Highlight
Functional characterization of the three classical CHASE domain-containing receptors from Physcomitrella patens reveals their key role in the moss cytokinin response.
Key words: Bryophyte, cytokinin, cytokinin receptor, evolution, moss, Physcomitrella patens, phytohormone, plant growth regulator, signaling, two-component system.
Abstract
While the molecular basis for cytokinin action is quite well understood in flowering plants, little is known about the cytokinin signal transduction in early diverging land plants. The genome of the bryophyte Physcomitrella patens (Hedw.) B.S. encodes three classical cytokinin receptors, the CHASE domain-containing histidine kinases, CHK1, CHK2, and CHK3. In a complementation assay with protoplasts of receptor-deficient Arabidopsis thaliana as well as in cytokinin binding assays, we found evidence that CHK1 and CHK2 receptors can function in cytokinin perception. Using gene targeting, we generated a collection of CHK knockout mutants comprising single (Δchk1, Δchk2, Δchk3), double (Δchk1,2, Δchk1,3, Δchk2,3), and triple (Δchk1,2,3) mutants. Mutants were characterized for their cytokinin response and differentiation capacities. While the wild type did not grow on high doses of cytokinin (1 µM benzyladenine), the Δchk1,2,3 mutant exhibited normal protonema growth. Bud induction assays showed that all three cytokinin receptors contribute to the triggering of budding, albeit to different extents. Furthermore, while the triple mutant showed no response in this bioassay, the remaining mutants displayed budding responses in a diverse manner to different types and concentrations of cytokinins. Determination of cytokinin levels in mutants showed no drastic changes for any of the cytokinins; thus, in contrast to Arabidopsis, revealing only small impacts of cytokinin signaling on homeostasis. In summary, our study provides a first insight into the molecular action of cytokinin in an early diverging land plant and demonstrates that CHK receptors play an essential role in bud induction and gametophore development.
Introduction
Phytohormones regulate many processes in plants such as the development of tissues and organs and the response to changes in the environment. One class of phytohormones, the cytokinins, is comprised of adenine derivatives carrying an isoprenoid or an aromatic side chain at the N 6-position (Mok and Mok, 2001). Cytokinin signaling is mediated via a multistep His-to-Asp phosphorelay system, a variant of the bacterial two-component system (TCS). While this type of signaling system is widespread in prokaryotes, it is unique to plants among multicellular eukaryotes (Heyl and Schmülling, 2003). For Arabidopsis thaliana, the current model of this signaling pathway predicts that the cytokinin ligand is bound by hybrid histidine kinase receptors via the cyclases/histidine kinases associated sensory extracellular (CHASE) domain (Anantharaman and Aravind, 2001; Mougel and Zhulin, 2001; Heyl et al., 2007). These CHASE domain-containing histidine kinases (CHKs) were shown to localize mainly to the endoplasmic reticulum (ER) (Caesar et al., 2011; Lomin et al., 2011; Wulfetange et al., 2011). The binding of the cytokinin ligand causes an autophosphorylation of the CHK receptor. After an intramolecular phosphotransfer, the signal is transmitted by phosphorylation to histidine phosphotransmitter proteins (HPTs), which shuttle between the cytoplasm and the nucleus (Punwani et al., 2010). In the nucleus, the HPTs activate type-B response regulators (RRBs), transcriptional regulators belonging to the class of Myb transcription factors via phosphorylation. Subsequently these transcription factors initiate the transcription of their target genes, one group of which are the type-A RRs (RRAs). RRA proteins have been shown to be involved in a negative feedback mechanism of the cytokinin signaling pathway (Hwang and Sheen, 2001; To et al., 2004). Most of the research on this signaling pathway has been done using the model plant Arabidopsis, but work in other plants species also contributed to the elucidation of the functioning of the pathway (Heyl et al., 2006a; Hellmann et al., 2010).
One of the open questions in cytokinin biology is the origin and evolution of this regulatory system and its contribution to the conquest of land by plants. Nevertheless, our knowledge of the cytokinin biology of algae and early diverging land plants is very limited (Tarakhovskaya et al., 2007; Pils and Heyl, 2009; von Schwartzenberg, 2009; Frebort et al., 2011; Spichal, 2012; Gruhn and Heyl, 2013). The streptophyta alga Klebsormidium flaccidum was recently shown to code for all parts of the TCS system in the evolution of the green lineage prior to the conquest of land (Hori et al., 2014). The moss Physcomitrella patens as an early divirging land plant also encodes all protein families involved in cytokinin biosynthesis, metabolism, and signaling (Pils and Heyl, 2009; Frébort et al., 2011; Spíchal, 2012; Gruhn and Heyl, 2013; Gruhn et al., 2014). Due to its simple developmental differentiation and its responsiveness to several plant hormones, P. patens is a long-standing model regarding hormonal action and homeostasis (Wang et al., 1981; Cove, 2005; Decker et al., 2006; von Schwartzenberg, 2006, 2009). Twenty different endogenous cytokinins were detected and quantified in P. patens, and the generation of cytokinin-deficient plants revealed the importance of extracellular cytokinins for bud formation (von Schwartzenberg et al., 2007). Furthermore, the apparent absence of adenylate isopentenyltransferases (IPTs), the key enzymes for cytokinin production in flowering plants, makes P. patens an interesting organism for studying cytokinin biology in general (Yevdakova and von Schwartzenberg, 2007; Yevdakova et al., 2008; Frébort et al., 2011; Patil and Nicander, 2013; Lindner et al., 2014). While cytokinin metabolism has a long tradition as a topic in P. patens research (reviewed by von Schwartzenberg, 2009), the signaling of this phytohormone has only recently attracted the attention of researchers (Pils and Heyl, 2009; Ishida et al., 2010). Last year a new subfamily of cytokinin receptors was described containing eight members from P. patens (Gruhn et al., 2014). This discovery makes this moss the only plant which encodes both classical and newly identified cytokinin receptors in its genome, and it raises the question of the biological role of both receptor subfamilies in P. patens.
Here we present the characterization of the three classical CHASE domain-containing histidine kinase cytokinin receptors from P. patens. Following the suggested nomenclature (Heyl et al., 2013), we refer to them as CHK1, CHK2, and CHK3 and describe their role in differentiation processes of the moss. Our results show that the proteins can function as cytokinin receptors in different assays, and analysis of single, double, and the triple mutants demonstrated that CHK1, CHK2, and CHK3 are necessary for cytokinin perception by the moss. The results highlight the importance of these receptors for the cytokinin response in this early diverging land plant species.
Materials and methods
CHK full-length cDNAs
For the functional assays it was essential to isolate the respective cDNA clone for each of the three receptor genes (genomic loci: CHK1, Pp1s50_141; CHK2, Pp1s194_72; CHK3, Pp1s252_49; see http://www.cosmoss.org; Lang et al., 2005). By using degenerated AHK4 primers (degAHK4 for, gcnathgaycargaracnttygc; and degAHK4 rev, tgngcngtytgngcrtartc) on wild-type protonemal cDNA, two 942bp fragments (CHK1–942 and CHK2–942) were amplified, subcloned, and sequenced. To retrieve the CHK1 sequence (accession no. KJ697768, 3123bp), the CHK1–942 fragment was used as a probe to isolate CHK1 from a P. patens λZAPII cDNA library (Strepp et al., 1998), according to standard procedures. Full-length CHK2 (accession no. KJ697769, 3249bp) was achieved by RACE (rapid amplification of cDNA ends) (SMART 5′ and 3′ RACE cDNA amplification kit; Clontech) with gene-specific primers (cre2 5′ RACE, gcagtagacggcgaaggtgaaca; and cre2 3′ RACE, tgccgtcatagcgaagtctcagt) on Δchk1 cDNA. To retrieve CHK3 (accession no. KJ697770, 3306bp), specific primers (chk3 for, atgagacaaagaaaacagttgatcaatcc; and chk3 rev, attcgcctggaagaaatgctttgcaacc) were used to amplify and subclone CHK3 from cDNA derived from 4-week-old gametophores. Partial sequencing served to prepare a complete cDNA by commercial gene synthesis (GenScript, Piscataway, NJ, USA).
Cytokinin binding assay
The cytokinin binding assay was performed as has been described previously (Romanov and Lomin, 2009). In brief, the respective cytokinin receptor (AHK4, CHK1, CHK2, and CHK3) was cloned into the pDEST15 vector (Invitrogen, Karlsruhe, Germany) and expressed using the Escherichia coli strain BL21 (DE) pLys. The empty pDEST15 vector was used as a negative control. Tritium-labeled trans-[3H]zeatin (tZ; 592 GBq mmol–1) was obtained from the Isotope Laboratory of the Institute of Experimental Botany (Prague, Czech Republic).
