Abstract
Background:
MiR-34a dysregulation has been implicated in tumorigenesis and progression of gastric cancer, but its role in prognosis of patients with gastric cancer remains unknown. The aim of this study was to investigate the expression and prognostic significance of miR-34a in gastric cancer patients after radical gastrectomy.
Methods:
Quantitative real-time polymerase chain reaction was performed to detect the expression of miR-34a in human gastric cancer cell lines and tissues in 76 patients with gastric adenocarcinoma from China. Results are assessed for association with clinical features and overall survival (OS) using Kaplan–Meier analysis. Prognostic values of miR-34a expression and clinical outcomes were evaluated by Cox regression analysis. A molecular prognostic stratification scheme incorporating miR-34a expression was determined using receiver operating characteristic analysis.
Results:
The results show that the expression level of miR-34a was decreased in human gastric cancer cell lines and tissues, and down-regulated expression of miR-34a was associated with Lauren classification (P = 0.034). Decreased miR-34a expression in gastric cancer tissues was positively correlated with poor OS of gastric cancer patients (P = 0.013). Further multivariate Cox regression analysis suggested that miR-34a expression was an independent prognostic indicator for gastric cancer (P = 0.027). Applying the prognostic value of miR-34a expression to tumor node metastasis (TNM) stage system showed a better prognostic value in patients with gastric cancer than miR-34a expression (P = 0.0435) or TNM stage (P = 0.0249) alone.
Conclusion:
The results reinforce the critical role for the down-regulated miR-34a expression in gastric cancer and suggest that miR-34a could be a prognostic indicator for this disease.
Keywords: Gastric Cancer, MiR-34a, Overall Survival, Prognostic Significance, Radical Gastrectomy
INTRODUCTION
Mortality associated with gastric cancer in many industrialized nations has decreased during recent decades.[1,2] However, China is still one of the countries with the highest incidence of gastric cancer and accounts for over 40% of all new cases worldwide.[3,4] Despite aggressive therapy, the prognosis of advanced gastric cancer in China tends to be dismal.[5] Because the heterogeneity of gastric cancer at the molecular level is present, selecting the patients who could benefit greatest from personalized treatment and targeted therapies is thought of as the best method to reduce the mortality rates of gastric cancer.[6,7] Therefore, the identification of molecular markers that are predictive of gastric cancer aggressiveness and prognosis of the patient has the potential to improve the ability to manage patients and provide important clinically relevant insights into disease treatment.
MicroRNAs (miRNAs) are small 21–23 nucleotides noncoding RNAs that regulate gene expression posttranscriptionally through base pairing with the 3’-untranslated region of target messenger RNAs (mRNAs).[8] In many human cancers, miRNAs can function as tumor suppressor genes or oncogenes to suppress translation or induce mRNA degradation depending on the nature of their targets.[9] The miR-34 family is a class of miRNAs those are highly evolutionarily conserved, and there are three miR-34 members, including miR-34a, miR-34b, and miR-34c.[10] They are widespread in arthropods, nematodes, and mammals. In most human tissues, the miR-34a expression level is much higher than that of miR-34b and miR-34c.[11,12] It has been demonstrated that the expression of miR-34a is altered in various types of cancer, including breast cancer,[13] lung cancer,[14] head and neck squamous cell carcinoma,[15] and prostate cancer.[16] Recently, Zhang et al. reported that down-regulation of miR-34a in gastric cancer is associated with high recurrence rate and poor overall survival (OS).[17] In addition, previous studies have also suggested that miR-34a could inhibit gastric cancer tumorigenesis by targeting platelet-derived growth factor receptor (PDGFR) and mesenchymal epithelial transition (MET) through the PI3K/Akt pathway[18] and regulate cisplatin-induce gastric cancer cell death by modulating PI3K/Akt/survivin pathway.[19] However, an extensive analysis of expression of miR-34a in correlated to the prognosis of gastric cancer patients has not been performed and awaits further elucidation.
In this research, we aim to determine the prognostic significance of expression of miR-34a in gastric cancer.
