases | HSCs | RRBS, WGBS | M. musculus | ||
| Organismal | Local increases and some local decreases | Small intestine, Colon, Lung, Liver, Spleen, Brain, Blood, Kidney, Muscle | Pyrosequencing, MCAM, Beadchip arrays | H. sapiens, M. musculus | ||
| Organismal | Increase (LINEs), local decreases and rare local increases | Sperm | Pyrosequencing (LINEs), Beadchip arrays | H. sapiens | ||
| Organismal | No global change | Cortex | WGBS | M. musculus, H. sapiens | ||
| 5-hmC | Transcriptional activation (?) | Organismal | Minor increase (SINEs, LTRs) | Cerebellum | Immunostaining, Chemical tagging/ sequencing | M. musculus |
| Organismal | Minor global decrease | HSCs | HPLC-MS | M. musculus | ||
| mCH (where H represents A,C,T) | (?) | Organismal | Minor global decrease | Cortex | WGBS | H. sapiens, M. musculus |
| *Histone H2A | Core chromatin component | Cellular (senescence) | Decrease | Fibroblasts | Western blot | H. sapiens |
| Organismal (replicative lifespan) | Decrease | Yeast cells | Western blot | S. cerevisiae | ||
| *Histone H2B | Core chromatin component | Cellular (senescence) | Decrease | Fibroblasts | Western blot | H. sapiens |
| Organismal | Decrease | Muscle stem cells | Transcriptional profiling | M. musculus | ||
| *Histone H3 | Core chromatin component | Cellular (senescence) | Decrease | Epidermis, Fibroblasts | Immunostaining, Western blot, SILAC-MS | H. sapiens |
| Organismal (replicative lifespan) | Decrease, changes in occupancy | Yeast cells | Western blot, SILAC-MS | S. cerevisiae | ||
| Organismal | Decrease | Soma, Whole male flies, Muscle stem cells | Western blot (normalization to tubulin), Transcriptional profiling | C. elegans, D. melanogaster, M. musculus | ||
| Organismal | No Change | Head | Western blot (normalization to total protein or DNA) | D. melanogaster | ||
| H4 | Core chromatin component | Cellular (senescence) | Decrease | Fibroblasts | Western blot, SILAC-MS | H. sapiens |
| macroH2A | Component of heterochromatin | Cellular (senescence) | Increase | Fibroblasts | Immunostaining, Western blot | H. sapiens |
| Organismal | Increase | Lung, Liver | Immunostaining, Western blot | M. musculus | ||
| *HP1 | Component of heterochromatin | Organismal | Remodeling | Head, Gut, Fat cells | Immunostaining, ChIP-chip | D. melanogaster |
| HP1α | Component of heterochromatin | Organismal (HGPS, WS) | Decrease | Fibroblasts, MSCs | Immunostaining, Western blot | H. sapiens |
| Organismal | Decrease | MSCs | Western blot | H. sapiens | ||
| HP1β | Component of heterochromatin | Cellular (senescence) | Increase | Fibroblasts | Immunostaining, Western blot | H. sapiens |
| HP1γ | Component of heterochromatin | Organismal (HGPS) | Decrease | Fibroblasts | Immunostaining | H. sapiens |
| Organismal | Decrease | Fibroblasts | Immunostaining | H. sapiens | ||
| H3K9me1 | Enriched in euchromatin along gene bodies, transcriptional repression | Cellular (senescence) | Increase | Fibroblasts | Western blot, SILAC-MS | H. sapiens |
| H3K9me2 | Enriched in euchromatin along gene bodies, transcriptional repression | Cellular (senescence) | Decrease | Fibroblasts | Western blot, SILAC-MS | H. sapiens |
| Organismal | Decrease | Whole male flies | Western blot | D. melanogaster | ||
| H3K9me3 | Enriched in heterochromatin regions, transcriptional silencing | Organismal (HGPS, WS) | Decrease | Fibroblasts, MSCs | Immunostaining, Western blot, ChIP-seq | H. sapiens |
| Cellular (senescence) | Decrease | Fibroblasts | SILAC-MS, Western blot | H. sapiens | ||
| Organismal | Increase, remodeling | Head | Western blot, ChIP-chip | D. melanogaster | ||
| Organismal | Decrease | Fibroblasts, Soma | Immunostaining, Western blot | H. sapiens, C. elegans | ||
| H3K27me3 | Enriched in euchromatin along gene bodies, transcriptional repression | Organismal (HGPS, WS) | Decrease, remodeling | Fibroblasts, MSCs | Immunostaining, ChIP-Seq | H. sapiens |
| Cellular (senescence) | Remodeling | Fibroblasts | ChIP-Seq | H. sapiens | ||
| Organismal | Decrease | Soma | Western blot | C. elegans | ||
| Organismal | Increase, remodeling | Brain, Muscle stem cells, HSCs | Immunostaining, ChIP-Seq | N. furzeri, M. musculus | ||
| H4K20me2 | DNA repair and genomic stability | Cellular (senescence) | Increase | Fibroblasts | Western blot, SILAC-MS | H. sapiens |
| H4K20me3 | Enriched in pericentric heterochromatin | Organismal (HGPS) | Increase | Fibroblasts | Western blot, Immunostaining | H. sapiens |
| Cellular (senescence) | Decrease | Fibroblasts | Western blot, SILAC-MS | H. sapiens | ||
| Organismal | Increase | Liver, Kidney | Liquid chromatography | R. norvegicus | ||
| H3K4me2 | Enriched in euchromatin along gene body, transcriptional activation | Organismal | Global increase, remodeling | Cortex | ChIP-Seq | M. mulatta |
| H3K4me3 | Enriched in euchromatin at promoters, transcriptional activation | Organismal (WS) | No global change | MSCs | ChIP-Seq | H. sapiens |
| Cellular (senescence) | Remodeling | Fibroblasts | ChIP-Seq | H. sapiens | ||
| Organismal | No change | Soma | Western blot | C. elegans | ||
| Organismal | Decrease, remodeling | Head | ChIP-chip | D. melanogaster | ||
| Organismal | Minor global increase, remodeling | Neurons, HSCs, Muscle stem cells | ChIP-Seq | H. sapiens, M. musculus | ||
| H3K36me3 | Enriched in euchromatin along gene body, transcriptional elongation | Organismal (replicative lifespan) | No global change, remodeling | Yeast cells | ChIP-Seq | S. cerevisiae |
| Organismal | Minor global decrease, remodeling | Soma, Head | Western blot, ChIP-chip, ChIP-Seq | C. elegans, D. melanogaster | ||
| H3K56ac | DNA replication, DNA damage response, nucleosome assembly | Cellular (senescence) | Decrease | Fibroblasts | Immunostaining, Western blot | H. sapiens |
| Organismal (replicative lifespan) | Decrease | Yeast cells | Western blot | S. cerevisiae, | ||
| H4K16ac | Regulation of telomere silencing, regulation of chromatin compaction (?) | Organismal (HGPS) | Decrease | Fibroblasts, Liver | Immunostaining, Western blot | M. musculus |
| Cellular (senescence) | Decrease | Fibroblasts | Western blot | H. sapiens | ||
| Organismal (replicative lifespan) | Increase | Yeast cells | Western blot | S. cerevisiae | ||
| Organismal | Decrease | Liver, Kidney | Western blot | H. sapiens, M. musculus | ||
| H4K12ac | Enriched in euchromatin along gene body, transcriptional elongation | Organismal | Decrease upon contextual fear conditioning | Hippocampus | Western blot | M. musculus |
see supplementary information S2 (table) for a fully referenced version of this table.
Note that the changes in chromatin reported for WS come from mesenchymal cells derived from an ESC model of WS. Chromatin changes reported for HGPS come from patient-derived cells or a genetic mouse model of the disease.
Reported studies of core histone changes correspond to all variants of a particular histone family (not assessed separately).
(?) Functional role of epigenetic mark unclear
see supplementary information S1 (box) to read more about targeted and global methods of epigenome mapping
Abbreviations: HGPS: Hutchinson-Gilford Progeria Syndrome; WS: Werner Syndrome; Soma: all body tissues except the germline cells; HSCs: hematopoietic stem cells; MSCs: mesenchymal stem cells; HPLC-MS: high performance liquid chromatography-mass spectrometry; RRBS: reduced representation bisulfite sequencing; WGBS: whole genome bisulfite sequencing; SILAC-MS: Stable isotope labeling by amino acids in cell culture- mass spectrometry; MCAM: methylated CpG island amplification microarrays; ChIP-Seq: Chromatin Immunoprecipitation followed by sequencing; ChIP-chip: Chromatin Immunoprecipitation followed by microarray hybridization; LINE: long interspersed nuclear element; SINE: short interspersed nuclear element; LTR: long tandem repeat.
Traditional model organisms for ageing are not well-suited for studying the role of 5-mC methylation in organismal lifespan, as this modification has not been observed in Saccharomyces cerevisiae29 or Caenorhabditis elegans30. However, DNA methylation is present in Drosophila melanogaster29, another invertebrate model for ageing. In D. melanogaster, overexpression of the putative DNA methyltransferase dDnmt2 increases longevity, whereas flies lacking a functional copy of the gene are short-lived31 (Table 2 and Supplementary information S3 (table)). Whether lifespan changes in dDnmt2 mutants result from changes in DNA methylation is still subject to debate. While an initial study identified a potential role for dDnmt2 in the methylation of CpG at transposons32, a subsequent study using bisulfite sequencing on wild-type and dDnmt2 mutant flies did not33. Additional evidence supports a role for dDNMT2 in tRNA methylation33. Thus, how CpG methylation impacts longevity remains unclear, but the relationship between DNA methylation and biological age in humans (Box 2) suggests that it could be used as a biomarker of ageing.
Table 2.
