Highlight
Expression of cell wall-related genes marks the onset of haustorium development in the parasitic plant Cuscuta. Action assays suggest a central role for xyloglucan endotransglucosylases/hydrolases in host plant infection.
Key words: Cell wall, Cuscuta, haustorial gene expression, haustorium development, parasitic plant, xyloglucan endotransglucosylase/hydrolase (XTH).
Abstract
Changes in cell walls have been previously observed in the mature infection organ, or haustorium, of the parasitic angiosperm Cuscuta, but are not equally well charted in young haustoria. In this study, we focused on the molecular processes in the early stages of developing haustoria; that is, before the parasite engages in a physiological contact with its host. We describe first the identification of differentially expressed genes in young haustoria whose development was induced by far-red light and tactile stimuli in the absence of a host plant by suppression subtractive hybridization. To improve sequence information and to aid in the identification of the obtained candidates, reference transcriptomes derived from two species of Cuscuta, C. gronovii and C. reflexa, were generated. Subsequent quantitative gene expression analysis with different tissues of C. reflexa revealed that among the genes that were up-regulated in young haustoria, two xyloglucan endotransglucosylase/hydrolase (XTH) genes were highly expressed almost exclusively at the onset of haustorium development. The same expression pattern was also found for the closest XTH homologues from C. gronovii. In situ assays for XTH-specific action suggested that xyloglucan endotransglucosylation was most pronounced in the cell walls of the swelling area of the haustorium facing the host plant, but was also detectable in later stages of haustoriogenesis. We propose that xyloglucan remodelling by Cuscuta XTHs prepares the parasite for host infection and possibly aids the invasive growth of the haustorium.
Introduction
The genus Cuscuta of the Convolvulaceae family (Solanales) includes ~200 species of thread-like parasitic plants with worldwide distribution (Dawson et al., 1994; Garcia et al., 2014). Being devoid of proper leaves and roots, and exhibiting very little to no photosynthetic activity, Cuscuta spp. are dependent on parasitizing a host plant to survive and reproduce. Host attachment and intrusion of stem and leaf tissue are mediated by specialized infection organs called haustoria that develop close to the apical stem tip. The initiation of haustorium differentiation is marked by site-specific cell elongation in areas where the parasite has contact with a host plant, and which is visible as a unilateral swelling of the parasite’s stem (Fig. 1). Subsequently, adhesive substances are secreted by the epidermis around the protruding haustorium (Vaughn, 2002), anchoring the parasite to the host and allowing the infection organ to grow into the host tissue using a combination of mechanical pressure and enzymatic digestion (Nagar et al., 1984; Johnsen et al., 2015; Kaiser et al., 2015) (Fig. 1). Upon reaching host xylem or phloem elements, searching hyphae emerge from the body of the haustorium and differentiate into these respective cell types, facilitating the transport of water and sugars from host to parasite (Vaughn, 2006). The successful connection to the host’s nutritional resources is visibly indicated by further swelling of the attached region, side shoot protrusion from the infection site, and restoration of apical tip growth, which typically ceases during the two previous stages (Fig. 1). Through the haustorium, a large variety of compounds are taken up by Cuscuta including small inorganic and organic molecules such as sugars, hormones, and amino acids, and macromolecules such as proteins and RNAs. These appear to serve both nutritional and regulatory purposes in this mutual interaction (Aly, 2013; Kim and Westwood, 2015). Having to share so many resources exposes the parasitized plants to a considerable amount of stress, and can result in reduced biomass or even host death in cases of severe attacks. In agricultural areas, the control of parasitic plants is difficult due to the physiological similarity and intimate connections of host and parasite. Thus, increased knowledge of the molecular mechanisms underlying haustorium development and host infection is required to develop effective strategies for combating the negative impacts of parasitic weeds (Aly, 2007; Alakonya et al., 2012; Jiang et al., 2013; Ichihashi et al., 2015). This is even more adamant when considering that many species of Cuscuta, in contrast to most other parasitic lineages, are generalists, being able to parasitize a large number of dicotyledonous plant species from different phylogenetic taxa. This variation in hosts can entail some molecular tuning of the infection process, for example by adapting the type of nutrient transfer cells in the mature haustoria to the type of host (Christensen et al., 2003). The onset of haustorium differentiation, however, does not seem to be influenced by different hosts, but rather depends on the presence of general signals initiating the infection process. The fact that the formation of Cuscuta haustoria can be stimulated in the absence of a host plant by applying a combination of far-red (FR) light and tactile stimuli corroborates this hypothesis (Tada et al., 1996). Haustoria induced in this way share morphological traits with haustoria developing in contact with a host plant, but have the advantage that the early stages of their development are more uniform and predictable than when induced by host attachment. In addition, in molecular studies, the consideration of RNA molecules transferred from the host can be neglected.
Fig. 1.
Cuscuta infecting the compatible host plant P. zonale. (A) C. reflexa and (B) C. gronovii parasitizing P. zonale. The infection of P. zonale by C. reflexa can be divided into three stages. (C and F) In the first swelling stage the parasite stem grows in width at the side facing the host (asterisk). (D and G) After attachment to the host surface (arrowhead in D), the haustorium (arrowhead in G) invades the host tissue (penetrating stage). (E and H) The mature stage is defined by established connections between the vascular systems of the two plants (arrowhead in H) and is recognized by apical shoot growth and the formation of additional side shoots (arrowheads in E). Cross-sections were made with respect to orientation of the parasite shoot axis in (F), and with respect to the host shoot axis in (G) and (H). Scale bars are 500 µm.
