Abstract
Background
MALAT-1 is a highly conserved nuclear long non-coding RNA (lncRNA). The overexpression of MALAT-1 has been reported in several types of cancers. This meta-analysis was conducted to further investigate its potential role as a prognostic indicator in various cancers.
Material/Methods
The meta-analysis was performed by use of systematic search terms in 13 databases for qualified papers on prognosis in cancer from inception to June 30, 2015. The combined hazard ratios (HRs) with 95% confidence interval (95% CI) were computed to demonstrate the effect of MALAT-1 on prognosis of cancers.
Results
A total of 590 papers were initially identified, and 17 studies were finally included in this paper. Meta-analysis was accomplished with a total of 1626 patients. Combined HRs and 95% CI were calculated by fixed-effects or random-effects models. The quality assessment of included studies was performed by the Newcastle-Ottawa scale (NOS). High expression of MALAT-1 was found to be an indicator of poor prognosis in overall survival (OS) (HR=1.84, 95% CI: 1.27–2.67) and disease-free survival (DFS) (HR=2.37, 95% CI: 1.55–3.62). In subgroups, the associations between MALAT-1 and survival were also apparent, for instance, in country subgroup: China (HR=1.85, 95% CI: 1.14–2.99).
Conclusions
The overexpression of MALAT-1 may be a potential prognostic indicator for various human cancers.
MeSH Keywords: Genes, Neoplasm; Meta-Analysis; Prognosis; RNA, Long Noncoding; Survival
Background
With incidence of all types of cancers on the rise, cancer is now the second leading cause of death worldwide [1]. As reported, although incidence rates of all cancers are almost 2 times higher in developed countries than in non-developed ones, mortality rates in developed countries are only 8% to 15% higher [2]. It is suggested that this difference in cancers is affected by detection methods and the availability of distinct therapeutic strategies.
Recent studies have shown that long non-coding RNAs (lncRNAs) greater than 200nt play important physiologic roles in cancers and normal tissue development [3]. Some lncRNAs have emerged as new members in cancer research and function as tumor suppressor genes, oncogenes, or both [4]. A nuclear lncRNA, metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1), also referred to as nuclear-enriched abundant transcript 2 (NEAT2), mascRNA, HCN, PRO2853, LINC00047, or NCRNA00047, encoded on chromosome 11q13, has recently emerged as a new focus in cancer research [5,6]. Several studies have shown that MALAT-1 is involved in cell cycle progression and tumorigenesis with a pro-proliferative function in regulating E2R1 and p53 [7,8]. The overexpression of MALAT-1 has been confirmed in several types of solid tumors, including glioma, lung, colorectal, bladder, laryngeal, pancreatic, gastric cancer, hepatocellular carcinoma, and multiple myeloma [9–17]. However, the effect of MALAT-1 on the outcome of cancer patients has been controversial, and no meta-analysis has been conducted on the correlation of MALAT-1 with the survival of cancer patients. Thus, the present meta-analysis is the first to evaluate the association between MALAT-1 expression and prognosis of all well-studied cancers.
Material and Methods
Literature search strategy
Studies in English were searched in PubMed, Web of Science, EMBASE, Science Direct, Wiley Online Library, Ovid, Cochrane Central Register of Controlled Trials, LILACS, and Google Scholar. Studies in Chinese were identified through CNKI, WanFang, ChongQing VIP, and China Biology Medicine disc. The search strategy was (“MALAT-1” or “MALAT-1” or “Metastasis-associated lung adenocarcinoma transcript” or “Nuclear-Enriched Abundant Transcript 2” or “NEAT2” or “mascRNA” or “HCN” or “PRO2853” or “LINC00047” or “NCRNA00047”) and (“prognos*” or “surviv*” or “follow-up” or “mortality” or “predict” or “outcome”). References of retrieved papers and reviews, as well as conference reports, were also read to identify additional relevant studies. The last update of searching time was June 30, 2015.