Modulation of lifespan by components and modifiers of chromatin¥
| Chromatin modification activity |
Protein (complex) |
Target histone and/or residue |
Chemical inhibitor |
Chemical activator |
Wild-type impact on health or lifespan |
Species |
|---|---|---|---|---|---|---|
| DNA methylation | dDNMT2 | 5-mC (?) | N/A | N/A | + | D. melanogaster |
| Histone Acetyltransferase | SAS2 | H4K16 | N/A | N/A | − | S. cerevisiae |
| GCN5 (SAGA complex) | H3, H3K14 | N/A | N/A | + | S. cerevisiae | |
| IKI3, SAS3 | H3K9, H3K14, H3K18 | spermidine | N/A | −/= | S. cerevisiae, D. melanogaster, C. elegans | |
| CBP | H4K5 | N/A | N/A | + | C. elegans | |
| Histone Deacetylase (Class I and II) | ? | Global effect | TrichostatinA, NaButyrate, PhenylButyrate, SAHA | N/A | + | D. melanogaster, C. elegans, M. musculus |
| RPD3 | H4K5, H4K12 | N/A | N/A | − | S. cerevisiae, D. melanogaster | |
| Histone Deacetylase (Class III) | SIR2(yeast), SIR2.1(worm), dSIR2 (fly) | H3K9, H3K14, H4K16 | Nicotinamide | N/A | + +/= +/= |
S. cerevisiae, D. melanogaster (?), C. elegans (?) |
| SIRT1 | H3K9, H3K14, H4K16 | Nicotinamide | SRT1720, SRT2104, SRT3025 | + | M. musculus | |
| SIRT6 | H3K9, H3K56 | Nicotinamide | N/A | + | M. musculus | |
| Histone Lysine methyltransferase | SET2 | H3K36 | N/A | N/A | +/− | S. cerevisiae |
| MET-1 | H3K36 (me3) | N/A | N/A | + | C. elegans | |
| SET-15 | ? | N/A | N/A | − | C. elegans | |
| SET-9, SET-26 | H3K4 (me1/2) | N/A | N/A | − | C. elegans | |
| SET1 | H3K4 (me3) | N/A | N/A | + | S. cerevisiae | |
| ASH-2, SET-2, WDR-5 (COMPASS complex) | H3K4 (me3) | N/A | N/A | − | C. elegans | |
| E(Z) (fly), MES-2 (worm) (Polycomb complex) | H3K27 | N/A | N/A | − | D. melanogaster, C. elegans | |
| Histone Lysine demethylase | RPH1 | H3K36 (me3) | N/A | N/A | − | S. cerevisiae |
| LSD-1 (T08D10.2 gene) | H3K4 (me2) | N/A | N/A | − | C. elegans | |
| RBR-2 (worm), LID (fly) | H3K4 (me3) | N/A | N/A | +/− +/= |
C. elegans, D. melanogaster | |
| JMJD-2 (worm), KDM4A(fly) | H3K9 | N/A | N/A | − −/= |
C. elegans, D. melanogaster | |
| UTX-1 | H3K27 | N/A | N/A | − | C. elegans | |
| Histone O-GlcNac regulation | OGT-1 | ? | N/A | N/A | + | C. elegans |
| OGA-1 | ? | N/A | N/A | −/= | C. elegans | |
| Histone monoubiquitinyl-transferase | RAD6, BRE1 (H2B monoubiquitination complex) | H2BK123 | N/A | N/A | + | S. cerevisiae |
| Nucleosome remodelers | ISW2 (ISWI complex) | N/A | N/A | N/A | − | S. cerevisiae, C. elegans |
| SWI/SNF | N/A | N/A | N/A | + | C. elegans | |
| CHD1 | N/A | N/A | N/A | − | S. cerevisiae | |
| LET-418 (worm), dMI2 (fly) (NurD complex) | N/A | N/A | N/A | − | C. elegans, D. melanogaster | |
| Histone chaperone | ASF1 | N/A | N/A | N/A | + | S. cerevisiae |
| HIR | N/A | N/A | N/A | − | S. cerevisiae | |
| Histone expression | H3/H4 | N/A | N/A | N/A | + | S. cerevisiae |
see supplemental information S3 (table) for a fully referenced version of this table.
indicates where the parameter is unknown or unconfirmed.
N/A:not applicable
The “wild-type impact on health or lifespan” column indicates what role the protein or complexhas on lifespan in physiological conditions based on experimental knock-down, mutation or overexpression results(‘−‘ to indicate that they normally restrict health or lifespan, ‘+’ to indicate that they normally promote health or lifespan, ‘=’ when no clear impact on lifespan was reported).
Core histone expression in ageing and longevity
The folding of cellular genomes into higher-order chromatin structure affects nearly all cellular processes that are linked to ageing, including transcription, DNA repair and DNA replication34. Age-dependent changes in chromatin structure, which are mediated in part by changes in histone expression, are linked to ageing phenotypes in mammalian cells and to lifespan regulation in yeast (Table 1 and Supplementary information S2 (table)). The expression of core histones is reduced during replicative ageing in yeast, as well as in other species and cell types35–39 (Table 1). In yeast, decreased histone expression is coupled to decreased nucleosome occupancy and aberrant upregulation of associated genes36. Interfering with complexes that control the exchange and deposition of histones onto chromatin also influences S. cerevisiae lifespan: the histone chaperone ASF1 (Anti-Silencing Function), which promotes histone deposition and stability, is required for normal replicative lifespan, whereas the HIR complex (Histone Information Regulator), which represses histone expression, limits yeast replicative lifespan40. Interestingly, increasing the cellular supply of histone H3 and H4, but not of histone H2A and H2B, increases the replicative lifespan of yeast by up to 50%40 (Table 2 and Supplementary information S3 (table)). The oligomerization of histones into chromatin is initiated by the deposition of H3–H4 dimers, which then allows the recruitment of H2A–H2B dimers41. Thus, an increased supply of H3–H4 dimers may promote nucleosome deposition in old cells and protect the genome against unwarranted genome activation. To our knowledge, whether histone expression levels limit metazoan longevity has not been investigated. However, because histone expression declines in some mammalian cells during ageing37, 38 (Table 1), this process may also impact ageing in mammals.
Histone methylation in ageing and longevity
Histone methylation is associated with active or repressed genome regions, depending on the residue affected and the level of methylation (Box 1), and it can be dynamically regulated by histone methyltransferases and histone demethylases (reviewed in42). The global level or genomic distribution of many histone methylations change in organismal and cellular models of ageing (Table 1). Furthermore, the manipulation of histone methyltransferases and histone demethylases can modulate longevity of model organisms (Table 2).
Data from both physiological and accelerated models of ageing generally support the so-called ‘heterochromatin loss model of ageing’43 (Table 1). This model postulates that decreased heterochromatin and/or the inappropriate redistribution of heterochromatin silencing proteins may cause cellular dysfunction with age. The mechanisms underlying heterochromatin remodeling are not fully understood, but may involve interactions between the chromatin machinery and nuclear periphery proteins such as nuclear lamins. Such interactions can establish nuclear microdomains that delineate regions of active and repressed gene expression44. For example, reducing Lamin B1 levels in proliferating fibroblasts can induce locus-specific remodeling of H3K4me3 and H3K27me3, which mimic alterations observed in senescent cells39. Interestingly, widespread alterations in heterochromatin organization are observed in mesenchymal stem cells derived from an embryonic stem cell (ESC) model of Werner syndrome, including a generalized reduction of H3K9me3 and decreased interactions with inner nuclear membrane proteins45. It will be important to determine the mechanisms that trigger large-scale changes in repressive chromatin with ageing.
Consistent with a model that loss of repressive chromatin is detrimental during ageing, the manipulation of chromatin regulators leading to increased ‘repressive’ histone methylation, such as H3K27me3, is often linked to increased longevity46–48 (Table 2). For example, reduced expression of the H3K27me3 demethylase utx-1 promotes C. elegans longevity46, 47. Interestingly, during worm ageing, a global decrease in somatic H3K27me347, 48 and an increase in utx-1 expression46 is observed, suggesting that utx-1 could limit worm longevity. However, results from several organisms indicate that the impact of H3K27me3 on ageing is likely more complex. Opposite to what could be expected from results with utx-1, reduction of mes-2, the C. elegans ortholog of the H3K27 trimethyltransferase component of Polycomb, extends the lifespan of sterile worms48. Furthermore, mutation in the Polycomb group protein E(Z) also increases lifespan in flies49. Finally, global levels of H3K27me3 increase in the muscle stem cells of old mice38 and in brains of old African killifish50, which contrasts with the age-related H3K27me3 decrease in C. elegans47, 48. That H3K27me3 levels can both be associated with lifespan extension and shortening suggests that different H3K27me3 regulators may influence lifespan through specific loci and/or in specific cells (for example, stem cells versus differentiated cells). It is also possible that some H3K27me3 regulators could exert their effect on lifespan via other, H3K27me3-independent effects.
Age-associated loss of heterochromatin is often coupled to remodeling of histone methylations associated with ‘active’ chromatin. Indeed, redistribution of the active histone modification H3K4me3 (a mark of accessible promoters) is observed during ageing and in cellular senescence39, 48, 51 (Table 1). For example, new widespread regions of H3K4me3 emerge in senescent human fibroblasts, even when normalized to the declining total histone H3 levels39. Spreading of H3K4me3 domains is also observed during ageing in mouse HSCs21, although it will be important to compare the magnitude of these changes to those in histone H3 levels. As broad H3K4me3 domains mark genes that are important for cell identity in many cell types52, the spreading of H3K4me3 during ageing could influence cell function. Targeted RNAi screens probing the effects of histone methyltransferases and demethylases on longevity in flies and worms have revealed that regulators of H3K4me3 can modulate lifespan46–48, 53 (Table 2).
Specifically, a longevity screen in fertile well-fed C. elegans hermaphrodites revealed that knock-down or mutation of genes encoding members of the COMPASS H3K4me3 methyltransferase complex54 (ASH-2, SET-2 - the worm homolog of SETD1A and SETD1B in humans - and WDR-5) increases lifespan, whereas knock-down of the gene encoding H3K4me3 demethylase RBR-2 shortens lifespan53. In addition, overexpressing RBR-2 in the germline extends lifespan53, supporting the idea that H3K4me3 demethylation in the germline promotes somatic maintenance. Consistently, male D. melanogaster deficient for the ortholog of RBR-2, LID, also exhibit shortened lifespan55, although female flies are unaffected. However, H3K4me3 regulators may have different effects depending on the H3K4me3 complex or conditions. For example, deficiency in Trx, which is part of another type of H3K4me3 methyltransferase complex in D. melanogaster54, does not significantly impact the lifespan of male flies49. In addition, several studies indicate that rbr-2 RNAi can actually also extend lifespan in worms under some experimental conditions48, 56, 57. These discrepancies may result from differences in H3K4me3 complexes and/or conditions, such as nutrient intake or reproductive status.
Maintaining the levels of another active histone methylation, H3K36me3, which is linked to transcriptional elongation, is required for healthy ageing in yeast and worms. Indeed, mutation of the yeast RPH1 gene, which encodes a H3K36 demethylase, extends yeast replicative lifespan, and yeast carrying H3 mutant forms that cannot be H3K36-methylated are short-lived58. In C. elegans, somatic levels of H3K36me3 moderately decrease with age48, and appear to be particularly reduced at genes that are deregulated with age51 (Table 1). Consistently, knock-down or mutation of the gene that encodes the putative C. elegans enzyme depositing the H3K36me3 mark, met-1, shortens worm lifespan51. These results suggest that the correct maintenance of H3K36me3 may be a critical process during ageing.
Thus, altering specific histone methyltransferases or demethylases modulates lifespan in yeast, worms, and flies. However, apart from studies in yeast, it is unclear whether changes in histone methylation directly impact ageing. Future studies should determine whether the role of histone methylation complexes in lifespan regulation is conserved throughout evolution.
Histone acetylation in ageing and lifespan regulation
Histone acetylation directly impacts the physical association of histones and DNA. Histone acetylation is a key, conserved player in longevity, and evidence suggests that its pattern changes during normal ageing (Table 1 and Table 2). In yeast, the global levels of H3K56ac decrease during replicative ageing, while that of H4K16ac increase, leading to de-silencing of telomeric repeats59. In contrast, the role of H4K16ac in mammalian ageing may be distinct from its functions in telomere maintenance. Global H4K16ac levels decrease during normal ageing and in a mouse model of HGPS and may be linked, at least in the progeroid model, to a decreased association of histone acetyltransferases with the nuclear periphery60. Following contextual fear conditioning, older mice fail to upregulate H4K12ac, a mark that promotes transcriptional elongation61, in their hippocampus, and this correlates with altered gene expression and learning impairment62. Changes in histone acetylation thus may be a consequence and a cause of the failure of older cells to transduce external stimuli to downstream transcriptional responses, a process that is particularly detrimental for rapid cell-to-cell signaling in the brain.
Both histone acetyltransferases (HAT) and deacetylases (HDAC) modulate lifespan and metabolic health (Table 2). For example, H4K16ac is deacetylated by the sirtuin SIR263, and increased SIR2 dosage extends yeast lifespan64 by limiting aberrant recombination at the ribosomal DNA (rDNA) locus. Although SIR2 and its orthologs also have non-histone substrates65, SIR2-mediated deacetylation of H4K16ac, which is counteracted by SAS2-mediated acetylation, is important in regulating yeast lifespan59. Indeed, the lifespan of yeast mutants carrying an H4 gene mimicking constitutive H4K16ac is not altered by SIR2 deletion, suggesting that SIR2 regulates lifespan through its activity on acetylated H4K1659.