Cell wall polymer profiling in infecting and non-infecting tissues of Cuscuta recently revealed such substantial differences in cell wall composition (Johnsen et al., 2015) that the initiation of changes to the cell wall must be assumed to occur early in haustoriogenesis. In the present study, gene expression in haustoria and stems of two Cuscuta species, C. reflexa and C. gronovii, was therefore analysed with the goal of identifying cell wall-related genes that mark the onset and progress of haustorium development. Cuscuta reflexa, a member of the subgenus Monogynella, possesses thick and yellow to green stems, while C. gronovii has thinner and more delicate stems of yellow to orange colour (Fig. 1A, B) and belongs to the subgenus Grammica (Costea et al., 2015). The distinct expression of xyloglucan endotransglucosylases/hydrolases (XTHs) in young haustorial tissues of both C. reflexa and C. gronovii was further substantiated by protein immunolocalization and xyloglucan endotransglucosylation (XET) assays. The potential function of XTHs in Cuscuta haustorium development and host plant infection will be discussed.
Materials and methods
Plant material
Cuscuta reflexa and C. gronovii were propagated on the compatible host Pelargonium zonale (Fig. 1) in a greenhouse at the Phytotron of the University of Tromsø, Norway, in 24h of light at 21 °C. For RNA sequencing and for the induction of haustorium development with the FR light system, filaments of Cuscuta that were distal to the host infection sites were used. For the analysis of host-induced haustorial stages, infection sites on P. zonale were visually inspected for the characteristics of the different stages: the swelling stage before penetration of the host that was defined as the starting point of haustoriogenesis (Fig. 1C, 1F); the penetrating stage after initial intrusion into the host tissue but before the parasite is able to feed (Fig. 1D, 1G); and the mature feeding stage that is recognized by further swelling of the attached region, protruding side shoots from the infection site, and restoration of apical tip growth (Fig. 1E, 1H). Other samples encompassed stems at least 5cm distal to infection sites and to shoot tips, elongating stem regions (an area of 3–5cm below the apical tip), and shoot tips (first apical centimetre of shoot).
FR light induction of haustoriogenesis
Host-free induction of haustorium development was carried out as described by Tada et al. (1996) with minor modifications. To facilitate tactile stimuli, apical shoot tips of ~10cm (harvested from the propagating culture) were placed between two plastic Petri dishes (Ø=13.5cm) that were lightly pressed and held together using adhesive tape. The pressed shoot tips were then placed in an upright position and irradiated with FR light (740nm) for 1h. After light treatment, the shoots were kept in darkness before further analysis. Tissue for RNA isolation was harvested from stem segments with and without haustoria, 3 d or 6 d after FR light induction, as indicated.
RNA isolation
All plant material used for RNA isolation was snap-frozen in liquid nitrogen and homogenized using a TissueLyser (Qiagen, Hilden, Germany). Total RNA was isolated using a combination of the hot borate method (Wan and Wilkins, 1994), and phenol–chloroform extraction in which pre-warmed (65 °C) borate buffer (200mM Borax, 30mM EDTA, 1% (w/v) SDS) and phenol were added to the frozen plant material to make up the first liquid–liquid extraction. Subsequently, one extraction with phenol:chloroform:isoamylalco hol (25:24:1) and two with chloroform:isoamylalcohol (24:1) were executed before total RNA was precipitated in 2M LiCl at 4 °C overnight. In order to remove residual DNA, the RNA samples were treated with DNase using the DNA-free kit (Ambion Inc., Austin, TX, USA). Removal of DNA and integrity of RNA were checked by agarose gel electrophoresis.
Construction of suppression subtractive hybridization (SSH) libraries and sequencing of subtracted cDNA clones
The two cDNA libraries used for generating differentially expressed SSH libraries were synthesized from 250ng of DNase-treated total RNA isolated from FR light-induced haustoria or stems 3 d after light treatment (for each RNA isolation, material was pooled from six induced shoots) using the SMARTer Pico PCR cDNA Synthesis kit (Clontech, Mountain View, CA, USA). SSH was carried out with the PCR-Select cDNA Subtraction Kit (Clontech). The Advantage 2 PCR Kit (Clontech) was used for all PCR amplifications. After amplification, the differentially expressed cDNAs were cloned into the pGEM-T Easy vector system (Promega, Madison, WI, USA). One Shot TOP10 Chemically Competent Escherichia coli cells (Invitrogen, Carlsbad, CA, USA) were transformed with the SSH libraries and incubated on LB/Carbenicillin/X-Gal/IPTG plates at 37 °C for blue/white screening. All procedures were carried out according to the manufacturers’ instructions. Plasmid DNA was isolated from white colonies by alkaline lysis (Birnboim and Doly, 1979) and sequenced with M13F primer (5′ GTAAAACGACGGCCAGT 3′) by Sanger sequencing (Macrogen Korea, Seoul, Korea).