METHODS
Cell culture
Immortalized normal human gastric epithelial cell line GES-1 and six human gastric cancer cell lines, including NCI-N87, AGS, MKN-45, MKN-28, BGC-823, and SGC7901, were obtained directly from Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China). All these cells were routinely grown and maintained in RPMI-1640 (Invitrogen, Carlsbad, CA, USA) culture medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) at 37°C in a humidified cell incubator with an atmosphere of 5% CO2.
Clinical specimens
Fresh gastric adenocarcinoma tissues were collected from patients with gastric cancer at The Second Affiliated Hospital of Xi’an Jiaotong University (Shaanxi province, China) from January 2008 to December 2012. Adjacent normal tissues were obtained at least 5 cm from the tumor at the same time. Samples were flash frozen in liquid nitrogen until use. Specimens were reassessed by two pathologists independently, and the stage of gastric cancer is classified according to the tumor node metastasis (TNM) stage system of the Union for International Cancer Control/American Joint Committee on Cancer.[20] The selected criteria were the patients harboring advanced gastric adenocarcinoma without distant metastases, and excluded criteria were patients with infiltration of adjacent structures (T4b), distant metastases, and gastric stump cancer. Also, patients were excluded if they had previously been exposed to any targeted therapy, radiotherapy, chemotherapy, and/or intervention therapy for gastric cancer. Routine chemotherapy based on 5-fluorouracil and oxaliplatin had been given to the patients with advanced-stage disease after operation, but no radiation treatment was done in any of the patients included in our study. This study was reviewed and approved by the Institutional Review Board of The Second Affiliated Hospital of Xi’an Jiaotong University (Shaanxi province, China). All study participants or their legal guardian provided informed written consent prior to study enrollment.
Quantitative real-time reverse transcriptase-polymerase chain reaction
Total RNA containing miRNA was extracted from cultured cells or tissues using miRNeasy Mini Kit (Qiagen, Germany). Complementary DNA was synthesized using miScript reverse transcription kit (Qiagen) following the manufacturer's instructions. Reverse transcription was undertaken using 50 ng total RNA with a primer specific for miR-34a, together with the SYBR Green miRNA reverse transcription kit. miRNAs were quantified using the SYBR Green miRNA quantitative real-time polymerase chain reaction (qRT-PCR) assay according to the manufacturer's protocol (Applied BioSystems, Foster City, CA, USA). The qRT-PCR reaction was carried out on a 7500 fast real-time system (Applied Biosystems, Foster City, CA, USA). All qRT-PCRs were performed in triplicate. The data were analyzed using an automated baseline. The threshold cycle (Ct) was defined as the fractional cycle number at which the fluorescence exceeded the given threshold. The data obtained from the qRT-PCR were analyzed using the ΔΔCt method (2ΔΔCt). The PCR primers sets used here for miR-34a was designed as follows: Mir-34a forward primer: 5’-UGGCAGUGUCUUAGCUGGUUGU-3’, and reverse primer: 5’-GUGCAGGGUCCAGGU-3’. U6 was used as an internal control and amplified with forward primer: 5’-GCTTCGGCAGCACATATACTAAAAT-3’ and reverse primer: 5’-CGCTTCACGAATTTGCGTGTCAT-3’.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA, USA) and SPSS 17.0 (SPSS, Chicago, IL, USA). Followup data were available for all patients. Pearson's Chi-square test was used to analyze the relationship between miR-34a expression and clinicopathological factors. Cumulative survival rates were calculated by Kaplan–Meier method and the differences between the subgroups were examined by the log-rank test. Numbers at risk were calculated for the beginning of each period. The prognostic value of miR-34a expression was determined by univariate and multivariate analysis. A prognostic model combining miR-34a expression with TNM stage system was constructed with the logistic regression. Receiver operating characteristic (ROC) analysis was performed to assess the prognostic value of the parameters. All P values were two-sided, and P < 0.05 was considered as statistically significant.
