Abstract
The essential oil (EO) from Salvia sclarea was shown to increase the susceptibility of methicillin resistant Staphylococcus epidermidis (MRSE) isolates to oxacillin. The purpose of this study was to investigate the effect of EO from S. sclarea on expression of mecA gene of MRSE carrying different types of staphylococcal chromosomal cassette (SCCmec) and to evaluate potential synergistic effect of EO with oxacillin. Using real-time PCR we found that EO alone inhibited the expression of the resistant genes mecA, mecR1, and mecI and blaZ, blaR1, and blaI. The use of the combination of EO with oxacillin resulted in significantly inhibited expression of mecA gene in all tested strains with different types of SCCmec. Using time-kill assay and checkerboard assay we confirmed synergistic effect of EO from S. sclarea and oxacillin in MRSE.
1. Introduction
The antibiotic resistance provides a great therapeutical and economic burden in the treatment of infectious diseases and it may threaten the success of antimicrobial chemotherapy. The disproportion between the slow development of new drugs and the fast emergence of resistant strains necessitates the development and research of new antimicrobial agents or resistance modifiers. One strategy employed to overcome resistance mechanisms is a combination therapy such as using clavulanic acid as inhibitor of β-lactamase in drugs sulbactam and tazobactam [1]. However, the frequent use of clavulanate has led to the emergence of resistant bacterial strains [2]. The promising strategy is the use of synergistic effects of natural compounds, products of plant secondary metabolism. Traditionally, medicinal plants have been used throughout the world for centuries for a range of medicinal complications. Plant drugs are considered to be less toxic and free of side effects than synthetic ones. Some work demonstrated that plants either contain antimicrobials that can operate in synergy with antibiotics or possess compounds that have no intrinsic antibacterial activity but are able to sensitize the pathogen to a previously ineffective antibiotic [3, 4]. Some plants can be sources of compounds that can potentiate the activity of antibiotics against resistant bacterial pathogens [5–9]. These compounds are believed to play a role in the plant's defense against infection by working in synergy with intrinsic antimicrobials. It has been suggested recently that such compounds can potentially be used to improve the efficacy of antibiotics against bacterial pathogens.
Salvia is an important genus widely cultivated and used in flavoring and folk medicines. Salvia species are used as traditional medicines all around the world, possessing antibacterial, antioxidant, anti-inflammatory, and analgesic properties [10]. S. sclarea, popularly known as clary sage, is a biennial or perennial herb with diverse biological activities manifested by different components, mainly of EO. Salvia and other plants such as thyme, lavender, sage, basil, coleus, hyssop, and skullcap belong to the large plant family Lamiaceae [11]. Plants from Lamiaceae family are known for high content of EOs.
Biological properties of EOs and their antimicrobial activity have been attributed to their main compounds such as mono-, di-, and sesquiterpenes and a variety of low molecular weight aliphatic hydrocarbons, acids, alcohols, aldehydes, acyclic esters or lactones, coumarins, and homologues of phenylpropanoids [12]. These compounds have hydrophobic characteristics and interact with different sites of microbial cell, namely, with cytoplasmic membrane. They affect the activities of membrane associated enzymes; certain components of EOs can act as uncouplers, which interfere with proton translocation over a membrane vesicle and subsequently interrupt ADP phosphorylation. Specific terpenoids with functional groups, for example, phenolic alcohols or aldehydes, also interfere with membrane integrated or associated proteins, stopping their production, or activity EOs are also able to inhibit the synthesis of DNA, RNA, proteins, and polysaccharides in fungal and bacterial cells [13, 14]. The effect of EO may be associated with the cell envelope by interfering with the rigidity and integrity of the membrane, thereby changing the expression or function of cell wall-related genes such as penicillin-binding proteins (PBPs) [15].
Thymol, a p-cymene derived compound primarily found in some EO, suppresses the toxic shock syndrome toxin (TSST-1) secretion in S. aureus [16] and decreases the production of α-hemolysin, SEA, and SEB enterotoxins in S. aureus [17]. Similar effects were described for perilla oil [18]. Diverse spectrum of EOs (S. officinalis, R. officinalis, A. alba, and E. caryophyllata) as well as some of their major compounds (limonene, eugenol, and eucalyptol) inhibited QS gene expression in S. aureus [19]. Extract from Rhus javanica showed the inhibition of the genetic expression of virulence factors such as sea, agrA, and sarA in methicillin resistant S. aureus [20], as well as mecA gene, which is responsible for staphylococcal resistance to β-lactam antibiotics.
Staphylococcus epidermidis was previously regarded as an innocuous commensal microorganism on the human skin and mucous membranes [21]. However, nowadays it is seen as an important opportunistic pathogen, one of the most prevalent causes of nosocomial infections associated with newborn, severely ill, and immunocompromised patients, and is also frequently isolated from postsurgical infections, especially in association with indwelling prosthetic devices [22]. It was found that approximately 70% of the S. epidermidis strains circulating in the hospital environment are resistant to methicillin and that the majority of them are also resistant to other antimicrobial classes [23]. Resistance to methicillin is at 75–90% among hospital isolates of S. epidermidis, which is even higher than the corresponding rate for S. aureus (40–60%) [24].
Resistance to β-lactam antibiotics in S. epidermidis, similarly as in S. aureus, is mediated by (i) production of β-lactamase which hydrolytically destroys the β-lactam antibiotics and (ii) the acquisition of mecA gene that produces alternative PBP2, which have low affinity to β-lactam antibiotics. Gene mecA is carried on a chromosomal genetic element designated as staphylococcal chromosomal cassette mec (SCCmec). Currently, there are several different types of SCCmec elements, with several subtypes, characterized by a unique combination of the mec and the recombinase-encoding ccr gene complexes [25]. Some reports suggest that, in coagulase negative staphylococci, namely, in S. epidermidis, SCCmec structures are more diverse and include either mec-ccr combinations not yet described for S. aureus [23] or more than one ccr allotype [26]. In S. epidermidis prevail SCCmec type IVa [27] and SCCmec type III [28]. Miragaia et al. [23] characterized 139 isolates of MRSE and found that 41% of the isolates harbored SCCmec type IV and 27% carried SCCmec type III, while SCCmec types V, I, and II were presented only in 6%, 4%, and 4% of the isolates, respectively. Of the 44 MRSE isolates recovered from the blood of patients with prosthetic valve endocarditis, 2% harbored SCCmec type I, 34% harbored type II, 28% harbored type III, and 36% harbored type IV [29].
