Abstract
Photolyases (PHRs) are DNA repair enzymes that revert UV-induced photoproducts, either cyclobutane pyrimidine dimers (CPD) or (6-4) photoproducts (PPs), into normal bases to maintain genetic integrity. (6-4) PHR must catalyze not only covalent bond cleavage, but also hydroxyl or amino group transfer, yielding a more complex mechanism than that postulated for CPD PHR. Previous mutation analysis revealed the importance of two histidines in the active center, H354 and H358 for Xenopus (6-4) PHR, whose mutations significantly lowered the enzymatic activity. Based upon highly sensitive FTIR analysis of the repair function, here we report that both H354A and H358A mutants of Xenopus (6-4) PHR still maintain their repair activity, although the efficiency is much lower than that of the wild type. Similar difference FTIR spectra between the wild type and mutant proteins suggest a common mechanism of repair in which (6-4) PP binds to the active center of each mutant, and is released after repair, as occurs in the wild type. Similar FTIR spectra also suggest that a decrease in volume by the H-to-A mutation is possibly compensated by the addition of water molecule( s). Such a modified environment is sufficient for the repair function that is probably controlled by proton-coupled electron transfer between the enzyme and substrate. On the other hand, two histidines must work in a concerted manner in the active center of the wild-type enzyme, which significantly raises the repair efficiency.
Keywords: DNA repair, Light-induced difference FTIR spectra, FAD, Hydrogen bond
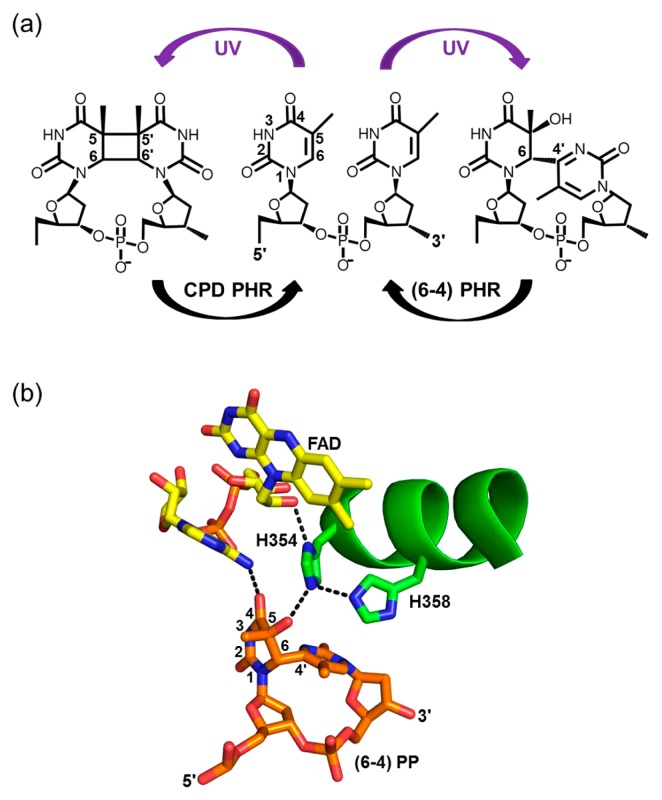
Most prokaryotes have a single photolyase (PHR) that repairs cyclobutane pyrimidine dimers (CPDs), while many eukaryotes possess an additional PHR that repairs pyrimidine-pyrimidone (6-4) photoproducts (PPs) (Fig. 1a) [1,2]. CPDs and (6-4) PPs both arise from UV-induced [2+2] cyclo-addition reactions between adjacent pyrimidines; yet the CPD formed between two C5-C6 double bonds is stable, whereas the PP of the C5-C6 bond with a C4 carbonyl group rapidly rearranges into (6-4) PP. Therefore, to maintain genetic integrity, (6-4) PHR must catalyze not only a covalent bond cleavage as in the CPD PHR reaction, but also the transfer of a hydroxyl or an amino group [3]. Thus, a more complex mechanism has been postulated for (6-4) PHR than for CPD PHR.
Figure 1.
(a) Molecular structures of two thymines (center) and UV-induced photoproducts, CPD (left) and (6-4) PP (right). (b) The (6-4) PP binding site of (6-4) PHR (structure from PDB: 3cvu). FAD chromophore, (6-4) PP and residues are shown as a stick drawing using yellow, orange and green, respectively. The hydrogen-network is shown as a dotted line. The number of amino acid residues corresponds to Xenopus (6-4) PHR.