De novo transcriptome sequencing and assembly
Poly(A)+ RNA purification, reverse transcription, size fractionation, titration, and sequencing were performed at the Norwegian High Throughput Sequencing Centre (NSC; Oslo, Norway) on a Roche GS FLX (454) sequencer. One-eighth of a run for both libraries was done as a test run, followed by re-titration and a full run with the same libraries. Together, both runs yielded 945 454 raw reads for C. reflexa and 914 135 raw reads for C. gronovii. Raw data were then processed using the ngs_backbone v.1.1.0 pipeline (Blanca et al., 2011). After trimming, filtering, and quality assessment, Mira (version 3.2.0) (Chevreux et al., 1999) was used to assemble the reads into contigs. The applied job options were ‘denovo, est’. The reliability of the assemblies was confirmed with an independent assembly strategy using the SeqClean Software for cleaning and the TGICL pipeline for the assembly (DFCI Gene Indices Software Tools, ftp://occams.dfci.harvard.edu/pub/bio/tgi/software/) (data not shown). Sequences were filtered using the Rfam (Griffiths-Jones et al., 2005) and the SILVA (Quast et al., 2013) databases. This Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession numbers GDKE00000000 (C. gronovii) and GDKD00000000 (C. reflexa). The versions described herein are the first versions, GDKE01000000 (C. gronovii) and GDKD01000000 (C. reflexa).
Analysis of de novo transcriptomes and SSH clones
For functional annotation, the transcriptome contigs were analysed by sequence-based and domain-based alignments. Sequence-based alignments were performed with BLAST (blastx) (Altschul et al., 1990) against the non-redundant (nr) protein database at NCBI (http://www.ncbi.nlm.nih.gov/protein) and the UniProt protein databases Swiss-Prot and UniRef90 (http://www.uniprot.org/). The E-value thresholds were set to 1e-15. Conserved protein domains were searched by using HMMER tools (HMMER 3.1, http://hmmer.janelia.org/) with the PFAM database (PFAM 27.0; Finn et al., 2014). The online tool Mercator (Lohse et al., 2014) was used to map all contigs to functional modules defined by the plant-specific ontology and pathway tool MapMan (Usadel et al., 2005).
Sequences of the SSH clones were trimmed by deleting vector backbone sequences (pGEM-T Easy) and poly(A) tails before comparison with the C. reflexa transcriptome using BLAST (blastn; E-value: 1e-99). The sequences which did not give a match with any of the contigs in the C. reflexa reference collection were aligned in Geneious Pro 5.6.6 (Biomatters Ltd, Auckland, New Zealand) with ClustalW (Li, 2003). Functional annotation was again conducted with Mercator. The minimum BLAST bit score was set to 50.
Phylogenetic analysis of XTHs
A rooted Neighbor–Joining tree was generated with PhyML on a ClustalW protein alignment of all Arabidopsis thaliana XTHs identified in the UniProt database (UniProt-Consortium, 2015), the two C. reflexa XTHs (Cr-XTH-1 and Cr-XTH-2) and a previously identified XTH, LeXTH1 (alias XTH1_Sly) from tomato, Solanum lycopersicum (Albert et al., 2004). Solanum lycopersicum Expansin A23 served as the root. The Jones–Taylor–Thornton (JTT) model was used for amino acid substitutions, and the phylogram was optimized for substitution rates. One thousand bootstrap replications were conducted. All calculations were performed with Geneious Pro 5.6.6 (Biomatters Ltd).
Reverse transcription quantitative real-time PCR (RT-qPCR)
SuperScript II Reverse Transcriptase (Invitrogen) and anchored oligo(dT)18 primers were used to synthesize cDNA from 1 µg of DNase-treated total RNA. Controls without reverse transcriptase were carried out for each target gene in order to verify the complete absence of contaminating DNA. Quantitative real-time PCR was performed in technical duplicates using the SsoFast EvaGreen Supermix (Bio-Rad, Oslo, Norway) according to the manufacturer’s specifications. Thermal cycling and fluorescence detection was carried out using a CFX96 Real-Time PCR Detection System (Bio-Rad) with the following cycling conditions: 95 °C for 30s followed by 40 cycles of 95 °C for 5s and 61 °C for 5s. After 40 cycles, melt curves were recorded by stepwise heating from 65 °C to 95 °C. The amplification efficiencies were taken into account when calculating the relative abundances of each target gene (Pfaffl, 2001). Relative abundances of C. reflexa (Cr)-ACTIN and Cr-SF2 were used to normalize the expression levels between C. reflexa samples. Cuscuta gronovii (Cg)-ACTIN and Cg-SF2 were used as reference genes for C. gronovii. In samples where a target transcript could not be detected in any of the technical duplicates [i.e. no Cq (quantification cycle) value], a Cq value was assigned by adding one cycle to the highest Cq in that run. Data were analysed with the CFX Manager Software 2.0 (Bio-Rad). Gene-specific primer sequences with their respective amplicon sizes and PCR efficiencies are listed in Supplementary Table S1 available at JXB online. Amplicon melt curve analyses and size separation on agarose gels are presented in Supplementary Figs S1 and S2, respectively.