Expression of mecA is inducible and can be controlled by either its cognate regulators MecI (DNA binding repressor protein) and MecR1 (sensor/signal transducer) or the structurally and functionally similar β-lactamase regulators BlaI and BlaR1, respectively [30]. The transcription of mecA and blaZ is corepressed by the regulators of the two regulons, MecI and BlaI. These regulators are almost identical and can replace each other [31] and both MecI and BlaI can bind as homodimers to the promoter/operator region of both mecA and blaZ [32]. In the presence of antibiotics, the sensing is mediated by signal transduction via the two transmembrane inducers, MecR1 or BlaR1, which will result in proteolytic autocleavage of the cytoplasmic domains of these proteins [33]. To perform this autocleavage, the transducers undergo acylation by the antibiotic that causes conformational changes in the molecule [34]. Autocleavage of the signal transducer is followed by cleavage of the cognate repressor, MecI or BlaI, and by subsequent induction of the transcription of mecA or blaZ [33].
In our previous work [35] we showed synergistic effects of oxacillin with plant extracts and EOs from several plant species of Lamiaceae family. The purpose of the present study was to determine the effect of EO from S. sclarea on the expression of mecA gene in strains S. epidermidis possessing different types of SCCmec and determine synergistic effect of oxacillin and EO in these strains.
2. Material and Methods
2.1. Plant Material and EO Isolation
The aerial parts of S. sclarea were harvested at the optimal growing and development stage. The EO was prepared in accordance with the European Pharmacopoeia [36]. The plant material was air-dried and submitted to hydrodistillation for 4 h. Isolated oil was diluted in n-hexane and dried over anhydrous sodium sulfate. The most frequent EO components were determined by the GC method.
2.2. Bacterial Strains
MRSE strains were obtained from clinical samples of patients with positive hemocultures from the University Teaching Hospital Old Town, Bratislava, Slovak Republic, and were kindly provided by Dr. Slobodníková from the Institute of Microbiology, Faculty of Medicine, Comenius University, in Bratislava, Slovak Republic.
2.3. Determination of Oxacillin Resistance and Genes from mec and bla Operons
The isolates were screened for their susceptibility towards oxacillin using disc diffusion method on Mueller-Hinton agar (HiMedia, India) in accordance with the Clinical and Laboratory Standard Institute Guidelines (CLSI) standards [37]. Suspension of the tested bacteria (0.1 mL of 108 cells/mL) was spread onto solid media plates. Antimicrobial susceptibility test discs (HiMedia, India) with ampicillin (10 μg) and oxacillin (1 μg) were placed on the incubated plates. These plates, after 2 h of maintenance at 4°C, were incubated for 24 h at 37°C and the diameters of the resulting zones of inhibition were measured in millimeters.
Extraction of genomic DNA was performed by using DNeasy Blood and Tissue Kit (Qiagen, Germany) according to the manufacturer's instructions. All the isolates were tested for genes of the mec and bla operon using the polymerase chain reaction method. Each PCR sample contained 2.5 μL of 10x Star Taq enzyme buffer (Gene Craft, Germany), 0.2 mM of each deoxynucleoside triphosphate (Gene Craft, Germany), 0.2 μM of each of the forward and reverse primers, 1.5 U Taq DNA polymerase (Gene Craft, Germany), and 2 μL template DNA. The PCR primers were designed by Primer3 and Primer-Blast and synthesized by Microsynth (Switzerland). Details of primer sequences are listed in Table 1. The cycling conditions were as follows: preheating for 5 minutes at 94°C, followed by 30 cycles of denaturation at 94°C/30 seconds, annealing at 55°C/40 seconds, extension at 72°C/30 seconds, and final extension for 5 minutes at 72°C. PCR amplicons were analyzed by 2% agarose gel electrophoresis and stained with ethidium bromide.
Table 1.
Primer sequences (5′ to 3′) used for PCR and real-time PCR.
| Primers | Sequences (5′-3′) | Product length (bp) |
|---|---|---|
| GAPDH | ||
| Forward | TCAACGATTTAACAGATGACGCA | 77 |
| Reverse | TTCGTCTTTGAAACGACCTTGTG | |
| mecA | ||
| Forward | TCCACCCTCAAACAGGTGAA | 139 |
| Reverse | TGGAACTTGTTGAGCAGAGGT | |
| mecI | ||
| Forward | TCATCTGCAGAATGGGAAGTT | 103 |
| Reverse | TTGGACTCCAGTCCTTTTGC | |
| mecR1 | ||
| Forward | AGCACCGTTACTATCTGCACA | 142 |
| Reverse | AGAATAAGCTTGCTCCCGTTCA | |
| blaZ | ||
| Forward | TCCTAAGGGCCAATCTGAACC | 105 |
| Reverse | ACACTCTTGGCGGTTTCACT | |
| blaI | ||
| Forward | ACTGTATGGAGGGGACATGAA | 89 |
| Reverse | TGTCTCGCAATTCTTCAATTTCTT | |
| blaR1 | ||
| Forward | GCCCTTACACAACGATTACCAA | 79 |
| Reverse | GCTGTACATGACGAAAGATCCAC |
2.4. SCCmec Typization
The SCCmec type was determined using the protocol and primers proposed by Zhang et al. [38]. Multiplex PCR was performed in 25 μL reactions with 2.5 μL of 10x Star Taq enzyme buffer (Gene Craft, Germany), 0.2 mM of each deoxynucleoside triphosphate (Gene Craft, Germany), various concentrations of the respective primers [38], 1.0 U of Taq DNA polymerase (Gene Craft, Germany), and 2 μL of template DNA. The amplification was performed using a Biometra Thermal Cycler (Germany) with an initial denaturation step at 94°C/5 min followed by 30 cycles of denaturation at 94°C/1 min, annealing at 55°C/1 min, extension at 72°C/2 min, and a final extension step at 72°C/5 min. PCR products were detected on a 2% agarose gel and stained with ethidium bromide.