In planta complementation assay
A protoplast transactivation assay (PTA) using protoplasts from the Arabidopsis ahk2,ahk3 double mutant was conducted as previously described (Choi et al., 2012). In brief, mesophyll protoplasts were isolated from 5- to 6-week-old Arabidopsis plants of the ahk2,ahk3 double cytokinin receptor knockout. The 350bp promoter fragment of the type-A response regulator ARR6 was used as a reporter construct and the type-B response regulator ARR2 as an effector. As an activator, the cDNAs of CHK1, CHK2, and CHK3 as well as that of AHK4 as a positive control were co-expressed with ARR2, respectively. The empty expression vector served as a negative control. The enzyme neuraminidase (NAN) was used as an internal control to standardize expression levels and to calculate relative expression levels (Kirby and Kavanagh, 2002). The details of the PTA protocol and the analysis of the results have been published previously (Ramireddy et al., 2013).
P. patens culture
The sequenced wild-type P. patens Hedw. Bruch & Schimp strain used in this study was collected from Gransden Wood, Huntingdonshire, UK in 1968 (Rensing et al., 2008). Standard growth conditions were 25 °C, in white light (100 µE m−2 s−1) for a light:dark cycle of 16:8h. For transformation and cytokinin profiling, liquid cultures were regularly disintegrated and grown in A′BCD(N)TV medium [0.356mM Ca(NO3)2, 1.01mM MgSO4, 1.84mM KH2PO4, 10mM KNO3, 0.044mM FeSO4 supplemented with Hoagland trace element solution (1ml l–1) and the vitamins nicotinic acid (8 µM), p-aminobenzoic acid (1.8 µM), and thiamine HCl (1.5 µM)] according to Wang et al. (1981). For phenotyping, budding assays, and quantitative real-time PCR, cultivation was performed on KNOP agar medium according to Hahn and Bopp (1968).
Generation of chk knockout mutants
A Δchk mutant collection comprising three single mutants, three double mutants, and one triple mutant was generated by sequential protoplast transformation with gene-disrupting vectors. The targeted loci, details on mutant generation [vector cloning, transformation protocol, and antibiotic selection Supplementary Table S1], as well as characterization [proof of insertion via PCR (see Supplementary Fig. S3) and absence of transcript (via RT–PCR (see Supplementary Fig. S4)], are given as the Supplementary data available at JXB online. For the Δchk1 mutants, a 300bp fragment has been deleted (scaffold_50:1326711..1326313), for Δchk2 77bp (scaffold_194:351,070..351,147), and for Δchk3 5016bp (scaffold_252:345,051..350,06 (5016bp).
Budding assay
Budding assays were performed as previously described by von Schwartzenberg et al. (2007) after cultivation of protonema for 10 d.
Cytokinin analysis by UHPLC-MS/MS
Liquid cultures of the wild type and the three double mutants (Δchk1,2; Δchk1,3; Δchk2,3) as well as the triple mutant were grown for 21 d and harvested as previously described (von Schwartzenberg et al., 2007). Three biological replicates were grown separately for the wild type and each mutant line. The extraction and purification was carried out in two technical replicates for each biological replicate. Samples (5mg DW) were homogenized under liquid nitrogen, extracted in modified Bieleski buffer (methanol/ water/formic acid, 15/4/1, v/v/v) (Novák et al., 2008), and then purified using two solid phase extraction columns, a C18 octadecylsilica-based column (500mg of sorbent, Applied Separations) and after that an MCX column (30mg of C18/SCX combined sorbent with cation-exchange properties, Waters) (Dobrev and Kaminek, 2002). Analytes were eluted by two-step elution using a 0.35M NH4OH aqueous solution and 0.35M NH4OH in 60% (v/v) MeOH solution. Cytokinin levels were determined using ultra high performance liquid chromatography-electrospray tandem mass spectrometry (UHPLC-MS/MS) with stable isotope-labeled internal standards as a reference (Svacinova et al., 2012).
RNA isolation and real-time PCR
RNA was extracted from the wild type as well as from the three double mutants and the triple mutant using the Trifast Reagent (Peqlab, Germany) according to the manufacturer. After DNaseI (Fermentas, Germany) treatment, cDNA was synthesized using peqGOLD M-MULV H plus (Peqlab). Real-time PCR was performed on a SteponePlus cycler (Applied Biosystems) using gene-specific primers and KAPA SYBR FAST Universal (Peqlab). Ribosomal protein L21 (Wang and Irving, 2011) was used as an endogenous control, and a primer efficiencies >95% were established for all targets (primers are given in Supplementary Table S2 at JXB online). Calculations were performed using the Stepone Software V. 2.3 with the ΔΔCt method.
Results
CHK1 and CHK2 bind trans-zeatin in an in vivo binding assay
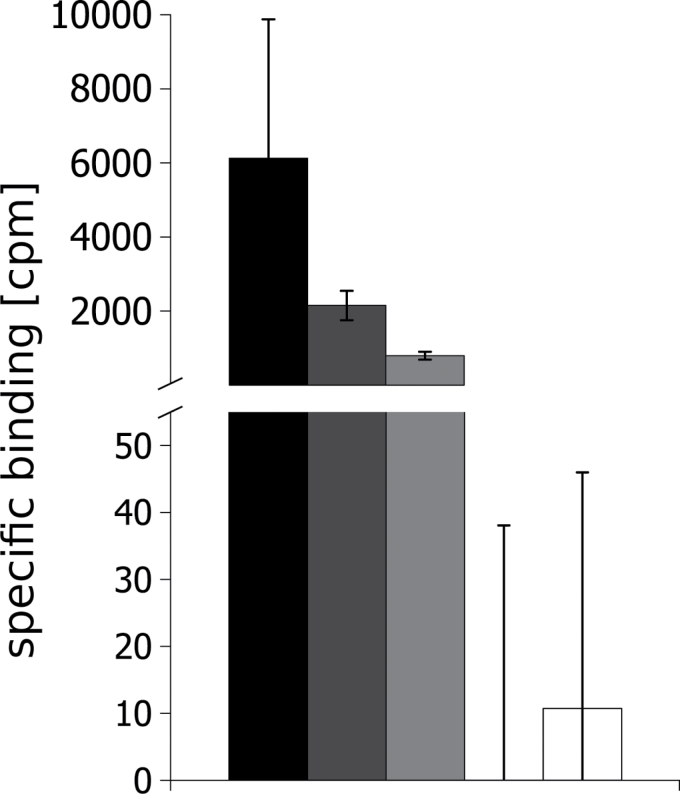
The sequences for the three CHK-coding sequences were retrieved by PCR cloning and submitted to the NCBI (CHK1, KJ697768; CHK2, KJ697769; and CHK3, KJ697770). In order to test the functionality of the three cytokinin receptors, we employed a cytokinin binding assay (Suzuki et al., 2001; Yamada et al., 2001; Romanov and Lomin, 2009). The cloned receptors and the respective controls were expressed in E. coli and the binding of radiolabeled tZ was tested. The assay was performed with AHK4 and the empty vector as positive and negative controls, respectively (Romanov et al., 2005; Heyl et al., 2007). CHK1 and CHK2 showed binding of tZ that was clearly above background (Fig. 1). Surprisingly, in this assay we did not detect any tZ binding for CHK3 although the protein was expressed in sufficient quantities (see Supplementary Fig. S1 at JXB online).
Fig. 1.
CHK1 and CHK2 bind tZ in an in vivo cytokinin binding assay. All receptors were expressed as GST fusion proteins in E. coli strain BL21 (DE) pLys. The specific binding to trans-[2-3H]zeatin was analyzed according to Romanov and Lomin (2009). Shown are biological replicates (n=3) and their SD (error bars). Expression of the different fusion proteins was confirmed by western blot (see Supplementary Fig. S1 at JXB online).