In 2001, Hitomi et al. reported that two conserved histidines play an important role for the repair function of (6-4) PHR [4]. The enzymatic activity of Xenopus (6-4) PHR, both in vitro and in vivo, was diminished in H354A and H358A, though H358A showed a tiny amount of activity. Binding affinities of (6-4) PP in H354A and H358A were identical to that in the wild type. These results indicate that H354 and H358 are a prerequisite for the repair function, but where H354 plays a principal role. X-ray crystallographic structures of (6-4) PHR support this idea [5,6]. Figure 1b shows the local structure of the active center of the Drosophila (6-4) PHR complexed with (6-4) PP, where H354 is in direct contact with the O-H group of C5 of the 5′ side, and H358 is within a hydrogen-bonding distance of H354. Based on these structure/function studies and additional data, various repair mechansims of (6-4) PP have been proposed [4–12], including the oxetane pathway, the concerted OH transfer pathway, the transient water pathway, among others. Various theoretical calculations have contributed to a deeper understanding of these repair mechanisms [9–12], but experimental data is limited. Although ultrafast spectroscopy detected an intermediate state [7], and flash photolysis implied a two-photon reaction mechansim [8], there are no experimental studies on structural dynamics.
Light-induced difference FTIR spectroscopy is a powerful, sensitive and informative method to study the structural dynamics of photoreceptive proteins [13,14]. This has been proven for photosynthetic reaction centers [15,16], animal and microbial rhodopsins [17–19], and various flavin-binding proteins [20,21]. To gain structural and chemical insights into the activation and repair processes of PHR, we recently established a method to measure light-induced FTIR spectra of CPD PHR [21–24] and (6-4) PHR [21,25–27]. In the latter case, the repair spectra clearly contain characteristic phosphate vibrations of the T(6-4)T dimer on the negative side (before repair) and normal DNA on the positive side (after repair) [25]. In addition, measurement of the 15N-labeled substrate clearly excluded the oxetane structure in the unphotolyzed state [26], which is consistent with the X-ray structure [5]. Thus, light-induced difference FTIR spectra provide information about structural dynamics in PHR function.
Here we studied the structural role of H354 and H358 in the photorepair process of (6-4) PHR by measuring difference FTIR spectra of H354A and H358A mutants. It is known that H354A has no repair activity while H358A has a tiny amount of repair activity [4]. Nonetheless, we believed that it was possible to detect the structural dynamics of H358A because light-induced difference FTIR spectroscopy is so sensitive. In fact, we can apply longer illumination to H358A; if a mutant exhibits 10-times lower repair activity, we can apply 10-times longer illumination, by which a similar repair signal can be expected. In this study, we were able to obtain the difference FTIR spectra of repair for H358A, which were compared to those of the wild type. This is what we expected. What was entirely unexpected was that we were also able to obtain the difference FTIR spectra of repair for H354A. Unlike the signal, which was very small, the repair activity by H354A is a solid result. The molecular mechanism of the repair reaction by (6-4) PHR will be discussed.
Materials and Methods
Sample Preparation
In the first paper for repair, Xenopus (6-4) PHR was expressed in E. coli as a fusion protein with glutathione-S-transferase (GST) at the N-terminus, which was cleaved with thrombin after purification by glutathione sepharose 4B resin (GE Healthcare) [25,26]. However, the amount of purified protein was low (0.26 mg of purified protein from 1 L of culture), which is disadvantageous for future studies using isotope-labeling and mutations. Therefore, we used a Histag system in this study as described previously [27]. Briefly, the wild-type and mutant genes of Xenopus (6-4) PHR containing a His-tag at the N-terminus were expressed in E. coli C41(DE3) strain, and purified by a TALON Metal Affinity Resin column (TAKARA) and a HiTrap Heparin HP column (GE Healthcare). The protein expressed in E. coli does not bind a second chromophore, such as MTHF [28]. A 5-liter culture of E. coli yielded 20 mg of (6-4) PHR, which is 15-times more than the previous preparation method using the GST-tag system.
(6-4) PP was synthesized and incorporated into double-stranded DNA according to a previously described method [29]. The damaged DNA substrate had the following sequences:
5′-CGCGAATTGCGCCC-3′
3′-GCGCTTAACGCGGG-5′
In FTIR measurements, we used redissolved samples, as established in previous studies [21,25–27]. First, we added 2 μL of the sample solution containing 1 mM (6-4) PHR in 50 mM Tris-HCl buffer (pH 8.0) and 200 mM NaCl on an IR window (BaF2). This was left to dry. We next gently mixed 1 μL of 2 mM (6-4) PP in aqueous solution with the sample solution of (6-4) PHR on the IR window, and then dried it. Finally we added 0.4 μL of H2O directly onto the dried film, and sandwiched it with another IR window. We used these re-dissolved samples for FTIR spectroscopy. In this study, the ratio of enzyme and substrate was 1:1. Of note, the oxidized form of (6-4) PHR can bind (6-4) PP [29], where >99% of (6-4) PHR binds (6-4) PP with a Ka value of 2.1×108 (M−1) [28] with either 5 mM (6-4) PHR or 5 mM (6-4) PP.