2.5. Determination of Minimal Inhibitory Concentration (MIC)
Minimal inhibitory concentrations (MIC) for oxacillin (Sigma-Aldrich) and EO (prepared as stated above) were determined by standard broth microdilution method using 96-well microtiter plates in accordance with CLSI [37] in Mueller-Hinton broth. The S. epidermidis suspension was adjusted to the 0.5 McFarland standard and diluted to obtain a final turbidity in wells approximately 1 × 106 CFU/mL. 0.1 mL of bacterial suspension was mixed with an equal volume of each dilution of oxacillin or EO, and optical density (OD600) values were determined after 24 hours of growth at 37°C using a 96-well plate reader Varioskan Flash (Thermo Fisher Scientific, Finland). The MIC was defined as the lowest concentration of antimicrobial agent that completely inhibits growth of the organism.
2.6. The Checkerboard Method
The study of the interaction between EO and oxacillin was done using the checkerboard method. Twofold serial dilutions of oxacillin prepared in horizontal rows of 96-well microtiter plate were subsequently cross-diluted vertically by twofold serial dilutions of EO. Microtiter plates were inoculated with test organism and incubated for 24 h at 37°C. MIC values of the combinations were determined as the lowest concentration that completely inhibited bacterial growth recorded as the optical density at 600 nm using Varioskan Flash (Thermo Fisher Scientific, Finland). Interaction between EO and oxacillin was then determined by calculating the fractional inhibitory concentration (FIC) indices. The FICI is defined as follows: MIC of substance A tested in combination/MIC of substance A tested alone + MIC of substance B tested in combination/MIC of substance B tested alone. The FICI is interpreted as follows: FICI < 0.5, synergistic effect; 0.5 < FIC < 1, additive effect; 1 < FICI < 4, indifferent effect; and FIC > 4, antagonistic effect [39]. The synergistic effect is shown graphically by applying isobole method. The shape of the isobologram curve can be convex, linear, or concave, which is indicative of the synergistic, indifferent, and antagonistic interactions, respectively [40].
2.7. The Time-Kill Assay
The time-kill assay was carried out in order to determine antibacterial and potential synergistic effects of EO when used singly and in combination with oxacillin. Bacteria (5 × 105 CFU/mL) were exposed to oxacillin and EO alone or in combination in concentrations 1/2 MIC and incubated at 37°C. Aliquots (0.1 mL) were taken at 0, 6, 10, and 24 h and diluted in normal saline as needed to enumerate 30–300 colonies. The diluted cultures were spread thoroughly on plates containing Mueller-Hinton agar. After incubating at 37°C for 24 h, the growing colonies were counted. The experiment was performed in triplicate; data are shown as mean ± standard deviation from three independent experiments.
2.8. RNA Isolation and Real-Time PCR
MRSE strains were treated with various concentrations of oxacillin and EO alone or with their combination for 30 minutes. Total RNA was isolated by using RNeasy Mini Kit (Qiagen, Germany) according to the manufacturer's instructions. DNA contamination from the total RNA preparations was removed with on-column RNase-Free DNase treatment (Qiagen, Germany). Measuring A260/A280 nm ratio assessed the nucleic acid purity. To generate cDNA, total RNA was reverse transcribed using the GoTaq 2-Step RT-qPCR System (Promega) with specific primers (Table 1). Real-time PCR was performed at least three times for each examined gene with an Applied Biosystem 7500 FAST Real-Time PCR System. The Ct values and the qPCR were normalized to the housekeeping gene for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the 2−ΔΔCt method [41].
2.9. Statistical Analysis
The experimental data are expressed as the mean ± standard deviation (±SD) from three independent experiments. Statistical analyses were performed using the t-test. P < 0.05 was considered to indicate a statistically significant difference.
3. Results and Discussion
3.1. Essential Oil
The essential oil obtained from aerial parts of S. sclarea was analyzed by gas chromatography (GC) and 12 compounds representing 75.2% of the essential oil were identified. The main components were linalyl acetate (38.67%), linalool (20.42%), germacrene (5.31%), geraniol (1.42%), and β-caryophyllene (1.80%).
3.2. Isolate Selection for Further Analysis
In our previous work we showed that addition of plant extracts and EOs from some species of Salvia plants increases the susceptibility of MRSE to oxacillin. EOs and oxacillin in combination showed synergistic effect, inhibited growth, and even killed the bacteria [35].
In this study we used 30 clinical isolates of MRSE for study of potential synergistic effects of EO from S. sclarea and oxacillin, and by using checkerboard method we confirmed this effect in 23 isolates (76.7%) (FICI from 0.125 to 0.381). The remaining 7 strains (23.3%) showed additive effect (FICI from 0.531 to 0.793). None of the strains showed antagonistic effect.
Based upon the phenotype analysis of methicillin resistance (MIC of oxacillin, Table 2), screening for presence or absence of mecA, mecI, mecR1, blaZ, blaI, and blaR1 gene using PCR (Figure 1), and analysis of mec class (Figure 2) and on SCCmec typization we selected four strains possessing different SCCmec types I, II, III, and IVa (Figure 3), which cover all SCCmec types and all variations of mec and bla operons present in collection of isolates.
Table 2.
Profile of mec and bla elements in S. epidermidis strains used in the work.
| Strain | mecA | mecI | mecR1 | blaZ | blaI | blaR1 | SCCmec type | Mec class | MIC oxa. (μg/mL) |
|---|---|---|---|---|---|---|---|---|---|
| R17 | + | − | − | + | + | + | I | B | 128 |
| R8 | + | + | + | + | + | + | II | A | 32 |
| R22 | + | + | + | − | − | − | III | A | 16 |
| R12 | + | − | − | + | + | + | IVa | B | 256 |
Figure 1.