RESULTS
Expression analysis of miR-34a in human gastric cancer cells
To ascertain the level of miR-34a expression in human gastric cancer cells, we first evaluated miR-34a expression by qRT-PCR in immortalized normal human gastric epithelial cell line GES-1 and six human gastric cancer cell lines, including NCI-N87, AGS, MKN-45, MKN-28, BGC-823, and SGC7901. As shown in Figure 1, the human gastric cancer cells expressed significantly lower levels of miR-34a than GES-1. This result indicated that decreased miR-34a expression may be related to the oncogenesis of gastric cancer.
Figure 1.
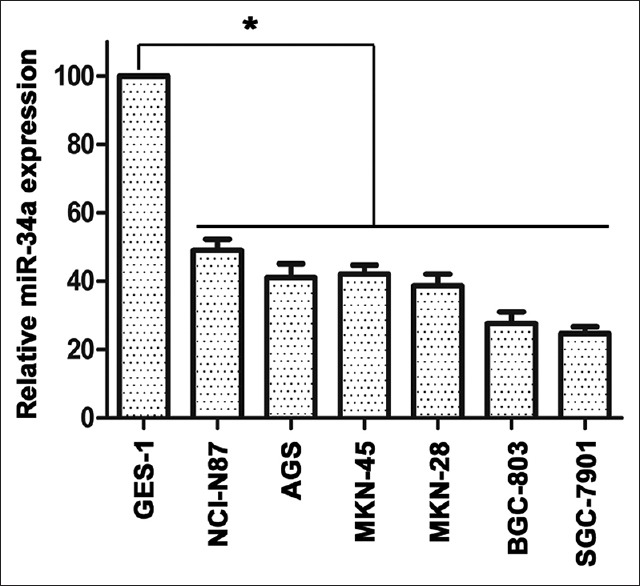
The expression of miR-34a in human gastric epithelial cell and gastric cancer cell lines. Quantitative real-time polymerase chain reaction analysis for miR-34a expression levels in immortalized normal human gastric epithelial cell line GES-1 and six human gastric cancer cell lines (NCI-N87, AGS, MKN-45, MKN-28, BGC-823, and SGC7901), *P < 0.05.
Associations between miR-34a expression and clinicopathologic factors
Table 1 summarizes the clinicopathological characteristics of patients enrolled in this study. Of 76 patients, most of the patients were female (69.7%) and had an intestinal type cancer (75.0%), middle or distal-located cancer (77.6%), TNM stage III cancer (61.8%), smaller tumor size (in cm, 52.6%), and poorly differentiation (59.2%). The expression levels of miR-34a were further measured to analyze their relationship with clinicopathologic factors. Patients were divided into high and low expression group according to the ratio of their normal/cancer tissue mean expression levels of miR-34a according to the results of qRT-PCR. And according to the criterion, approximately 47.4% (36 of 76) tumors were scored as low miR-34a expression (normal/cancer ratio <1.0). As shown in Table 1, the expression of miR-34a was significantly correlated with Lauren classification (P = 0.034). Besides, there are no other differences significantly between the high and low group of miR-34a expression.
Table 1.
