Abstract
Due to the lack of annotated data sets, there are few studies on machine learning based approaches to extract named entities (NEs) in clinical text. The 2009 i2b2 NLP challenge is a task to extract six types of medication related NEs, including medication names, dosage, mode, frequency, duration, and reason from hospital discharge summaries. Several machine learning based systems have been developed and showed good performance in the challenge. Those systems often involve two steps: 1) recognition of medication related entities; and 2) determination of the relation between a medication name and its modifiers (e.g., dosage). A few machine learning algorithms including Conditional Random Field (CRF) and Maximum Entropy have been applied to the Named Entity Recognition (NER) task at the first step. In this study, we developed a Support Vector Machine (SVM) based method to recognize medication related entities. In addition, we systematically investigated various types of features for NER in clinical text. Evaluation on 268 manually annotated discharge summaries from i2b2 challenge showed that the SVM-based NER system achieved the best F-score of 90.05% (93.20% Precision, 87.12% Recall), when semantic features generated from a rule-based system were included.
1 Introduction
Named Entity Recognition (NER) is an important step in natural language processing (NLP). It has many applications in general language domain such as identifying person names, locations, and organizations. NER is crucial for biomedical literature mining as well (Hirschman, Morgan, & Yeh, 2002; Krauthammer & Nenadic, 2004) and many studies have focused on biomedical entities, such as gene/protein names. There are mainly two types of approaches to identify biomedical entities: rule-based and machine learning based approaches. While rule-based approaches use existing biomedical knowledge/resources, machine learning (ML) based approaches rely much on annotated training data. The advantage of rule-based approaches is that they usually can achieve stable performance across different data sets due to the verified resources, while machine learning approaches often report better results when the training data are good enough. In order to harness the advantages of both approaches, the combination of them, called the hybrid approach, has often been used as well. CRF and SVM are two common machine learning algorithms that have been widely used in biomedical NER (Takeuchi & Collier, 2003; Kazama, Makino, Ohta, & Tsujii, 2002; Yamamoto, Kudo, Konagaya, & Matsumoto, 2003; Torii, Hu, Wu, & Liu, 2009; Li, Savova, & Kipper-Schuler, 2008). Some studies reported better results using CRF (Li, Savova, & Kipper-Schuler, 2008), while others showed that the SVM was better (Tsochantaridis, Joachims, & Hofmann, 2005) in NER. Keerthi & Sundararajan (Keerthi & Sunda-rarajan, 2007) conducted some experiments and demonstrated that CRF and SVM were quite close in performance, when identical feature functions were used.
2 Background
There has been large ongoing effort on processing clinical text in Electronic Medical Records (EMRs). Many clinical NLP systems have been developed, including MedLEE (Friedman, Alderson, Austin, Cimino, & Johnson, 1994), SymTex (Haug et al., 1997), Meta-Map (Aronson, 2001). Most of those systems recognize clinical named entities such as diseases, medications, and labs, using rule-based methods such as lexicon lookup, mainly because of two reasons: 1) there are very rich knowledge bases and vocabularies of clinical entities, such as the Unified Medical Language System (UMLS) (Lindberg, Humphreys, & McCray, 1993), which includes over 100 controlled bio-medical vocabularies, such as RxNorm, SNOMED, and ICD-9-CM; 2) very few annotated data sets of clinical text are available for machine learning based approaches.
Medication is one of the most important types of information in clinical text. Several studies have worked on extracting drug names from clinical notes. Evans et al. (Evans, Brownlow, Hersh, & Campbell, 1996) showed that drug and dosage phrases in discharge summaries could be identified by the CLARIT system with an accuracy of 80%. Chhieng et al. (Chhieng, Day, Gordon, & Hicks, 2007) reported a precision of 83% when using a string matching method to identify drug names in clinical records. Levin et al. (Levin, Krol, Doshi, & Reich, 2007) developed an effective rule-based system to extract drug names from anesthesia records and map to RxNorm concepts with 92.2% sensitivity and 95.7% specificity. Sirohi and Peissig (Sirohi & Peissig, 2005) studied the effect of lexicon sources on drug extraction. Recently, Xu et al. (Xu et al., 2010) developed a rule-based system for medication information extraction, called MedEx, and reported F-scores over 90% on extracting drug names, dose, route, and frequency from discharge summaries.