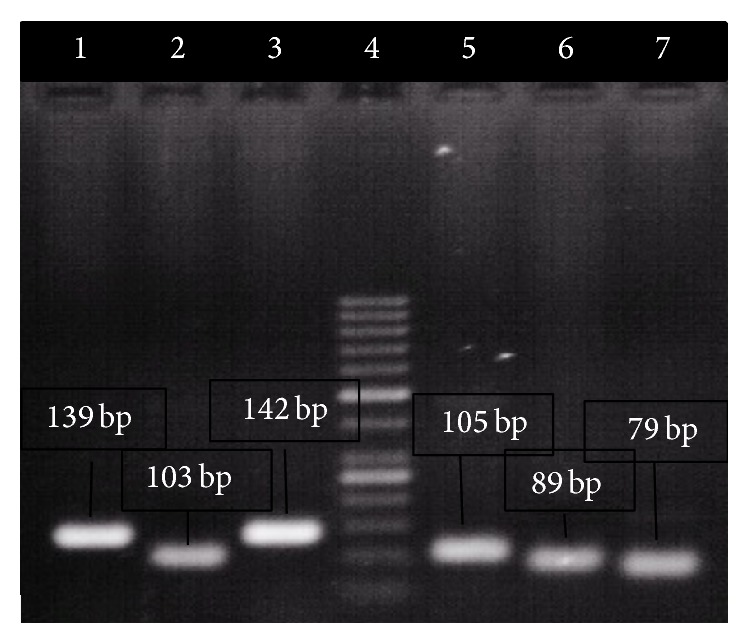
Detection of genes from mec and bla operons in strain R8. Lane 1: mecA, lane 2: mecI, lane 3: mecR1, lane 4: 100 bp marker, lane 5: blaZ, lane 6: blaI, and lane 7: blaR1.
Figure 2.
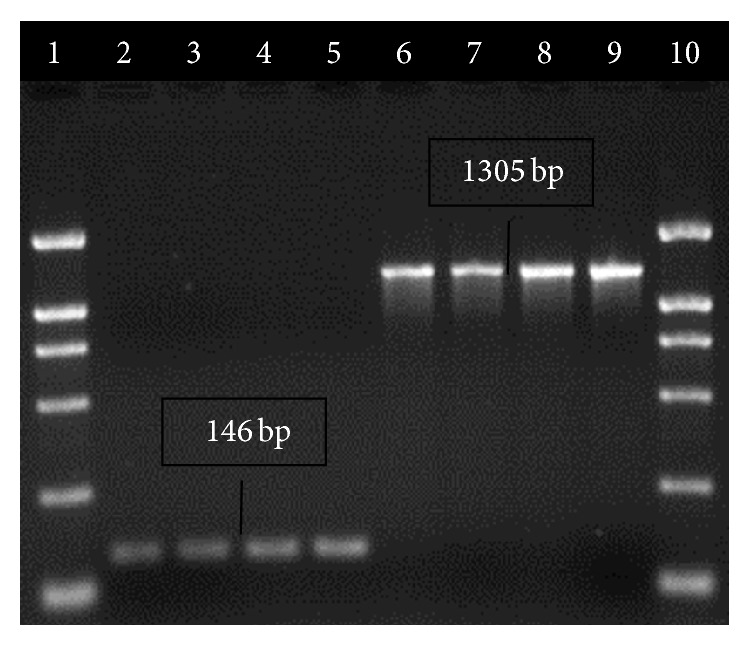
Detection of mec class. Lane 1: 2000 bp marker, lanes 2, 3: strain R8, lanes 4, 5: strain R22 (class A), lanes 6, 7: strain R17, lanes 8, 9: strain R12 (class B), and lane 10: 2000 bp marker.
Figure 3.
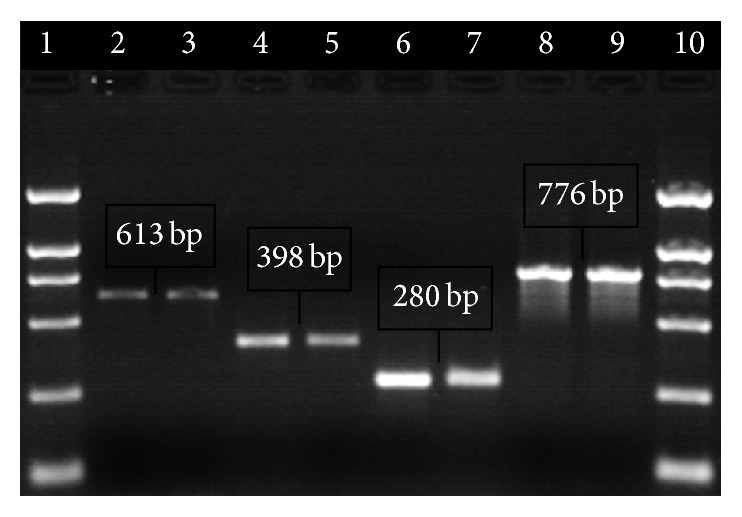
Typization of SCCmec in selected strains. Lane 1: 2000 bp marker, lanes 2, 3: SCCmec type I (strain R17), lanes 4, 5: SCCmec type II (strain R8), lanes 6, 7: SCCmec type III (strain R22), lanes 8, 9: SCCmec type IVa (strain R12), and lane 10: 2000 bp marker.
3.3. Real-Time PCR Analysis of Expression of mecA Gene
We have attempted to assess if EO alone or in combination with oxacillin influences the expression of mecA gene. Firstly, we characterize the mecA/mecI/mecR1 and blaZ/blaI/blaR1 regions of our strains and determine the effect of presence or absence of mec and bla elements on the MIC of oxacillin. The characteristics of these strains are in Table 2. Isolates R17 and R12 which possess SCCmec types I and IVa, respectively, and class B mec have genotypes +blaI/+blaR1 and −mecI/mutmecR1 (absent mecI and truncation of mecR1 by insertion of IS1272). These strains showed high resistance, correlating with the fact that mecI deletion can lead to increased resistance [42]. Isolate R8 (SCCmec type II) has both complete repressor/sensor systems +mecI/+mecR1 and +blaI/+blaR1. Isolate R22 (SCCmec type III) has +mecI/+mecR1 system but this isolate was the only one in which we were unable to confirm the presence of the bla operon. This isolate has the lowest MIC of oxacillin from four isolates. Hackbarth et al. described that interruption of blaR1 results in constitutive repression and therefore decreased resistance [43].