While the role of class III HDACs (also known as ‘sirtuins’) in lifespan modulation in metazoans has been challenged66, new evidence supports a role of SIR2 orthologs, such as SIRT1 in mice, in promoting health and lifespan under standard conditions or at least in the context of dietary restriction (DR)-mediated longevity and health67 (Table 2). The role of mammalian SIRT6 in regulating longevity is clearer. SIRT6 can deacetylate H3K9ac68 and H3K56ac69. SIRT6 is the only sirtuin for which a deficiency induces a progeroid phenotype in mice70. Conversely, Sirt6 transgenic male mice are long-lived71. The mechanism by which SIRT6 promotes longevity is unclear, but may stem from its role in supporting genomic stability70, 72, partly by recruiting chromatin remodelers at sites of DNA-damage69. More generally, sirtuins may have a pro-longevity role by promoting increased genomic stability69, 70, 72–74 (see also section below).
In general, deregulation of both HATs and HDACs, through genetic manipulation or targeting by chemicals or drugs, has been associated with significant changes in longevity across taxa (Table 2). However, in most cases, whether modulation of these enzymes regulates lifespan by acting solely on chromatin or also by regulating non-histone substrates warrants further studies.
Enzymes regulating other histone modifications
Regulators of other types of histone modifications could also impact ageing, although this has been much less studied (Table 2). For instance, O-N-acetyl-glucosamine (O-GlcNac) can be deposited on histones H2A, H2B and H4 and is enriched at promoters of genes that are important for ageing and stress response in C. elegans, notably targets of insulin-signaling75. Specifically, mutation of the gene encoding the enzyme that deposits O-GlcNac (ogt-1) shortens worm lifespan75, 76, and mutation of the gene encoding the enzyme that removes O-GlcNac (oga-1) can extend lifespan in worms depending on the genetic background75, 76. However, O-GlcNac can also modify metabolic enzymes in addition to histones76. Thus, the exact mechanism by which O-GlcNac cycling impacts lifespan remains to be explored.
Role of nucleosome remodelers
Nucleosome remodelers also modify chromatin and their perturbation can regulate lifespan in yeast and worms (Table 2). Chromatin remodelers can modify nucleosome positioning, chromatin higher-order structure and overall nuclear organization (reviewed in77). Changes to heterochromatin in ageing models have been linked to activity of nucleosome remodelers, such as the NuRD complex78. NuRD components RBBP4 and RBBP7 are downregulated in fibroblasts isolated from HGPS patients and healthy aged donors, and depletion of NuRD by RNAi in HeLa cells recapitulates the loss of heterochromatin in HGPS cells78. Inactivation of the chromatin remodeling ISWI complex has recently been shown to promote lifespan in S. cerevisiae and C. elegans79. Indeed, deletion of ISW2, a gene encoding a subunit of the ISWI complex, in yeast leads to shifts in nucleosome positioning at thousands of stress-response genes and to increased replicative lifespan79. Disruption of the homologous complex in C. elegans through another subunit leads to increased longevity79. On the contrary, SWI/SNF complexes are crucial for promoting longevity in the context of the Insulin–FOXO pathway in C. elegans80. Indeed, both the transcriptional changes and the lifespan extension promoted by DAF-16 (the worm FOXO transcription factor), require functional SWI/SNF80. These studies suggest that the regulation of nucleosome positioning and chromatin compaction by remodelers can both impact pro-ageing or pro-longevity genes. However, how these complexes are targeted to specific genes, how their different activities compete, and whether they have a role in the physiological regulation of ageing remains to be explored.
Environment and age-related epigenetics
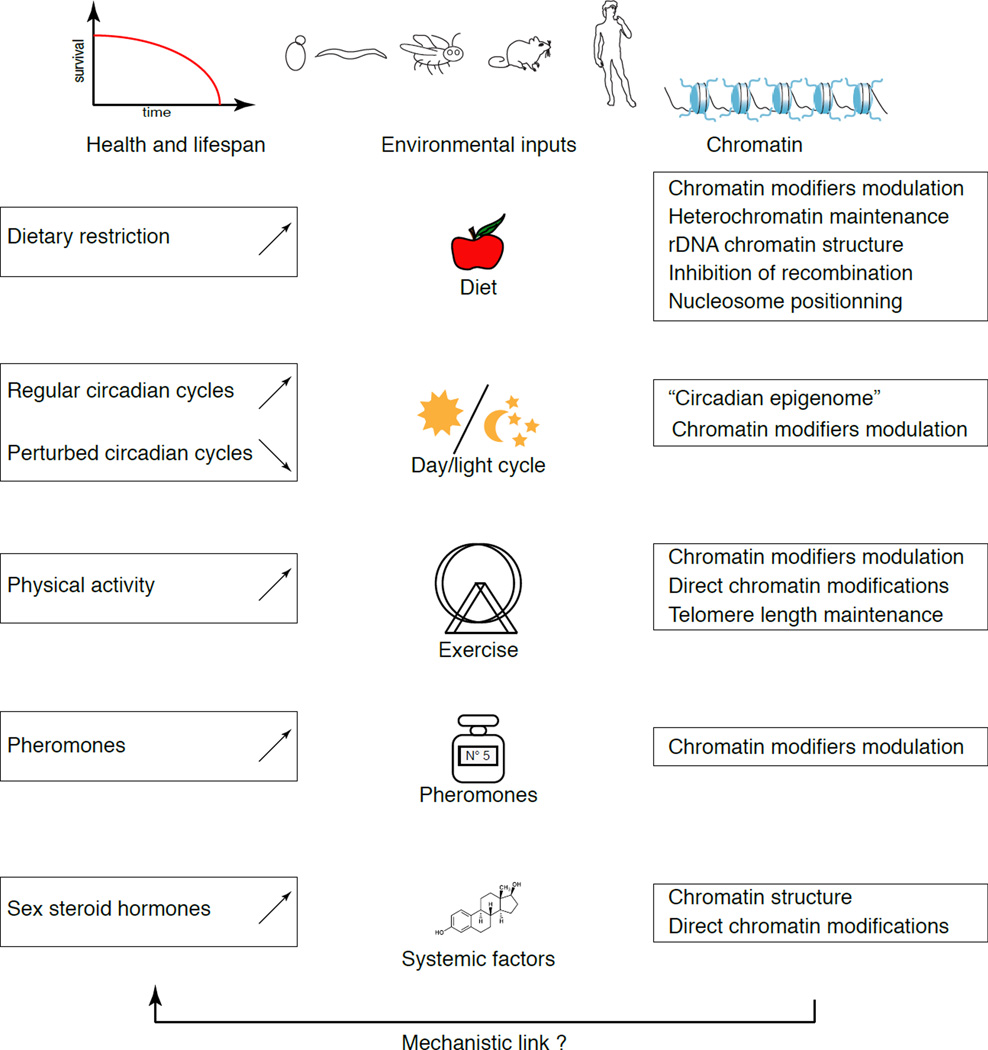
Emerging evidence highlights the importance of non-genetic cues in the regulation of ageing and longevity, including diet3, exercise81, sexual stimuli82, and circadian rhythms83 (Figure 1). Many proteins that directly or indirectly induce epigenetic remodeling are also modulated by environmental cues. However, causal evidence linearly linking external cues to specific chromatin changes, and subsequently to alterations in ageing and longevity, is still missing. We discuss a subset of environmental signals that may act, at least in part, through changes in chromatin to promote healthspan and lifespan.
Figure 1. Environmental inputs that impact longevity can also affect the chromatin landscape.
Environmental signals that modulate lifespan might do so by modulating chromatin. Dietary restriction increases lifespan in a range of organisms, and DR has also been linked to changes in the chromatin landscape. Those include changes in the expression of chromatin modifiers, for example increased expression of several sirtuins, or an increased maintenance of heterochromatin. Robust circadian light cycles also promote health and lifespan and are linked to circadian epigenomic changes (for example, periodic increases in H3K14 acetylation of circadian promoters by the CLOCK protein) and the modulation of the activity of chromatin modifiers such as SIRT1. Physical activity is also beneficial to health and lifespan, and has been associated with changes in chromatin modifications (for example, increased H3K36ac in human skeletal muscle) and with the regulation of chromatin modifiers (for example, induced nuclear exclusion of HDAC4 and HDAC5 in human skeletal muscle). Recent work has shown that, in C. elegans, pheromones may increase lifespan, through a mechanism requiring chromatin-modifying enzymes (for example, SIR-2.1 is required for lifespan extension following exposure to ascarosides #2 or #3). Finally, in women, the strong decrease in production of sex steroid hormones with age, such as estrogens, contributes to age-related diseases, and estrogens can directly remodel chromatin at target genes through their receptor. Whether there is a linear pathway from the environmental output to the changes in chromatin and, ultimately, to lifespan extension, remains untested.
Nutrient intake, ageing and chromatin
Dietary restriction (DR) – a reduction in food intake without malnutrition – extends lifespan and delays signs of ageing in many organisms3. DR induces changes in gene expression across tissues (reviewed in10), including the expression of genes regulating metabolism, stress responses, DNA repair, and chromatin structure. These DR-induced transcriptional changes are consistent with a global preservation of genome integrity and chromatin structure84. Thus, the age-delaying effect of DR may be due to increased genomic stability10, 84. DR can also impact the chromatin landscape (reviewed in10). For instance, DR induces positional shifts of nucleosomes at thousands of genes in yeast, which may potentiate a stress response at the transcriptional level79. Interestingly, shifts at partially overlapping locations are observed in long-lived ISW2 deletion mutants in yeast, and the effects of DR and ISW2 mutation on longevity are epistatic79. Thus, nucleosome rearrangements may partly underlie DR-induced longevity in yeast79.
DR may also affect the global chromatin landscape in metazoans. A study using a position variegation effect reporter suggests that DR may delay the age-related loss of facultative heterochromatin in flies85. Diet-switch experiments between an unrestricted diet and DR suggest that diet-regulated changes in heterochromatin may be rapid (within 3 days) and reversible85, which is suggestive of short-term adaptations. High nutrient intake in humans, using increased body mass index as a proxy, also seems to induce an ‘aged-like’ DNA methylation profile in liver86. Thus, nutrient intake has important ties to the regulation of both longevity and chromatin structure. However, it is unclear whether the observed changes are directly responsible for ageing and longevity changes. An important step in understanding how diet impacts longevity will be to establish the molecular link between changes in chromatin that are induced by diet and organismal ageing.
Energy and nutrient sensing and chromatin modulation
Consistent with the widespread anti-ageing effect of DR, many proteins that modulate longevity are linked to nutrient sensing and metabolic regulation. These proteins may integrate metabolic signals into chromatin responses. The detection of DR can occur through the systemic sensing of nutrient availability (for example, via signaling pathways) and/or the direct sensing of cellular energy levels that are reflected by changes in NADH:NAD+ or ATP:ADP:AMP ratios (reviewed in10). Although these pathways also have well-known non-chromatin substrates, their impact on ageing and longevity may be partly mediated through chromatin.
The insulin and insulin-like growth factor (IGF) signaling pathways are well characterized nutrient-sensing pathways that regulate lifespan across evolution1. Critical effectors of insulin signaling are FOXO transcription factors. In high nutrient conditions, FOXO transcription factors (FOXOs) are phosphorylated and excluded from the nucleus; in low nutrient conditions they re-localize to the nucleus87. Although FOXOs are not classical chromatin modifiers, FOXO1 can directly decrease chromatin compaction of the mouse Igfbp1 promoter88. Furthermore, DAF-16, the worm FOXO transcription factor, directly recruits the SWI/SNF remodeling complex to target genes, and this complex is required for FOXO-mediated longevity80.