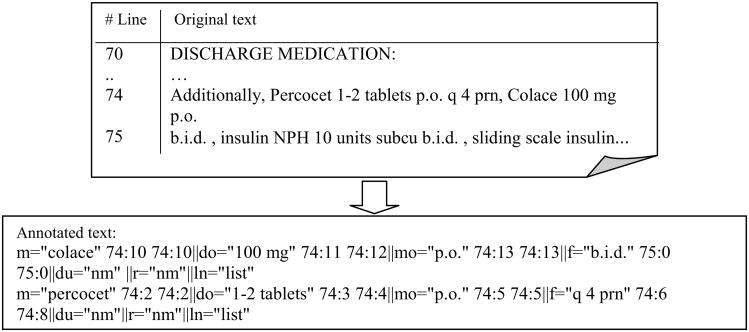
Starting 2007, Informatics for Integrating Biology and the Bedside (i2b2), an NIH-funded National Center for Biomedical Computing (NCBC) based at Partners Healthcare System in Boston, organized a series of shared tasks of NLP in clinical text. The 2009 i2b2 NLP challenge was to extract medication names, as well as their corresponding signature information including dosage, mode, frequency, duration, and reason from de-identified hospital discharge summaries (Uzüner, Solti, & Cadag, 2009). At the beginning of the challenge, a training set of 696 notes were provided by the organizers. Among them, 17 notes were annotated by the i2b2 organizers, based on an annotation guideline (see Table 1 for examples of medication information in the guideline), and the rest were un-annotated notes. Participating teams would develop their systems based on the training set, and they were allowed to annotate additional notes in the training set. The test data set included 547clinical notes, from which 251 notes were randomly picked by the organizers. Those 251 notes were then annotated by participating teams, as well as the organizers, and they served as the gold standard for evaluating the performance of systems submitted by participating teams. An example of original text and annotated text were shown in Figure 1.
Table 1.
Number of classes and descriptions with examples in i2b2 2009 dataset.
| Class | # | Example | Description |
|---|---|---|---|
| Medication | 12773 | “Lasix”, “Caltrate plus D”, “fluoci-nonide 0.5% cream”, “TYLENOL (ACETAMINOPHEN)” | Prescription substances, biological substances, over-the-counter drugs, excluding diet, allergy, lab/test, alcohol. |
| Dosage | 4791 | “1 TAB”, “One tablet”, “0.4 mg” “0.5 m.g.”, “100 MG”, “100 mg × 2 tablets” | The amount of a single medication used in each administration. |
| Mode | 3552 | “Orally”, “Intravenous”, “Topical”, “Sublingual” | Describes the method for administering the medication. |
| Frequency | 4342 | “Prn”, “As needed”, “Three times a day as needed”, “As needed three times a day”, “×3 before meal”, “×3 a day after meal as needed” | Terms, phrases, or abbreviations that describe how often each dose of the medication should be taken. |
| Duration | 597 | “×10 days”, “10-day course”, “For ten days”, “For a month”, “During spring break”, “Until the symptom disappears”, “As long as needed” | Expressions that indicate for how long the medication is to be administered. |
| Reason | 1534 | “Dizziness”, “Dizzy”, “Fever”, “Diabetes”, “frequent PVCs”, “rare angina” | The medical reason for which the medication is stated to be given. |
Figure. 1.
An example of the i2b2 data, ‘m’ is for MED NAME, ‘do’ is for DOSE, ‘mo’ is for MODE, ‘f’ is for FREQ, ‘du’ is for DURATION, ‘r’ is for REASON, ‘ln’ is for “list/narrative.”
The results of systems submitted by the participating teams were presented at the i2b2 workshop and short papers describing each system were available at i2b2 web site with protected passwords. Among top 10 systems which achieved the best performance, there were 6 rule-based, 2 machine learning based, and 2 hybrid systems. The best system, which used a machine learning based approach, reached the highest F-score of 85.7% (Patrick & Li, 2009). The second best system, which was a rule-based system using the existing MedEx tool, reported an F-score of 82.1% (Doan, Bastarache L., Klimkowski S., Denny J.C., & Xu, 2009). The difference between those two systems was statistically significant. However, this finding was not very surprising, as the machine learning based system utilized additional 147 annotated notes by the participating team, while the rule-based system mainly used 17 annotated training data to customize the system.