Relation between intratumoral miR-34a expression and clinical characteristics in patients with gastric cancer (n=76)
| Items | Patients, n (%) | miR-34a expression | P | |
|---|---|---|---|---|
| Low | High | |||
| All patients | 76 (100) | 36 | 40 | |
| Age* | 0.646 | |||
| ≤65 years | 38 (50.0) | 19 | 19 | |
| >65 years | 38 (50.0) | 17 | 21 | |
| Gender | 0.580 | |||
| Female | 53 (69.7) | 24 | 29 | |
| Male | 23 (30.3) | 12 | 11 | |
| Localization | 0.506 | |||
| Proximal | 17 (22.4) | 8 | 9 | |
| Middle | 30 (39.5) | 12 | 18 | |
| Distal | 29 (38.2) | 16 | 13 | |
| Differentiation | 0.276 | |||
| Well | 12 (15.8) | 6 | 6 | |
| Moderately | 19 (25.0) | 6 | 13 | |
| Poorly | 45 (59.2) | 24 | 21 | |
| Lauren classification | 0.034 | |||
| Intestinal type | 57 (75.0) | 23 | 34 | |
| Diffusetype | 19 (25.0) | 13 | 6 | |
| T stage | 0.146 | |||
| T1 | 7 (9.2) | 2 | 5 | |
| T2 | 6 (7.9) | 1 | 5 | |
| T3 | 10 (13.2) | 7 | 3 | |
| T4 | 53 (69.7) | 26 | 27 | |
| N stage | 0.254 | |||
| N0 | 25 (32.9) | 11 | 14 | |
| N1 | 8 (10.5) | 2 | 6 | |
| N2 | 17 (22.4) | 7 | 10 | |
| N3 | 26 (34.2) | 16 | 10 | |
| TNM stage | 0.357 | |||
| I | 10 (13.2) | 3 | 7 | |
| II | 19 (25.0) | 8 | 11 | |
| III | 47 (61.8) | 25 | 22 | |
| Tumor size* | 0.981 | |||
| <3.5 cm | 40 (52.6) | 19 | 21 | |
| ≥3.5 cm | 36 (47.4) | 17 | 19 | |
TNM: Tumor node metastasis. *Split at median.
Prognostic value of miR-34a expression in patients with gastric cancer
Kaplan–Meier survival analysis was performed to investigate the prognostic value of miR-34a expression. As shown in Figure 2, patients with low miR-34a expression showed significantly shorter OS than those high ones [P = 0.013; Figure 2a], which indicated a vital impact of miR-34a expression on clinical outcome in gastric cancer patients. Also, the prognosis of gastric cancer patients with TNM stage III was also poorer compared with TNM stage I + II significantly [P = 0.011; Figure 2b].
Figure 2.
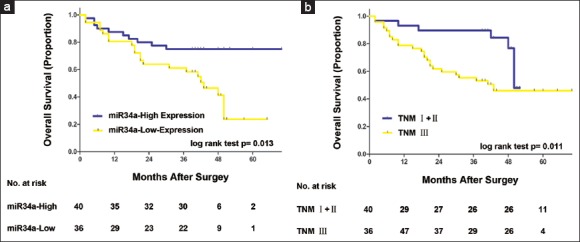
Analyses of overall survival according to the expression of miR-34a and tumor node metastasis stage in gastric cancer patients. Kaplan–Meier analyses of overall survival according to miR-34a expression (a) and tumor node metastasis stage (b) in patients with gastric cancer. P value was calculated by log-rank test.
Univariate and multivariate analyses for OS in this study population exhibited that T stage (P = 0.027), N stage (P = 0.012), TNM stage (P = 0.015), and miR-34a expression (P = 0.018) significantly affected the survival of gastric cancer [Table 2]. In addition, as shown in Table 3, TNM stage (hazard ratio [HR], 1.615; 95% confidence interval [CI], 1.071–2.436; P = 0.022) and miR-34a expression (HR, 2.327; 95% CI, 1.099–4.927; P = 0.027) were recognized as independent and significant prognostic parameters.
Table 2.