CHK1 and CHK2 function as cytokinin receptors in an in planta complementation assay
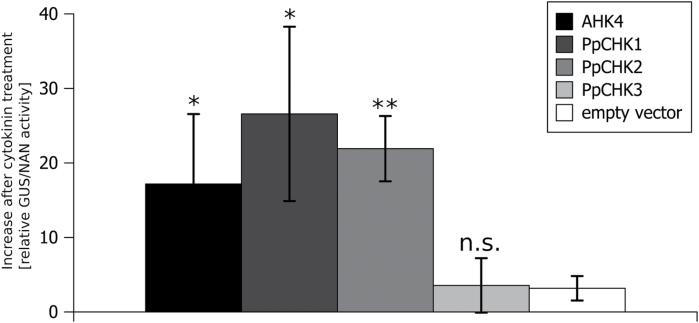
In order to test the in vivo functionality of the three cytokinin receptors, we employed an in planta complementation assay in which a candidate receptor is expressed in protoplasts from Arabidopsis plants in which two of the three cytokinin receptors are mutated (ahk2,ahk3) (Choi et al., 2012). The complementation was quantified using a β-glucuronidase (GUS) reporter gene. All genes were individually expressed under the control of the 35S promoter and the cells were treated with tZ. Using the cytokinin receptor AHK4 from Arabidopsis as a positive control, ahk2,ahk3 was complemented as described previously, while the empty vector as negative control showed only a weak activation of the reporter gene (Fig. 2). Of the three cytokinin receptors, only CHK1 and CHK2 showed a complementation in the double mutant. In fact, they complemented the ahk2,ahk3 double mutant even better than AHK4. However, no receptor activity was detected in this assay for CHK3 as compared with the controls (Fig. 2).
Fig. 2.
P. patens cytokinin receptors PpCHK1 and PpCHK2 activate the cytokinin-dependent TCS in ahk2-5,ahk3-7 double knockout mutant of Arabidopsis. Cytokinin perception-deficient protoplasts (ahk2-5,ahk3-7) (Riefler et al., 2006) were co-transformed with the cytokinin-responsive ARR2 (effector), the ARR6 promoter fused to β-glucuronidase (reporter), 35S::NAN (internal reference), and the indicated cytokinin receptor (activator) under the control of the 35S promoter. Protoplasts were incubated overnight with and without trans-zeatin; subsequently ARR6 promoter trans-activation was measured. Results were normalized by the internal reference, and the specific activity upon cytokinin treatment was calculated (normalized reporter activity with cytokinin minus normalized reporter activity without cytokinin). Depicted results are mean values of three biological replicates, and whiskers represent the SD (n=3, mean ±SD, t-test different from vector control, *P<0.05; **P<0.005; n.s., not significant).
Taking the results of the binding and complementation assay together, it was shown that at least two of the three classical CHK proteins fulfill the requirements to function as a cytokinin receptor.
Generation of Δchk knockout mutants
In order to characterize the in planta function of the CHK1, CHK2, and CHK3 receptors in P. patens, a mutant collection comprising single (Δchk1, Δchk2, and Δchk3), double (Δchk1,2; Δchk1,3; and Δchk2,3), and triple (Δchk1,2,3) mutants was generated by protoplast transformation using gene targeting constructs for each locus. Detailed information of the generation and characterization of this collection are provided in Supplementary Fig. S2 at JXB online. Each of the constructs harbored a different resistance cassette (for selection on G418, hygromycin B, and zeocin, respectively), thus enabling selection of plants with up to three CHK loci targeted. Mutants were analyzed by detailed PCR-based characterization of genomic DNA (see Supplementary Fig. S3) and cDNA (see Supplementary Fig. S4) which proved that (i) the respective CHK loci were targeted and (ii) the corresponding transcripts were no longer detectable. Furthermore, it was confirmed that the mutants had maintained the haploid status using flow cytometry (not shown).
Phenotype of CHK knockout mutants: protonema and gametophore development
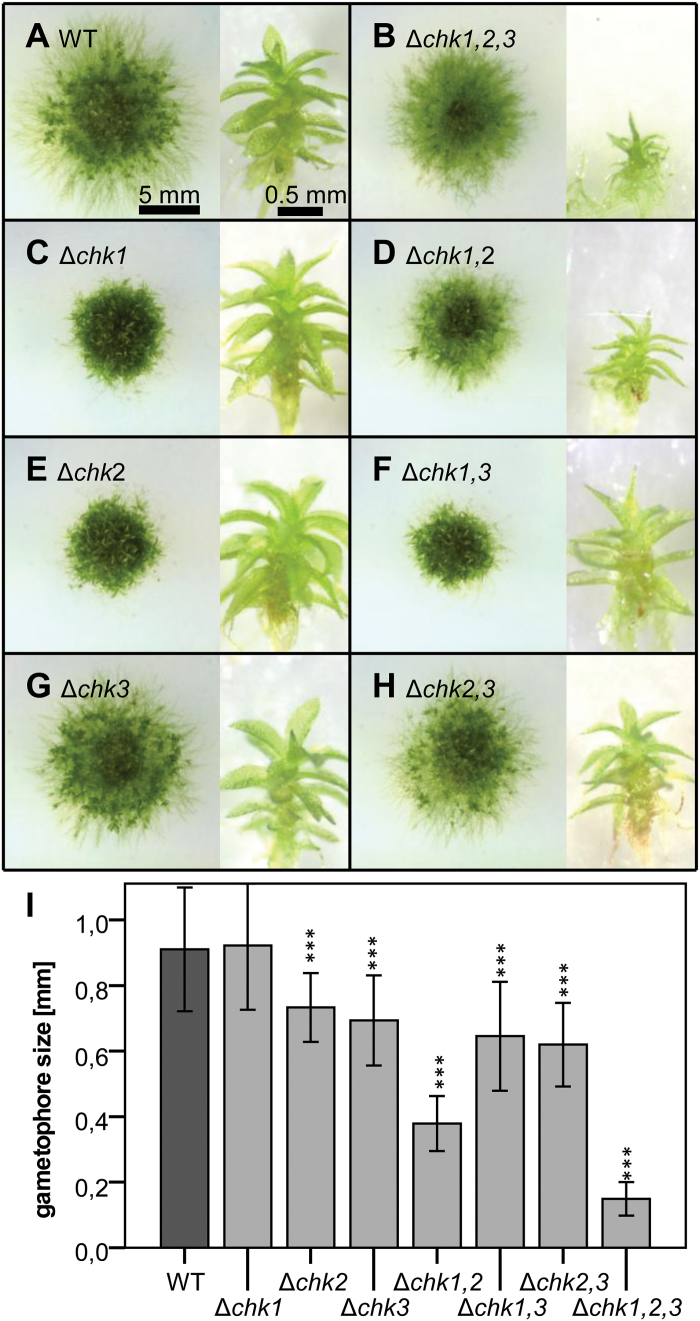
Knockout of a single receptor in Δchk1 or Δchk2 altered the growth morphology of moss grown on agar medium. Wild-type colonies showed a large area with undifferentiated protonema in the outer parts, and displayed bud and gametophore formation in the inner parts. The Δchk1 and Δchk2 single mutants had a smaller colony diameter and fewer protruding protonema. In contrast, Δchk3 did not exhibit a reduction of the colony size (Fig. 3C, E, G). For the double mutants, the colony size was most strongly reduced for Δchk1,3 (Fig. 3D, F, H). Detailed data on colony diameter over 6 weeks are given in Supplementary Fig. S5A at JXB online).
Fig. 3.
(A–H) Phenotypes of single, double, and triple mutants of CHK1, CHK2, and CHK3. Each panel shows moss colonies (7 weeks old) on the left and isolated gametophores (14 weeks old) on the right. Cultures were grown on KNOP agar medium. Corresponding pictures were taken at the same magnification; scale bars are given in (A). (I) Mean size of gametophores after 14 weeks [mean ±SD, t-test different from the wild type (WT), ***P<0.001].
While the gametophores formed by Δchk1 mutants had an average size comparable with the wild type, the size of gametophores of Δchk2 and Δchk3 was reduced. The size of the gametophores formed by Δchk1,2 was drastically reduced compared with the wild type and the single mutants (Fig. 3D). Gametophores of the Δchk1,3 and the Δchk2,3 mutants were smaller compared with the wild type, although they were larger than gametophores of Δchk1,2. This indicates the relevance of CHK1 and CHK2 for gametophore development. The development of gametophores in the double mutants occurred as for the wild type within 2 weeks of culture (Supplementary Fig. S5B at JXB online). All three CHK double mutants eventually developed antheridia and archegonia, produced a sporophyte after water-mediated fertilization, and finally completed the entire life cycle with the germination of haploid spores.
The triple receptor mutant Δchk1,2,3 showed a minor reduction in colony diameter compared with the wild type. However, the number of gametophores per colony was reduced as the colony consisted mainly of protonema. Gametophore formation was delayed by ~1 week (Supplementary Fig. S5B at JXB online). Furthermore, even after 14 weeks, the size of the gametophores was <20% of the size of the wild type, indicating the importance of all three receptors not only for bud initiation but also for gametophore development (Fig. 3B). This result functionally links two essential processes in the development of the moss, namely the onset of budding and gametophore formation, to the cytokinin receptors. Moreover, no antheridia and archegonia were observed for the triple mutants which arrested their life cycle at the gametophore stage, further highlighting the importance of the classical cytokinin receptors for the life cycle of P. patens.