Interestingly, two machine learning systems in the top ten systems achieved very different performance, one (Patrick et al., 2009) achieved an F-score of 85.7%, ranked the first; while another (Li et al., 2009) achieved an F-score of 76.4%, ranked the 10th on the final evaluation. Both systems used CRF for NER, on the equivalent number of training data (145 and 147 notes respectively). The large difference in F-score of those two systems could be due to: the quality of training set, and feature sets using for classification. More recently, i2b2 organizers also reported a Maximum Entropy (ME) based approach for the 2009 challenge (Halgrim, Xia, Solti, Cadag, & Uzuner, 2010). Using the same annotated data set as in (Patrick et al., 2009), they reported an F-score of 84.1%, when combined features such as unigram, word bigrams/trigrams, and label of previous words were used. These results indicated the importance of feature sets used in machine learning algorithms in this task.
For supervised machine learning based systems in the i2b2 challenge, the task was usually divided into two steps: 1) NER of six medication related findings; and 2) determination of the relation between detected medication names and other entities. It is obvious that NER is the first crucial step and it affects the performance of the whole system. However, short papers presented at the i2b2 workshop did not show much detailed evaluation on NER components in machine learning based systems. The variation in performance of different machine learning based systems also motivated us to further investigate the effect of different types of features on recognizing medication related entities.
In this study, we developed an SVM-based NER system for recognizing medication related entities, which is a sub-task of the i2b2 challenge. We systematically investigated the effects of typical local contextual features that have been reported in many biomedical NER studies. Our studies provided some valuable insights to NER tasks of medical entities in clinical text.
3 Methods
A total of 268 annotated discharge summaries (17 from training set and 251 from test set) from i2b2 challenge were used in this study. This annotated corpus contains 9,689 sentences, 326,474 words, and 27,589 entities. Annotated notes were converted into a BIO format and different types of feature sets were used in an SVM classifier for NER. Performance of the NER system was evaluated using precision, recall, and F-score, based on 10-fold cross validation.
3.1 Preprocessing
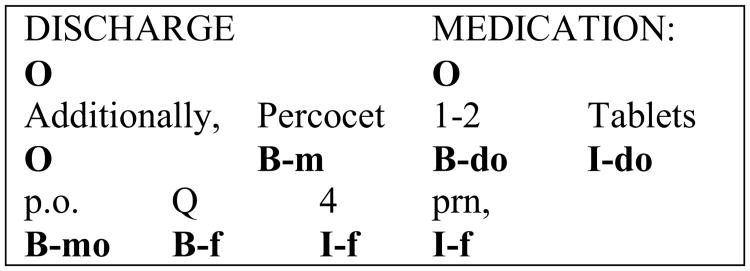
The annotated corpus was converted into a BIO format (see an example in Figure 2). Specifically, it assigned each word into a class as follows: B means beginning of an entity, I means inside an entity, and O means outside of an entity. As we have six types of entities, we have six different B classes and six different I classes. For example, for medication names, we define the B class as “B-m”, and the I class as “I-m”. Therefore, we had total 13 possible classes to each word (including O class).
Figure 2.
An example of the BIO representation of annotated clinical text (Where m as medication, do as dose, mo as mode, and f as frequency).
After preprocessing, the NER problem now can be considered as a classification problem, which is to assigns one of the 13 class labels to each word.
3.2 SVM
Support Vector Machine (SVM) is a machine learning method that is widely used in many NLP tasks such as chunking, POS, and NER. Essentially, it constructs a binary classifier using labeled training samples. Given a set of training samples, the SVM training phrase tries to find the optimal hyperplane, which maximizes the distance of training sample nearest to it (called support vectors). SVM takes an input as a vector and maps it into a feature space using a kernel function.
In this paper we used TinySVM1 along with Yamcha2 developed at NAIST (Kudo & Matsu-moto, 2000; Kudo & Matsumoto, 2001). We used a polynomial kernel function with the degree of kernel as 2, context window as +/-2, and the strategy for multiple classification as pair-wise (one-against-one). Pairwise strategy means it will build K(K-1)/2 binary classifiers in which K is the number of classes (in this case K=13). Each binary classifier will determine whether the sample should be classified as one of the two classes. Each binary classifier has one vote and the final output is the class with the maximum votes. These parameters were used in many biomedical NER tasks such as (Takeuchi & Collier, 2003; Kazama et al., 2002; Yamamoto et al., 2003).
3.3 Features sets
In this study, we investigated different types of features for the SVM-based NER system for medication related entities, including 1) words; 2) Part-of-Speech (POS) tags; 3) morphological clues; 4) orthographies of words; 5) previous history features; 6) semantic tags determined by MedEx, a rule based medication extraction system. Details of those features are described below:
Words features: Words only. We referred it as a baseline method in this study.