We used real-time PCR to investigate the expression of genes from mec and bla operons and to determine the influence of EO on their expression. Quantification data for all genes were normalized to the reference gene for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). In Figure 4 is relative expression of genes in strain R8, which have complete both operons. Regardless of constitutive or inducible expression of genes our data show that 30 min treatment of culture with subinhibitory concentrations of EO from S. sclarea leads to decreasing of expression of all genes from both operons. It is known that EOs influence membranes and proteins in cytoplasmic membrane [44]; therefore we can assume that they can influence conformation of MecR1, or BlaR1, which can have impact on transcription of both operons. While EO showed similar and dose dependent effect on single genes from mec operon, in case of bla genes the effect of EO on blaI and blaR1 gene expression was weaker. However the expression of gene blaZ was decreased the most from all genes. Similar results, the inhibition of expression of resistant genes mecA, mecI, and mecR1, against methicillin resistant S. aureus by hexane and chloroform fractions of Salvia miltiorrhiza Bunge, were described by Lee et al. [45].
Figure 4.
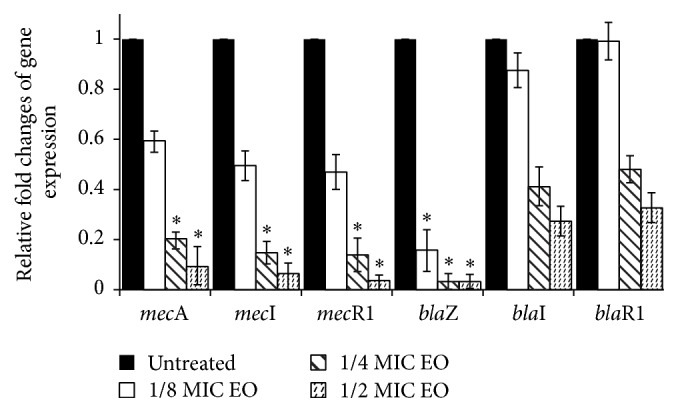
Relative expression of genes of mec and bla operons. S. epidermidis R8 was treated with subinhibitory concentrations of EO from S. sclarea for 30 min. Transcript levels were monitored by real-time PCR as described in the text. Using the 2−ΔΔCt method, the data are presented as the fold change in gene expression normalized to an endogenous reference gene (GAPDH) and relative to the untreated control (value 1). Values represent the mean ± SD for three independent experiments. ∗ P < 0.05.
One isolate from each SCCmec type was exposed to increasing amount of EO and as is shown in Figure 5, EO reduced the expression of mecA gene in all strains. Regarding SCCmec type and genetic background of mec and bla operons, strains R8 and R22 have mec A class with all three mec genes, while strains R17 and R12 have mec B class (absent mecI and truncation of mecR1 by insertion of IS1272). As is obvious from Figure 5, we did not find significant differences in expression of mecA gene between strains with different mec class after treatment with EO at concentrations 1/4 and 1/2 of MIC; however in concentration 1/8 MIC there was statistically significant (P < 0.05) difference between strains of both classes of mec complex, when EO decreased the expression of mecA gene more in strains R17 and R12 in the comparison with strains R8 and R22. We estimated the expression of mecA gene in the presence of EO also in three random selected strains with IVa type of SCCmec and the results were similar (data not shown).
Figure 5.
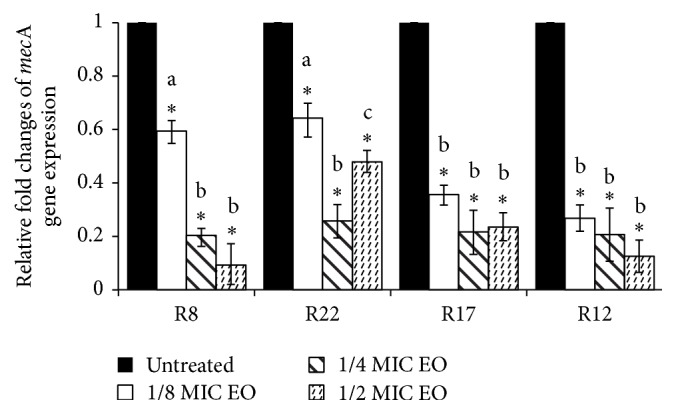
Relative expression of mecA gene in S. epidermidis strains with different types of SCCmec. Strains were treated with subinhibitory concentrations of EO from S. sclarea for 30 min. Using the 2−ΔΔCt method, the data are presented as the fold change in gene expression normalized to an endogenous reference gene (GAPDH) and relative to the untreated control (value 1). Values represent the mean ± SD for three independent experiments. ∗ P < 0.05. Different letters signify statistical differences between values (P < 0.05).
Finally we examined the expression of mecA gene after 30 min treatment with oxacillin and EO. Oxacillin alone induced higher expression of mecA gene (data not shown). The addition of EO not only inhibited the induction of expression, but also reduced the expression of mecA gene in the comparison with untreated control (Figure 6). The combination of 1/4 MIC of both compounds was more effective, probably due to lower concentration of oxacillin and lower induction of expression of mecA gene. We found similar trend as in the case of EO alone, that is, higher reduction of expression in strains R17 and R12 in the comparison with strains R8 and R22.
Figure 6.