Sirtuins rely on NAD+ as a cofactor and are inhibited by nicotinamide; thus, they are direct sensors of the cellular metabolic state63. Sirtuins are important mediators of DR-induced longevity across species89, 90 (Table 2). Although they also regulate non-histone substrates, sirtuins have a strong functional link to chromatin regulation, which is required for their effect on lifespan, at least in yeast59. In mice, SIRT6 can recruit the SWI/SNI subunit SNF2H to DNA break sites and prevent genomic instability through chromatin remodeling and local deacetylation of H3K56ac69. Whether, as a rule, sirtuins promote DR-induced longevity by directly acting on chromatin remains unclear.
The energy-sensor AMP-activated protein kinase (AMPK) is necessary for longevity in response to several DR regimens in worms91 and flies92. In addition, metformin, an AMPK activator, extends mouse lifespan93, although metformin may have additional targets. AMPK can modify several chromatin regulators. In yeast, AMPK promotes histone acetylation by controlling the genomic occupancy and activity of the HAT GCN594 (Table 2). In mammals, AMPK can also inhibit histone deacetylation by phosphorylating HDAC4, HDAC5 or HDAC795, 96. Interestingly, AMPK has several other substrates that are directly or indirectly implicated in chromatin regulation and genomic stability, including histone H2B itself97, SIRT198, and O-GlcNAc transferase (OGT)99. Whether AMPK promotes longevity by modifying chromatin targets or its non-chromatin substrates remains unknown.
Metabolites and metabolic enzymes may also directly regulate age-related epigenetics. Glycolytic enzymes, such as GAPDH (glyceraldehyde-3-phosphate dehydrogenase) or HK (Hexokinase), when mutated, can extend replicative lifespan in yeast100, 101. These enzymes can also modulate transcription in the nucleus in a metabolism-dependent manner (reviewed in10). For instance, GAPDH directly promotes the transcription of H2B genes during S-phase102. The nuclear functions of these enzymes may be important to integrate metabolic input and chromatin regulation. Metabolites generated by mitochondrial respiration have also been implicated in longevity and chromatin regulation. For instance, the Krebs cycle intermediate citrate can be converted to acetyl-CoA in mammalian cells, and this process can in turn modulate global levels of histone acetylation103. Several other Krebs cycle intermediates may also impact the global levels of DNA and histone methylation because they can act as obligatory co-substrates (such as α-ketoglutarate) or potent inhibitors (such as succinate) of the Ten Eleven Translocation (TET) enzymes (which are involved in DNA demethylation) and the KDM2/7 histone demethylases104. Thus, perturbation of the Krebs cycle during ageing may induce stochastic or aberrant chromatin remodeling. Interestingly, supplementation with Krebs cycle metabolites (such as oxaloacetate, malate, fumarate and α-ketoglutarate) can extend worm lifespan through the genetic pathways that mediate DR-induced longevity105–107. Thus, strong molecular ties link DR, metabolism, longevity and the regulation of chromatin states. Future studies should establish whether these links are fortuitous, or whether DR requires chromatin remodeling to have a lasting impact on longevity.
The circadian clock and ageing
The circadian clock controls many physiological and behavioral systems and is highly linked to energy metabolism108 and chromatin83. Disrupting circadian rhythms negatively impacts health and longevity across species (reviewed in108). Conversely, restoring a functional circadian clock in ageing animals improves health and/or lifespan109, 110. Furthermore, isogenic mice with innate circadian periods closest to 24h live longer than their littermates with shorter or longer innate circadian periods5, further highlighting that a robust circadian clock is key to longevity.
At a molecular level, the circadian clock regulates a circadian epigenome and gene program, which requires the rhythmic recruitment of protein complexes to chromatin111. CLOCK, a primary molecular component of the clock, has H3K9 and H3K14 HAT activity112, 113. The circadian epigenome can be influenced by metabolic cues, and several energy sensors that mediate DR-induced longevity also modulate the circadian machinery and the chromatin state of circadian promoters. For instance, SIRT1 deacetylates the circadian clock components PER2 and BMAL1113, 114. Activation of SIRT1 leads to decreased H3 acetylation, and thus repression of circadian genes in a time-specific manner115. Other energy sensors that modulate the clock and have links to chromatin include AMPK116. Thus, although the mechanism linking circadian rhythms to health and longevity is elusive, the circadian clock may promote longevity by modulating the epigenome.
Impact of physical exercise on ageing and chromatin
Physical activity promotes healthy ageing in mammals. For example, voluntary exercise promotes neurogenesis and healthy cognitive ageing in mice117, 118. Exercise may also prevent cognitive decline in humans119, and regular physical activity is associated with longevity, a 30% reduction in all-cause mortality, and a dose-response improvement in overall health in humans120.
Evidence suggests that exercise may influence chromatin dynamics, though the underlying mechanisms are unknown. In rat skeletal muscle, ageing is associated with decreased SIRT1 activity, and exercise can counteract this effect121. In humans, exercise is associated with enhanced AMPK activity, nuclear exclusion of HDAC4 and HDAC5, and increased H3K36 acetylation in skeletal muscle tissue122. The molecular mechanisms underlying the beneficial effects of exercise during ageing and whether they involve chromatin changes are still unresolved questions, but future work is likely to explore this connection.
Longevity modulation by pheromones
The longevity of worms and flies can be modulated by the opposite sex82, 123, 124, a process that can involve pheromone production and/or sensing82, 124. In the presence of males in C. elegans, hermaphrodites are short-lived with signs of premature ageing, and the expression of the H3K27 demethylase utx-1 is highly upregulated82. Interestingly, knock-down of utx-1 in hermaphrodites extends longevity even in the presence of males82, suggesting that the effect of males on hermaphrodite longevity may involve chromatin remodeling. As a portion of the premature demise induced by the presence of males is dependent on the sensing of male pheromones by hermaphrodites82, it is possible that pheromones may influence chromatin states to regulate premature ageing.
Pheromones are not only involved in sexual sensing, but also in sensing of animal density (‘crowding’) in C. elegans125. Interestingly, exposure to pheromones involved in crowding (ascaroside #2 or ascaroside #3) is sufficient to extend C. elegans lifespan126. This pheromone-induced longevity requires the histone deacetylase gene sir2.1, but is independent of insulin-signaling126. Thus, chromatin modifiers may mediate the effect of pheromones, although whether pheromones directly impact chromatin marks or the overall chromatin structure is unknown. While ascarosides are nematode-specific molecules, their chemical structures resemble that of metabolites and they are part of a class of molecules termed ‘secondary metabolites’. As secondary metabolites are present across evolution, the potential link between pheromonal signaling, chromatin, and ageing deserves further exploration in other species.
Systemic regulation of ageing: an epigenetic connection?
Sex steroid hormones (for example, estrogens and androgens) are systemic factors that decline with age in humans. Indeed, the cessation of ovarian lifespan (menopause in humans) ends its endocrine activity and shuts down the bulk synthesis of female sex steroid hormones. Estrogens act mainly through nuclear receptors (for example, the estrogen receptor (ER)), which can impact transcription by directly modulating chromatin states127 and higher order chromatin structure128. Interestingly, global histone acetylation levels decrease with age in rat brains, and estradiol promotes histone acetylation in ex-vivo brain slices from young, but not old, rats129. Estrogens are thought to limit the prevalence or age-of-onset of osteoporosis, sarcopenia, cardiovascular diseases, immune decline and neurodegeneration130. In reproductively senescent women, age-related decrease in estrogens may induce ageing phenotypes, partly due to decreased activity of estrogen receptors on chromatin. More studies are needed to understand the mechanistic impact of sex steroid hormones on human ageing and the chromatin changes induced by the age-induced drop in estrogen synthesis.
Environmental factors, thus, seem to modulate the activity of chromatin modifiers and organismal longevity. However, whether longevity is achieved through chromatin changes or via other mechanisms is still very much unexplored.
Epigenetics and genomic stability in ageing
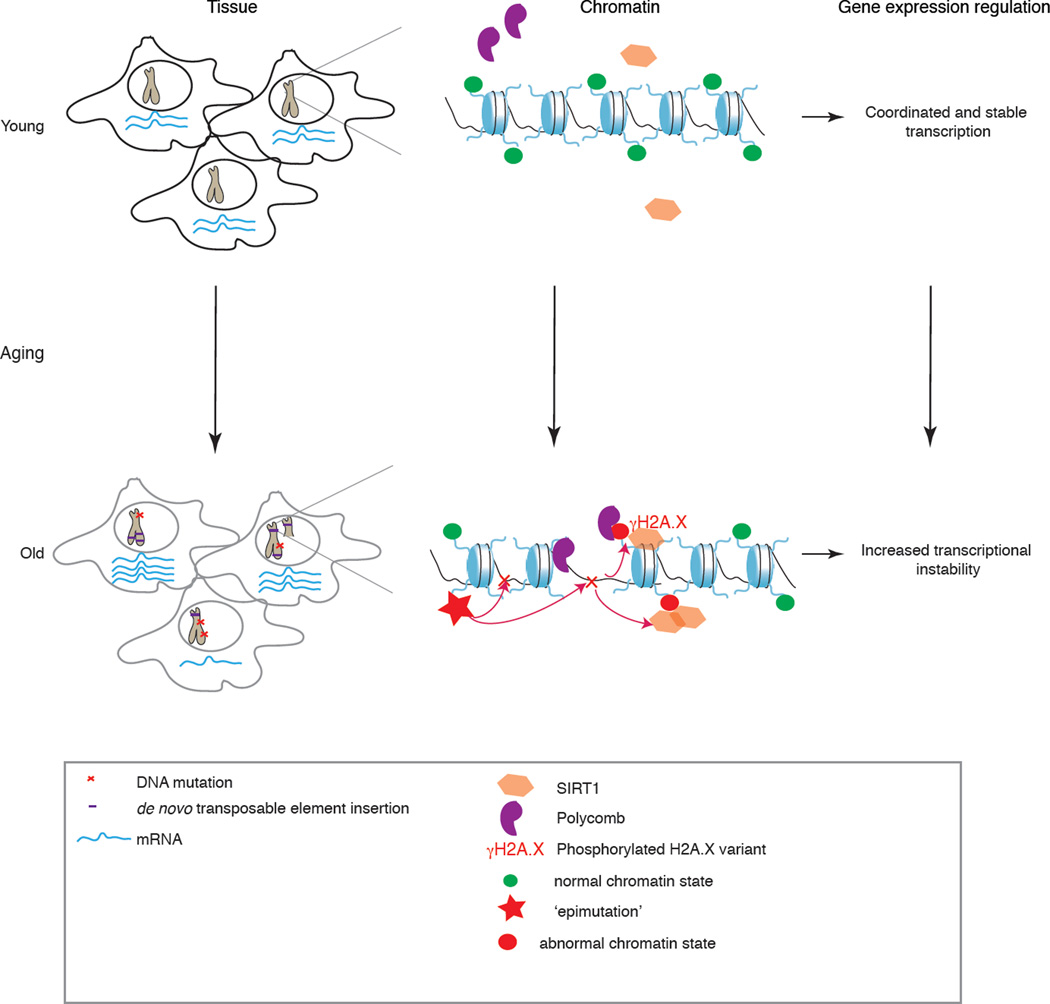
Epigenomic changes during ageing can generate instability at two levels; age-dependent changes in chromatin may increase the susceptibility of the genome to mutation and may reduce transcriptional precision. Epigenomic states can themselves be unstable; indeed, ‘epimutations’, which correspond to aberrant or atypical changes in epigenetic states131, appear to stochastically increase throughout life132. Thus, accumulated epimutations during ageing may induce further genomic instability (Figure 2).
Figure 2. Model for the possible cross-talk between chromatin changes and transcriptional and genomic instability during ageing.