CHK mutants display an altered cytokinin tolerance and response
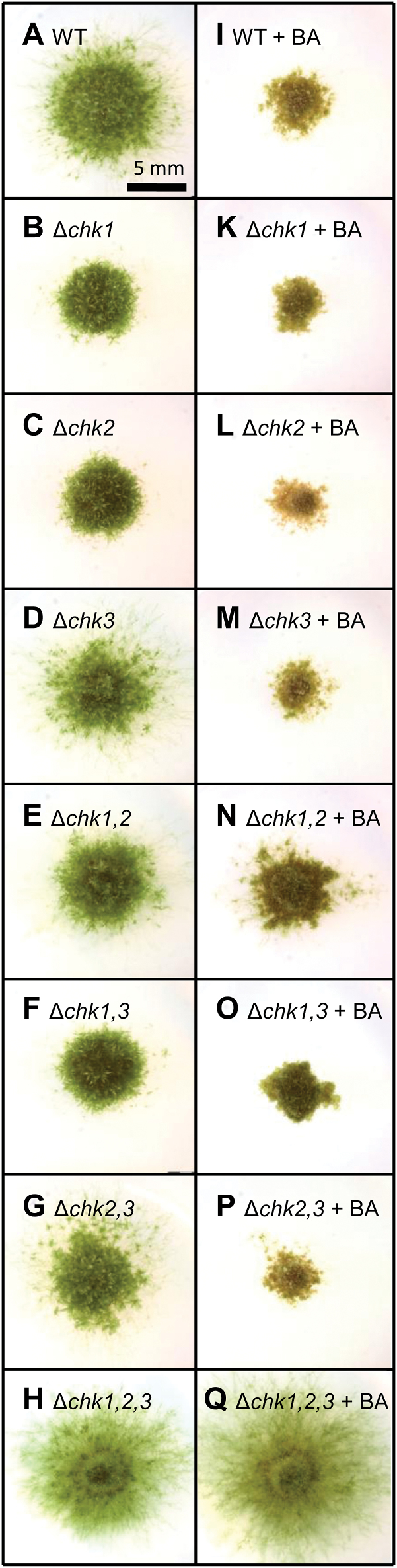
Next, we investigated the effect of cytokinin treatment on the different mutant lines in a cytokinin tolerance assay. Plants of the CHK mutant collection were inoculated on KNOP agar medium supplemented with 1 µM benzyladenine (BA), representing a concentration far beyond the range measured for endogenously produced cytokinins in P. patens (von Schwartzenberg et al., 2007). We have further chosen BA for this experiment as it is less prone to degradation by cytokinin oxidase/dehydrogenase compared with isoprenoid cytokinins (Avalbaev et al., 2012). At a concentration of 1 µM BA, the protonemal growth of the wild type was strongly inhibited, and malformed buds developed. In contrast, the high dose of BA did not lead to growth reduction and bud formation in the Δchk1,2,3 triple mutant, indicating a strong cytokinin insensitivity of this mutant (Fig. 4).
Fig. 4.
Tolerance of single, double, and triple mutants of CHK1, CHK2, and CHK3 to a high dose of cytokinin in comparison with the wild type. Seven-week-old cultures grown on KNOP agar plates. (A–H) No exogenous cytokinin, (I–Q) 1 µM benzyladenine (BA). All pictures are at the same scale; the scale bar is given in (A).
The exposure of the other members of the chk mutant collection to high doses of BA showed that the presence of a single CHK receptor is sufficient to confer sensitivity to an excess of cytokinin (Fig. 4). While all single and double mutant genotypes showed a brown or pale color, the protonema of the triple Δchk1,2,3 mutant was not visibly affected in pigmentation by the BA overdose. The tolerance assay showed that all three classical CHK receptors are involved in growth inhibition as well as in the formation of malformed buds, which are typical responses of P. patens to a high dose of cytokinin (von Schwartzenberg, 2009).
Budding bioassay reveals differences in the biological roles of CHK1, CHK2, and CHK3
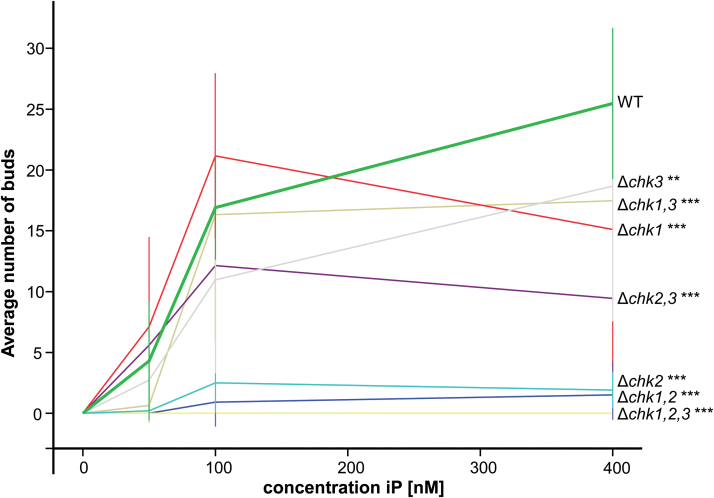
Cytokinins affect many aspects in the development of mosses (for a review, see von Schwartzenberg, 2009), with the induction of buds being the most striking. In order to establish whether one or more of the three CHKs under investigation are involved in this process, the CHK mutant collection was tested in a dose-dependent budding assay (Hahn and Bopp, 1968). The number of buds was counted after 10 d of growth on different concentrations of isopentenyladenine (iP; 50, 100, and 400nM). For the genotypes Δchk1, Δchk3, Δchk1,3, and Δchk2,3, a slightly reduced budding was observed (Fig. 5); however, the high variability of these bioassays results should be taken into account. For the genotypes Δchk2 and Δchk1,2 only minimal bud formation was recorded. These genotypes only responded to concentrations of iP >50nM (Supplementary Table S3 at JXB online), whereas in the wild type and most of the other mutants bud induction was already clearly detectable at 50nM iP. Strikingly, in this bioassay, the triple mutant Δchk1,2,3 did not exhibit any budding response.
Fig. 5.
Dose-dependent budding response to isopentenyladenine (iP) in the wild type (WT), and single, double, and triple Δchk mutants. Equal amounts of protonema were suspended on KNOP agar medium containing 0–400nM iP, and bud formation was analyzed microscopically after 10 d under standard conditions. The average number of buds corresponds to one microscopic view field (3.8mm2). At least two different biological replicates were counted in 5–10 view fields (mean ±SD, t-test different from the WT at 400nM, **P<0.01, ***P<0.001; the complete data set is given in Supplementary Table S3 at JXB online.
From the results for the different double mutant combinations tested in the budding assay it can be deduced that CHK1 and CHK2 alone are capable of mediating budding as a response to increased iP concentrations. The response to iP in the presence of CHK1 alone (in Δchk2,3) was slightly weaker than in the presence of CHK2 alone (in Δchk1,3). Only a very low budding response was observable in the double mutant Δchk1,2 mediated by CHK3 alone. Noticeably, the CHK3 receptor in the absence of CHK2 and CHK1 was insufficient to transduce the iP signal in order to result in significant budding (Fig. 5). In summary all three receptors participate in the budding response in this short-term assay. The absence of all three receptors leads to a complete lack of cytokinin-dependent bud induction, thus indicating an essential role for the CHKs in this developmental transition.
Differential budding in response to distinct cytokinins
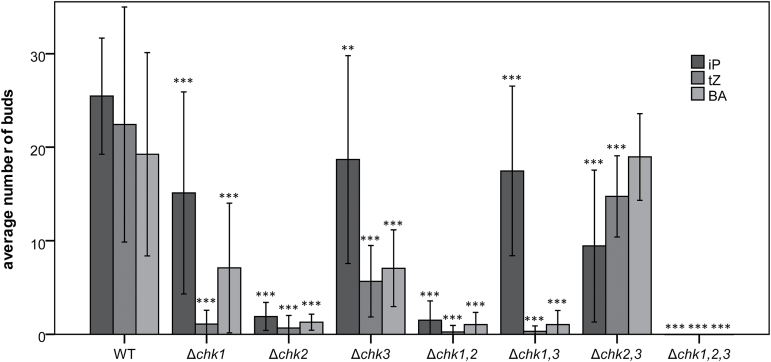
In order to investigate how the three CHK receptors differ in their response to different cytokinins, we performed the budding assay with the three double mutants as well as the triple mutant using the cytokinins iP, tZ, and BA (each at 400nM, Fig. 6) known to be the most active in this assay (von Schwartzenberg et al., 2007). As determined in the Δchk2,3 mutant background, the receptor CHK1 alone is capable of mediating a budding response to all applied cytokinins. In Δchk1,3, where only the CHK2 receptor is present, the budding response was high with iP but strongly impaired for tZ and BA—indicating a preference for iP. A strongly reduced response for all the three cytokinin bases was noted for the Δchk1,2 double mutant, indicating that CHK3 alone is not very active—at least in the protonemal stage. No budding response at all was found for the Δchk1,2,3 triple mutant no matter which of the three cytokinins was applied (Fig. 6).