Univariate analysis of factors for OS in 76 patients with gastric cancer
| Items | Patients, n | Events, n | OS (univariate) Hazard radio (95% CI) | P |
|---|---|---|---|---|
| All patients | 76 | 32 | ||
| Age* | 0.410 | |||
| ≤65 years | 38 | 13 | 1.000 (reference) | |
| >65 years | 38 | 19 | 1.348 (0.662–2.746) | |
| Gender | 0.290 | |||
| Female | 53 | 20 | 1.000 (reference) | |
| Male | 23 | 12 | 0.679 (0.332–1.390) | |
| Localization | 0.074 | |||
| Proximal + middle | 47 | 23 | 1.000 (reference) | |
| Distal | 29 | 9 | 0.490 (0.223–1.073) | |
| Differentiation | 0.052 | |||
| Well + moderately | 31 | 9 | 1.000 (reference) | |
| Poorly | 45 | 23 | 2.152 (0.993–4.664) | |
| Lauren classification | 0.792 | |||
| Intestinal type | 57 | 25 | 1.000 (reference) | |
| Diffuse-type | 19 | 7 | 1.086 (0.589–1.999) | |
| T stage | 0.027 | |||
| T1 + T2 | 13 | 1 | 1.000 (reference) | |
| T3 + T4 | 63 | 31 | 3.096 (1.140–8.409) | |
| N stage | 0.012 | |||
| N0 + N1 | 33 | 9 | 1.000 (reference) | |
| N2 + N3 | 43 | 23 | 2.690 (1.239–5.840) | |
| TNM stage | 0.015 | |||
| I + II | 29 | 8 | 1.000 (reference) | |
| III | 47 | 24 | 1.655 (1.101–2.487) | |
| Tumor size* | 0.078 | |||
| <3.5 cm | 40 | 12 | 1.000 (reference) | |
| ≥3.5 cm | 36 | 20 | 1.906 (0.931–3.902) | |
| miR-34a expression | 0.018 | |||
| High | 40 | 10 | 1.000 (reference) | |
| Low | 36 | 22 | 2.469 (1.166–5.228) |
*Split at median. OS: Overall survival; 95% CI: 95% confidence interval.
Table 3.
Multivariate analysis of factors for OS in 76 patients with gastric cancer
| Items | Patients, n | Events, n | OS (multivariate) Hazard radio (95% CI) | P |
|---|---|---|---|---|
| All patients | 76 | 32 | ||
| TNM stage | 0.022 | |||
| I + II | 29 | 8 | 1.000 (reference) | |
| III | 47 | 24 | 1.615 (1.071–2.436) | |
| miR-34a expression | 0.027 | |||
| High | 40 | 10 | 1.000 (reference) | |
| Low | 36 | 22 | 2.327 (1.099–4.927) |
OS: Overall survival; 95% CI: 95% confidence interval; TNM: Tumor node metastasis.
Prognostic model based on tumor node metastasis stage and miR-34a expression
In order to develop a more sensitive predictive method for patients with gastric cancer, a prognostic model combining TNM stage and miR-34a expression was constructed. ROC analysis was performed to compare its prognostic ability. As shown in Figure 3, prognostic model of TNM stage plus miR-34a expression (area under the curve [AUC] = 0.729) exhibited a better prognostic value than miR-34a expression (AUC = 0.685, P = 0.0435) or TNM stage (AUC = 0.614, P = 0.0249) alone.
Figure 3.
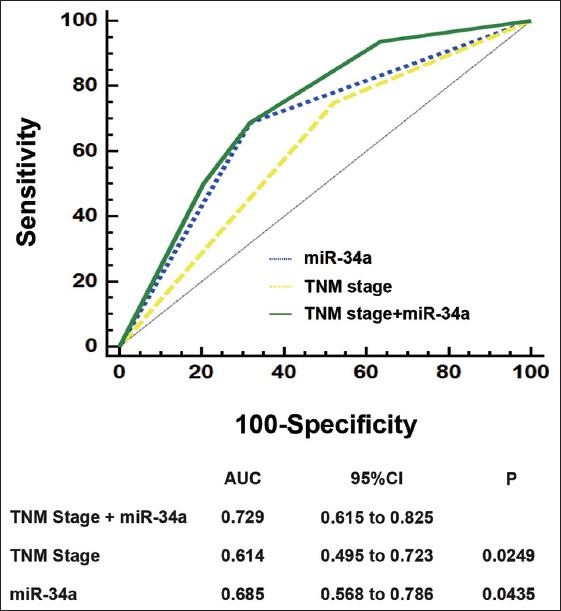
Receiver operating characteristic analysis for the prediction of overall survival in patients with gastric cancer. Receiver operating characteristic analysis of the sensitivity and specificity for the prediction of overall survival by the combined miR-34a expression and tumor node metastasis stage model, the tumor node metastasis stage model, and the miR-34a expression model. P values show the area under the receiver operating characteristic curves (area under the curve) of the combined miR-34a expression and tumor node metastasis stage model versus area under the curves of the tumor node metastasis stage model or the miR-34a expression model.