A possible model is the following: in cells from young organisms (top left), transcriptional programs are robustly defined and precise between cells (depicted by consistent mRNA levels). The genomic integrity is maintained (depicted by intact chromosomes), because mutations are rare or correctly repaired. As a result, ‘normal’ chromatin states are found throughout the genome. With increased age (bottom left), transcriptional instability is increased between cells of a tissue (depicted by variable mRNA levels between cells). Genomic instability is also a hallmark of ageing and is increased at both a macro level (for example, aneuploidies depicted by partial chromosome duplication), increased transposable element insertions and, more locally by DNA mutations in the form of single nucleotide mutations, small insertions or deletions (indels). DNA damage can trigger the recruitment of chromatin modifiers, and the acquisition of abnormal chromatin states. Thus, genomic instability could modify the epigenetic landscape of old cells. Reciprocally, aberrant changes in epigenetic marks, known as ‘epimutations’, can further promote the accumulation of DNA mutations in a feedback loop mechanism. The epigenetic changes acquired during ageing could also decrease the transcriptional precision of neighboring genes.
Epigenetic instability and DNA mutations
The accumulation of mutations and epimutations with age can result from errors in DNA repair and failure to correctly replicate the genome and epigenome133, 134. While the extent and importance of mutations during ageing is debated, persistent DNA-damage signaling during ageing may drive local changes in chromatin structure and epigenetic modifications132, 134. DNA-damage signaling may also promote specific chromatin alterations, such as recruitment of chromatin modifiers, including Polycomb, SIRT1, SIRT6, and DNA-methyltransferases (reviewed in134). The resulting aberrant chromatin environment may in turn further increase susceptibility of DNA to damage134. Consistent with the idea that DNA damage can globally impact chromatin structure, deficiency in the WRN DNA repair helicase in mesenchymal stem cells leads to global loss of chromatin compaction45, decreased levels of the heterochromatin marks H3K9me3 and H3K27me3, and increased phosphorylation of the H2A.X variant at centromeric loci45.
Different types of genomic instability, such as single nucleotide mutations, aneuploidy, and transposition events increase with age and might contribute to ageing phenotypes (reviewed in133). Age-dependent genomic instability is due in part to the progressive failure of DNA repair pathways135. The importance of accumulated DNA damage in ageing is illustrated by several progeroid phenotypes, which are induced by mutations in DNA repair enzymes136, although it is unclear whether increased DNA repair itself can extend lifespan. Interestingly, metabolites linked to the regulation of longevity, such as NAD or α-ketoglutarate, modulate the activity of proteins involved in both DNA repair and chromatin remodeling137. For example, the recruitment of NAD-dependent deacetylases SIRT1 and SIRT6 may directly promote genomic stability69, 70, 73, 74. By triggering chromatin remodeling, the accumulation of mutations with age may also induce changes in gene expression, driving some aspects of age-related functional decline.
Epigenetic instability and transposition
Another source of genomic instability during ageing comes from the activation of endogenous mobile genetic elements (transposable elements (TE)). In eukaryotes, 30–80% of the genome is composed of TEs138, which are transcribed and can regulate the expression of nearby genes. Active TEs induce extensive genomic instability139 and they are normally kept in check, especially in the germline, by heterochromatin marks such as H3K9me3140. Interestingly, transposition can increase during ageing in several species72, 139, 141–144, and TE activation correlates with neurodegenerative disorders in humans145. Conversely, DR may counteract the age-linked derepression of TEs in the liver and skeletal muscle of aged mice144. TE reactivation during ageing may result at least in part from heterochromatin loss. It may also be due to loss of SIRT6 specifically at TEs, as SIRT6 normally promotes heterochromatin formation and transcriptional repression in fibroblasts, heart, liver and brain from young mice72. Consistent with the idea that TE activation may negatively impact organismal lifespan, flies that are deficient for the Argonaute gene Ago2 exhibit exacerbated transposition and a shortened lifespan143. This result suggests that activation of TEs may cause certain aspects of ageing, although the shortening of lifespan could also result from defects in small RNA pathways due to Ago2 deficiency. Finally, in senescent human fibroblasts, chromatin accessibility increases at retrotransposon insertion sites, leading to increased TE transcription and transposition146. Together, these observations suggest that increased transposition during ageing is partially associated with aberrant chromatin remodeling and may promote ageing-phenotypes.
Epigenetics and transcriptional instability
Age-related epimutations could also impact ageing by causing stereotypical changes in gene expression7, 21, 38, as well as by adversely affecting transcriptional precision. The integrity of transcriptional networks seems to decline with age in C. elegans147 and in mice8, as does coordinated gene expression within cells of a tissue148. Interestingly, an increase in cell-to-cell transcriptional noise was observed at 11 out of 15 tested genes in cardiomyocytes from old mice149. However, transcriptional noise was not increased, at least in the 6 assayed genes, in HSCs from old mice150. Because these pioneering studies were limited to only a few genes and cell types, whether transcriptional noise increases globally or at subsets of genes during ageing remains unclear. Genome-wide studies are needed to assess the significance and impact of alterations to transcriptional noise during ageing.
While transcriptional instability may be a byproduct of genomic instability149, 151, it could also result from the accumulation of epimutations over the lifetime of an organism. Aspects of transcriptional precision may be affected by age-dependent changes in chromatin modifiers. For example, age-dependent fluctuations in gene expression are accompanied by decreases in H3K36me3 levels in worms. Maintenance of H3K36me3 levels restricts these fluctuations in gene expression and promotes longevity51. In yeast, sustained H3K36me3 levels are required to suppress cryptic transcription, and deletion of the H3K36me3 demethylase promote longer lifespan58. Transcriptional precision may also be affected by other chromatin regulators, including HDACs152, and modifiers of H3K4me3 breadth52, and it will be interesting to determine if these chromatin modifiers are themselves altered with ageing. Future studies will need to systematically elucidate the link between changes in transcriptional precision and in the ageing chromatin landscape to determine whether chromatin regulates transcriptional precision during ageing.
Transgenerational regulation of ageing
Because the germline propagates intact genetic material throughout generations153, 154, there has been interest in understanding the differential mechanisms of chromatin regulation in somatic versus germline cells during ageing (Box 3). Fertilization, which is mimicked to some degree by in vitro reprogramming, requires the resetting of age-dependent perturbations such as protein aggregates, chromatin disorganization, and mitochondrial dysfunction, and this reset may not always be complete. Intriguingly, some age-related phenotypes, including changes in lifespan and fertility, may be inherited through generations in model organisms. Thus, while the bulk of the epigenome is reset between generations, select epigenomic loci escape reprogramming or are re-established through unidentified triggers in subsequent generations. For example, in S. pombe, C. elegans, and mice, various chromatin marks (H3K9me3, H3K27me3, and DNA methylation) may be transmitted through mitosis and meiosis to the next generation155–157. We discuss examples of transgenerational inheritance of ageing phenotypes that are triggered by perturbations in chromatin regulators. As the molecular nature of the inherited signals for ageing phenotypes remains unknown, it is important to note that the mechanisms underlying this transgenerational inheritance could be independent of chromatin and could even involve cryptic genetic or microbiome-based transmission.
Box 3. Epigenetics of the ‘mortal soma’ and ‘immortal germline’.
An emerging question in the field of ageing epigenetics is whether different epigenetic mechanisms act in somatic cells and germ cells during ageing. Regulatory mechanisms that maintain germ cells may be more robust187 and a number of studies (reviewed in188) have explored the hypothesis that aged somatic cells could ‘rejuvenate’ through reprograming, the process by which differentiated cells are converted back to a germ or embryonic stem cell (ESC)-like state via the re-expression of pluripotency regulators189. Indeed, both senescent and aged fibroblasts undergo some aspects of chromatin rejuvenation upon reprogramming and the subsequent re-differentiation into fibroblasts, including restoration of DNA methylation patterns at the loci of the pluripotency genes OCT4 and NANOG and disappearance of senescence-associated heterochromatin foci188. Aged HSCs, which normally show persistent age-dependent gene expression changes, produce induced pluripotent stem cells (iPSCs) with similar transcriptional profiles to both normal ESCs and iPSCs clones derived from young HSCs, suggesting that chromatin modifications that influence transcriptional states may have been reset by reprogramming190. The differentiation potential of iPSCs derived from aged HSCs, both in vitro and through embryos, is similar to lines derived from young cells190, consistent with a stable re-setting of epigenetic states rather than a masking of defects in the pluripotent state. However, it cannot be excluded that seemingly rejuvenated properties of iPSCs generated from aged donor cells results from the selection of cells with more ‘youthful’ chromatin states during reprogramming rather than an active reset process. Nevertheless, together, these studies highlight that the pluripotent state possesses unique properties for the erasure and re-establishment of epigenetic marks.
Despite their unique modes of chromatin regulation, germ cells may also undergo epigenetic alterations with ageing. Longitudinal studies of DNA methylation changes in human sperm reveal global hypermethylation at repetitive elements and site-specific CpG hypomethylation with age, particularly at the regions retaining canonical nucleosomes (as opposed to smaller protamines that normally replace bulky nucleosomes in mature sperm chromatin to promote compaction)191. Oocytes isolated from aged mice also display disordered chromatin and changes in the levels of certain histone modifications, including decreased histone methylation levels192, 193. Moreover, transcriptional profiling of oocytes from young and old mice reveal that several genes encoding chromatin modifiers are down-regulated with age, such as Polycomb genes Ezh2 and Bmi1 and DNA methyltransferase genes Dnmt1 and Dnmt3l194. These studies suggest that the deregulation of chromatin states in ageing germ cells may be more common than previously thought. Delineating which epigenetic alterations are reset, and which are fixed and transmittable, will be invaluable for understanding how lifespan trajectories evolved, and whether certain mechanisms can be harnessed to induce ‘erasure’ of ageing signs.
Progressive germline mortality
The germline is normally ‘immortal’ (Box 3). However, there are examples of progressive sterility over generations from worms to mammals, a phenomenon called ‘germline mortality’. Understanding progressive germline mortality could give insights into the transmission of chromatin changes to the next generation, which could in turn affect somatic maintenance. Recently, epigenetic mechanisms, including small RNAs and chromatin modifiers, were shown to contribute to germline mortality in C. elegans (reviewed in158). Mutations of spr-5, one of the worm homologs of the H3K4me2 demethylase LSD1, increase H3K4me2 levels in whole worms and primordial germ cells and induce sterility by generation 20159, 160. A predicted null mutation of the rbr-2 H3K4me3 demethylase gene, as well as simultaneous loss of spr-5 and rbr-2, result in progressive sterility at higher temperatures57, as does loss of function of the set-2 H3K4me2/3 methyltransferase161. Germline maintenance throughout generations may partly depend on the coordinated control of H3K4 methylation and repressed chromatin states. Accordingly, deletion of a predicted H3K9-demethylase in C. elegans can suppress the fertility defects of spr-5 mutants, whereas loss of H3K9-methyltransferases leads to loss of fertility at earlier generations160, 162. These results suggest that the presence of H3K4me2 in H3K9-methylated regions promotes inappropriate gene activation that is detrimental for germ cells160, 163. Whether the marks themselves or small RNA intermediates cause the transgenerational phenotype is unknown, but it is interesting that feedback loops exist between chromatin and small RNAs164, 165. It will be important to determine the role of chromatin states in germline function across generations in other species, including mammals.