Fig. 6.
Budding response of the wild type (WT), and double and triple Δchk mutants to iP, tZ, and BA. The budding response of the different genotypes was assessed after 10 d on KNOP agar medium supplemented with 400nM iP, tZ, and BA, respectively. Equal amounts of protonema were suspended on KNOP agar medium and bud formation was analyzed microscopically after 10 d under standard conditions. The number of buds corresponds to one microscopic view field (3.8mm2). At least two different biological replicates were counted in 5–10 view fields (mean ±SD, t-test different from the WT for each individual cytokinin, **P< 0.01, ***P<0,001); the complete data set is given in Supplementary Table S3 at JXB online).
Relative expression of CHK genes in Δchk mutant backgrounds
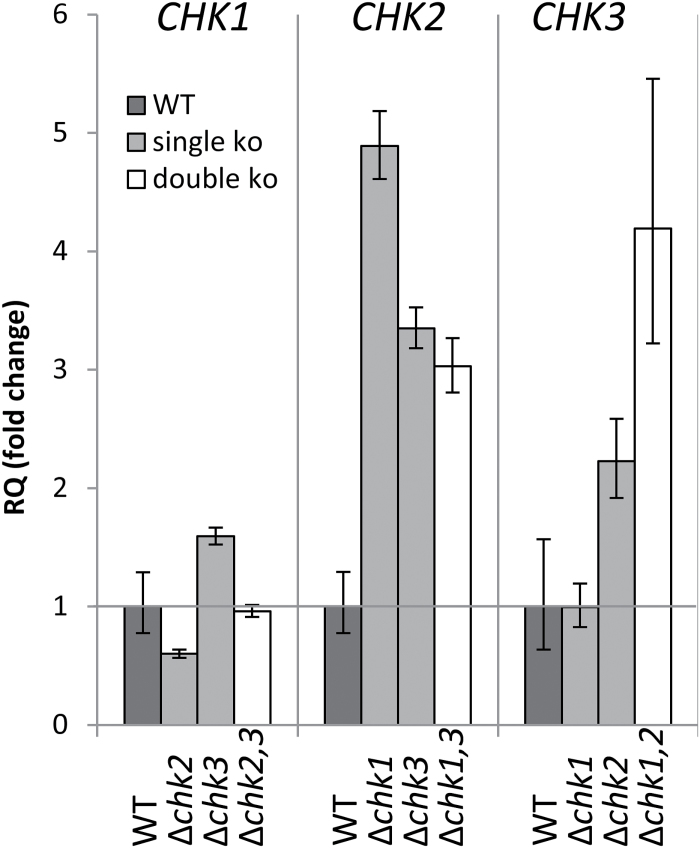
In order to investigate compensatory interaction between the different receptors on the transcript level, quantitative real-time-based analysis of CHK gene expression in the mutants and wild type was carried out. This analysis revealed that CHK1 expression is not or is only slightly affected by the knockout of CHK2, CHK3, or both receptors. However, expression of CHK2 seems to be 3- to 5-fold up-regulated in the single, and the double mutants of CHK1 and CHK3 compared with the expression level measured for the wild type. The expression of CHK3 was found to be up-regulated in Δchk2 and Δchk1,2, but not in Δchk1 (Fig. 7). Thus while the expression of CHK1 is quite stable regardless of the genetic background, the transcript level of both CHK2 and CHK3 increased in most receptor mutant backgrounds.
Fig. 7.
Relative expression of CHK1, CHK2, and chk3 in the respective single and double mutants. Three biological replicates were measured (7-day-old plants grown on KNOP agar, under standard conditions) with three technical replicates. 60S ribosomal protein L21 (Wang and Irving, 2011) served as the endogenous control, and gene expression was normalized to wild-type (WT) levels. Analysis of RQ (relative quantities) was performed with StepOne Plus software (Life Technonolgies®). Mean ±SD from technical replicates. t-test for differences from the WT for each target among biological replicates yielded no significant differences.
Cytokinin profiles of Δchk mutants
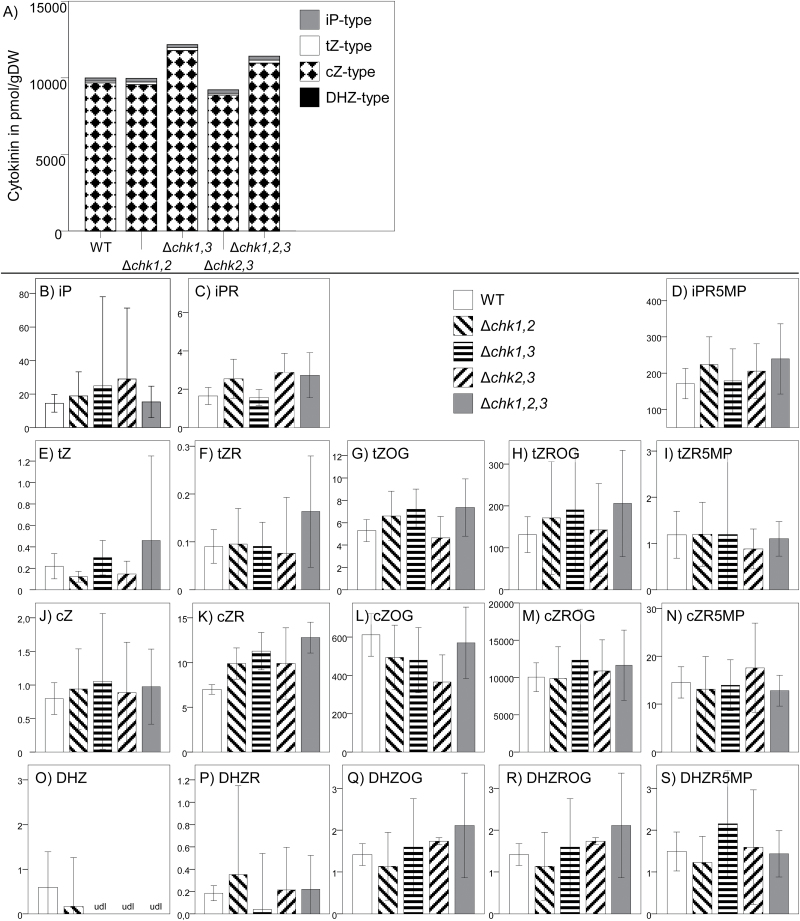
The transcriptional response of CHK2 and CHK3 to a receptor deficiency led to the question of whether there is a connection between cytokinin signaling and metabolism in P. patens. Previously, it has been shown in Arabidopsis that deficiencies in cytokinin receptors can result in changes of cytokinin homeostasis (Riefler et al., 2006). Thus we established the cytokinin profiles of the three double mutants (Δchk1,2, Δchk1,3, and Δchk2,3) and the triple mutant Δchk1,2,3 using UHPLC-MS/MS measurements and compared them with the profile of the wild type (Fig. 8). Each of the genotypes was cultured three times independently as a protonemal culture in liquid medium, and two technical replicates were made for each extract. Although there were individual changes, a general increase of all types of cytokinins, as described for Arabidopsis (Riefler et al., 2006), was not found for the moss mutants. cis-Zeatin riboside O-glucoside (cZROG), which is by far the most abundant cytokinin in P. patens (von Schwartzenberg et al., 2007), was only found at slightly higher levels in the Δchk1,3 mutant. However no significant changes in cytokinin levels were measured in the cytokinin receptor mutants when compared with the wild type.
Fig. 8.