Selection criteria
Titles and abstracts of studies were checked first by 4 authors (Ruixue Tang, Mengtong Jiang, Lu Liang, and Dandan Xiong), and a study was extracted if it tested the expression of MALAT-1 in cancer tissues and analyzed the relationship between the MALAT-1 expression and survival of patients. After the duplicates were removed and the titles and abstracts were checked, eligible papers were analyzed with full-text if they met the following inclusion criteria: (1) The expression level of MALAT-1 was examined in cancer tissues; (2) MALAT-1 expression level was comparatively analyzed with patient survival time; (3) Studies were written in English or Chinese; (4) HR for survival time was reported or could be calculated from the reported data; and (5) The detection methods of MALAT-1 were restricted to reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) and fluorescence in-situ hybridization (FISH). Papers not fitting the above inclusion criteria, reviews, animal studies, or case report were excluded. If research on the same cohort was published in more than 1 study, only the most recent was selected. Differences among authors were resolved by deliberation with a fifth investigator (Gang Chen).
Data extraction
Data extraction from eligible studies was performed independently by 4 authors (Ruixue Tang, Mengtong Jiang, Lu Liang, and Dandan Xiong). The following data were extracted from eligible studies: first author’s name; study publication year; regions of study; language of study; sample size; cancer type; follow-up time; method of MALAT-1 testing; percentage of MALAT-1-positive cases; survival results (overall survival [OS], disease-free survival [DFS], progression-free survival [PFS]), cut-off value, HR, and 95% CI. The cut-off value depended on MALAT-1/GAPDH ratio or median/mean value of MALAT-1 levels, which was determined by authors of the original study. Differences of opinion were discussed with a fifth investigator (Gang Chen) in conference.
Quality assessment
The quality of eligible papers was assessed by Newcastle–Ottawa quality assessment scale (NOS). Assessed items included selection, outcome and comparability, with a score range of 0–9. Four reviewers (Ruixue Tang, Mengtong Jiang, Lu Liang, and Dandan Xiong) evaluated each paper independently and then compared the results. Disagreements about quality were resolved with a fifth investigator (Yiwu Dang) via discussion.
Statistical analysis
HR and its 95% CI were used to indicate the effect of MALAT-1 expression on the survival of cancer patients. HR and its 95% CI were extracted directly if they were reported in a study, and if not, they were calculated with available data, such as the exact patient numbers and Kaplan-Meier (K-M) curve by the method from Parmar et al. [18]. An HR>1 implied that MALAT-1 had a negative effect on survival outcome and a 95% CI of HR not overlapping 1 was considered statistically significant.
Statistical heterogeneity among the studies was tested by inconsistency (I2) and Q test (chi-squared test; χ2). If heterogeneity was significant (Q test p<0.05, I2>50%), the random-effects model was used to estimate the pooled HR, and if not, the fixed-effects model was used.
Publication bias was tested with Begg’s and Egger’s tests [19,20]. STATA 12.0 (STATA Corp., College Station, TX) was used to perform statistical analysis. All p values were 2-sided, and p<0.05 was considered as statistically significant.
Results
Characteristics of Included Studies
As shown in the flow diagram (Figure 1), 590 studies were found in 13 databases, including PubMed, Web of Science, EMBASE, Science Direct, Wiley Online Library, Ovid, Cochrane Central Register of Controlled Trials, LILACS and Google Scholar, CNKI, WanFang, ChongQing VIP, and China Biology Medicine disc. Thirty-six duplicated papers were removed. After the titles and abstracts were reviewed, 534 irrelevant articles were excluded. Then, after a more cautious consideration with full texts, 5 ineligible papers were eliminated due to the different methods of expression detection (only RT-qPCR and FISH were allowed), insufficient survival data, or language other than English and Chinese. Thus, 15 eligible studies remained. Additionally, 2 qualified research datasets from our own group were accepted into the analysis, which were named as A1 and A2. Finally, 17 studies [9,10,14–17,21–29] were included in the present meta-analysis. The principle characteristics of included studies are displayed in Table 1 and details about NOS score of eligible studies are shown in Table 2.
Figure 1.