Epigenetic memory of longevity
Several phenotypes associated with somatic maintenance, including lifespan and stress resistance, can be inherited in a transgenerational manner. In C. elegans, wild-type descendants from ancestors deficient for members of the H3K4 trimethyltransferase complex have a lifespan extended by up to four generations166. This extension can be reverted by transient inactivation of the rbr-2 demethylase166. The mechanisms of lifespan extension are unclear, since global levels of H3K4me3 are unchanged in worms descended from ancestors with deficiencies in H3K4me3 modifiers166. However, a small number of genes are aberrantly expressed in genetically wild-type descendants from mutant ancestors in the fourth, but not the fifth, generation166. Whether specific changes to H3K4me3-marked loci escape the reprogramming of germ cells during meiosis to influence the expression of the associated genes is unknown. It is noteworthy that transgenerationally-regulated genes are enriched in genes involved in metabolic pathways166, suggesting that loss of H3K4me methyltransferases in the parental generation may alter diet and/or trigger a metabolic cascade that affects lifespan independently of H3K4me3.
Consistent with the possibility that metabolic states could be inherited and thereby affect lifespan, a recent study indicated that worms whose grandparents were starved display increased organismal lifespan up to the third generation, although the extent of lifespan extension varies between replicates (22–70% extension)167. The transgenerational inheritance of metabolic states has also been observed in mammals (reviewed in154) and one hypothesis to explain this, at least in mice, is that metabolic changes in the parental generation influence chromatin states in germ cells, and that this permits the transmission of a previous environmental state to offspring157, 168.
A phenotype associated with longevity is stress resistance. In C. elegans, exposure of the parental generation to high glucose promotes resistance to oxidative stress and can reduce neurodegeneration in the F1 progeny169. The transmission of stress-resistance requires intact sir-2.1 and genes encoding COMPASS H3K4 methyltransferase complex components WDR-5 and SET-2, although changes in global H3K4me3 levels are not inherited along with stress resistance169. In flies, exposure to heat shock or osmotic stress induces heterochromatin changes that persist over several generations in the absence of stress11. In another model, chemical stress in flies induces the derepression of Polycomb-targeted genes, a subset of which remains derepressed in the absence of the stressor over multiple generations170. Thus, the memory of environmental exposures that affect lifespan might alter chromatin states in a manner that could be maintained through generations.
Many of these phenotypes revert after a certain number of generations, raising the questions of why and how the stimulus ceases to be passed on. Future research should identify the mechanisms of inheritance and establish whether chromatin directly mediates the inheritance of longevity phenotypes.
Concluding remarks
Epigenetic marks are remodeled during ageing and experimental perturbation of chromatin modifiers can directly impact lifespan. Thus, a compelling hypothesis is that the observed epigenomic changes throughout lifespan, and in the context of life-extending regimens, may actively modulate ageing. Throughout lifespan, environmental insults cause stochastic epimutations, which could contribute to functional decline at different scales, from cells to organs. These events can have systemic effects on ageing at the organismal level. While the sequence of events leading to genomic and transcriptional instability during ageing is unclear, genome surveillance and epigenetic remodeling demonstrably influence each other. Thus, we propose that that environment-induced epigenomic instability throughout life is a major driver of the ageing process, which is supported by the accuracy of the DNA-methylation clock in humans (Box 2).
In general, the plasticity of ageing chromatin states, notably upon environmental changes or cellular reprogramming, suggests that chromatin-regulating enzymes may be important therapeutic targets in the pursuit of healthy ageing. A variety of drugs that target chromatin modifiers have been identified171, 172, and a number of these increase the lifespan of model organisms (see Table 2). Improving the specificity of such drugs and testing their efficacy in different cellular contexts could help treat age-related diseases, as proposed for Alzheimer’s disease173. A major goal for drug design will be the specificity of delivery given their differential effects in various cells and tissues.
Despite recent progress, a full understanding of how epigenetic information integrates environmental input in the context of a genetic background during ageing is still in its infancy. This is partly due to the technical challenge of definitively studying how alterations in chromatin state may cause in ageing phenotypes. Among these challenges, the high copy number of histone genes in metazoan genomes prohibits simple ‘gene-swap’ experiments to study the function of specific residues in the regulation of longevity and ageing. Moreover, when perturbing chromatin modifying complexes we need to disentangle how to direct the activity of the enzyme to a particular histone residue and how to target the inhibition or activation of a mark at a subset of loci of interest rather than genome-wide. Finally, it will be important to further develop techniques to probe the chromatin landscape at the single cell and single molecule levels174, 175. This will help distinguish between three biological scenarios: the consistent chromatin remodeling in all ageing cells within a population, the emergence or disappearance of specific subpopulations during ageing, or the allele-specific epigenetic regulation changes during ageing in humans or outbred populations.
Reference epigenomes have transformed our understanding of transcription and highlighted key differences in chromatin states between tissues in young individuals. To better understand the integrative changes that occur during ageing, epigenome maps throughout lifespan would be particularly helpful. Such an ageing roadmap should help define better strategies for delaying or reversing aspects of ageing and age-related diseases.
Supplementary Material
Key points.
-
-
Widespread epigenomic remodeling, at the level of DNA or histone protein modification, has been observed during ageing across species and cell types. Some epigenetic states can serve as “molecular clocks”.
-
-
The experimental perturbation of chromatin modifiers can influence the lifespan of model organisms.
-
-
Environmental inputs, such as diet, physical activity, hormones or pheromones, have been linked to remodeling of the epigenome as well as to changes in lifespan. Chromatin may thus act as a molecular integrator of environmental exposures.
-
-
Random epigenetic changes, or epimutations, throughout life may trigger increases in transcriptional and genomic instability.
-
-
The unique regulation of chromatin in germline cells might protect these cells more than somatic cells during ageing.
-
-
Some ageing or age-related phenotypes, such as lifespan, fertility, and stress-resistance, may be inherited through successive generations in model organisms, through non-genetic mechanisms.
-
-
The use of epigenetic drugs or epigenome-editing technologies may be a promising avenue for age-related therapeutics.
Acknowledgments
The authors apologize to those whose work could not be cited owing to space constraints. B.A.B., E.A.P and A.B. wrote the review together. We thank Aaron Daugherty, Elizabeth Schroder, and Kévin Contrepois, as well as the anonymous reviewers for helpful comments on the manuscript. B.A.B. is supported by K99 AG049934. E.A.P. is supported by NSF GRFP and NIH F31 AG043232 graduate fellowships. A.B. is supported by NIH DP1 AG044848, P01 AG036695, and the Glenn Laboratories for the Biology of Ageing.
Glossary
- Lifespan
refers to the time elapsed between birth and death in time units. In yeast, lifespan can be measured in two ways: chronological lifespan, which corresponds to the length of time a non-dividing yeast cell can survive; replicative lifespan, which corresponds to the number of mitotic cell divisions a mother cell has undergone.
- Healthspan
corresponds to the duration of disease-free physiological health within the lifespan of an individual. In humans, for instance, this corresponds to the period with high cognitive abilities, immune competence, and peak physical condition.
- Isogenic
characterized by essentially identical genetic material. Highly inbred populations are usually considered to be isogenic.
- Epigenetic
the broad definition of the term corresponds to modes of genomic regulation not directly encoded in DNA. We use it specifically to refer to chromatin-level regulation. In its strictest definition, epigenetic encompasses strictly heritable changes without changes to the underlying gene sequence.
- Chromatin
a complex between DNA and histone proteins that packages genetic information. Chromatin can be broadly stratified in two states: euchromatin, a loose and transcription-permissive compartment, and heterochromatin, a dense and compact compartment that harbors repressed DNA.
- Epigenome
corresponds to the epigenetic landscape of a cell, including the genome-wide distributions of chromatin marks, such as histone modifications or DNA methylation.
- Replicative senescence
a limitation in the number of time a cell can divide. Replicative senescence has sometimes been used as an in vitro proxy of ageing for dividing mammalian cells such as fibroblasts or stem cells.
- CpG islands
clusters of CpG dinucleotides, usually found in the promoter regions of some genes. They are typically defined by a minimum length (~200bp–500bp) and an unusual enrichment of CpG (>60%). They are usually unmethylated in the germline or pluripotent state.
- Chronological age
the time elapsed since an individual’s birth. Chronological age is often compared to biological age.
- Biological age
is used as a proxy for individual health. It is expressed as the age of the average population that is most similar to the individual under observation. Younger biological ages are linked to high performance and health, while older biological ages correlate with disease onset.
- Histone chaperones
proteins that aid the folding or dimerization of histones, as well as their loading onto the chromatin fiber. Different histones have different chaperones.
- Sirtuin
the family of class III histone deacetylases (HDACs), the founding member of which is the yeast Sir2p. The specificity of these HDACs is their dependency on NAD+ as a co-factor to catalyze deacetylation or other enzymatic reactions, including ADP-ribosylation.
- Dietary restriction
a reduction in food intake without malnutrition. There are many types of DR-regimens, such as intermittent-fasting, global caloric restriction or specific reduction of a nutrient type.
- Position Variegation Effect
variability in the activation state of a gene depending on its proximity to a heterochromatin domain.
- Insulin and insulin-like growth factor (IGF) signaling pathways
important conserved signaling pathways that are sensitive to nutrient levels and have been linked to longevity and stress resistance across metazoans. Primary components relevant to ageing include the insulin and IGF1 receptors, the activation of which triggers a phosphorylation cascade involving PI3-Kinase, AKT and ultimately the FOXO transcription factors.
- Pheromones
secreted chemical factors that can trigger a response in other members of a given species. Pheromone signaling has been linked to sexual behavior, feeding and stress.
- Epimutations
aberrant or atypical changes in epigenetic states, often stochastic.
- Transposable Elements
endogenous DNA elements that change their position or amplify their copy number within a host genome. There are two main types of transposable elements: Type I elements, which function through an RNA intermediate (LINEs, LTRs; copy–paste mechanism) and Type II elements, which function through a DNA intermediate (cut–paste mechanism). They usually occupy a large portion of eukaryotic genomes.
- Genomic instability
random loss of integrity of genetic material, whether through large-scale rearrangements (such as chromosomal translocations, large inversions and deletions, and so on), site-specific changes (such as single nucleotide changes) or transposon insertions. Genomic instability may be a driver of ageing and age-related diseases, such as cancer.
Biographies
Bérénice A. Benayoun is a post-doctoral fellow in Anne Brunet’s lab in the Department of Genetics at Stanford University. She performed her graduate work in Reiner Veitia’s lab at the Jacques Monod Institute (France), studying ovarian ageing. She joined the Brunet laboratory to study the genetics and epigenetics of ageing.
Elizabeth A. Pollina is a graduate student in the Cancer Biology Program at Stanford University. She joined Anne Brunet’s lab in the Department of Genetics to study the role of chromatin in ageing, with particular interest in the central nervous system.
Anne Brunet is a professor of Genetics at Stanford University and the co-director of the Glenn Laboratories for Ageing Biology at Stanford. Her lab is interested in the mechanisms of ageing and longevity.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 2.Cournil A, Kirkwood TB. If you would live long, choose your parents well. Trends Genet. 2001;17:233–235. doi: 10.1016/s0168-9525(01)02306-x. [DOI] [PubMed] [Google Scholar]
- 3.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Remolina SC, Hughes KA. Evolution and mechanisms of long life and high fertility in queen honey bees. Age. 2008;30:177–185. doi: 10.1007/s11357-008-9061-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Libert S, Bonkowski MS, Pointer K, Pletcher SD, Guarente L. Deviation of innate circadian period from 24 h reduces longevity in mice. Aging cell. 2012;11:794–800. doi: 10.1111/j.1474-9726.2012.00846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waddington CH. The Epigenotype. International Journal of Epidemiology. 2012;41:10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 7.Zahn JM, et al. AGEMAP: a gene expression database for aging in mice. PLoS genetics. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Southworth LK, Owen AB, Kim SK. Aging mice show a decreasing correlation of gene expression within genetic modules. PLoS genetics. 2009;5:e1000776. doi: 10.1371/journal.pgen.1000776. First genome-wide investigation of the effect of ageing on the robustness of transcriptional networks in mice.