Average level of isoprene-type cytokinins in tissue of P. patens wild type and the cytokinin receptor mutants Δchk1,2,3 (triple) and Δchk1,2; Δchk1,3 and Δchk2,3 (double). Each genotype was cultured three times independently for 21 d, sampled at three independent time points, and measured by UHPLC-MS/MS (n=9–24). Data present mean values with 95% confidence intervals. Cytokinin content in the mutants was compared with that in the wild type by independent samples Kruskal–Wallis test (confidence interval 95%, significance level 0.5); no significant changes compared in all samples with the wild type. Values are given in pmol g–1 DW. (A) Sum of all measured cytokinins. (B–S) Levels of each individual cytokinin; columns are the same as indicated in (A). tZ, trans-zeatin; tZOG, trans-zeatin O-glucoside; tZR, trans-zeatin riboside; tZRMP, trans-zeatin riboside-5′-monophosphate; tZROG, trans-zeatin riboside O-glucoside cZ, cis-zeatin; cZOG, cis-zeatin O-glucoside; cZR, cis-zeatin riboside; cZRMP, cis-zeatin riboside-5′-monophosphate; cZROG, cis-zeatin riboside O-glucoside; DHZ, dihydrozeatin; DHZOG, dihydrozeatin O-glucoside; DHZR, dihydrozeatin riboside; DHZRMP, dihydrozeatin riboside-5′-monophosphate; DHZROG, dihydrozeatin riboside O-glucoside; iP, N 6-isopentenyladenine; iPR, N 6-isopentenyladenosine; iPRMP, N 6-isopentenyladenosine-5′-monophosphate. udl, under the detection limit.
Discussion
The aim of this study was to characterize the properties and biological roles of the three classical cytokinin receptors of P. patens. In order to test the functionality of the three cloned receptors CHK1, CHK2, and CHK3, we used two well-established cytokinin receptor assays, the cytokinin binding assay and the in planta complementation assay (Mizuno and Yamashino, 2010; Choi et al., 2012). The assays confirmed the activity of CHK1 and CHK2 in hormone binding (Fig. 1), as well as their translation into downstream signaling, at least in Arabidopsis (Fig. 2). To our surprise, for CHK3 no activity was confirmed in either of these assays (Fig. 1, 2). However, this does not mean that the third receptor is not functional in P. patens. In fact the analysis of the knockout lines clearly shows that CHK3 has a role as a cytokinin receptor in the moss.
CHK receptors are functional in planta
To confirm the function of CHKs as cytokinin receptors in P. patens, different cytokinin-dependent assays using a collection of receptor knockout mutants were conducted. In flowering plants it is known that high concentrations of cytokinins can induce senescence and programmed cell death (Carimi et al., 2003; Vescovi et al., 2012). Our experiments confirmed the growth-inhibiting effect of 1 µM BA in P. patens (Thelander et al., 2005). We clearly demonstrated that this cytokinin-dependent growth inhibition is mediated via the CHK receptors, as the Δchk1,2,3 triple mutant was not affected by a high dose of BA. The cytokinin tolerance assay (Fig. 4) confirmed a role for CHK3 in cytokinin perception as the Δchk1,2 double mutant, with only CHK3 left as a functional receptor, was more affected by BA than plants with a simultaneous knockout of all three classical receptors (Δchk1,2,3). We deduce from the in planta experiments that CHK3 is capable of reacting to BA. The absence of a tZ-mediated response in the complementation and binding assays could be explained by either an incorrect protein processing in a heterologous system or a general low functionality of CHK3. While a tZ binding and a tZ-dependent response of CHK3 remains unclear, it can be stated that CHK3 mediates a clear iP response in the budding assay (Fig. 6) and a clear BA response in the tolerance assay (Fig. 4).
This experiment also indicates that apart from CHK1, CHK2, and CHK3, no other receptor is necessary to confer sensitivity to a cytokinin overdose, at least at this growth stage of the moss (Fig. 4).
A more detailed analysis using the well characterized cytokinin-dependent budding response (Hahn and Bopp, 1968) revealed that each CHK receptor, including CHK3, mediates a cytokinin response in planta as all three double mutants respond to cytokinins. CHK2 has a prominent role in this developmental process as all mutants in which this cytokinin receptor was missing showed a much weaker cytokinin response and (almost) no response to low levels of iP (50nM; Fig. 5; Supplementary Table S3 at JXB online) when compared with those missing CHK1 or CHK3. In contrast, the mutants Δchk1, Δchk3, Δchk1,3, and Δchk2,3 were less affected in budding frequency, also at high cytokinin concentrations (Fig. 5). In order to check if these effects depend on the type of cytokinin used, the assay was extended using three different cytokinins at 400nM (Fig. 6). These experiments showed weak budding when only CHK3 was present regardless of the cytokinin used, which indicates that this receptor is functional, but does not play a critical role for the transition from protonema to gametophore during the moss life cycle. In contrast, CHK1 responded to a broader cytokinin spectrum as all three tested cytokinins led to bud formation. Only CHK2 showed a preference for one particular cytokinin, iP, as this cytokinin had a far stronger bud-inducing effect than tZ or BA in the Δchk1,3 mutant (Fig. 6). We conclude from the in planta cytokinin response assays that all three receptors mediate a cytokinin-dependent signal independently from each other. Despite its responsiveness towards high doses of cytokinin, CHK3 seems to be of only minor importance for bud formation. CHK1 and CHK2 play a major role in triggering this developmental process, however with differences in the cytokinin preference.
The differences and redundancies among the investigated cytokinin receptors were further highlighted by the changes in their expression in the different mutant backgrounds. The peculiar finding of a higher sensitivity of the Δchk2,3 mutant compared with the Δchk2 mutant to iP can be explained by taking into account the expression level of CHK1. In Δchk2, but not in the Δchk2,3 mutant, the CHK1 level is reduced, which could contribute to the reduced sensitivity of Δchk2. However, in what way and to what extent the regulation and redundancies among the receptors are realized remain to be investigated. The CHK2 and CHK3 transcript levels were elevated in mutants still expressing the respective receptor. Thus it seems possible that the relatively small loss of bud formation observed in Δchk1,3 (Fig. 5) was partly due to an up-regulated CHK2 expression in this mutant. The expression levels of CHK1 either remained constant or increased slightly in Δchk2 mutants (Fig. 7).
P. patens development depends on functional CHK receptors
The phenotypic characterization of colony shape and gametophore size revealed differences between the mutants and wild type. Single mutants expressed minor phenotypic changes (reduced colony diameter of Δchk1 and Δchk2) or were even indistinguishable from the wild type (Δchk3), thus indicating again that CHK3 is not a major player for cytokinin perception in P. patens. The apparent minor role of CHK3 was further corroborated by the similar phenotype of the Δchk1,2 double mutant and the triple mutant Δchk1,2,3, which both exhibited reduced differentiation and a reduction of gametophore size. However, the CHK3 receptor possesses enough activity to provide basal functions in cytokinin activity as the Δchk1,2 double mutant like the Δchk1,3 and Δchk2,3 mutants, is able to undergo its entire life cycle including the formation of gametangia, sporophyte, and viable spores (not shown).
The importance of the cytokinin receptors for sexual reproduction was further emphasized by the inability of the triple mutant to form sporophytes. A similar phenotype was also observed when the cytokinin oxidase gene AtCKX2 was overexpressed in P. patens. These plants with a lowered content of cytokinins also showed a reduced budding response and absence of sporophyte formation (von Schwartzenberg et al., 2007). The life cycle of P. patens is apparently dependent on a functionality in the level of cytokinin homeostasis as well as of cytokinin perception.
A detailed developmental analysis of vegetative and generative stages in CHK mutants employing approaches such as presented by Coudert et al. (2015) and Landberg et al. (2013) is the subject of ongoing studies.
Classical CHKs play a key role in cytokinin perception and moss development
The experiments of this study clearly demonstrate the crucial role of CHK1, CHK2, and CHK3 for cytokinin perception and especially for the cytokinin-triggered formation of buds in moss. Recently, an additional subfamily of cytokinin receptors has been discovered (Gruhn et al., 2014). While sharing the same overall domain structure with the classical CHKs, their CHASE domain shows a lower conservation compared with the classical cytokinin receptors (Gruhn et al., 2014). One of the eight members of the new P. patens CHK subfamily, CHK4, has been characterized, and cytokinin binding and cytokinin-dependent activation of a two-component signaling chain was shown. The results of the analysis presented in this study of the classical Δchk mutants raise the question of the biological role of the receptors of the new subfamily. In particular, the facts that the triple mutant was completely resistant to the applied cytokinins in the tolerance assay and that bud formation was strongly delayed seem to indicate that these new receptors might not be critical in the cytokinin biology of the moss. However, one has to consider that budding is not the only developmental process regulated by cytokinin in P. patens and that small gametophores did form eventually in the Δchk1,2,3 triple mutant. Furthermore the transition from chloronema to caulonema, the formation of secondary chloronema, and the development of brachycytes and tnema cells, amongst other processes, are influenced by cytokinin (von Schwartzenberg, 2009). It will be interesting to test if those processes are also affected in the Δchk1,2,3 mutant or if the newly identified receptors function in one or more of these developmental processes. While the biological function of these ‘novel’ CHKs in not yet clear, their evolutionary origin is clearly different from those of the other cytokinin receptors (Gruhn et al., 2014). Interestingly, sequences from charophyceae algae [i.e. EST (expressed sequence tag) evidence from Spirogyra pratensis and the sequenced genome of Klebsormidium flaccidum] clustered between both clades of CHKs and those might be ancestral to both (Gruhn et al., 2014; Hori et al., 2014; E. Kaltenegger and A. Heyl, unpublished data). It is conceivable that they represent an ancestral type of cytokinin receptor that evolved into the classical CHK receptors found in land plants (e.g. PpCHK1, AtAHK4, and others).