POS features: Part-of-Speech tags of words. To obtain POS information, we used a POS tagger in the NLTK package3.
Morphologic features: suffix/prefix of up to 3 characters within a word.
Orthographic features: information about if a word contains capital letters, digits, special characters etc. We used orthographic features described in (Collier, Nobata, & Tsujii, 2000) and modified some as for medication information such as “digit and percent”. We had totally 21 labels for orthographic features.
Previous history features: Class assignments of preceding words, by the NER system itself.
Semantic tag features: semantic categories of words. Typical NER systems use dictionary lookup methods to determine semantic categories of a word (e.g., gene names in a dictionary). In this study, we used MedEx, the best rule-based medication extraction system in the i2b2 challenge, to assign medication specific categories into words.
MedEx was originally developed at Vanderbilt University, for extracting medication information from clinical text (Xu et al., 2010). MedEx labels medication related entities with a pre-defined semantic categories, which has overlap with the six entities defined in the i2b2 challenge, but not exactly same. For example, MedEx breaks the phrase “fluocinonide 0 5% cream” into drug name: “fluocinonide”, strength: “0.5%”, and form: “cream”; while i2b2 labels the whole phrase as a medication name. There are a total of 11 pre-defined semantic categories which are listed in (Xu et al., 2010c). When the Vanderbilt team applied MedEx to the i2b2 challenge, they customized and extended MedEx to label medication related entities as required by i2b2. Those customizations included:
- Customized Rules to combine entities recognized by MedEx into i2b2 entities, such as combine drug name: “fluocinonide”, strength: “0.5%”, and form: “cream” into one medication name “fluocinonide 0.5% cream”.
- A new Section Tagger to filter some drug names in sections such as “allergy” and “labs”.
- A new Spell Checker to check whether a word can be a misspelled drug names.
In a summary, the MedEx system will produce two sets of semantic tags: 1) initial tags that are identified by the original MedEx system; 2) final tags that are identified by the customized MedEx system for the i2b2 challenge. The initial tagger will be equivalent to some simple dictionary look up methods used in many NER systems. The final tagger is a more advanced method that integrates other level of information such as sections and spellings. The outputs of initial tag include 11 pre-defined semantic tags in MedEx, and outputs of final tags consist of 6 types of NEs as in the i2b2 requirements. Therefore, it is interesting to us to study effects of both types of tags from MedEx in this study. These semantic tags were also converted into the BIO format when they were used as features.
4 Results and Discussions
In this study, we measured Precision, Recall, and F-score using the CoNLL evaluation script4. Precision is the ratio between the number of correctly identified NE chunks by the system and the total number of NE chunks found by the system; Recall is the ratio between the number of correctly identified NE chunks by the system and the total number of NE chunks in the gold standard. Experiments were run in a Linux machine with 16GB RAM and 8 cores of Intel Xeon 2.0GHz processor. The performance of different types of feature sets was evaluated using 10-fold cross-validation.
Table 2 shows the precision, recall, and F-score of the SVM-based NER system for all six types of entities, when different combinations of feature sets were used. Among them, the best F-score of 90.05% was achieved, when all feature sets were used. A number of interesting findings can be concluded from those results. First, the contribution of different types of features to the system's performance varies. For example, the “previous history feature” and the “morphology feature” improved the performance substantially (F-score from 81.76% to 83.83%, and from 83.81% to 86.06% respectively). These findings were consistent with previous reported results on protein/gene NER (Kazama et al., 2002; Takeuchi and Collier, 2003; Yamamoto et al., 2003). However, “POS” and “orthographic” features contributed very little, not as much as in protein/gene names recognition tasks. This could be related to the differences between gene/protein phrases and medication phrases – more orthographic clues are observed in gene/protein names. Second, the “semantic tags” features alone, even just using the original tagger in Me-dEx, improved the performance dramatically (from 81.76% to 86.51% or 89.47%). This indicates that the knowledge bases in the biomedical domain are crucial to biomedical NER. Third, the customized final semantic tagger in MedEx had much better performance than the original tagger, which indicated that advanced semantic tagging methods that integrate other levels of linguistic information (e.g., sections) were more useful than simple dictionary lookup methods.
Table 2.