FTIR Spectroscopy
IR spectra of the re-dissolved sample were measured using an FTS-7000 (DIGILAB) spectrophotometer [14,18,21]. Samples were placed in an Oxford Optistat-DN cryostat mounted in the spectrophotometer, which was also equipped with a temperature controller (ITC-4, Oxford). The illumination source was a 300 W xenon lamp (MAX-302, ASAHI SPECTRA), and illumination at >450 nm with a yellow filter (VY-45, Toshiba) was used to obtain light-induced difference FTIR spectra for activation (the reduced-minus-oxidized spectra) [25,27]. After activation was complete, illumination at >390 nm without filters was used to obtain light-induced difference FTIR spectra for repair (after-minus-before repair) [25]. FTIR spectra were constructed from 128 interferograms with a spectral resolution of 2 cm−1. The difference spectrum was calculated by subtracting the spectrum recorded before illumination from the spectrum recorded after illumination. Four to six difference spectra obtained in this way were averaged for each difference spectrum.
Results
Photorepair Spectra of the Wild-Type and Mutant (6-4) PHR
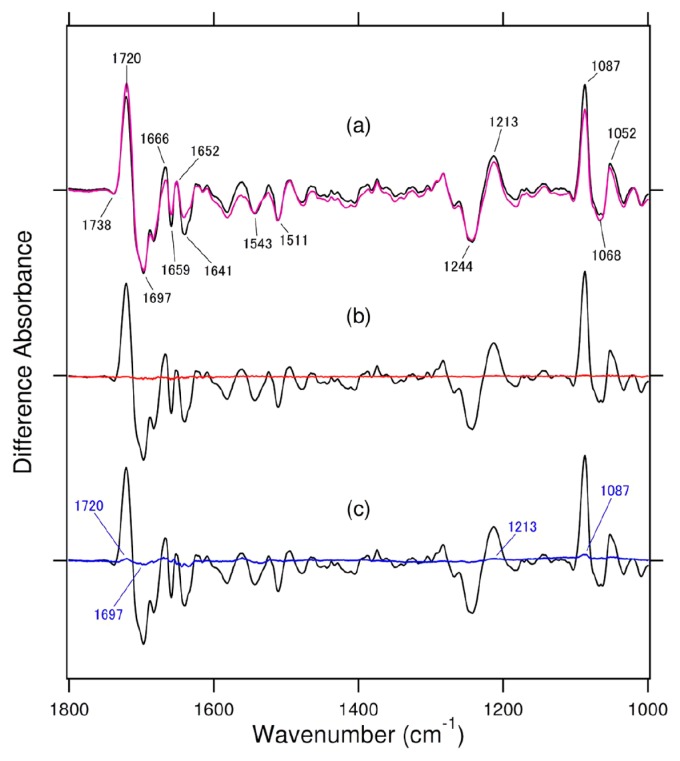
Xenopus (6-4) PHR was originally expressed in E. coli as a fusion protein with GST at the N-terminus, which was cleaved with thrombin after the purification [25,26]. To obtain more protein, we used the His-tag system [27]. The difference FTIR spectra for activation (FADH− minus FADox) were very similar between the two sample preparations [27]. Figure 2a compares the difference FTIR spectra for repair between the two sample preparations, where the ratio of enzyme and substrate was 1:1. Similar FTIR spectra were also observed for repair, indicating no significant dependence on sample preparation. Consequently, we used His-tagged Xenopus (6-4) PHR in the present study.
Figure 2.
(a) The photorepair difference FTIR spectra of Xenopus (6-4) PHR; His-tagged sample in this study (black line) and GST-tagged sample in a previous study (purple line). The purple spectrum is reproduced from Zhang et al. [16]. (b) Black and red lines represent the spectra with the wild type and the H354A mutant of (6-4) PHR under 2-min illumination, respectively. (c) Black and blue lines represent the spectra with the wild type and the H358A mutant of (6-4) PHR under 2-min illumination, respectively. One division of the y-axis corresponds to 0.008 absorbance units.