Comparison of cytokinin signaling between P. patens and Arabidopsis
Given the importance of cytokinin as a regulator of plant growth and development (Hwang et al., 2012; Kieber and Schaller, 2014), it is surprising that A. thaliana is the only species in which cytokinin receptors have been analyzed systematically.
On the protein level, the classical CHK receptors from Arabidopsis and P. patens show a high degree of similarity with respect to their domain architecture of the whole protein and on the sequence level, also within the CHASE domain (Gruhn et al., 2014, 2015). These structural similarities also translate to functional similarities as we were able to complement ahk2,ahk3 deficiency in Arabidopsis protoplasts by transient expression of PpCHK1 and PpCHK2 (Fig. 1).
This study investigated the classical cytokinin receptors of the moss P. patens in detail, thus allowing phenotypic comparisons of cytokinin receptor mutants of an early diverging and a flowering plant. In both cases, the single mutants showed weak or no phenotypes, indicating a high level of redundancy among the receptors. While in Arabidopsis the simultaneous knockout of all cytokinin receptors (ahk2,ahk3,ahk4) leads to a severe dwarf plant (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006), the respective P. patens Δchk1,2,3 mutant showed a protonemal growth area that is comparable with that of the wild type. However, gametophores which appeared delayed in the Δchk1,2,3 triple mutant when compared with the wild type also exhibited a strong dwarf phenotype (Fig. 3, Supplementary Fig. S5B at JXB online). Both the Arabidopsis triple mutant shoot and the P. patens triple mutant gametophore are strongly impaired in growth and development. Further, both plants are highly resistant to exogenous cytokinin treatment. In both plant species, different mutant combinations show a distinct response to certain cytokinins, indicating different biological roles for the respective receptors (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006; Stolz et al., 2011). Despite the similarities in the cytokinin signaling mechanisms of flowering plants and bryophytes, we uncovered differences in the impacts of cytokinin signaling on the homeostasis of these hormones. In Arabidopsis, it has been shown that deficiency in cytokinin perception results in drastic changes of cytokinin homeostasis; notably, with increasing number of deleted receptors, a significantly increased concentration of numerous cytokinin species was found. In the Arabidopsis triple receptor mutant, there was, for example, a 15-fold increase of tZ compared with the wild type (Riefler et al., 2006). The levels of active cytokinins in P. patens [iP, iPR, tZ, tZR, and dihydrozeatin (DHZ) (von Schwartzenberg et al., 2007)] were not significantly altered in the analyzed Δchk mutants (Fig. 8). Thus the relatively small reduction in budding response for Δchk1, Δchk3, or Δchk1,3, for example (Fig. 5), cannot be explained by an increased production of active cytokinins to compensate the loss of function in the receptor system. In summary, in the moss P. patens—in contrast to flowering plants—there is only a minor contribution of the CHK1, CHK2, and CHK3 receptors to cytokinin homeostasis.
Conclusions
The study presented reveals that at the evolutionary stage of bryophytes, cytokinin signaling is fully established and uses classical receptors of the CHK gene family. The different experiments highlight the common and different properties of the receptors and their roles in developmental processes such as bud formation and gametophore development. The results of this study demonstrate the functionality of the classical PpCHK receptors, which are crucial for key steps in the life cycle of P. patens. Currently studies are under way to investigate the impact of the CHK receptors on multiple physiological and developmental aspects, and we expect that this and the presented research will contribute to the understanding of how hormonal regulation was established at the level of bryophytes.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Expression control of the fusion proteins analyzed in Fig. 1.
Supplementary Fig. S2. Overview of the chk knockout plants generated by transformation with the listed vectors.
Supplementary Fig. S3. PCR screening of knockout mutants.
Supplementary Fig. S4. RT–PCR screen of the different mutant lines.
Supplementary Fig. S5. (A) Average colony radius; (B) time course of gametophore frequency.
Supplementary Table S1. Generation of the chk mutant collection.
Supplementary Table S2. Primer sequences.Supplementary Table S3. Data for the budding assay (Figs 5, 6).
Acknowledgements
The authors thank Vera Schwekendiek (University of Hamburg) for excellent assistance related to moss culturing and bioassays, and Hana Martínková and Michaela Glosová (Palacký University, Olomouc) for sample purification and hormone analyses. The contributions of Florian Gelhaar and Niels Wegner (University of Hamburg) in an early phase of the project are acknowledged; furthermore, Frank Woithe (University of Hamburg) is acknowledged. Susanne Bringe (University Hamburg), who sadly passed away in 2014, deserves special recognition for her technical contributions to the project. NG and AH are grateful for the financial support of the Volkswagen Foundation to NG (Az I/83 477). AL appreciates support from the Estuary and Wetland Research Graduate School Hamburg (ESTRADE) as a member of the State Excellence Initiative (LExI) funded by the Hamburg Science and Research Foundation. JŠ, ON, and MS acknowledge the Ministry of Education, Youth and Sports of the Czech Republic (the National Program for Sustainability I, grant no. LO1204) and the Internal Grant Agency of Palacký University (IGA_PrF_2015_024 and IGA_PrF_2015_021).
References
- Anantharaman V, Aravind L. 2001. The CHASE domain: a predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends in Biochemical Science 26, 579–582. [DOI] [PubMed] [Google Scholar]
- Avalbaev AM, Somov KA, Yuldashev RA, Shakirova FM. 2012. Cytokinin oxidase is key enzyme of cytokinin degradation. Biochemistry (Moscow) 77, 1354–1361. [DOI] [PubMed] [Google Scholar]
- Caesar K, Thamm AM, Witthoft J, Elgass K, Huppenberger P, Grefen C, Horak J, Harter K. 2011. Evidence for the localization of the Arabidopsis cytokinin receptors AHK3 and AHK4 in the endoplasmic reticulum. Journal of Experimental Botany 62, 5571–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carimi F, Zottini M, Formentin E, Terzi M, Lo Schiavo F. 2003. Cytokinins: new apoptotic inducers in plants. Planta 216, 413–421. [DOI] [PubMed] [Google Scholar]
- Choi J, Lee J, Kim K, Cho M, Ryu H, An G, Hwang I. 2012. Functional identification of OsHk6 as a homotypic cytokinin receptor in rice with preferential affinity for iP. Plant and Cell Physiology 53, 1334–1343. [DOI] [PubMed] [Google Scholar]
- Coudert Y, Palubicki W, Ljung K, Novak O, Leyser O, Harrison CJ. 2015. Three ancient hormonal cues co-ordinate shoot branching in a moss. Elife 4, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove D. 2005. The moss Physcomitrella patens . Annual Review of Genetics 39, 339–358. [DOI] [PubMed] [Google Scholar]
- Decker EL, Frank W, Sarnighausen E, Reski R. 2006. Moss systems biology en route: phytohormones in Physcomitrella development. Plant Biology 8, 397–405. [DOI] [PubMed] [Google Scholar]
- Dobrev PI, Kaminek M. 2002. Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. Journal of Chromatography A 950, 21–29. [DOI] [PubMed] [Google Scholar]
- Frébort I, Kowalska M, Hluska T, Frébortova J, Galuszka P. 2011. Evolution of cytokinin biosynthesis and degradation. Journal of Experimental Botany 62, 2431–2452. [DOI] [PubMed] [Google Scholar]
- Gruhn N, Halawa M, Snel B, Seidl MF, Heyl A. 2014. A subfamily of putative cytokinin receptors is revealed by an analysis of the evolution of the two-component signaling system of plants. Plant Physiology 165, 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhn N, Heyl A. 2013. Updates on the model and the evolution of cytokinin signaling. Current Opinion in Plant Biology 16, 569–574. [DOI] [PubMed] [Google Scholar]
- Gruhn N, Seidl MF, Halawa M, Heyl A. 2015. Members of a recently discovered subfamily of cytokinin receptors display differences and similarities to their classical counterparts. Plant Signaling and Behavior 10, e984512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn H, Bopp M. 1968. A cytokinin test with high specificity. Planta 83, 115–118. [DOI] [PubMed] [Google Scholar]
- Hellmann E, Gruhn N, Heyl A. 2010. The more, the merrier: cytokinin signaling beyond Arabidopsis. Plant Signaling and Behavior 5, 1384–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Brault M, Frugier F, Kuderova A, Lindner AC, Motyka V, Rashotte AM, Schwartzenberg KV, Vankova R, Schaller GE. 2013. Nomenclature for members of the two-component signaling pathway of plants. Plant Physiology 161, 1063–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Schmülling T. 2003. Cytokinin signal perception and transduction. Current Opinion in Plant Biology 6, 480–488. [DOI] [PubMed] [Google Scholar]