Immunolocalization of XTHs
Cross-sections (70 µm) of haustoria 3 d after FR light induction were prepared using a Leica VT1000E vibratome (Leica Biosystems, Nussloch GmbH, Nussloch, Germany). Free binding sites were blocked for 30min with 5% (w/v) non-fat milk powder in standard phosphate-buffered saline buffer (1× PBS) (blocking buffer). After washing with PBS, 1:20 dilutions of polyclonal anti-XTH rabbit IgGs or IgGs from the pre-immune serum (both provided by Dr E. Labrador and her group at the University of Salamanca, Spain) in blocking buffer were applied to the sections for 2h followed by three 5min washes with PBS. The sections were then incubated for 1h in the dark with Alexa Fluor 555 Goat Anti-Rabbit IgG (Life Technologies, Carlsbad, CA, USA) (1:200 in blocking buffer) and subsequently washed with PBS. Labelled sections were stained with toluidine blue O in order to quench autofluorescence. Micrographs were taken with a SteREO Lumar V12 equipped with an AxioCam MRc5 camera and the Lumar 43 filter set (all from Carl Zeiss, Jena, Germany).
XET action assays
XET test papers were prepared as described by Fry (1997). A piece of Whatman No. 1 filter paper was passed over the surface of 1% (w/v) Tamarind xyloglucan (Megazyme, Ireland) dissolved in 0.5% (w/v) chlorobutanol and left to dry. The dry xyloglucan-coated paper was dipped in 5 µM sulphorhodamine-labelled xyloglucan oligosaccharides (XyGO-SR) dissolved in 75% (v/v) acetone and dried before use. XyGO-SR were prepared by conjugation of xyloglucan oligosaccharides with sulphorhodamine carried out as described by Kosik and Farkas (2008). Xyloglucan-coated papers without XyGO-SR were used as control papers. Tissue prints were made by placing hand-cut 0.5–1.0mm thick pieces of Cuscuta cross-sections on XET test papers or control papers soaked in 50mM Na-acetate, 300mM NaCl (pH 5.5) followed by incubation between two sheets of acetate for 1h. The background on printed papers was removed by washing in water:ethanol:formic acid (1:1:1) for 2h with gentle agitation followed by rinsing in distilled water. Fluorescence micrographs were taken of dried de-stained papers using a SteREO Lumar V12 equipped as described above.
Results
Identification of haustorium-specific gene transcripts
Tada et al. (1996) reported that the formation of haustoria in C. japonica can be induced through the synergistic effect of FR light and tactile stimuli without the presence of a host plant. In the present study, the same signals were applied to activate haustorium differentiation in C. reflexa and C. gronovii. While host plant infection by Cuscuta progressed with varying speed, the development of FR light-induced haustoria proceeded in a very predictable manner: 3 d after treatment with FR light the parasite stem was noticeably swollen at one side, and after 6 d the haustorial body could be discerned (shown for C. reflexa in Fig. 2). Cross-sections of these young haustorial stages (Fig. 2C, D) resembled those during early host infection (Fig. 1F). The predictability of haustorial onset is, of course, a distinctive advantage when searching for developmentally regulated genes in the Cuscuta haustorium. Moreover, the intimate connection between parasite and infected host, and the translocation of transcripts from host to parasite through the haustorium (Leblanc et al., 2012; Kim et al., 2014), makes it difficult to avoid contamination of the RNA from haustorial infection sites with host-encoded transcripts. The concentration of host mRNA in Cuscuta is generally increased towards the host–parasite interface (Leblanc et al., 2013), which would lead to an enrichment of host transcripts during the SSH procedure and consequently mask the differences in parasitic gene expression (Diatchenko et al., 1996). Therefore, haustoria and stem tissue from FR light-induced shoot tips of C. reflexa were used for the generation of differentially expressed SSH libraries (Fig. 2E).
Fig. 2.
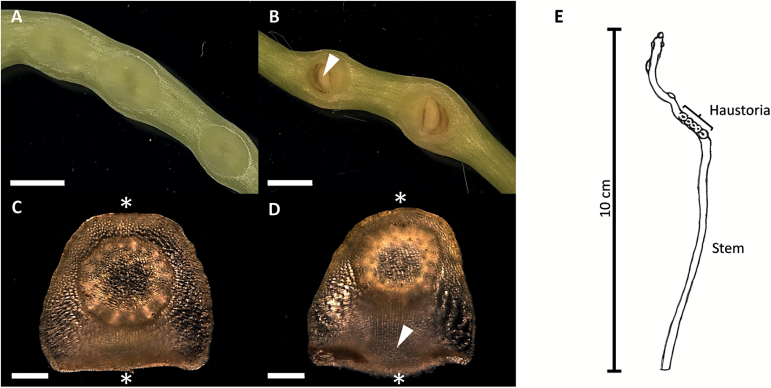
Haustorium development of FR light-induced C. reflexa shoots. (A and C) Initial stages of haustoriogenesis were visible 3 d after treatment with FR light. (B and D) After 6 d, the haustorial body (arrowheads in B and D) could be discerned. Asterisks indicate sites of contact with the plastic surface of the Petri dish. (E) Schematic overview of FR light-induced C. reflexa shoots indicating the young haustoria and stem areas used for SSH. Cross-sections were made with respect to the orientation of the parasite shoot axis. Scale bars are 2000 µm (A and B) and 500 µm (C and D).