Performance of the SVM-based NER system for different feature combinations.
| Features | Pre | Rec | F-score |
|---|---|---|---|
| Words (Baseline) | 87.09 | 77.05 | 81.76 |
| Words + History | 90.34 | 78.17 | 83.81 |
| Words + History + Morphology | 91.72 | 81.08 | 86.06 |
| Words + History + Morphology + POS | 91.81 | 81.06 | 86.10 |
| Words + History + Morphology + POS + Orthographies | 91.78 | 81.29 | 86.22 |
| Words + Semantic Tags (Original MedEx) | 90.15 | 83.17 | 86.51 |
| Words + Semantic Tags (Customized MedEx) | 92.38 | 86.73 | 89.47 |
| Words + History + Morphology + POS + Orthographies + Semantic Tags (Original MedEx) | 91.43 | 84.2 | 87.66 |
| Words + History + Morphology + POS + Orthographies + Semantic Tags (Customized MedEx) | 93.2 | 87.12 | 90.05 |
Table 3 shows the precision, recall, and F-score for each type of entity, from the MedEx alone, and the baseline and the best runs of the SVM-based NER system. As we can see, the best SVM-based NER system that combines all types of features (including inputs from MedEx) was much better than the MedEx system alone (90.05% vs. 85.86%). This suggested that the combination of rule-based systems with machine learning approaches could yield the most optimized performance in biomedical NER tasks.
Table 3.
Comparison between a rule based system and the SVM based system.
| Entity | MedEx only | SVM (Baseline) | SVM (Best) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Pre | Rec | F-score | Pre | Rec | F-score | Pre | Rec | F-score | |
| ALL | 87.85 | 83.97 | 85.86 | 87.09 | 77.05 | 81.76 | 93.2 | 87.12 | 90.05 |
| Medication | 87.25 | 90.21 | 88.71 | 88.38 | 75.03 | 81.16 | 93.3 | 91.35 | 92.31 |
| Dosage | 92.79 | 83.94 | 88.14 | 89.43 | 83.65 | 86.41 | 94.38 | 90.99 | 92.65 |
| Mode | 95.86 | 90.06 | 92.87 | 96.18 | 93.30 | 94.70 | 97.12 | 93.8 | 95.41 |
| Frequency | 92.67 | 89.00 | 90.80 | 90.33 | 87.60 | 88.94 | 95.88 | 93.04 | 94.43 |
| Duration | 42.65 | 40.15 | 41.36 | 24.16 | 19.62 | 21.45 | 65.18 | 40.16 | 49.57 |
| Reason | 54.23 | 36.72 | 43.79 | 48.40 | 25.51 | 33.30 | 69.21 | 37.39 | 48.4 |
Among six types of medication entities, we noticed that four types of entities (medication names, dosage, mode, and frequency) got very high F-scores (over 92%); while two others (duration and reason) had low F-scores (up to 50%). This finding was consistent with results from i2b2 challenge. Duration and reason are more difficult to identify because they do not have well-formed patterns and few knowledge bases exist for duration and reasons.
This study only focused on the first step of the i2b2 medication extraction challenge – NER. Our next plan is to work on the second step of determining relations between medication names and other entities, thus allowing us to compare our results with those reported in the i2b2 challenge. In addition, we will also evaluate and compare the performance of other ML algorithms such as CRF and ME on the same NER task.
5 Conclusions
In this study, we developed an SVM-based NER system for medication related entities. We systematically investigated different types of features and our results showed that by combining semantic features from a rule-based system, the ML-based NER system could achieve the best F-score of 90.05% in recognizing medication related entities, using the i2b2 annotated data set. The experiments also showed that optimized usage of external knowledge bases were crucial to high performance ML based NER systems for medical entities such as drug names.
Acknowledgments
Authors would like to thank i2b2 organizers for organizing the 2009 i2b2 challenge and providing dataset for research studies. This study was in part supported by NCI grant R01CA141307-01.
Footnotes
Available at http://chasen.org/∼taku/software/TinySVM/
Available at http://chasen.org/∼taku/software/YamCha/
Available at http://www.cnts.ua.ac.be/conll2002/ner/bin/conlleval.txt
Contributor Information
Son Doan, Email: Son.Doan@Vanderbilt.edu.
Hua Xu, Email: Hua.Xu@Vanderbilt.edu.