We prepared re-dissolved samples of H354A and H358A mutants according to the same method for the wild type, and attempted to measure light-induced difference FTIR spectra for repair. Figure 2b compares the difference FTIR spectra for the wild type (black line) and H354A mutant (red line) under 2-min illumination. Unlike the wild type, the difference FTIR spectrum of H354A coincides with the zero line, indicating no repair. This observation is consistent with a previous in vitro and in vivo study [4], and the present FTIR study also indicates the important role of His354 in the repair reaction of (6-4) PHR.
Figure 2c compares the difference FTIR spectra for the wild type (black line) and H358A mutant (blue line) under 2-min illumination. Unlike the wild type, the difference FTIR spectrum of H358A is close to the zero line, which is also consistent with a previous in vitro and in vivo study [4]. However, the blue spectrum shows small peaks at 1720 (+), 1697 (−), 1213 (+), and 1087 (+) cm−1, which are characteristic of the repair spectrum of the wild-type (6-4) PHR (black line in Fig. 2). It should be noted that a previous mutation study reported H358A to possess limited repair activity, unlike H354A [4]. The present FTIR result suggests possible monitoring of the repair reaction by H358A (6-4) PHR. If this is the case, difference FTIR spectroscopy is able to detect structural changes upon photorepair. To achieve this aim, we applied a longer period of illumination for H358A.
Photorepair Spectra of H358A (6-4) PHR
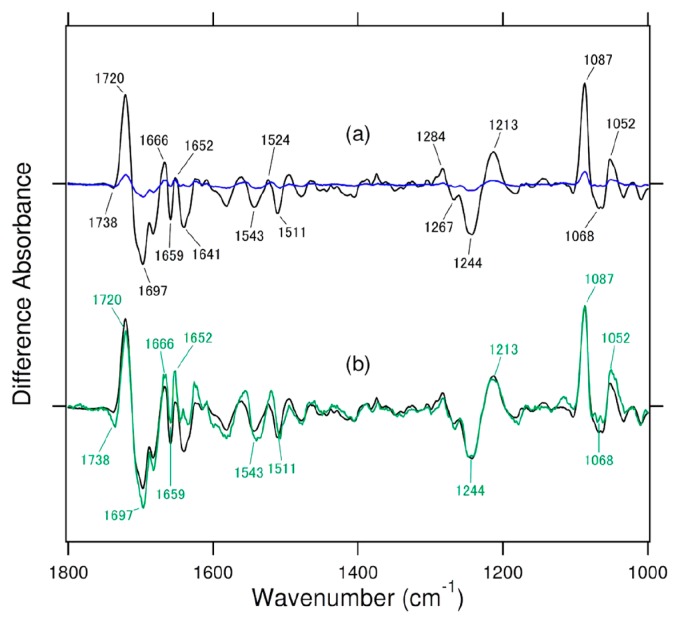
Figure 3a compares the difference FTIR spectra for the wild type (black line) under 2-min illumination and H358A mutant (blue line) under 10-min illumination. Longer illumination clearly allowed a difference FTIR spectrum to be detected, where short and long illuminations yielded similar difference spectra. The green line in Figure 3b represents the 8-times expanded spectrum of the blue spectrum in Figure 3a. The green spectrum coincides with the black spectrum. In particular, phosphate vibrations [30–32] such as 1244 (−), 1213 (+), 1087 (+), 1068 (−), and 1052 (+) cm−1 that are characteristic of repair were similarly observed between H358A and the wild type. This fact indicates that H358A can repair (6-4) PP in the same manner as the wild type. It should be noted that contamination of the wild type protein never happens in the measurements of H358A, because we expressed H358A (6-4) PHR exclusively. On the other hand, repair efficiency is very low. Previous in vitro and in vivo repair measurements reported a small repair activity of H358A [4] but where the repair mechanism remained unclear. The present FTIR spectroscopy provided almost identical FTIR spectra between H358A and the wild type, from which we are able to conclude identical structural changes upon repair.
Figure 3.
Comparison of the light-induced difference FTIR spectra of Xenopus (6-4) PHR for the wild type (black line) under 2-min illumination, the H358A mutant (blue line) under 10-min illumination (a) and the H358A mutant spectrum (8-fold expansion of the blue spectrum in (a) (green line) (b). One division of the y-axis corresponds to 0.01 absorbance units.