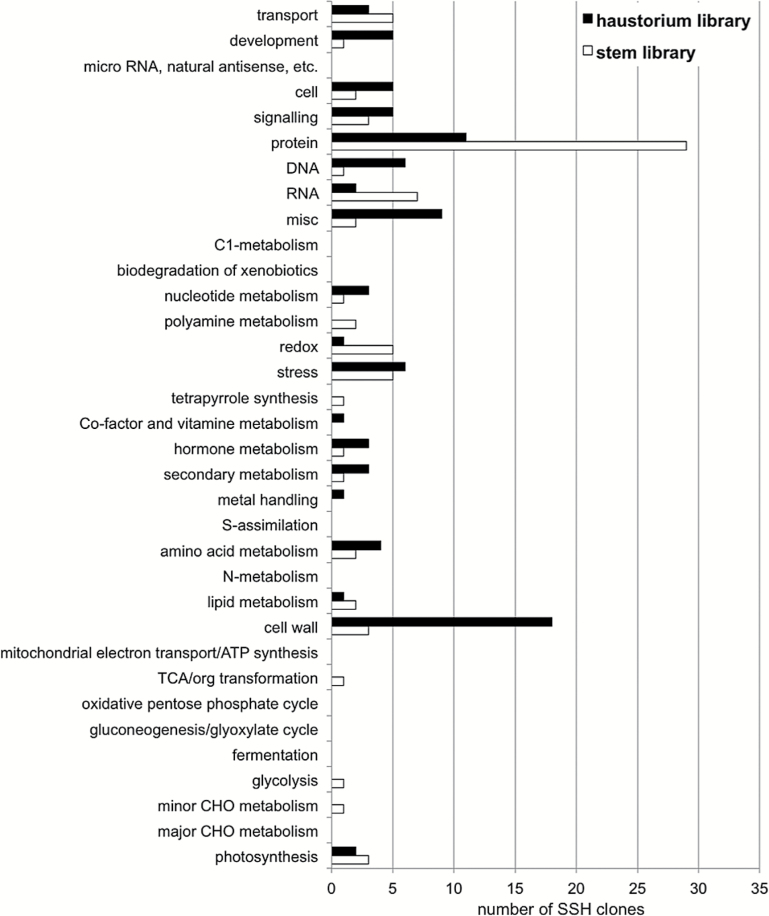
The SSH procedure is directional, only enriching transcripts that are present in higher abundance in one sample (tester) compared with another (driver). In this study, the SSH was carried out in both directions, leading to the construction of two libraries: the haustorium library enriched for sequences that are more abundant in young (3-day-old) haustoria than in the stem; and the reciprocal stem library enriched for sequences with higher expression in the stem tissue. Clones from both libraries were randomly picked and sequenced, yielding 182 and 179 readable sequences from the stem- and haustorium-specific libraries, respectively, which were subjected to an analysis by the functional annotation tool Mercator (Lohse et al., 2014). Mercator combines sequence similarity searches to a variety of plant genomes as well as information derived from InterProScan, KOG, and cdd searches, and assigns them to so-called bins representing different functional categories. Seventy-nine stem-specific sequences and 89 haustorium-specific sequences could be confidently assigned to a MapMan category. The quantitative distribution of assigned functions showed that particularly transcripts associated with the bins ‘cell wall’, ‘DNA’, ‘development’, and ‘misc’ were frequent in the haustorium-specific library but not in the stem-specific library (Fig. 3). The stem library, on the other hand, contained more transcripts for genes associated with the bins ‘protein’, ‘RNA’, and ‘redox’.
Fig. 3.
MapMan binning of C. reflexa SSH clones. The clones were mapped and functionally classified using the online tool Mercator. The minimum BLAST bit score was set to 50.
De novo sequencing of transcriptomes of two Cuscuta species
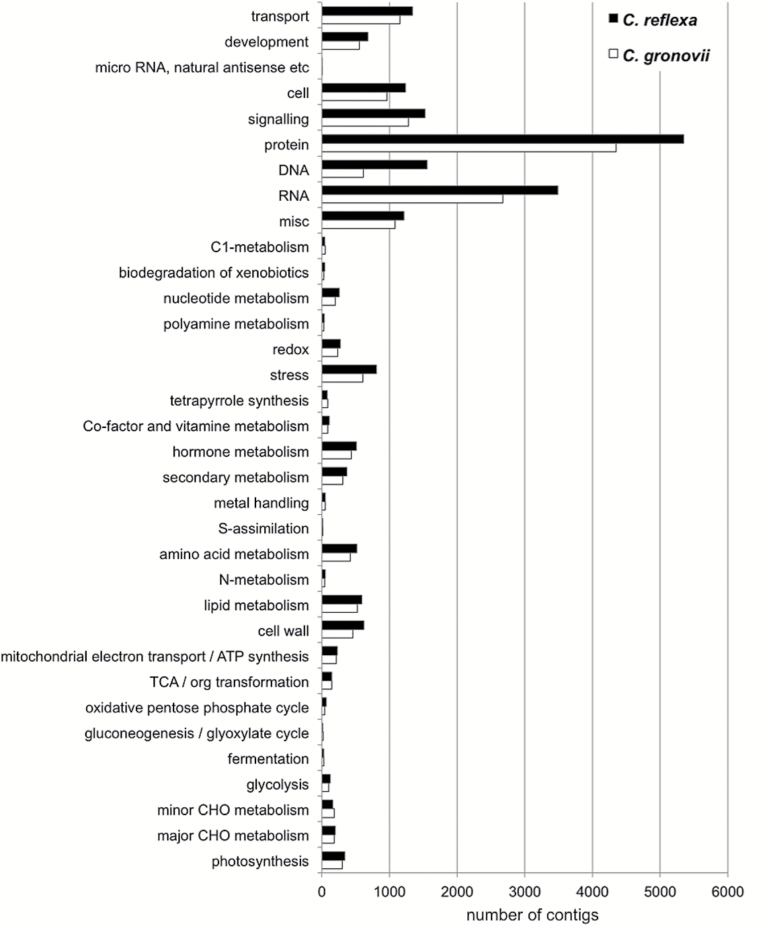
To create a ‘baseline’ for the relative sizes of MapMan categories in Cuscuta, the representation of MapMan bins in entire transcriptomes was investigated. For that purpose, 454-based de novo sequencing of the mRNAs from pooled vegetative tissue of C. reflexa and a second species, C. gronovii, was performed. These reference transcriptomes were also expected to improve the sequence information for the transcripts of interest obtained by the SSH screen. One and one-eighth 454 runs (see the Materials and methods) yielded just under 1×106 raw reads per species, which resulted in a total of 42 103 and 31 685 contigs for C. reflexa and C. gronovii, respectively (Table 1). Both the average and largest contig sizes were only marginally larger in C. reflexa than in C. gronovii. To predict the function of the Cuscuta contigs, their sequences were compared with different databases. The NCBI nr database, the UniProt Swiss-Prot and the UniRef90 databases individually yielded hits with up to 96% of the sequences for both species (Table 2). All approaches combined (i.e. nr, Swiss-Prot, and PFAM) allowed the annotation of 98% of the contigs of C. reflexa and of C. gronovii (Table 2).