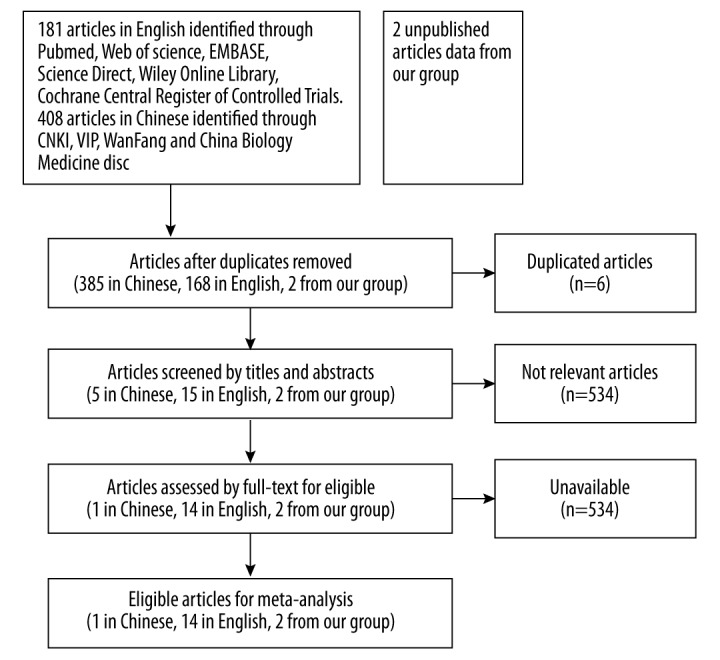
The flow diagram of the meta-analysis.
Table 1.
Characteristics of patients in different studies.
| Author | Year | Country | Sample size | Cancer type | TNM stage (I/II vs. III/IV) | Follow-up (mon) | Cutoff (high/low) | Method | Variance Analysis | Outcomes | HR statistics | Quality score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hirata | 2015 | Japan | 50 | CCRCC | NR | 47 (total) | NR | RT-qPCR | Multivariate | OS | Survival curve | 6 |
| Zhang | 2014 | China | 106 | CCRCC | 62/55 | NR | 46/60 | RT-qPCR | Univariate | OS | Reported | 6 |
| Shen | 2014 | China | 78 | NSCLC | NR | NR | 24/54 | RT-qPCR | Univariate | DFS | Survival curve | 6 |
| Ji | 2003 | Germany | 70 | NSCLC | NR | NR | 28/22 | RT-qPCR | Univariate | OS | Survival curve | 6 |
| Schmidt | 2011 | Germany | 222 | NSCLC | 184/38 | 56 (total) | 83/139 | RT-qPCR | Univariate | OS | Reported | 6 |
| Mu | 2013 | China | 76 | NSCLC | 29/15 | 80 (total) | 32/44 | RT-PCR | Multivariate analysis | OS | Reported | 5 |
| Pang | 2014 | China | 126 | PC | 41/85 | 60 (total) | 63/63 | RT-qPCR | Univariate | OS | Reported | 7 |
| Liu | 2014 | China | 45 | PC | 24/21 | 47 (total) | 26/19 | RT-qPCR | Multivariate | DFS | Reported | 7 |
| Lai | 2012 | China | 60 | HCC | NR | 18.6 (median) | 33/27 | RT-qPCR | Multivariate | OS | Reported | 6 |
| Fan | 2014 | China | 95 | BC | NR | NR | 45/50 | RT-qPCR | Multivariate | OS | Survival curve | 6 |
| Okugawa | 2014 | Japan | 150 | GC | 48/102 | 79 (total | 88/62 | RT-qPCR | Univariate | OS | Reported | 6 |
| Zheng | 2014 | China | 146 | CC | 23/91 | 56.2 (median) | 73/73 | RT-qPCR | Multivariate | OS DFS | Reported | 6 |
| Ma | 2014 | China | 118 | Glioma | NR | NR | NR | RT-qPCR | Univariate | OS | Reported | 6 |
| Cho | 2014 | China | 45 | MM | 17/19 | 48 (total) | 20/16 | RT-qPCR | Multivariate | OS PFS | Survival curve | 7 |
| Dong | 2014 | China | 19 | Osteosar-coma | NR | 60 (total) | 14/5 | RT-qPCR | Univariate | OS | Data in paper | 6 |
| A1 | 2015 | China | 125 | NSCLC | 54/71 | 13 (median) | 66/59 | RT-qPCR | Univariate | OS | Original data | |
| A2 | 2015 | China | 95 | HCC | 22/73 | 32.8 (median) | 52/43 | RT-qPCR | Univariate | PFS | Original data |
Table 2.