Common peaks were observed not only for the phosphate vibrations, but also for other vibrations. The most prominent peak pair at 1720 (+) and 1697 (−) cm−1 of the wild type was assigned as the C=O stretch of DNA [25,26,33], which was reproduced for H358A. In addition to the peaks of the substrate, spectral changes were observed in the frequency region of amide-I vibration which is a signature of secondary structural alterations. In the case of the wild type, peaks were observed at 1666 (+), 1659 (−), 1652 (+) and 1641 (−) cm−1 (Fig. 2a and 3a). It should be however noted that amide-I vibrations are generally H/D-insensitive, and a previous study showed that the bands at 1666 (+) and 1641 (−) cm−1 downshifted in D2O, suggesting different origins [25]. In contrast, the bands at 1659 (−) and 1652 (+) cm−1 were H/D-insensitive, and from the frequency, we are able to identify the amide-I vibrations of the α-helix [34]. Figure 3b shows that H358A possesses peaks at 1666 (+), 1659 (−) and 1652 (+) cm−1. Although the negative 1641-cm−1 band is largely reduced, it is still observable.
The present FTIR analysis revealed that H358A exhibits very similar light-induced difference FTIR spectra to those of the wild type. (6-4) PP probably binds to the active center of H358A and the wild type similarly, and the protein structures are also similar after the release of the repaired DNA. A decrease in volume caused by the H-to-A mutation may be occupied by water molecule(s) near the 358 position, by which a similar structure is maintained. Despite this similar structure, the enzymatic activity of H358A is significantly lowered. Assuming a linear increase of DNA repair with illumination period, the efficiency of H358A is about 40-times lower than that of the wild type. This value is consistent with a previous report [4]. Figure 1b shows that H358 is not in direct contact with (6-4) PP, but forms a hydrogen bond with the side chain of H354. Therefore, we infer that H354 is directly involved in a catalytic reaction where H358 regulates the pKa of the imidazole group of H354.
Photorepair Spectra of H354A (6-4) PHR
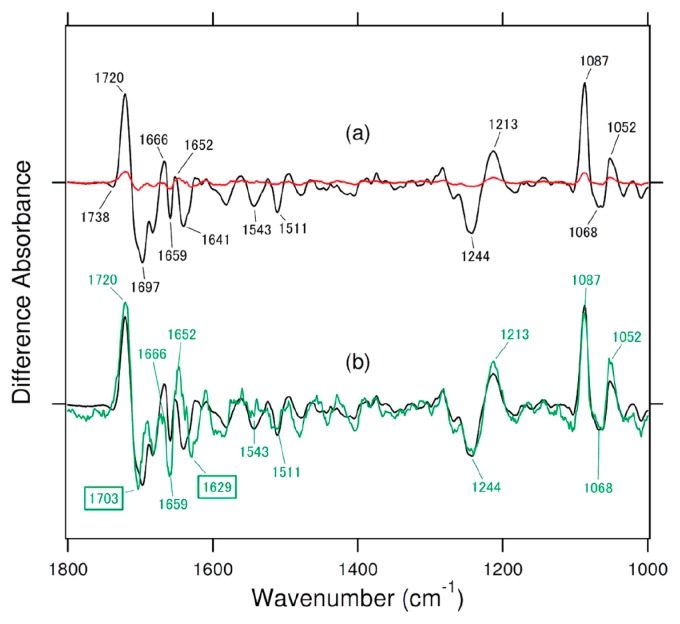
We then attempted a similar analysis for H354A. Figure 4a compares the difference FTIR spectra for the wild type (black line) under 2-min illumination and the H354A mutant (red line) under 30-min illumination. A 15-fold longer illumination of H354A provided some difference FTIR spectra, and we normalized the red and black spectra. The green line in Figure 4b represents the 10-times expanded spectrum of the red spectrum in Figure 4a. The expanded green spectrum is noisy, but phosphate-specific vibrations such as 1244 (−), 1213 (+), 1087 (+), 1068 (−), and 1052 (+) cm−1 were clearly reproduced in H354A. In addition, the C=O stretching vibrations of DNA at 1720 (+) and 1697 (−) cm−1 of the wild type were similarly observed for H354A. However, it should be noted that the positive 1720-cm−1 band has an identical frequency in H354A and the wild type, but the negative peak is slightly upshifted for H354A (1703 cm−1) relative to the wild type (1697 cm−1). An identical positive peak frequency is reasonable because it originates from the C=O stretch of the repaired DNA after the release from (6-4) PHR. In contrast, the higher C=O stretching frequency of (6-4) PP in H354A indicates a weaker hydrogen bond of the C=O groups than in the wild type. Figure 1b shows that H354 forms a complex hydrogen-bonding network, including the OH group of the 2′-hydroxyl group of FAD and the 5′-OH group of (6-4) PP. Replacement of H354 by Ala presumably causes local structural perturbation, leading to a shift of the C=O group(s).