DISCUSSION
Despite aggressive therapy, the prognosis of advanced gastric cancer in China tends to be dismal.[5,21] Identification of patients who could benefit greatest from personalized treatment and targeted therapies is thought of as the best method to reduce the mortality rates of gastric cancer. Recently, research concerning the relationship between miRNAs and gastric cancer has gradually gained attention from researchers in the general surgery field, which has become an emerging hotspot in the field of gastric cancer research.
The miR-34 family is a class of miRNAs that are highly evolutionarily conserved, and they are widespread in arthropods, nematodes, and mammals.[10,11] Through investigating the miRNA expression profile in gastric cancer, Yao et al. determined that miR-34 was displaying a greater than 2-fold expression difference between gastric cancer and normal gastric tissues.[22] In virtually all vertebrates, there are three miR-34 members, including miR-34a, miR-34b, and miR-34c. In most human tissues, the miR-34a expression level is much higher than that of miR-34b and miR-34c.[11] It has been demonstrated that the dysregulation of miR-34a is present in various types of cancer, including breast cancer,[13] lung cancer,[14] head and neck squamous cell carcinoma,[15] and prostate cancer.[16] Also, Shen et al. observed that the miR-34a expression is down-regulated in laryngeal squamous cell carcinoma.[23] The above-mentioned studies confirmed that miR-34a might play an important role that is similar to tumor suppressor miRNAs.
Recently, Zhang et al. reported that down-regulation of miR-34a in gastric cancer is associated with high recurrence rate and poor OS, and low miR-34a expression level was associated with lymph node involvement, advanced TNM stage, and poor tumor differentiation.[17] Their results are in accordance with ours. In the present research, we also investigated the expression level of miR-34a in gastric cancer patients and proved that demonstrated that high miR-34a expression as an independent poor prognostic factor for OS of patients with gastric cancer after radical gastrectomy. In addition, incorporation of the level of miR-34a expression into TNM stage system improved the prognostic value of traditional staging system. These results reinforce the critical role for the down-regulated miR-34a expression in gastric cancer and suggest that miR-34a could be a prognostic indicator for this disease. However, the potential changing of clinical practice should be validated in a randomized controlled trial in the future.
Like many other miRNAs, miR-34a could antagonize various processes of cancer cells through regulating different genes and influencing their functions in many cellular pathways.[24] The control of cellular proliferation is a major function of miR-34a and ectopic expression of miR-34a could lead to the arrest of G1/G2 in cancer cells and result in the decreasing of cell doubling times.[25] In addition, it has been demonstrated that ectopic miR-34a could induce apoptosis when reintroduced into some cancer cells because it represses SIRT1, YY1, and Bcl-2, in which, miR-34a was identified as a p53 target, and miR-34a mediated apoptosis may be suppressed by the inactivation of p53.[15,26,27] MiR-34a was down-regulated in many cancers, including gastric cancer.[28] Bhatt et al. showed that restoration of tumor suppressor miR-34 inhibits tumorspheres of gastric cancer of p53-mutant.[29] Peng et al. demonstrated that miR-34a could inhibit gastric cancer tumourigenesis by targeting PDGFR and MET through the PI3K/Akt pathway,[18] and Cao et al. proved that miR-34a could regulate cisplatin-induce gastric cancer cell death by modulating PI3K/Akt/survivin pathway.[19] In this research, we found that the human gastric cancer cells, including NCI-N87, AGS, MKN-45, MKN-28, BGC-823, and SGC7901, expressed significantly lower levels of miR-34a than GES-1, which is an immortalized normal human gastric epithelial cell line. This result indicated that abnormal miR-34a expression may be related to the oncogenesis of gastric cancer. Also, we identified the expression of miR-34a was significantly correlated with Lauren classification. There are fewer intestinal-type gastric cancer patients with high miR-34a expression than diffuse-type patients. However, clarifying the underlying mechanism of differential expression of miR-34a between the intestinal and diffuse type progression of gastric cancer awaits further investigation.