Details about NOS score of eligible studies.
| NOS author | Hirata | Zhang | Shen | Ji | Schmidt | Mu | Pang | liu | Lai | Fan | Okugawa | Zheng | Ma | Cho | Dong |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Is the case definition adequate? | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Representativeness of the cases | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Selection of controls | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Definition of controls | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Comparability of cases and controls on the basis of the design or analysis | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Ascertainment of exposure | 1 | 1 | 1 | 1 | 1 | – | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 |
| Non-response rate | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Total score | 6 | 6 | 6 | 6 | 6 | 5 | 7 | 7 | 6 | 6 | 6 | 6 | 6 | 7 | 6 |
In this meta-analysis, all essential data were collected from the 17 studies, including a total sample of 1626 patients from China (13), Germany (2), and Japan (2). Thirteen studies were published in English, and 1 was in Chinese. The 2 datasets (A1 and A2) were on file from our research group. All of the studies involved the prognostic value of MALAT-1 on survival with 10 classes of cancer: non-small cell lung cancer (NSCLC) (5) [10,25,27,28], clear cell renal cell carcinoma (CCRCC) (2) [14,22], hepatocellular carcinoma (HCC) (2) [16], pancreatic cancer (PC) (2) [9,23], gastric cancer (GC) (1) [15], colorectal cancer (CC) (1) [26], bladder cancer (BC) (1) [21], multiple myeloma (MM) (1) [17], glioma (1) [24], and osteosarcoma (1) [29]. RT-qPCR or FISH was applied for MALAT-1 level evaluation in various tissue samples. Among the 17 studies, 9 directly reported HRs, 5 showed the survival curves, and 3 (Dong et al. [29] and A1 and A2) were calculated from original data. For prognostic indicators (OS, DFS, PFS), 1 [26] provided OS and DFS, and 1 [17] had OS and PFS. The other 15 studies only showed single indicators (OS or DFS or PFS).
Meta-analysis between MALAT-1 and survival in 10 malignancies
Based on different prognostic indicators, all the studies were divided into 3 groups to avoid data reuse: OS (14) [9,10,15–17, 21,22,24,26–29], DFS (3) [23,25,26], and PFS group (2) [17].
In the OS group, 14 studies including 1378 cases assessed the effect of up-expression of MALAT-1 on OS (HR=1.84, 95% CI: 1.27–2.67; random-effects model), suggesting that high expression of MALAT-1 was a predicator of poor prognosis among the included cancers. Due to the existence of heterogeneity (I2=80.3%, p<0.001), subgroups were analyzed based on country, type of cancer, sample number, and analysis methods with the random-effects model. A significant relationship was observed between overexpressed MALAT-1 and poor OS in the studies in Germany (HR=1.75, 95% CI: 1.08–2.82) and China (HR=1.85, 95% CI: 1.14–2.99), but not in Japan (HR=2.34, 95% CI: 0.49–11.22). In addition, the heterogeneity only presented in the subgroup of China (I2=85%, p<0.001). In various types of cancers, the relevance between MALAT-1 and poor OS was only found in CCRCC (HR=3.76, 95% CI: 1.84–7.67), and there was no heterogeneity (I2=0%, p=0.466). With 100 patients as a threshold, the sample size was divided into 2 categories. The negative effect of overexpressed MALAT-1 in predicting poor prognosis was shown in studies with larger patient sample sizes (HR=2.38, 95% CI: 1.81–3.13) with no heterogeneity (I2=31.4%, p=0.200). In different analysis methods, the association of MALAT-1 with OS was found in univariate analysis (HR=1.78, 95% CI: 1.16–2.73), and heterogeneity existed (I2=83.6%, p<0.001) (Table 3).
Table 3.
Meta-analysis results of subgroups in the OS group.
| Subgroup | No. of study | HR (95%CI) | Heterogeneity | ||
|---|---|---|---|---|---|
| I2 | P | ||||
| OS | 14 | 1.84 (1.27–2.67) | 80.3% | <0.001 | |
| Country | Germany | 2 | 1.75 (1.08–2.82) | 0.0% | 0.779 |
| China | 10 | 1.85 (1.14–2.99) | 85.0% | <0.001 | |
| Japan | 2 | 2.34 (0.49–11.22) | 39.2% | 0.200 | |
| Type of Cancers | CCRCC | 2 | 3.76 (1.84–7.67) | 0.0% | 0.466 |
| NSCLC | 5 | 1.08 (0.71–1.65) | 63.1% | 0.043 | |
| Sample Number | <100 | 8 | 1.25 (0.73–2.15) | 61.2% | 0.012 |
| >100 | 6 | 2.38 (1.81–3.13) | 31.4% | 0.200 | |
| Analysis Methods | Univariate | 8 | 1.78 (1.16–2.73) | 83.6% | <0.001 |
| Multivariate | 6 | 2.13 (0.83–5.44) | 75.2% | 0.001 | |
CCRCC – clear cell renal cell carcinoma; NSCLC – non-small cell lung cancer.