Figure 4.
Comparison of the light-induced difference FTIR spectra of Xenopus (6-4) PHR for the wild type (black line) under 2-min illumination, the H354A mutant (red line) under 30-min illumination (a) and the H354A mutant spectrum (10-fold expansion of the blue spectrum in (a) (green line) (b). One division of the y-axis corresponds to 0.01 absorbance units.
Different bands were observed at 1641 and 1629 cm−1 for the wild type and H354A, respectively. Note that the band at 1641 (−) cm−1 for the wild type does not originate from the amide-I vibration, because it shows a prominent deuterium effect [25]. Therefore, the vibrational origin of the 1641 (−) cm−1 band presumably comes from an H/D-exchangeable side chain. The green spectrum in Figure 4b also shows that the amide-I vibration of the α-helix at 1659 (−) and 1652 (+) cm−1 in the wild type was similarly observed for H354A. This observation suggests that structural changes of the α-helix occur even in the absence of H354, and such changes are coupled to the repair reaction. On the other hand, the peak pair at 1659 (−) and 1652 (+) cm−1 seems to be intensified in H354A in comparison to the wild type. This may indicate that the observed helical structural alteration originates from the helix that involves H354 and H358 (Fig. 1b). The lack of the side chain of H354, which is important for the catalytic reaction, may yield a larger helical structural rearrangement in H354A.
The present FTIR analysis revealed that H354A is able to repair (6-4) PP. Similar FTIR spectra show that (6-4) PP binds to the active center of H354A and the wild type in a similar manner, and that the protein structures are also similar after the release of the repaired DNA. A decrease in volume caused by the H-to-A mutation may be occupied by water molecule(s) near the 354 position, by which a similar structure is maintained. Despite this similar structure, the enzymatic activity of H354A is significantly lowered, but is more than that of H358A. Assuming a linear increase of DNA repair illumination period, the efficiency of H354A is about 150-times lower than that of the wild type. It is reasonable that such low efficient repair cannot be measured by the previous in vitro and in vivo assay [4]. In other words, highly sensitive FTIR measurements were utilized to show the repair activity of H354A in the present study.
Discussion
Previous mutation analysis revealed the importance of two histidines in the active center, H354 and H358 for Xenopus (6-4) PHR, whose mutations significantly lowered the enzymatic activity. This paper reports the difference FTIR spectra of (6-4) PHR mutants, H354A and H358A, bound to (6-4) PP in double-stranded DNA upon photorepair. The present spectroscopic study clearly shows that both H354A and H358A mutants of Xenopus (6-4) PHR still maintain their repair activity, although the efficiency is much lower than that of the wild type. Similar difference FTIR spectra between the wild type and mutant proteins suggest a common mechanism of repair in which (6-4) PP binds to the active center of each mutant and is released after repair, as occurs in the wild type. Similar FTIR spectra also suggest that the decrease in volume caused by the H-to-A mutation is possibly compensated by the addition of water molecule(s), and that such a modified environment is essentially sufficient for the repair function that is probably controlled by proton-coupled electron transfer between the enzyme and substrate. On the other hand, two histidines must work in a concerted manner in the active center of the wild-type enzyme to significantly raise the repair efficiency.
While this study provides important structural information on the repair process of (6-4) PHR, this does not deny any models at present. Theoretical calculations may favor the concerted OH transfer pathway because of lower activation energy than other pathways [9,11]. Nevertheless, the oxetane and transient water pathways are also possible. It should be noted that these models are based on the prerequisite role of H354 in general. However, as shown in the present study, removal of H354 still retains the essential repair ability of (6-4) PHR, which is also the case for H358. Therefore, theoretical calculations should explain the mechanism of histidine mutants. From an experimental perspective, structural dynamics of the reaction intermediates need to be elucidated. Previous light-induced difference FTIR spectroscopy of (6-4) PHR clearly excluded the oxetane structure in the unphotolyzed state [26], which is consistent with the X-ray structure [5]. This does not deny the formation of oxetane during the repair reaction. We will study the reaction intermediates by using low-temperature light-induced difference FTIR spectroscopy in the future.
Acknowledgment
This work was supported by a grant from the Japanese Ministry of Education, Culture, Sports, Science and Technology to H.K. (25104009).
Footnotes
Conflicts of Interest
All authors declare that they have no conflict of interest.