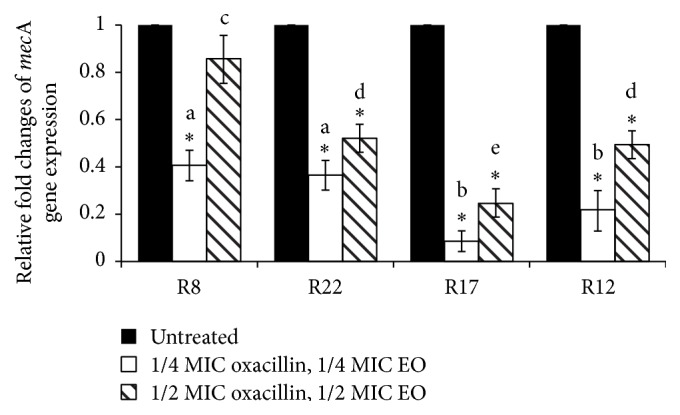
Relative expression of mecA gene in S. epidermidis strains with different types of SCCmec after treatment with EO from S. sclarea and oxacillin. Using the 2−ΔΔCt method, the data are presented as the fold change in gene expression normalized to an endogenous reference gene (GAPDH) and relative to the untreated control (value 1). Values represent the mean ± SD for three independent experiments. ∗ P < 0.05. Different letters signify statistical differences between values (P < 0.05).
3.4. Modulatory Study
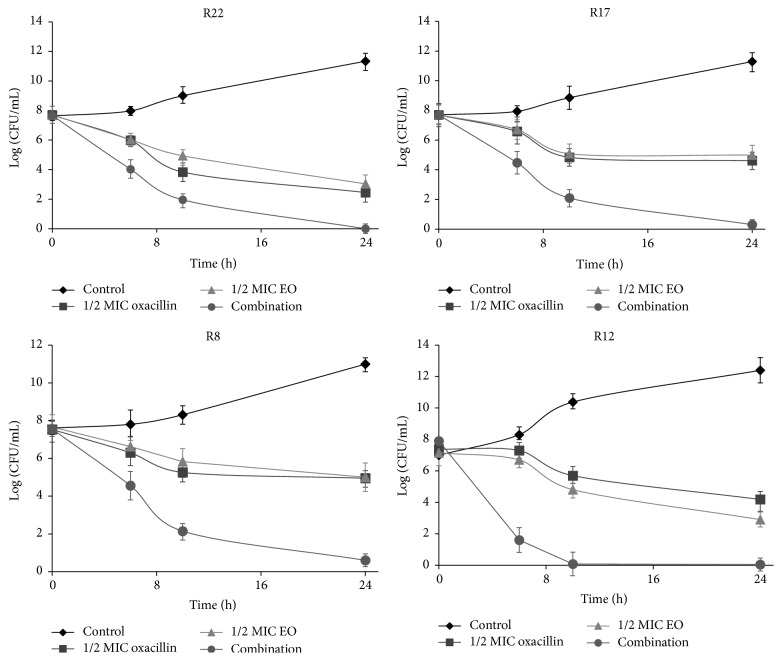
The viable cell counts for four isolates of MRSE differing in type of SCCmec after exposure to 1/2 MIC of oxacillin and EO alone and in combination at different times are shown in Figure 7. The 1/2 MIC values were specific for each particular strain. The combination of 1/2 MIC EO + 1/2 MIC oxacillin completely inhibited the growth of all four strains after 24 h. The same combination caused an over 3 log10-fold reduction in the bacterial count yet after 10 h (strains R17 and R8) in comparison with the most active compound alone. In strain R12 the reduction was more than 5 log10-fold in 6 h and from 10 h the growth of this strain was completely inhibited.
Figure 7.
Time-kill curves of strains after treatment with 1/2 MIC of oxacillin and 1/2 MIC of EO alone or in combination.
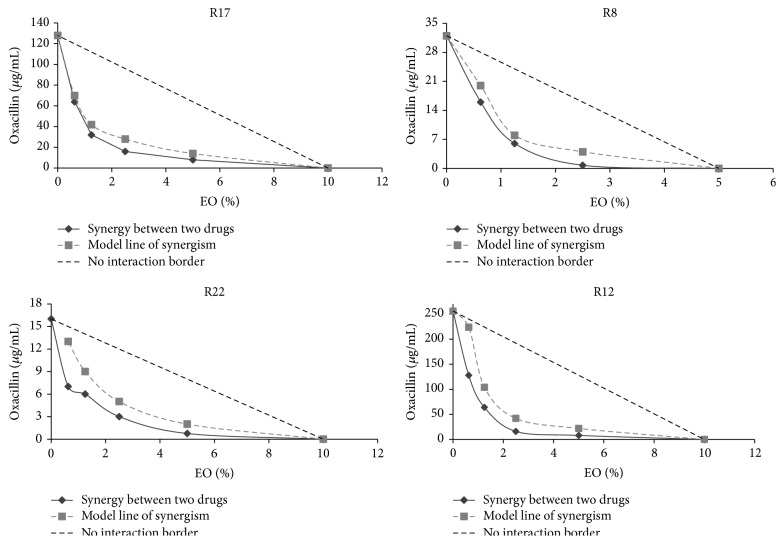
To get a better understanding of the modulatory effect of EO we examined the interaction between EO and oxacillin via the checkerboard method and described it in terms of fractional inhibitory concentration (FIC) indices. The FIC indices of EO in combination with oxacillin were 0.381 for strain R17, 0.156 for R8, 0.125 for R22, and 0.376 for R12, which according to Pillai et al. [39] indicate synergistic interactions. Combinatorial profiles are presented graphically in Figure 8. The synergistic interaction can be read according to the curve indicating borderline synergy according to FIC 0.5. Isobolograms of all strains have convex shape beyond the borderline of synergisms and clearly show the potentiating effect of S. sclarea EO on oxacillin susceptibility in these strains.
Figure 8.
Isobole curves revealing the synergistic effect of S. sclarea EO with oxacillin against S. epidermidis strains with different types of SCCmec.
4. Conclusion
Our data reveal the potential of EO from S. sclarea to be candidate for combination therapy against MRSE, because it has synergistic effect with oxacillin in all tested strains. The killing effect of the combinatorial treatment is connected with reduction of expression of mecA gene and other genes participating in staphylococcal resistance to β-lactams antibiotics. Lower expression of these genes reduces the two major bacterial defense mechanisms against β-lactam antibiotics. Although synergistic effect of EO may be caused by several mechanisms, observed different levels of the reduction of mecA expression in strains with different types SCCmec and different genetic background in mec and bla operons confirmed that reduction of mecA gene expression can be one of major mechanisms.