To date, there is an increasing interest with respect to the miRNA responses to lifestyle and environmental exposures. Stánitz et al. had been investigated that whether living conditions, lifestyle behaviors, and social status, such as alcohol consumption and cigarette smoking, are associated with specific miRNA expression patterns of gastric cancer.[30] Their study showed that down-expression of miR-34a was observed in gastric cancer samples of patients from western countries, which is in accordance with our results in China. Also, gastric cancer of the same histopathology from different geographic region's population could show differential miRNAs and protein expression patterns, which might result from the existence of different risk factors of carcinogenesis.
In summary, the results of this study demonstrate that miR-34a is a promising marker to predict the OS of patients with gastric cancer after radical gastrectomy. In addition, incorporation of the level miR-34a expression into TNM stage system improved the prognostic value of traditional staging system, and the gastric cancer patients with low miR-34a expression might need closer follow-up and aggressive postoperative treatment.
Financial support and sponsorship
This work was supported by a grant of National “12th 5-Year” Plan for Science and Technology Support (No. 2012BAJ8B03).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Yuan-Yuan Ji
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Anderson WF, Camargo MC, Fraumeni JF, Jr, Correa P, Rosenberg PS, Rabkin CS. Age-specific trends in incidence of noncardia gastric cancer in US adults. JAMA. 2010;303:1723–8. doi: 10.1001/jama.2010.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen WQ, Zheng RS, Zhang SW, Zeng HM, Zou XN. The incidences and mortalities of major cancers in China, 2010. Chin J Cancer. 2014;33:402–5. doi: 10.5732/cjc.014.10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17–20. doi: 10.3748/wjg.v12.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thrumurthy SG, Chaudry MA, Hochhauser D, Mughal M. The diagnosis and management of gastric cancer. BMJ. 2013;347:f6367. doi: 10.1136/bmj.f6367. [DOI] [PubMed] [Google Scholar]
- 6.Figueiredo C, Garcia-Gonzalez MA, Machado JC. Molecular pathogenesis of gastric cancer. Helicobacter. 2013;18(Suppl 1):28–33. doi: 10.1111/hel.12083. [DOI] [PubMed] [Google Scholar]
- 7.Stadtländer CT, Waterbor JW. Molecular epidemiology, pathogenesis and prevention of gastric cancer. Carcinogenesis. 1999;20:2195–208. doi: 10.1093/carcin/20.12.2195. [DOI] [PubMed] [Google Scholar]
- 8.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–15. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 10.Wong MY, Yu Y, Walsh WR, Yang JL. microRNA-34 family and treatment of cancers with mutant or wild-type p53 (Review) Int J Oncol. 2011;38:1189–95. doi: 10.3892/ijo.2011.970. [DOI] [PubMed] [Google Scholar]
- 11.Tscherner A, Gilchrist G, Smith N, Blondin P, Gillis D, LaMarre J. MicroRNA-34 family expression in bovine gametes and preimplantation embryos. Reprod Biol Endocrinol. 2014;12:85. doi: 10.1186/1477-7827-12-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maroof H, Salajegheh A, Smith RA, Lam AK. MicroRNA-34 family, mechanisms of action in cancer: A review. Curr Cancer Drug Targets. 2014;14:737–51. doi: 10.2174/1568009614666141020100337. [DOI] [PubMed] [Google Scholar]
- 13.Yang S, Li Y, Gao J, Zhang T, Li S, Luo A, et al. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2013;32:4294–303. doi: 10.1038/onc.2012.432. [DOI] [PubMed] [Google Scholar]