Table 1.
Summary of 454 sequencing data and contig assembly
| C. gronovii | C. reflexa | |
|---|---|---|
| Number of raw sequences | 914 135 | 945 254 |
| Number of contigs | 31 685 | 42 103 |
| Mean contig size | 800.4 bp | 843.9 bp |
| Largest contig | 5 965 bp | 6 735 bp |
| N25 | 1 318 bp | 1 391 bp |
| N50 | 867 bp | 906 bp |
| N75 | 617 bp | 642 bp |
Table 2.
Annotation of Cuscuta contigs
The number of contigs with hits to annotated sequences in public databases (blastx thresholds E-value: 1e-15) and with no annotation are shown (percentages in parentheses).
| nr (NCBI) | SwissProt (UniProt) | UniRef90 (UniProt) | PFAM 27.0 (EMBL-EBI) | No annotation | |
|---|---|---|---|---|---|
| C. gronovii | 29 052 (92%) | 30 105 (95%) | 30 347 (96%) | 13 780 (43%) | 555 (2%) |
| C. reflexa | 38 944 (92%) | 39 465 (94%) | 39 927 (95%) | 18 648 (44%) | 984 (2%) |
Using Mercator (Lohse et al., 2014), 52% of the C. reflexa contigs and 55% of the C. gronovii contigs could be assigned to bins. The distribution chart in Fig. 4 shows that most cellular functions are covered by the contig collections of both Cuscuta species and that the relative distribution of contigs to most of the bins is overall similar. Cell wall-related transcripts were not over-represented in the total transcriptomes (Fig. 4), substantiating the suspicion that their abundance in the haustorium-specific SSH cDNA library (Fig. 3) reflects elevated cell wall remodelling activities in this tissue.
Fig. 4.
MapMan binning of Cuscuta transcriptome contigs. The contigs were mapped and functionally classified using the online tool Mercator. The minimum BLAST bit score was set to 50.
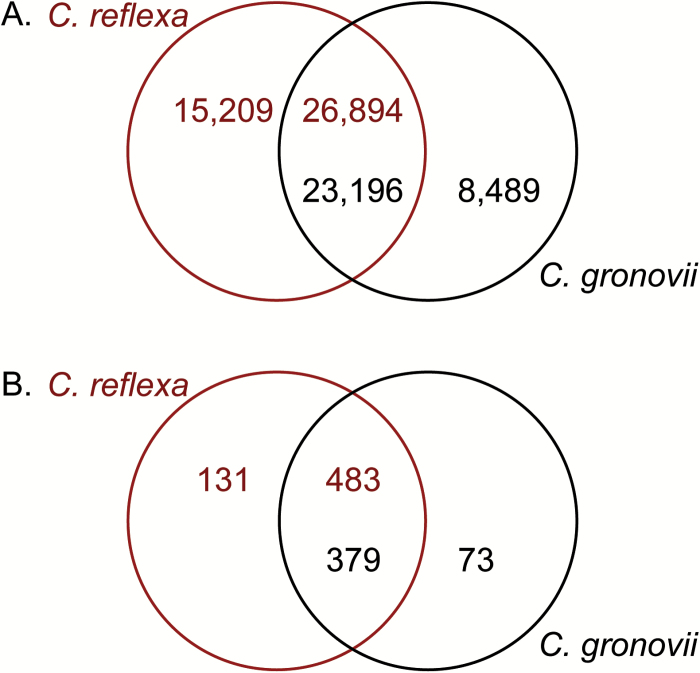
A direct sequence homology comparison between the two Cuscuta species showed that 73% (23 196) of the C. gronovii contigs had homologues in the C. reflexa transcriptome, while 64% (26 894) of the C. reflexa transcripts were also retrieved in C. gronovii (Fig. 5A). The difference in the percentages is at least in part due to the lower number of contigs assembled for C. gronovii. For contigs belonging to the cell wall-related bin, congruencies were even a bit higher. Of the 452 (C. gronovii) and 614 (C. reflexa) contigs that were sorted to this bin, 379 (84%) and 483 (79%), respectively, were also found in the other Cuscuta species (Fig. 5B).