Acknowledgment
This study was supported by Grant VEGA 1/0287/15 of the Ministry of Education, Science, Research and Sport of the Slovak Republic.
Conflict of Interests
The authors do not have any conflict of interests regarding the content of the paper.
References
- 1.Lee N., Yuen K.-Y., Kumana C. R. Clinical role of β-lactam/β-lactamase inhibitor combinations. Drugs. 2003;63(14):1511–1524. doi: 10.2165/00003495-200363140-00006. [DOI] [PubMed] [Google Scholar]
- 2.Enright M. C., Robinson D. A., Randle G., Feil E. J., Grundmann H., Spratt B. G. The evolutionary history of methicillin resistant Staphylococcus aureus (MRSA) Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stermitz F. R., Lorenz P., Tawara J. N., Zenewicz L. A., Lewis K. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(4):1433–1437. doi: 10.1073/pnas.030540597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betoni J. E. C., Mantovani R. P., Barbosa L. N., Di Stasi L. C., Fernandes A., Jr. Synergism between plant extract and antimicrobial drugs used on Staphylococcus aureus diseases. Memórias do Instituto Oswaldo Cruz. 2006;101(4):387–390. doi: 10.1590/s0074-02762006000400007. [DOI] [PubMed] [Google Scholar]
- 5.Smith E. C. J., Williamson E. M., Wareham N., Kaatz G. W., Gibbons S. Antibacterials and modulators of bacterial resistance from the immature cones of Chamaecyparis lawsoniana . Phytochemistry. 2007;68(2):210–217. doi: 10.1016/j.phytochem.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Fadli M., Saad A., Sayadi S., et al. Antibacterial activity of Thymus maroccanus and Thymus broussonetii essential oils against nosocomial infection—Bacteria and their synergistic potential with antibiotics. Phytomedicine. 2012;19(5):464–471. doi: 10.1016/j.phymed.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Taylor P. W. Alternative natural sources for a new generation of antibacterial agents. International Journal of Antimicrobial Agents. 2013;42(3):195–201. doi: 10.1016/j.ijantimicag.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Cragg G. M., Newman D. J. Natural products: a continuing source of novel drug leads. Biochimica et Biophysica Acta—General Subjects. 2013;1830(6):3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cushnie T. P. T., Cushnie B., Lamb A. J. Alkaloids: an overview of their antibacterial, antibiotic-enhancing and antivirulence activities. International Journal of Antimicrobial Agents. 2014;44(5):377–386. doi: 10.1016/j.ijantimicag.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Ibrahim T. A. Chemical composition and biological activity of extracts from Salvia bicolor desf. growing in Egypt. Molecules. 2012;17(10):11315–11334. doi: 10.3390/molecules171011315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iijima Y., Davidovich-Rikanati R., Fridman E., et al. The biochemical and molecular basis for the divergent patterns in the biosynthesis of terpenes and phenylpropenes in the peltate glands of three cultivars of basil. Plant physiology. 2004;136(3):3724–3736. doi: 10.1104/pp.104.051318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorman H. J. D., Deans S. G. Antimicrobial agents from plants: antibacterial activity of plant volatile oils. Journal of Applied Microbiology. 2000;88(2):308–316. doi: 10.1046/j.1365-2672.2000.00969.x. [DOI] [PubMed] [Google Scholar]
- 13.Kalemba D., Kunicka A. Antibacterial and antifungal properties of essential oils. Current Medicinal Chemistry. 2003;10(10):813–829. doi: 10.2174/0929867033457719. [DOI] [PubMed] [Google Scholar]
- 14.Burt S. Essential oils: their antibacterial properties and potential applications in foods—a review. International Journal of Food Microbiology. 2004;94(3):223–253. doi: 10.1016/j.ijfoodmicro.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 15.Bonde M., Højland D. H., Kolmos H. J., Kallipolitis B. H., Klitgaard J. K. Thioridazine affects transcription of genes involved in cell wall biosynthesis in methicillin-resistant Staphylococcus aureus . FEMS Microbiology Letters. 2011;318(2):168–176. doi: 10.1111/j.1574-6968.2011.02255.x. [DOI] [PubMed] [Google Scholar]
- 16.Nostro A., Blanco A. R., Cannatelli M. A., et al. Susceptibility of methicillin-resistant Staphylococci to oregano essential oil, carvacrol and thymol. FEMS Microbiology Letters. 2004;230(2):191–195. doi: 10.1016/s0378-1097(03)00890-5. [DOI] [PubMed] [Google Scholar]
- 17.Qiu J., Wang D., Xiang H., et al. Subinhibitory concentrations of thymol reduce enterotoxins A and B and α-hemolysin production in Staphylococcus aureus isolates. PLoS ONE. 2010;5(3) doi: 10.1371/journal.pone.0009736.e9736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu J., Zhang X., Luo M., et al. Subinhibitory concentrations of perilla oil affect the expression of secreted virulence factor genes in Staphylococcus aureus . PLoS ONE. 2011;6(1) doi: 10.1371/journal.pone.0016160.e16160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saviuc C., Cotar A. I., Holban A. M., Banu O., Grumezescu A. M., Chifiriuc M. C. Phenotypic and molecular evaluation of Pseudomonas aeruginosa and Staphylococcus aureus virulence patterns in the presence of some essential oils and their major compounds. Letters in Applied NanoBioScience. 2013;2(1):91–96. [Google Scholar]
- 20.You Y.-O., Choi N.-Y., Kang S.-Y., Kim K.-J. Antibacterial activity of Rhus javanica against methicillin-resistant Staphylococcus aureus . Evidence-Based Complementary and Alternative Medicine. 2013;2013:8. doi: 10.1155/2013/549207.549207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohanty S. S., Kay P. R. Infection in total joint replacements. Why we screen MRSA when MRSE is the problem? The Journal of Bone & Joint Surgery—British Volume. 2004;86(2):266–268. doi: 10.1302/0301-620x.86b2.14129. [DOI] [PubMed] [Google Scholar]