References
- Aronson AR. Effective mapping of biomed-ical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- Chhieng D, Day T, Gordon G, Hicks J. Use of natural language programming to extract medication from unstructured electronic medical records. AMIA Annu Symp Proc. 2007;908 [PubMed] [Google Scholar]
- Collier N, Nobata C, Tsujii J. Extracting the names of genes and gene products with a hidden Markov model. Proc of the 18th Conf on Computational linguistics. 2000;1:201–207. [Google Scholar]
- Doan S, Bastarache L, Klimkowski S, Denny JC, Xu H. Vanderbilt's System for Medication Extraction. Proc of 2009 i2b2 workshop 2009 [Google Scholar]
- Evans DA, Brownlow ND, Hersh WR, Campbell EM. Automating concept identification in the electronic medical record: an experiment in extracting dosage information. Proc AMIA Annu Fall Symp. 1996:388–392. [PMC free article] [PubMed] [Google Scholar]
- Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgrim S, Xia F, Solti I, Cadag E, Uzuner O. Statistical Extraction of Medication Infor- mation from Clinical Records. AMIA Summit on Translational Bioinformatics. 2010:10–12. [Google Scholar]
- Haug PJ, Christensen L, Gundersen M, Clemons B, Koehler S, Bauer K. A natural language parsing system for encoding admitting diagnoses. Proc AMIA Annu Fall Symp. 1997:814–818. [PMC free article] [PubMed] [Google Scholar]
- Hirschman L, Morgan AA, Yeh AS. Rutabaga by any other name: extracting biological names. J Biomed Inform. 2002;35:247–259. doi: 10.1016/s1532-0464(03)00014-5. [DOI] [PubMed] [Google Scholar]
- Kazama J, Makino T, Ohta Y, Tsujii T. Tuning Support Vector Machines for Biomedical Named Entity Recognition. Proceedings of the ACL-02 Workshop on Natural Language Processing in the Biomedical Domain. 2002:1–8. [Google Scholar]
- Keerthi S, Sundararajan S. CRF versus SVM-struct for sequence labeling. Yahoo Research Technical Report 2007 [Google Scholar]
- Krauthammer M, Nenadic G. Term identification in the biomedical literature. J Biomed Inform. 2004;37:512–526. doi: 10.1016/j.jbi.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Kudo T, Matsumoto Y. Use of Support Vector Learning for Chunk Identification. Proc of CoNLL-2000 2000 [Google Scholar]
- Kudo T, Matsumoto Y. Chunking with Support Vector Machines. Proc of NAACL 2001 2001 [Google Scholar]
- Levin MA, Krol M, Doshi AM, Reich DL. Extraction and mapping of drug names from free text to a standardized nomenclature. AMIA Annu Symp Proc. 2007:438–442. [PMC free article] [PubMed] [Google Scholar]
- Li D, Savova G, Kipper-Schuler K. Conditional random fields and support vector machines for disorder named entity recognition in clinical texts. Proceedings of the workshop on current trends in biomedical natural language processing (BioNLP'08) 2008:94–95. [Google Scholar]
- Li Z, Cao Y, Antieau L, Agarwal S, Zhang Z, Yu H. A Hybrid Approach to Extracting Medication Information from Medical Discharge Summaries. Proc of 2009 i2b2 workshop 2009 [Google Scholar]
- Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993;32:281–291. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick J, Li M. A Cascade Approach to Extract Medication Event (i2b2 challenge 2009) Proc of 2009 i2b2 workshop 2009 [Google Scholar]
- Sirohi E, Peissig P. Study of effect of drug lexicons on medication extraction from electronic medical records. Pac Symp Biocomput. 2005:308–318. doi: 10.1142/9789812702456_0029. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Collier N. Bio-medical entity extraction using Support Vector Machines. Proceedings of the ACL 2003 workshop on Natural language processing in biomedicine. 2003:57–64. [Google Scholar]
- Torii M, Hu Z, Wu CH, Liu H. Bio-Tagger-GM: a gene/protein name recognition system. J Am Med Inform Assoc. 2009;16:247–255. doi: 10.1197/jamia.M2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsochantaridis I, Joachims T, Hofmann T. Large margin methods for structured and interdependent output variables. Journal of Machine Learning Research. 2005;6:1453–1484. [Google Scholar]
- Uzüner O, Solti I, Cadag E. The third 2009 i2b2 challenge. 2009 https://www.i2b2.org/NLP/Medication/
- Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17:19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Kudo T, Konagaya A, Matsumoto Y. Protein name tagging for biomedical annotation in text. Proceedings of ACL 2003 Workshop on Natural Language Processing inBiomedi-cine, 2003. 2003;13:65–72. [Google Scholar]