In the other 2 groups with DFS or PFS information, high level of MALAT-1 was observed to be a negative factor for DFS (HR=2.37, 95% CI: 1.55–3.62) with no heterogeneity (I2=0%, p=0.552, fixed-effects model) (Figure 2A), but not for PFS (HR=1.09, 95% CI: 0.57–2.06, fixed-effects model) (Figure 2B). Due to the limited number of studies, subgroup analysis for DFS and PFS were not performed.
Figure 2.
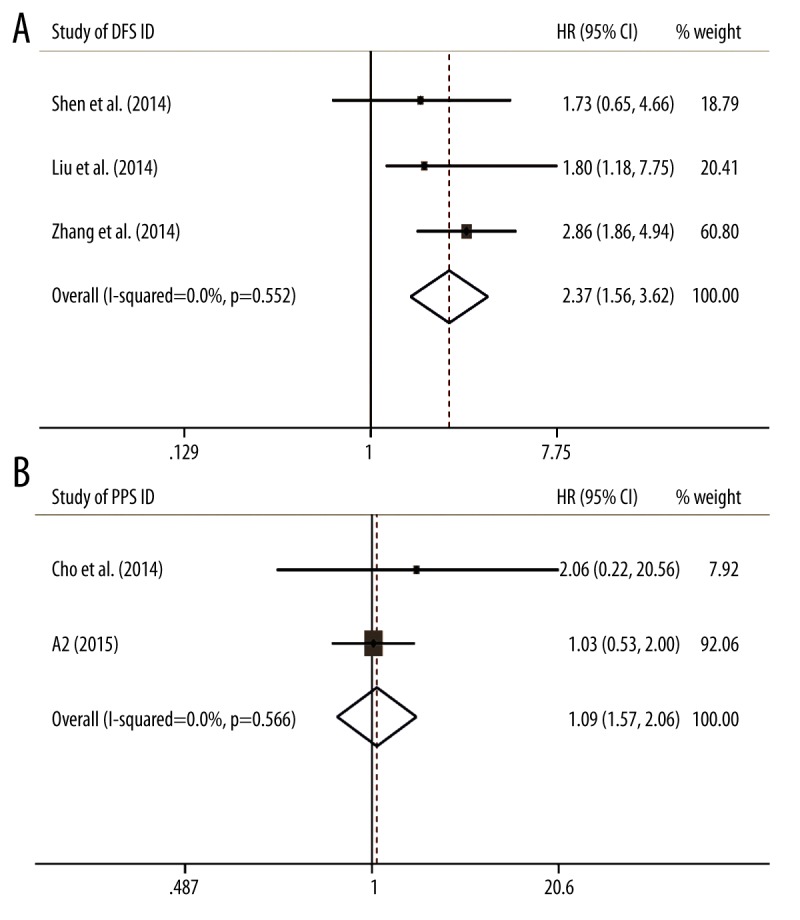
Meta-analysis of combined HRs with increased MALAT-1 expression. (A) The combined HRs by factor of DFS. (B) The combined HRs by factor of PFS.
Sensitivity analysis
Sensitivity analyses demonstrated that the studies by Mu et al. [28] and A1 were the top 2 with heterogeneity in the OS group, and their removal changed the results into more significant ones with no heterogeneity, such as for OS (HR=2.33, 95% CI: 1.79–3.04; I2=27.1%, p=0.178), country subgroup (China) (HR=2.73, 95% CI: 2.01–3.69; I2=18.3%, p=0.285), cancer subgroup (NSCLC) (HR=1.75, 95% CI: 1.08–2.82; I2=0%, p=0.779), smaller patient sample size (HR=1.88, 95% CI: 0.85–4.14; I2=35.4%, p=0.171), and analysis method subgroup (multivariate) (HR=3.34, 95% CI: 2.01–5.55; I2=0%, p=0.787).
Bias assessment
The Begg’s funnel plot and the Egger’s test were performed to assess publication bias. In the OS group, neither Begg’s funnel plot nor Egger’s test displayed obvious publication bias for the HR evaluations of OS (Begg’s test, Z=0.31, p=0.760; Egger’s test, t=−0.21, p=0.840; Figure 3). Due to the small number of studies, publication bias was not analyzed in the DFS and PFS groups.