Fig. 5.
Contig overlap between the two Cuscuta species (A) for all contigs and (B) for contigs related to cell wall functions. The Venn diagrams show how many contigs in the C. reflexa (top row) and C. gronovii (bottom row) transcriptomes have homologues in the other species (blastn threshold E-value: 1e-10). (This figure is available in colour at JXB online.)
Validation of differential expression by RT-qPCR
The reference transcriptomes described above provided information on complete or almost complete cDNA sequences for most of the partial cDNA clones that were recovered from the SSH libraries. With this additional sequence information, RT-qPCR primers for validation of the differential transcript accumulation indicated by the SSH were generated. To this end, the relative expression levels of 10 genes from the haustorium library and two genes from the stem library putatively related to cell wall functions were quantified in the RNA samples used for the SSH by RT-qPCR. All genes displayed fold changes that reflected the differential expression suggested by the SSH; the two stem-specific genes (Cr-GH5 and Cr-GT20) were less expressed in young haustoria compared with the stem, whereas the transcript abundances of the 10 haustorium-specific genes (Cr-GH1, Cr-GH17, Cr-GH31, Cr-PAE, Cr-PX-1, Cr-PX-2, Cr-PX-3, Cr-RGP, Cr-XTH-1, and Cr-XTH-2) were higher in the haustorial tissue (Table 3).
Table 3.
Validation of SSH-identified differentially expressed cell wall-related genes by RT-qPCR
Gene designations, the corresponding SSH library, and fold changes ±SD of technical duplicates are shown.
| SSH library | Gene | Fold change (H:S)a |
|---|---|---|
| Haustorium | Cr-GH1 | 15±2.3 |
| Cr-GH17 | 2.2±0.54 | |
| Cr-GH31 | 25±3.6 | |
| Cr-PAE | 11±2.8 | |
| Cr-PX-1 | 1.9±0.33 | |
| Cr-PX-2 | 781b | |
| Cr-PX-3 | 8.2±1.2 | |
| Cr-RGP | 13±1.9 | |
| Cr-XTH-1 | 129±21 | |
| Cr-XTH-2 | 195±29 | |
| Stem | Cr-GH5 | –1.7±0.25 |
| Cr-GT20 | –16±4.8 |
a Values are means of technical duplicates in the normalized transcript abundances of the RNA sample used to generate the tester cDNA for the haustorium library (H) compared with the RNA sample used to generate the tester cDNA for the stem library (S). Reference genes: Cr-ACTIN and Cr-SF2.
b No SD because target transcript could not be detected in the stem RNA sample.
Expression mapping of differentially expressed genes
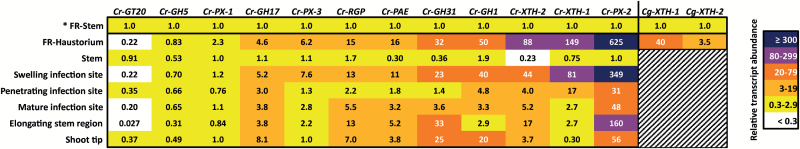
In order to investigate if these tentative haustorium-associated genes were also expressed during host-induced haustoriogenesis and if their expression is maintained during later stages of haustorium development or not, the gene-specific transcript abundances were quantified in swelling, penetrating, and mature stages of C. reflexa infecting P. zonale. All genes displayed similar expression levels in the early swelling stage of the host infection to those in the FR light-induced haustoria (Fig. 6). The expression in the stems was also similar, indicating that FR light irradiation in itself had no severe effect on the expression of these genes. Interestingly, Cr-RGP, Cr-PAE, two glycoside hydrolase members (Cr-GH31 and Cr-GH1), the two XTH genes (Cr-XTH-1 and Cr-XTH-2), and the peroxidase genes Cr-PX-2 and Cr-PX-3 showed a considerable decrease in expression levels in the more advanced stages of haustorium development (Fig. 6). To examine further if these genes were expressed in young haustoria only as a result of the high rates of cell division and cell elongation in these organs, the expression levels were also quantified in growing shoot tips, where the apical meristem is located, and in the stem region just below the tip that displays high rates of elongation (see Supplementary Fig. S3 at JXB online). While some genes indeed displayed an expression behaviour that suggests a general association with either cell division, cell elongation, or both (e.g. Cr-GH17, Cr-RGP, and Cr-GH31), the two Cr-XTH genes and Cr-PX-2 showed a more differentiated expression pattern with a clear increase in young haustoria (Fig. 6). These three genes displayed the highest differential expression levels, with changes of ≥80-fold in both our experiments (Table 3; Fig. 6). Moreover, the closest homologues to Cr-XTH-1 and Cr-XTH-2 in C. gronovii, Cg-XTH-1 and Cg-XTH-2, were both more highly expressed in young FR light-induced haustoria than in stems of this species (Fig. 6). The numerical values of all gene expression levels are presented in Supplementary Tables S2 and S3.
Fig. 6.