- 14.Stahlhut C, Slack FJ. Combinatorial Action of MicroRNAs let-7 and miR-34 Effectively Synergizes with Erlotinib to Suppress Non-small Cell Lung Cancer Cell Proliferation. Cell Cycle. 2015;14:2171–80. doi: 10.1080/15384101.2014.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar B, Yadav A, Lang J, Teknos TN, Kumar P. Dysregulation of microRNA-34a expression in head and neck squamous cell carcinoma promotes tumor growth and tumor angiogenesis. PLoS One. 2012;7:e37601. doi: 10.1371/journal.pone.0037601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rokhlin OW, Scheinker VS, Taghiyev AF, Bumcrot D, Glover RA, Cohen MB. MicroRNA-34 mediates AR-dependent p53-induced apoptosis in prostate cancer. Cancer Biol Ther. 2008;7:1288–96. doi: 10.4161/cbt.7.8.6284. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Li S, Yang J, Liu S, Gong X, Yu X. The prognostic value of miR-34a expression in completely resected gastric cancer: Tumor recurrence and overall survival. Int J Clin Exp Med. 2015;8:2635–41. [PMC free article] [PubMed] [Google Scholar]
- 18.Peng Y, Guo JJ, Liu YM, Wu XL. MicroRNA-34A inhibits the growth, invasion and metastasis of gastric cancer by targeting PDGFR and MET expression. Biosci Rep. 2014;34 doi: 10.1042/BSR20140020. pii: E00112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Cao W, Yang W, Fan R, Li H, Jiang J, Geng M, et al. miR-34a regulates cisplatin-induce gastric cancer cell death by modulating PI3K/AKT/survivin pathway. Tumour Biol. 2014;35:1287–95. doi: 10.1007/s13277-013-1171-7. [DOI] [PubMed] [Google Scholar]
- 20.Washington K. 7th edition of the AJCC cancer staging manual: Stomach. Ann Surg Oncol. 2010;17:3077–9. doi: 10.1245/s10434-010-1362-z. [DOI] [PubMed] [Google Scholar]
- 21.Shen L, Shan YS, Hu HM, Price TJ, Sirohi B, Yeh KH, et al. Management of gastric cancer in Asia: Resource-stratified guidelines. Lancet Oncol. 2013;14:e535–47. doi: 10.1016/S1470-2045(13)70436-4. [DOI] [PubMed] [Google Scholar]
- 22.Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L, et al. MicroRNA profiling of human gastric cancer. Mol Med Rep. 2009;2:963–70. doi: 10.3892/mmr_00000199. [DOI] [PubMed] [Google Scholar]
- 23.Shen Z, Zhan G, Ye D, Ren Y, Cheng L, Wu Z, et al. MicroRNA-34a affects the occurrence of laryngeal squamous cell carcinoma by targeting the antiapoptotic gene survivin. Med Oncol. 2012;29:2473–80. doi: 10.1007/s12032-011-0156-x. [DOI] [PubMed] [Google Scholar]
- 24.Bader AG. miR-34 – A microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatani F, Ferracin M, Manara MC, Ventura S, Del Monaco V, Ferrari S, et al. miR-34a predicts survival of Ewing's sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J Pathol. 2012;226:796–805. doi: 10.1002/path.3007. [DOI] [PubMed] [Google Scholar]
- 26.Kofman AV, Letson C, Dupart E, Bao Y, Newcomb WW, Schiff D, et al. The p53-microRNA-34a axis regulates cellular entry receptors for tumor-associated human herpes viruses. Med Hypotheses. 2013;81:62–7. doi: 10.1016/j.mehy.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luan S, Sun L, Huang F. MicroRNA-34a: A novel tumor suppressor in p53-mutant glioma cell line U251. Arch Med Res. 2010;41:67–74. doi: 10.1016/j.arcmed.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Zhang S, Chen P, Huang Z, Hu X, Chen M, Hu S, et al. Sirt7 promotes gastric cancer growth and inhibits apoptosis by epigenetically inhibiting miR-34a. Sci Rep. 2015;5:9787. doi: 10.1038/srep09787. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Bhatt K, Zhou L, Mi QS, Huang S, She JX, Dong Z. MicroRNA-34a is induced via p53 during cisplatin nephrotoxicity and contributes to cell survival. Mol Med. 2010;16:409–16. doi: 10.2119/molmed.2010.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stánitz E, Juhász K, Tóth C, Gombos K, Natali PG, Ember I. Evaluation of MicroRNA expression pattern of gastric adenocarcinoma associated with socioeconomic, environmental and lifestyle factors in northwestern Hungary. Anticancer Res. 2013;33:3195–200. [PubMed] [Google Scholar]