- Heyl A, Werner T, Schmülling T. 2006. Cytokinin metabolism and signal transduction. In: Annual Plant Reviews Volume 24: Plant Hormone Signaling. Oxford: Blackwell Publishing, 93–123. [Google Scholar]
- Heyl A, Wulfetange K, Pils B, Nielsen N, Romanov GA, Schmülling T. 2007. Evolutionary proteomics identifies amino acids essential for ligand-binding of the cytokinin receptor CHASE domain. BMC Evolutionary Biology 7, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mahonen AP, et al. 2004. In planta functions of the Arabidopsis cytokinin receptor family. Proceedings of the National Academy of Sciences, USA 101, 8821–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, et al. 2014. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nature Communications 5, 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. 2001. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B. 2012. Cytokinin signaling networks. Annual Review of Plant Biology 63, 353–380. [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Nakanishi H, Mizuno T. 2010. Classification of the genes involved in the two-component system of the moss Physcomitrella patens . Bioscience, Biotechnology, and Biochemistry 74, 2542–2545. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE. 2014. Cytokinins. Arabidopsis Book 12, e0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby J, Kavanagh TA. 2002. NAN fusions: a synthetic sialidase reporter gene as a sensitive and versatile partner for GUS. The Plant Journal 32, 391–400. [DOI] [PubMed] [Google Scholar]
- Landberg K, Pederson ER, Viaene T, Bozorg B, Friml J, Jonsson H, Thelander M, Sundberg E. 2013. The moss Physcomitrella patens reproductive organ development is highly organized, affected by the two SHI/STY genes and by the level of active auxin in the SHI/STY expression domain. Plant Physiology 162, 1406–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Eisinger J, Reski R, Rensing SA. 2005. Representation and high-quality annotation of the Physcomitrella patens transcriptome demonstrates a high proportion of proteins involved in metabolism in mosses. Plant Biology 7, 238–250. [DOI] [PubMed] [Google Scholar]
- Lindner AC, Lang D, Seifert M, Podlesakova K, Novak O, Strnad M, Reski R, von Schwartzenberg K. 2014. Isopentenyltransferase-1 (IPT1) knockout in Physcomitrella together with phylogenetic analyses of IPTs provide insights into evolution of plant cytokinin biosynthesis. Journal of Experimental Botany 65, 2533–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomin SN, Yonekura-Sakakibara K, Romanov GA, Sakakibara H. 2011. Ligand-binding properties and subcellular localization of maize cytokinin receptors. Journal of Experimental Botany 62, 5149–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Yamashino T. 2010. Biochemical characterization of plant hormone cytokinin-receptor histidine kinases using microorganisms. Methods in Enzymology 471, 335–356. [DOI] [PubMed] [Google Scholar]
- Mok DWS, Mok MC. 2001. Cytokinin metabolism and action. Annual Review of Plant Physiology and Plant Molecular Biology 52, 89–118. [DOI] [PubMed] [Google Scholar]
- Mougel C, Zhulin IB. 2001. CHASE: an extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends in Biochemial Science 26, 582–584. [DOI] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. 2004. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. The Plant Cell 16, 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O, Hauserova E, Amakorová P, Doležal K, Strnad M. 2008. Cytokinin profiling in plant tissues using ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69, 2214–2224. [DOI] [PubMed] [Google Scholar]
- Patil G, Nicander B. 2013. Identification of two additional members of the tRNA isopentenyltransferase family in Physcomitrella patens . Plant Molecular Biology 82, 417–426. [DOI] [PubMed] [Google Scholar]
- Pils B, Heyl A. 2009. Unraveling the evolution of cytokinin signaling. Plant Physiology 151, 782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punwani JA, Hutchison CE, Schaller GE, Kieber JJ. 2010. The subcellular distribution of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin signaling. The Plant Journal 62, 473–482. [DOI] [PubMed] [Google Scholar]
- Ramireddy E, Brenner WG, Pfeifer A, Heyl A, Schmulling T. 2013. In planta analysis of a cis-regulatory cytokinin response motif in Arabidopsis and Identification of a novel enhancer sequence. Plant and Cell Physiology 54, 1079–1092. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, et al. 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. 2006. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, flowering size, germination, root development, and cytokinin metabolism. The Plant Cell 18, 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov GA, Lomin SN. 2009. Hormone-binding assay using living bacteria expressing eukaryotic receptors. Methods in Molecular Biology 495, 111–120. [DOI] [PubMed] [Google Scholar]
- Romanov GA, Spichal L, Lomin SN, Strnad M, Schmulling T. 2005. A live cell hormone-binding assay on transgenic bacteria expressing a eukaryotic receptor protein. Analytical Biochemistry 347, 129–134. [DOI] [PubMed] [Google Scholar]
- Spíchal L. 2012. Cytokinins—recent news and views of evolutionally old molecules. Functional Plant Biology 39, 267–284. [DOI] [PubMed] [Google Scholar]
- Stolz A, Riefler M, Lomin SN, Achazi K, Romanov GA, Schmülling T. 2011. The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. The Plant Journal 67, 157–168. [DOI] [PubMed] [Google Scholar]
- Strepp R, Scholz S, Kruse S, Speth V, Reski R. 1998. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proceedings of the National Academy of Sciences, USA 95, 4368–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T. 2001. The Arabidopsis sensor His-kinase, AHk4, can respond to cytokinins. Plant and Cell Physiology 42, 107–113. [DOI] [PubMed] [Google Scholar]
- Svacinova J, Novak O, Plackova L, Lenobel R, Holik J, Strnad M, Dolezal K. 2012. A new approach for cytokinin isolation from Arabidopsis tissues using miniaturized purification: pipette tip solid-phase extraction. Plant Methods 8, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarakhovskaya ER, Maslov Yu I, Shishova MF. 2007. Phytohormones in algae. Russian Journal of Plant Physiology 54, 163–170. [Google Scholar]
- Thelander M, Olsson T, Ronne H. 2005. Effect of the energy supply on filamentous growth and development in Physcomitrella patens . Journal of Experimental Botany 56, 653–662. [DOI] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. 2004. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. The Plant Cell 16, 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vescovi M, Riefler M, Gessuti M, Novák O, Schmülling T, Lo Schiavo F. 2012. Programmed cell death induced by high levels of cytokinin in Arabidopsis cultured cells is mediated by the cytokinin receptor CRE1/AHK4. Journal of Experimental Botany 63, 2825–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwartzenberg K. 2006. Moss biology and phytohormones—cytokinins in Physcomitrella . Plant biol 8, 382–388. [DOI] [PubMed] [Google Scholar]
- von Schwartzenberg K. 2009. Hormonal regulation of development by auxin and cytokinin in moss. In: Knight C, Perroud P-F, Cove D, eds. Annual Plant Reviews: The Moss Physcomitrella patens , Vol. 36 Wiley-Blackwell, 246–281. [Google Scholar]
- von Schwartzenberg K, Fernandez Núnez MF, Blaschke H, Dobrev PI, Novák O, Motyka V, Strnad M. 2007. Cytokinins in the bryophyte Physcomitrella patens: analyses of activity, distribution, and cytokinin oxidase/dehydrogenase overexpression reveal the role of extracellular cytokinins. Plant Physiology 145, 786–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TL, Beutelmann P, Cove DJ. 1981. Cytokinin biosynthesis in mutants of the moss Physcomitrella patens . Plant Physiology 68, 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Irving HR. 2011. Developing a model of plant hormone interactions. Plant Signaling and Behavior 6, 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfetange K, Lomin SN, Romanov GA, Stolz A, Heyl A, Schmülling T. 2011. The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiology 156, 1808–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T. 2001. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant and Cell Physiology 42, 1017–1023. [DOI] [PubMed] [Google Scholar]
- Yevdakova NA, Motyka V, Malbeck J, Trávnícková A, Novák O, Strnad M, von Schwartzenberg K. 2008. Evidence for importance of tRNA-dependent cytokinin biosynthetic pathway in the moss Physcomitrella patens . Journal of Plant Growth Regulation 27, 271–281. [Google Scholar]
- Yevdakova NA, von Schwartzenberg K. 2007. Characterisation of a prokaryote-type tRNA-isopentenyltransferase gene from the moss Physcomitrella patens . Planta 226, 683–695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.