- 9.Gut P, Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature. 2013;502:489–498. doi: 10.1038/nature12752. [DOI] [PubMed] [Google Scholar]
- 10.Vaquero A, Reinberg D. Calorie restriction and the exercise of chromatin. Genes & development. 2009;23:1849–1869. doi: 10.1101/gad.1807009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seong KH, Li D, Shimizu H, Nakamura R, Ishii S. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell. 2011;145:1049–1061. doi: 10.1016/j.cell.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nature biotechnology. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruickshanks HA, et al. Senescent cells harbour features of the cancer epigenome. Nat Cell Biol. 2013;15:1495–1506. doi: 10.1038/ncb2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 16.Day K, et al. Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome Biol. 2013;14:R102. doi: 10.1186/gb-2013-14-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maegawa S, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teschendorff AE, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou J, Lippert C, Heckerman D, Aryee M, Listgarten J. Epigenome-wide association studies without the need for cell-type composition. Nat Methods. 2014;11:309–311. doi: 10.1038/nmeth.2815. [DOI] [PubMed] [Google Scholar]
- 20.Beerman I, et al. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell. 2013;12:413–425. doi: 10.1016/j.stem.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun D, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14:673–688. doi: 10.1016/j.stem.2014.03.002. First extensive study of the epigenomic landscape (several histone marks and DNA-methylation) in purified stem cells (hematopoietic stem cells) from young and old mice.
- 22.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greer EL, et al. DNA Methylation on N6-Adenine in C. elegans. Cell. 2015;161:868–878. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y, et al. N6-methyldeoxyadenosine marks active transcription start sites in chlamydomonas. Cell. 2015;161:879–892. doi: 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang G, et al. N6-methyladenine DNA modification in Drosophila. Cell. 2015;161:893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 26.Weidner CI, et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014;15:R24. doi: 10.1186/gb-2014-15-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. Most extensive demonstration that DNA-methylation levels at specific loci are predictive of age across a variety of cells and tissues.
- 29.Capuano F, Mulleder M, Kok R, Blom HJ, Ralser M. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Analytical chemistry. 2014;86:3697–3702. doi: 10.1021/ac500447w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson VJ, Johnson TE, Hammen RF. Caenorhabditis elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic Acids Res. 1986;14:6711–6719. doi: 10.1093/nar/14.16.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin MJ, Tang LY, Reddy MN, Shen CK. DNA methyltransferase gene dDnmt2 and longevity of Drosophila. The Journal of biological chemistry. 2005;280:861–864. doi: 10.1074/jbc.C400477200. [DOI] [PubMed] [Google Scholar]
- 32.Phalke S, et al. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nature genetics. 2009;41:696–702. doi: 10.1038/ng.360. [DOI] [PubMed] [Google Scholar]
- 33.Schaefer M, Lyko F. Lack of evidence for DNA methylation of Invader4 retroelements in Drosophila and implications for Dnmt2-mediated epigenetic regulation. Nature genetics. 2010;42:920–921. doi: 10.1038/ng1110-920. author reply 921. [DOI] [PubMed] [Google Scholar]
- 34.Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feser J, Tyler J. Chromatin structure as a mediator of aging. FEBS Lett. 2011;585:2041–2048. doi: 10.1016/j.febslet.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Z, et al. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 2014;28:396–408. doi: 10.1101/gad.233221.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu L, et al. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. First genome-wide study of age-dependent changes in histone modifications in purified adult stem cells.
- 39.Shah PP, et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 2013;27:1787–1799. doi: 10.1101/gad.223834.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Feser J, et al. Elevated histone expression promotes life span extension. Mol Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. This study provided the first evidence that the expression level of core histones may directly impact lifespan.
- 41.Das C, Tyler JK. Histone exchange and histone modifications during transcription and aging. Biochim Biophys Acta. 2013;1819:332–342. doi: 10.1016/j.bbagrm.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nature reviews. Genetics. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsurumi A, Li WX. Global heterochromatin loss: a unifying theory of aging? Epigenetics. 2012;7:680–688. doi: 10.4161/epi.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dechat T, et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W, et al. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015 doi: 10.1126/science.aaa1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin C, et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;14:161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 47.Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10:980–990. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ni Z, Ebata A, Alipanahiramandi E, Lee SS. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell. 2012;11:315–325. doi: 10.1111/j.1474-9726.2011.00785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siebold AP, et al. Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:169–174. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baumgart M, et al. RNA-seq of the aging brain in the short-lived fish N. furzeri - conserved pathways and novel genes associated with neurogenesis. Aging Cell. 2014;13:965–974. doi: 10.1111/acel.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pu M, et al. Trimethylation of Lys36 on H3 restricts gene expression change during aging and impacts life span. Genes & development. 2015;29:718–731. doi: 10.1101/gad.254144.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benayoun BA, et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158:673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Greer EL, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. This study linked histone methylation to lifespan extension for the first time in a metazoan.
- 54.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L, Greer C, Eisenman RN, Secombe J. Essential functions of the histone demethylase lid. PLoS genetics. 2010;6:e1001221. doi: 10.1371/journal.pgen.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 57.Alvares SM, Mayberry GA, Joyner EY, Lakowski B, Ahmed S. H3K4 demethylase activities repress proliferative and postmitotic aging. Aging Cell. 2014;13:245–253. doi: 10.1111/acel.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sen P, et al. H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Genes Dev. 2015;29:1362–1376. doi: 10.1101/gad.263707.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. The first experimental swap of histone residues showed that H4K16 itself plays a role in yeast replicative ageing.
- 60.Krishnan V, et al. Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc Natl Acad Sci U S A. 2011;108:12325–12330. doi: 10.1073/pnas.1102789108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peleg S, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 63. Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. First demonstration that longevity-promoting sirtuins are histone deacetylases.
- 64.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kugel S, Mostoslavsky R. Chromatin and beyond: the multitasking roles for SIRT6. Trends in biochemical sciences. 2014;39:72–81. doi: 10.1016/j.tibs.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burnett C, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Satoh A, et al. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell metabolism. 2013;18:416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toiber D, et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Molecular cell. 2013;51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 71. Kanfi Y, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. Studies 70 and 71 were the first to link a member of the Sirtuin family with ageing and lifespan in mammals.
- 72.Van Meter M, et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang RH, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Love DC, et al. Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7413–7418. doi: 10.1073/pnas.0911857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rahman MM, et al. Intracellular protein glycosylation modulates insulin mediated lifespan in C.elegans. Aging. 2010;2:678–690. doi: 10.18632/aging.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annual review of biochemistry. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pegoraro G, et al. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dang W, et al. Inactivation of Yeast Isw2 Chromatin Remodeling Enzyme Mimics Longevity Effect of Calorie Restriction via Induction of Genotoxic Stress Response. Cell metabolism. 2014;19:952–966. doi: 10.1016/j.cmet.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Riedel CG, et al. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat Cell Biol. 2013;15:491–501. doi: 10.1038/ncb2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Janssen I, Carson V, Lee IM, Katzmarzyk PT, Blair SN. Years of life gained due to leisure-time physical activity in the U.S. American journal of preventive medicine. 2013;44:23–29. doi: 10.1016/j.amepre.2012.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Maures TJ, et al. Males shorten the life span of C. elegans hermaphrodites via secreted compounds. Science. 2014;343:541–544. doi: 10.1126/science.1244160. Together with references 123 and 124, this study was the first to investigate the mechanistic bases of sex interactions on lifespan, and this study identified a link with a chromatin modifier UTX-1.
- 83.Orozco-Solis R, Sassone-Corsi P. Circadian clock: linking epigenetics to aging. Current opinion in genetics & development. 2014;26C:66–72. doi: 10.1016/j.gde.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heydari AR, Unnikrishnan A, Lucente LV, Richardson A. Caloric restriction and genomic stability. Nucleic acids research. 2007;35:7485–7496. doi: 10.1093/nar/gkm860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang N, et al. Dietary and genetic effects on age-related loss of gene silencing reveal epigenetic plasticity of chromatin repression during aging. Aging. 2013;5:813–824. doi: 10.18632/aging.100614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Horvath S, et al. Obesity accelerates epigenetic aging of human liver. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 88.Hatta M, Liu F, Cirillo LA. Acetylation curtails nucleosome binding, not stable nucleosome remodeling, by FoxO1. Biochemical and biophysical research communications. 2009;379:1005–1008. doi: 10.1016/j.bbrc.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 89.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boily G, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PloS one. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stenesen D, et al. Adenosine nucleotide biosynthesis and AMPK regulate adult life span and mediate the longevity benefit of caloric restriction in flies. Cell metabolism. 2013;17:101–112. doi: 10.1016/j.cmet.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martin-Montalvo A, et al. Metformin improves healthspan and lifespan in mice. Nature communications. 2013;4:2192. doi: 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Abate G, et al. Snf1/AMPK regulates Gcn5 occupancy, H3 acetylation and chromatin remodelling at S. cerevisiae ADY2 promoter. Biochimica et biophysica acta. 2012;1819:419–427. doi: 10.1016/j.bbagrm.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McGee SL, et al. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57:860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- 96.Mihaylova MM, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bungard D, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lau AW, Liu P, Inuzuka H, Gao D. SIRT1 phosphorylation by AMP-activated protein kinase regulates p53 acetylation. American journal of cancer research. 2014;4:245–255. [PMC free article] [PubMed] [Google Scholar]
- 99.Xu Q, et al. AMPK regulates histone H2B O-GlcNAcylation. Nucleic acids research. 2014;42:5594–5604. doi: 10.1093/nar/gku236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS biology. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hachinohe M, et al. A reduction in age-enhanced gluconeogenesis extends lifespan. PloS one. 2013;8:e54011. doi: 10.1371/journal.pone.0054011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zheng L, Roeder RG, Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
- 103.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Salminen A, Kauppinen A, Hiltunen M, Kaarniranta K. Krebs cycle intermediates regulate DNA and histone methylation: Epigenetic impact on the aging process. Ageing research reviews. 2014 doi: 10.1016/j.arr.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 105.Williams DS, Cash A, Hamadani L, Diemer T. Oxaloacetate supplementation increases lifespan in Caenorhabditis elegans through an AMPK/FOXO-dependent pathway. Aging cell. 2009;8:765–768. doi: 10.1111/j.1474-9726.2009.00527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Edwards CB, Copes N, Brito AG, Canfield J, Bradshaw PC. Malate and fumarate extend lifespan in Caenorhabditis elegans. PloS one. 2013;8:e58345. doi: 10.1371/journal.pone.0058345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chin RM, et al. The metabolite alpha-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature. 2014;509:397–401. doi: 10.1038/nature13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Froy O. Circadian rhythms, aging, and life span in mammals. Physiology. 2011;26:225–235. doi: 10.1152/physiol.00012.2011. [DOI] [PubMed] [Google Scholar]
- 109.Hurd MW, Ralph MR. The significance of circadian organization for longevity in the golden hamster. Journal of biological rhythms. 1998;13:430–436. doi: 10.1177/074873098129000255. [DOI] [PubMed] [Google Scholar]
- 110.Rakshit K, Giebultowicz JM. Cryptochrome restores dampened circadian rhythms and promotes healthspan in aging Drosophila. Aging cell. 2013;12:752–762. doi: 10.1111/acel.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aguilar-Arnal L, Sassone-Corsi P. The circadian epigenome: how metabolism talks to chromatin remodeling. Current opinion in cell biology. 2013;25:170–176. doi: 10.1016/j.ceb.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. Demonstration that the molecular circadian clock is an epigenetic regulator.