Figure 3.
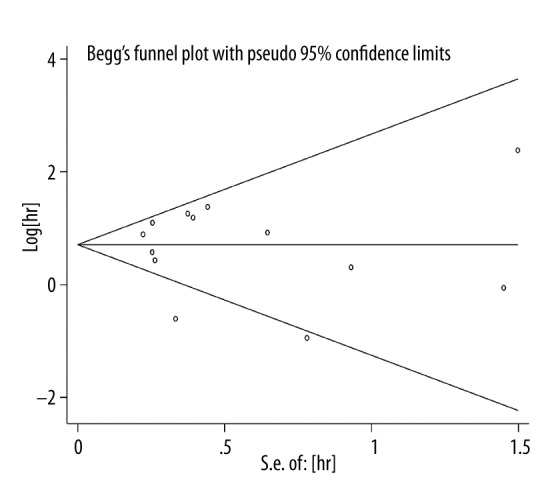
Begg’s funnel plot for the evaluation of potential publication bias for OS estimation.
Discussion
MALAT-1 was dysregulated in several tumors. However, to the best of our knowledge, no meta-analysis has been performed to evaluate MALAT-1 expression level as a prognostic indicator in published studies. Thus, we performed the first meta-analysis to explore the prognostic values of MALAT-1 in various tumors.
It is well known that invasion/metastasis of solid tumors is one of main causes of death for patients, which is an extremely complex process. Previously, some studies on aberrant expression of MALAT-1 of cancers have been published, revealing that the overexpressed MALAT-1 can promote cancer cell proliferation and migration, which suggests its important role in malignant tumor development [30]. Thus, it is important to find specific and sensitive biomarkers for early tumor prognostic prediction, for example: MALAT-1.
In hepatocellular carcinoma (HCC) [16] and multiple myeloma [17], MALAT1 is an independent prognostic factor for predicting disease progression. In CCRCC [22], it was reported that MALAT1 silencing decreased cell proliferation and invasion, and increased apoptosis of cancer cells. Similarly, cell proliferation and invasion were inhibited and metastasis was suppressed when MALAT-1was knocked down in osteosarcoma [29]. Based on these studies, MALAT1 may be a predicting factor for prognosis in general.
In the present meta-analysis, 17 studies comprising 1626 patients were screened by a series of selection criteria concerned with survival on the 10 classes of malignancies. The results demonstrated that overexpression of MALAT-1 was concerned with poorer survival in reported cancers. As aforementioned, studies were classified into 3 groups (OS group, DFS group, and PFS group) to avoid reusing data.
In the OS group, the combined HR was 1.84 (95% CI: 1.27–2.67). Among the subgroups, the followed results confirmed the significant negative role of overexpressed MALAT-1 on OS: country (Germany and China), type of cancer (CCRCC), sample size (>100 cases), and analysis method (univariate). On the contrary, there was no significant effect of MALAT-1 on OS in Japan, smaller sample size (<100 cases), or multivariate analyses. The statistical results revealed that the outcomes of MALAT-1 on cancer development might to some extent be influenced by racial or environmental factors. After the sensitivity analysis, 2 main sources of heterogeneity were found: Mu et al. [28] and A1. The existing heterogeneity might be related to the sample size and sample selection of these 2 studies. Additionally, MALAT-1 showed an obvious impact on poor OS, except in NSCLC. In 5 NSCLC studies, Mu et al. [28] reported that MALAT-1 was downregulated in tumor cells, and MALAT-1 was considered as a tumor suppressor. However, the other 4 studies regarded MALAT-1 as a tumor-promoting indicator. Due to the existence of different conclusions, we analyzed the data as comprehensively as possible in this meta-analysis.
Moreover, the overexpression of MALAT-1 was also highly associated with shorter survival in DFS. According to the results, MALAT1 can affect the progression and prognosis of different tumors in a variety of subgroups; however, the mechanisms remain unclear.
As a meta-analysis, the present study was limited in several aspects that should be further discussed. Firstly, potential biases might exist if negative results papers were less frequently published. Secondly, as studies included were published in Chinese and English only, the bias of selection and language might exist. Thirdly, the inaccuracy might add the potential bias when calculating HRs from the survival curves. Finally, the number of included studies and the total sample size were relatively small and 10 cancers cannot represent all types of malignancies. Therefore, our conclusions should be interpreted with caution.