- 22.Arciola C. R., Campocciaa D., Gamberinia S., et al. Antibiotic resistance in exopolysaccharide—forming Staphylococcus epidermidis clinical isolates from orthopedic implant infections. Biomaterials. 2005;26:6530–6535. doi: 10.1016/j.biomaterials.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 23.Miragaia M., Thomas J. C., Couto I., Enright M. C., De Lencastre H. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. Journal of Bacteriology. 2007;189(6):2540–2552. doi: 10.1128/jb.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diekema D. J., Pfaller M. A., Schmitz F. J., et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clinical Infectious Diseases. 2001;32(supplement 2):S114–S132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 25.Xiao X. M., Ito T., Tiensasitorn C., et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrobial Agents and Chemotherapy. 2002;46(4):1147–1152. doi: 10.1128/aac.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanssen A.-M., Sollid J. U. E. Multiple staphylococcal cassette chromosomes and allelic variants of cassette chromosome recombinases in Staphylococcus aureus and coagulase-negative staphylococci from Norway. Antimicrobial Agents and Chemotherapy. 2007;51(5):1671–1677. doi: 10.1128/aac.00978-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jamaluddin T. Z. M. T., Kuwahara-Arai K., Hisata K., et al. Extreme genetic diversity of methicillin-resistant Staphylococcus epidermidis strains disseminated among healthy Japanese children. Journal of Clinical Microbiology. 2008;46(11):3778–3783. doi: 10.1128/jcm.02262-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machado A. B. M. P., Reiter K. C., Paiva R. M., Barth A. L. Distribution of staphylococcal cassette chromosome mec (SCCmec) types I, II, III and IV in coagulase-negative staphylococci from patients attending a tertiary hospital in southern Brazil. Journal of Medical Microbiology. 2007;56(10):1328–1333. doi: 10.1099/jmm.0.47294-0. [DOI] [PubMed] [Google Scholar]
- 29.Wisplinghoff H., Rosato A. E., Enright M. C., Noto M., Craig W., Archer G. L. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrobial Agents and Chemotherapy. 2003;47(11):3574–3579. doi: 10.1128/aac.47.11.3574-3579.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ender M., McCallum N., Berger-Bächi B. Impact of mecA promoter mutations on mecA expression and β-lactam resistance levels. International Journal of Medical Microbiology. 2008;298(7-8):607–617. doi: 10.1016/j.ijmm.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 31.Lewis R. A., Dyke K. G. H. MecI represses synthesis from the β-lactamase operon of Staphylococcus aureus . Journal of Antimicrobial Chemotherapy. 2000;45(2):139–144. doi: 10.1093/jac/45.2.139. [DOI] [PubMed] [Google Scholar]
- 32.Sharma V. K., Hackbarth C. J., Dickinson T. M., Archer G. L. Interaction of native and mutant MecI repressors with sequences that regulate mecA, the gene encoding penicillin binding protein 2a in methicillin-resistant staphylococci. Journal of Bacteriology. 1998;180(8):2160–2166. doi: 10.1128/jb.180.8.2160-2166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H. Z., Hackbarth C. J., Chansky K. M., Chambers H. F. A proteolytic transmembrane signaling pathway and resistance to β-lactams in staphylococci. Science. 2001;291(5510):1962–1965. doi: 10.1126/science.1055144. [DOI] [PubMed] [Google Scholar]
- 34.Thumanu K., Cha J., Fisher J. F., Perrins R., Mobashery S., Wharton C. Discrete steps in sensing of β-lactam antibiotics by the BlaR1 protein of the methicillin-resistant Staphylococcus aureus bacterium. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(28):10630–10635. doi: 10.1073/pnas.0601971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chovanová R., Mikulášová M., Vaverková Š. In vitro antibacterial and antibiotic resistance modifying effect of bioactive plant extracts on methicillin-resistant Staphylococcus epidermidis . International Journal of Microbiology. 2013;2013:7. doi: 10.1155/2013/760969.760969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Council of Europe. European Pharmacopoeia, Supplement. 3rd 2001. [Google Scholar]
- 37.CLSI. CLSI Document. M100-S21. Wayne, Pa, USA: Clinical and Laboratory Standards Institute; 2011. Performance standards for antimicrobial susceptibility testing; Twenty-first informational supplement. [Google Scholar]
- 38.Zhang K., McClure J.-A., Elsayed S., Louie T., Conly J. M. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus . Journal of Clinical Microbiology. 2005;43(10):5026–5033. doi: 10.1128/jcm.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pillai S. K., Moellering R. C., Eliopoulos G. M. Antimicrobial combinations. In: Lorian V., editor. Antibiotics in Laboratory Medicine. 5th. 2005. pp. 365–440. [Google Scholar]
- 40.Din W. M., Jin K. T., Ramli R., Khaithir T. M. N., Wiart C. Antibacterial effects of ellagitannins from Acalypha wilkesiana var. macafeana hort.: surface morphology analysis with environmental scanning electron microcopy and synergy with antibiotics. Phytotherapy Research. 2013;27(9):1313–1320. doi: 10.1002/ptr.4876. [DOI] [PubMed] [Google Scholar]
- 41.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Katayama Y., Zhang H.-Z., Chambers H. F. PBP 2a mutations producing very-high-level resistance to beta-lactams. Antimicrobial Agents and Chemotherapy. 2004;48(2):453–459. doi: 10.1128/aac.48.2.453-459.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hackbarth C. J., Miick C., Chambers H. F. Altered production of penicillin-binding protein 2a can affect phenotypic expression of methicillin resistance in Staphylococcus aureus . Antimicrobial Agents and Chemotherapy. 1994;38(11):2568–2571. doi: 10.1128/aac.38.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nazzaro F., Fratianni F., De Martino L., Coppola R., De Feo V. Effect of essential oils on pathogenic bacteria. Pharmaceuticals. 2013;6(12):1451–1474. doi: 10.3390/ph6121451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J.-W., Ji Y.-J., Lee S.-O., Lee I.-S. Effect of Saliva miltiorrhiza Bunge on antimicrobial activity and resistant gene regulation against methicillin resistant Staphylococcus aureus (MRSA) Journal of Microbiology. 2007;45(4):350–357. [PubMed] [Google Scholar]