Author Contributions
H. K. directed the research, and wrote the manuscript. D. Y. expressed and purified proteins, and performed FTIR experiments. J. Y. and S. I. synthesized substrates. T. I. helps all experiments, and K. H., T. T. and E. D. G. helped to organize the project.
References
- 1.Sinha RP.&Häder DP. UV-induced DNA damage and repair: a review. Photochem Photobiol Sci. 2002;1:225–236. doi: 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg EC, Walker GC, Siede Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2nd edition. ASM Press; 2006. [Google Scholar]
- 3.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–2238. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 4.Hitomi K, Nakamura H, Kim ST, Mizukoshi T, Ishikawa T, Iwai S, et al. Role of two histidines in the (6-4) photolyase reaction. J Biol Chem. 2001;276:10103–10109. doi: 10.1074/jbc.M008828200. [DOI] [PubMed] [Google Scholar]
- 5.Maul MJ, Barends TRM, Glas AF, Cryle MJ, Domratcheva T, Schneider S, et al. Crystal structure and mechanism of a DNA (6-4) photolyase. Angew Chem Int Ed. 2008;47:10076–10080. doi: 10.1002/anie.200804268. [DOI] [PubMed] [Google Scholar]
- 6.Hitomi K, DiTacchio L, Arvai AS, Yamamoto J, Kim ST, Todo T, et al. Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes. Proc Natl Acad Sci USA. 2009;106:6962–6967. doi: 10.1073/pnas.0809180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Liu Z, Tan C, Guo X, Wang L, Sancar A, et al. Dynamics and mechanism of repair of ultraviolet-induced (6-4) photoproduct by photolyase. Nature. 2010;466:887–890. doi: 10.1038/nature09192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto J, Martin R, Iwai S, Plaza P.&Brettel K. Repair of the (6-4) photoproduct by DNA photolyase requires two photons. Angew Chem Int Ed. 2013;52:7432–7436. doi: 10.1002/anie.201301567. [DOI] [PubMed] [Google Scholar]
- 9.Faraji S, Groenhof G.&Dreuw A. Combined QM/MM investigation on the light-driven electron-induced repair of the (6-4) thymine dimer catalyzed by DNA photolyase. J Phys Chem B. 2013;117:10071–10079. doi: 10.1021/jp401662z. [DOI] [PubMed] [Google Scholar]
- 10.Sadeghian K, Bocola M, Merz T.&Schütz M. Theoretical study on the repair mechanism of the (6-4) photolesion by the (6-4) photolyase. J Am Chem Soc. 2010;132:16285–16295. doi: 10.1021/ja108336t. [DOI] [PubMed] [Google Scholar]
- 11.Domratcheva T. Neutral histidine and photoinduced electron transfer in DNA photolyases. J Am Chem Soc. 2011;133:18172–18182. doi: 10.1021/ja203964d. [DOI] [PubMed] [Google Scholar]
- 12.Dreuw A.&Faraji S. A quantum chemical perspective on (6-4) photolesion repair by photolyases. Phys Chem Chem Phys. 2013;15:19957–19969. doi: 10.1039/c3cp53313a. [DOI] [PubMed] [Google Scholar]
- 13.Gerwert K. Molecular reaction mechanisms of proteins monitored by time-resolved FTIR-spectroscopy. Biol Chem. 1999;380:931–935. doi: 10.1515/BC.1999.115. [DOI] [PubMed] [Google Scholar]
- 14.Kandori H. Role of internal water molecules in bacteriorhodopsin. Biochim Biophys Acta. 2000;1460:177–191. doi: 10.1016/s0005-2728(00)00138-9. [DOI] [PubMed] [Google Scholar]
- 15.Breton J. Fourier transform infrared spectroscopy of primary electron donors in type I photosynthetic reaction centers. Biochim Biophys Acta. 2001;1507:180–193. doi: 10.1016/s0005-2728(01)00206-7. [DOI] [PubMed] [Google Scholar]
- 16.Noguchi T. FTIR detection of water reactions in the oxygen-evolving centre of photosystem II. Philos Trans R Soc Lond, B, Biol Sci. 2008;363:1189–1194. doi: 10.1098/rstb.2007.2214. discussion 1194–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siebert F. Infrared-spectroscopy applied to biochemical and biological problems. Meth Enzymol. 1995;246:501–526. doi: 10.1016/0076-6879(95)46022-5. [DOI] [PubMed] [Google Scholar]