Conclusions
In summary, despite of the limitations mentioned above, this meta-analysis for the first time demonstrates that overexpression of MALAT-1 is related with poor prognosis in malignant tumors. The expression of MALAT-1 might be a prognostic factor for the aforementioned cancers, especially NSCLC.
Footnotes
Conflicts of interest
The authors declare there are no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Source of support: The study was partly supported by the Fund of Guangxi Natural Scientific Research (No. 2014GXNSFBA118167) and the Fund of National Natural Science Foundation of China (NSFC 81360327). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript
References
- 1.Tanday S. Global cancer cases on the rise. Lancet Oncol. 2015;16(7):e317. doi: 10.1016/S1470-2045(15)00022-4. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Zhou S, Wang J, Zhang Z. An emerging understanding of long noncoding RNAs in kidney cancer. J Cancer Res Clin Oncol. 2014;140:1989–95. doi: 10.1007/s00432-014-1699-y. [DOI] [PubMed] [Google Scholar]
- 5.Ishizone S, Maruta F, Saito H, et al. Efficacy of S-1 for patients with peritoneal metastasis of gastric cancer. Chemotherapy. 2006;52:301–7. doi: 10.1159/000096002. [DOI] [PubMed] [Google Scholar]
- 6.Thomassen I, van Gestel YR, van Ramshorst B, et al. Peritoneal carcinomatosis of gastric origin: a population-based study on incidence, survival and risk factors. Int J Cancer. 2014;134:622–28. doi: 10.1002/ijc.28373. [DOI] [PubMed] [Google Scholar]
- 7.Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS genetics. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, Lin C, Liu W, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–88. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015;36:2403–7. doi: 10.1007/s13277-014-2850-8. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt LH, Spieker T, Koschmieder S, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–92. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 11.Xu C, Yang M, Tian J, et al. MALAT-1: a long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–75. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 12.Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–89. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malek R, Ajili F, Assaad-Khalil SH, et al. Similar glucose control with basal-bolus regimen of insulin detemir plus insulin aspart and thrice-daily biphasic insulin aspart 30 in insulin-naive patients with type 2 diabetes: Results of a 50-week randomized clinical trial of stepwise insulin intensification. Diabetes Metab. 2015;41:223–30. doi: 10.1016/j.diabet.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Zhang HM, Yang FQ, Chen SJ, et al. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36:2947–55. doi: 10.1007/s13277-014-2925-6. [DOI] [PubMed] [Google Scholar]
- 15.Okugawa Y, Toiyama Y, Hur K, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–39. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai MC, Yang Z, Zhou L, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Medical Oncol. 2012;29:1810–16. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 17.Cho SF, Chang YC, Chang CS, et al. MALAT1 long non-coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer. 2014;14:809. doi: 10.1186/1471-2407-14-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815–34. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 19.Peters JL, Sutton AJ, Jones DR, et al. Contour-enhanced meta-analysis funnel plots help distinguish publication bias from other causes of asymmetry. J Clin Epidemiol. 2008;61:991–96. doi: 10.1016/j.jclinepi.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan Y, Shen B, Tan M, et al. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20:1531–41. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 22.Hirata H, Hinoda Y, Shahryari V, et al. Long Noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–31. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu JH, Chen G, Dang YW, et al. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014;15:2971–77. doi: 10.7314/apjcp.2014.15.7.2971. [DOI] [PubMed] [Google Scholar]
- 24.Ma KX, Wang HJ, Li XR, et al. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 2015;36:3355–59. doi: 10.1007/s13277-014-2969-7. [DOI] [PubMed] [Google Scholar]
- 25.Shen L, Chen L, Wang Y, et al. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol. 2015;121:101–8. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 26.Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:3174–81. [PMC free article] [PubMed] [Google Scholar]
- 27.Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 28.Mu YY, Wang HJ, Zhu H, et al. Expression of the long noncoding MALAT-1 RNA in human lung cancer tissues and its clinical significance. Chinese Journal of Practical Medicine. 2013;40:1–4. [Google Scholar]
- 29.Dong YQ, Liang GJ, Yuan B, et al. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumor Biol. 2014;36:1477–86. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 30.Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med. 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]