- 113.Nakahata Y, et al. The NAD+dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 115.Bellet MM, et al. Pharmacological modulation of circadian rhythms by synthetic activators of the deacetylase SIRT1. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3333–3338. doi: 10.1073/pnas.1214266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu CW, et al. Exercise enhances the proliferation of neural stem cells and neurite growth and survival of neuronal progenitor cells in dentate gyrus of middle-aged mice. Journal of applied physiology. 2008;105:1585–1594. doi: 10.1152/japplphysiol.90775.2008. [DOI] [PubMed] [Google Scholar]
- 118.Lugert S, et al. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell stem cell. 2010;6:445–456. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 119.Li L, et al. Acute Aerobic Exercise Increases Cortical Activity during Working Memory: A Functional MRI Study in Female College Students. PloS one. 2014;9:e99222. doi: 10.1371/journal.pone.0099222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gremeaux V, et al. Exercise and longevity. Maturitas. 2012;73:312–317. doi: 10.1016/j.maturitas.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 121.Koltai E, et al. Exercise alters SIRT1, SIRT6, NAD and NAMPT levels in skeletal muscle of aged rats. Mechanisms of ageing and development. 2010;131:21–28. doi: 10.1016/j.mad.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McGee SL, Fairlie E, Garnham AP, Hargreaves M. Exercise-induced histone modifications in human skeletal muscle. The Journal of physiology. 2009;587:5951–5958. doi: 10.1113/jphysiol.2009.181065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shi C, Murphy CT. Mating induces shrinking and death in Caenorhabditis mothers. Science. 2014;343:536–540. doi: 10.1126/science.1242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gendron CM, et al. Drosophila life span and physiology are modulated by sexual perception and reward. Science. 2014;343:544–548. doi: 10.1126/science.1243339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Edison AS. Caenorhabditis elegans pheromones regulate multiple complex behaviors. Curr Opin Neurobiol. 2009;19:378–388. doi: 10.1016/j.conb.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ludewig AH, et al. Pheromone sensing regulates Caenorhabditis elegans lifespan and stress resistance via the deacetylase SIR-2.1. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5522–5527. doi: 10.1073/pnas.1214467110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wiench M, Miranda TB, Hager GL. Control of nuclear receptor function by local chromatin structure. The FEBS journal. 2011;278:2211–2230. doi: 10.1111/j.1742-4658.2011.08126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Osmanbeyoglu HU, et al. Estrogen represses gene expression through reconfiguring chromatin structures. Nucleic acids research. 2013;41:8061–8071. doi: 10.1093/nar/gkt586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kanungo MS, Thakur MK. Modulation of acetylation of histones and transcription of chromatin by butyric acid and 17beta-estradiol in the brain of rats of various ages. Biochemical and biophysical research communications. 1979;87:266–271. doi: 10.1016/0006-291x(79)91675-9. [DOI] [PubMed] [Google Scholar]
- 130.Deroo BJ, Korach KS. Estrogen receptors and human disease. The Journal of clinical investigation. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chong S, Youngson NA, Whitelaw E. Heritable germline epimutation is not the same as transgenerational epigenetic inheritance. Nature genetics. 2007;39:574–575. doi: 10.1038/ng0507-574. author reply 575–6. [DOI] [PubMed] [Google Scholar]
- 132.Holliday R. The inheritance of epigenetic defects. Science. 1987;238:163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- 133.Vijg J, Suh Y. Genome instability and aging. Annual review of physiology. 2013;75:645–668. doi: 10.1146/annurev-physiol-030212-183715. [DOI] [PubMed] [Google Scholar]
- 134.Burgess RC, Misteli T, Oberdoerffer P. DNA damage, chromatin, and transcription: the trinity of aging. Current opinion in cell biology. 2012;24:724–730. doi: 10.1016/j.ceb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vyjayanti VN, Rao KS. DNA double strand break repair in brain: reduced NHEJ activity in aging rat neurons. Neuroscience letters. 2006;393:18–22. doi: 10.1016/j.neulet.2005.09.053. [DOI] [PubMed] [Google Scholar]
- 136.Burtner CR, Kennedy BK. Progeria syndromes and ageing: what is the connection? Nature reviews. Molecular cell biology. 2010;11:567–578. doi: 10.1038/nrm2944. [DOI] [PubMed] [Google Scholar]
- 137.Liu J, Kim J, Oberdoerffer P. Metabolic modulation of chromatin: implications for DNA repair and genomic integrity. Frontiers in genetics. 2013;4:182. doi: 10.3389/fgene.2013.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kidwell MG. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 2002;115:49–63. doi: 10.1023/a:1016072014259. [DOI] [PubMed] [Google Scholar]
- 139.Maxwell PH, Burhans WC, Curcio MJ. Retrotransposition is associated with genome instability during chronological aging. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20376–20381. doi: 10.1073/pnas.1100271108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505:353–359. doi: 10.1038/nature12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wood JG, Helfand SL. Chromatin structure and transposable elements in organismal aging. Frontiers in genetics. 2013;4:274. doi: 10.3389/fgene.2013.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dennis S, Sheth U, Feldman JL, English KA, Priess JR. C. elegans germ cells show temperature and age-dependent expression of Cer1, a Gypsy/Ty3-related retrotransposon. PLoS pathogens. 2012;8:e1002591. doi: 10.1371/journal.ppat.1002591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li W, et al. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat Neurosci. 2013;16:529–531. doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.De Cecco M, et al. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY) 2013;5:867–883. doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Reilly MT, Faulkner GJ, Dubnau J, Ponomarev I, Gage FH. The role of transposable elements in health and diseases of the central nervous system. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:17577–17586. doi: 10.1523/JNEUROSCI.3369-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.De Cecco M, et al. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging cell. 2013;12:247–256. doi: 10.1111/acel.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vinuela A, Snoek LB, Riksen JA, Kammenga JE. Genome-wide gene expression regulation as a function of genotype and age in C. elegans. Genome research. 2010;20:929–937. doi: 10.1101/gr.102160.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang Q, et al. The spatial association of gene expression evolves from synchrony to asynchrony and stochasticity with age. PloS one. 2011;6:e24076. doi: 10.1371/journal.pone.0024076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bahar R, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 150. Warren LA, et al. Transcriptional instability is not a universal attribute of aging. Aging cell. 2007;6:775–782. doi: 10.1111/j.1474-9726.2007.00337.x. With reference 149, these pioneering studies investigated the impact of ageing on cell-to-cell noise at select genes.
- 151.Busuttil RA, et al. Intra-organ variation in age-related mutation accumulation in the mouse. PloS one. 2007;2:e876. doi: 10.1371/journal.pone.0000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Weinberger L, et al. Expression noise and acetylation profiles distinguish HDAC functions. Molecular cell. 2012;47:193–202. doi: 10.1016/j.molcel.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jones DL. Aging and the germ line: where mortality and immortality meet. Stem Cell Rev. 2007;3:192–200. doi: 10.1007/s12015-007-0009-3. [DOI] [PubMed] [Google Scholar]
- 154.Lim JP, Brunet A. Bridging the transgenerational gap with epigenetic memory. Trends Genet. 2013;29:176–186. doi: 10.1016/j.tig.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ragunathan K, Jih G, Moazed D. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science. 2015;348:1258699. doi: 10.1126/science.1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gaydos LJ, Wang W, Strome S. Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science. 2014;345:1515–1518. doi: 10.1126/science.1255023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Radford EJ, et al. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science. 2014;345:1255903. doi: 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kelly WG. Transgenerational epigenetics in the germline cycle of Caenorhabditis elegans. Epigenetics Chromatin. 2014;7:6. doi: 10.1186/1756-8935-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Katz DJ, Edwards TM, Reinke V, Kelly WG. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Greer EL, et al. A histone methylation network regulates transgenerational epigenetic memory in C. elegans. Cell Rep. 2014;7:113–126. doi: 10.1016/j.celrep.2014.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xiao Y, et al. Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proc Natl Acad Sci U S A. 2011;108:8305–8310. doi: 10.1073/pnas.1019290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kerr SC, Ruppersburg CC, Francis JW, Katz DJ. SPR-5 and MET-2 function cooperatively to reestablish an epigenetic ground state during passage through the germ line. Proc Natl Acad Sci U S A. 2014;111:9509–9514. doi: 10.1073/pnas.1321843111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Robert VJ, et al. The SET-2/SET1 Histone H3K4 Methyltransferase Maintains Pluripotency in the Caenorhabditis elegans Germline. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 164.Buckley BA, et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Simon M, et al. Reduced insulin/IGF-1 signaling restores germ cell immortality to caenorhabditis elegans Piwi mutants. Cell Rep. 2014;7:762–773. doi: 10.1016/j.celrep.2014.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Greer EL, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rechavi O, et al. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Carone BR, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tauffenberger A, Parker JA. Heritable transmission of stress resistance by high dietary glucose in Caenorhabditis elegans. PLoS Genet. 2014;10:e1004346. doi: 10.1371/journal.pgen.1004346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Stern S, Fridmann-Sirkis Y, Braun E, Soen Y. Epigenetically heritable alteration of fly development in response to toxic challenge. Cell Rep. 2012;1:528–542. doi: 10.1016/j.celrep.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 171.Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nature reviews. Drug discovery. 2013;12:917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 172.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nature reviews. Drug discovery. 2014;13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 173.Graff J, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015 doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cusanovich DA, et al. Epigenetics. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 177.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 178.Maze I, Noh KM, Soshnev AA, Allis CD. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nature reviews. Genetics. 2014;15:259–271. doi: 10.1038/nrg3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nature structural & molecular biology. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 180.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 181.Bocklandt S, et al. Epigenetic predictor of age. PLoS One. 2011;6:e14821. doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Horvath S, et al. Accelerated epigenetic aging in Down syndrome. Aging Cell. 2015 doi: 10.1111/acel.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Marioni RE, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tsygankov D, Liu Y, Sanoff HK, Sharpless NE, Elston TC. A quantitative model for age-dependent expression of the p16INK4a tumor suppressor. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16562–16567. doi: 10.1073/pnas.0904405106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Epel ES, et al. Accelerated telomere shortening in response to life stress. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17312–17315. doi: 10.1073/pnas.0407162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kellinger MW, et al. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol. 2012;19:831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kirkwood TB, Holliday R. The evolution of ageing and longevity. Proc R Soc Lond B Biol Sci. 1979;205:531–546. doi: 10.1098/rspb.1979.0083. [DOI] [PubMed] [Google Scholar]
- 188.Mahmoudi S, Brunet A. Aging and reprogramming: a two-way street. Curr Opin Cell Biol. 2012;24:744–756. doi: 10.1016/j.ceb.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 190.Wahlestedt M, et al. An epigenetic component of hematopoietic stem cell aging amenable to reprogramming into a young state. Blood. 2013;121:4257–4264. doi: 10.1182/blood-2012-11-469080. [DOI] [PubMed] [Google Scholar]
- 191.Jenkins TG, Aston KI, Pflueger C, Cairns BR, Carrell DT. Age-associated sperm DNA methylation alterations: possible implications in offspring disease susceptibility. PLoS Genet. 2014;10:e1004458. doi: 10.1371/journal.pgen.1004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shao GB, et al. Aging alters histone H3 lysine 4 methylation in mouse germinal vesicle stage oocytes. Reprod Fertil Dev. 2014 doi: 10.1071/RD13293. [DOI] [PubMed] [Google Scholar]
- 193.Manosalva I, Gonzalez A. Aging changes the chromatin configuration and histone methylation of mouse oocytes at germinal vesicle stage. Theriogenology. 2010;74:1539–1547. doi: 10.1016/j.theriogenology.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 194.Hamatani T, et al. Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Genet. 2004;13:2263–2278. doi: 10.1093/hmg/ddh241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.