- 18.Furutani Y.&Kandori H. Hydrogen-bonding changes of internal water molecules upon the actions of microbial rhodopsins studied by FTIR spectroscopy. Biochim Biophys Acta. 2014;1837:598–605. doi: 10.1016/j.bbabio.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Lórenz-Fonfría VA.&Heberle J. Channelrhodopsin unchained: structure and mechanism of a light-gated cation channel. Biochim Biophys Acta. 2014;1837:626–642. doi: 10.1016/j.bbabio.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Kottke T, Hegemann P, Dick B.&Heberle J. The photochemistry of the light-, oxygen-, and voltage-sensitive domains in the algal blue light receptor phot. Biopolymers. 2006;82:373–378. doi: 10.1002/bip.20510. [DOI] [PubMed] [Google Scholar]
- 21.Yamada D.&Kandori H. FTIR spectroscopy of flavin-binding photoreceptors. Methods Mol Biol. 2014;1146:361–376. doi: 10.1007/978-1-4939-0452-5_14. [DOI] [PubMed] [Google Scholar]
- 22.Wijaya IMM, Zhang Y, Iwata T, Yamamoto J, Hitomi K, Iwai S, et al. Detection of distinct α-helical rearrangements of cyclobutane pyrimidine dimer photolyase upon substrate binding by Fourier transform infrared spectroscopy. Biochemistry. 2013;52:1019–1027. doi: 10.1021/bi3016179. [DOI] [PubMed] [Google Scholar]
- 23.Wijaya IMM, Iwata T, Yamamoto J, Hitomi K, Iwai S, Getzoff ED, et al. FAD chromophore charge controls the conformation of CPD-photolyase α-helices. Biochemistry. 2014;53:5864–5875. doi: 10.1021/bi500638b. [DOI] [PubMed] [Google Scholar]
- 24.Wijaya IMM, Iwata T, Yamamoto J, Hitomi K, Iwai S, Getzoff ED, et al. FTIR study of CPD photolyase with substrate in single strand DNA. Biophysics. 2015;11:39–45. doi: 10.2142/biophysics.11.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Iwata T, Yamamoto J, Hitomi K, Iwai S, Todo T, et al. FTIR study of light-dependent activation and DNA repair processes of (6-4) photolyase. Biochemistry. 2011;50:3591–3598. doi: 10.1021/bi1019397. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Yamamoto J, Yamada D, Iwata T, Hitomi K, Todo T, et al. Substrate assignment of the (6-4) photolyase reaction by FTIR spectroscopy. J Phys Chem Lett. 2011;2:2774–2777. [Google Scholar]
- 27.Yamada D, Zhang Y, Iwata T, Hitomi K, Getzoff ED.&Kandori H. Fourier-transform infrared study of the photoactivation process of Xenopus (6-4) photolyase. Biochemistry. 2012;51:5774–5783. doi: 10.1021/bi300530x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hitomi K, Kim ST, Iwai S, Harima N, Otoshi E, Ikenaga M, et al. Binding and catalytic properties of Xenopus (6-4) photolyase. J Biol Chem. 1997;272:32591–32598. doi: 10.1074/jbc.272.51.32591. [DOI] [PubMed] [Google Scholar]
- 29.Iwai S, Shimizu M, Kamiya H.&Ohtsuka E. Synthesis of a phosphoramidite coupling unit of the pyrimidine (6-4) pyrimidone photoproduct and its incorporation into oligode-oxynucleotides. J Am Chem Soc. 1996;118:7642–7643. [Google Scholar]
- 30.Schleicher E, Hessling B, Illarionova V, Bacher A, Weber S, Richter G, et al. Light-induced reactions of Escherichia coli DNA photolyase monitored by Fourier transform infrared spectroscopy. FEBS J. 2005;272:1855–1866. doi: 10.1111/j.1742-4658.2005.04617.x. [DOI] [PubMed] [Google Scholar]
- 31.Taillandier E.&Liquier J. Infrared spectroscopy of DNA. Meth Enzymol. 1992;211:307–335. doi: 10.1016/0076-6879(92)11018-e. [DOI] [PubMed] [Google Scholar]
- 32.Klähn M, Schlitter J.&Gerwert K. Theoretical IR spectroscopy based on QM/MM calculations provides changes in charge distribution, bond lengths, and bond angles of the GTP ligand induced by the Ras-protein. Biophys J. 2005;6:3829–3844. doi: 10.1529/biophysj.104.058644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowak MJ. IR matrix isolation studies of nucleic acid constituents: the spectrum of monomeric thymine. J Mol Struct. 1989;193:35–49. [Google Scholar]
- 34.Krimm S.&Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv Protein Chem. 1986;38:181–364. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]