Expression of cell wall-related genes in tissues of Cuscuta. Transcript abundances in relation to the transcript abundances in ‘FR-Stem’ (* set to 1) are presented in a heat map with colours ranging from light (<0.3) to dark (≥300). All transcript abundances in C. reflexa and C. gronovii are normalized to the abundances of Cr-ACTIN and Cr-SF2 transcripts, and the abundances of Cg-ACTIN and Cg-SF2 transcripts, respectively. Values are the mean of three biological replicates. (This figure is available in colour at JXB online.)
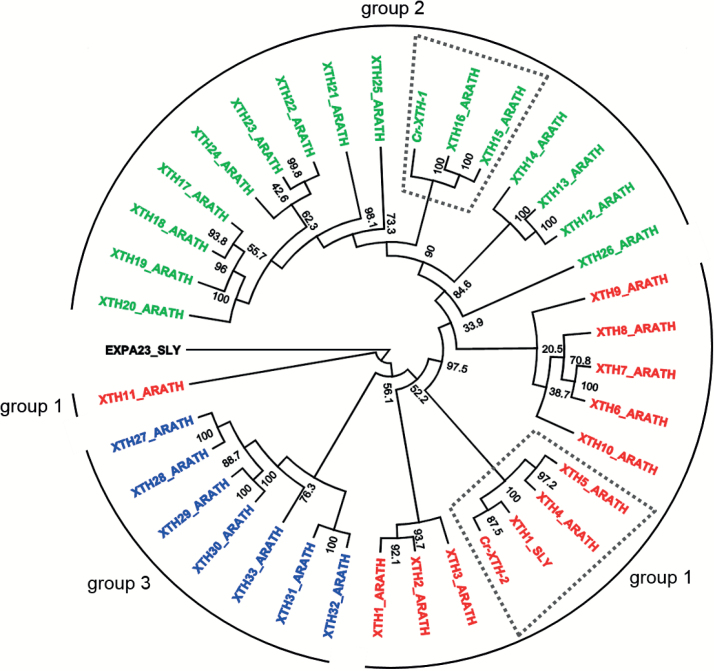
Interestingly, the increased expression of a tomato gene encoding an XTH, Lycopersicum esculentum LeXTH1 (here referred to as S. lycopersicum XTH1_SLY), has been described as a possible defence reaction of an incompatible tomato plant being attacked by C. reflexa potentially by tightening the cell walls (Albert et al., 2004). A comparison of the XTH1_SLY protein sequence and the Cuscuta XTHs showed that the tomato protein is very similar to Cr-XTH-2 and that both cluster with the A. thaliana XTH4 and XTH5 proteins in group 1 (Rose et al., 2002) (Fig. 7). In contrast, Cr-XTH-1 clusters with A. thaliana XTH15 and XTH16 within group 2 of this protein family.
Fig. 7.
Neighbor–Joining tree showing the relationship between C. reflexa XTH-1 and XTH-2 and Solanum lycopersicum XTH1 (XTH1_SLY, formerly LeXTH1) based on their phylogenetic placement within the Arabidopsis thaliana XTH gene family tree. The tree was rooted with S. lycopersicum Expansin A23 (EXPA23_SLY). The phylogram was optimized for substitution rates. Numbers next to the nodes represent bootstrap proportion values from 1000 replications. Designation of the subgroups 1–3 of the XTH protein family of A. thaliana are adopted from Rose et al. (2002). Clusters that include Cr-XTH-1, Cr-XTH-2, and XTH1_SLY are accentuated by a surrounding broken line. (This figure is available in colour at JXB online.)
Presence and distribution of XTH proteins and of XET action
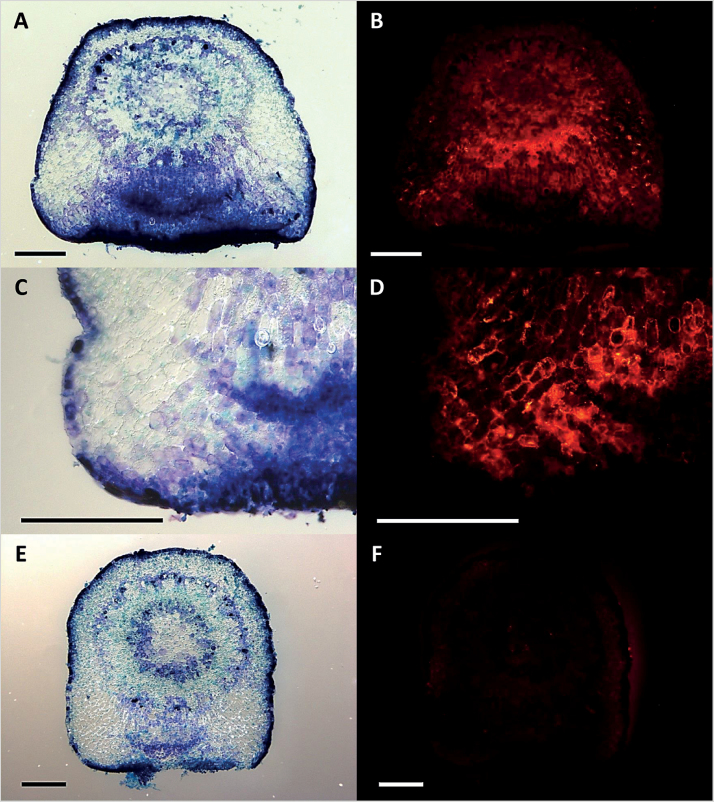
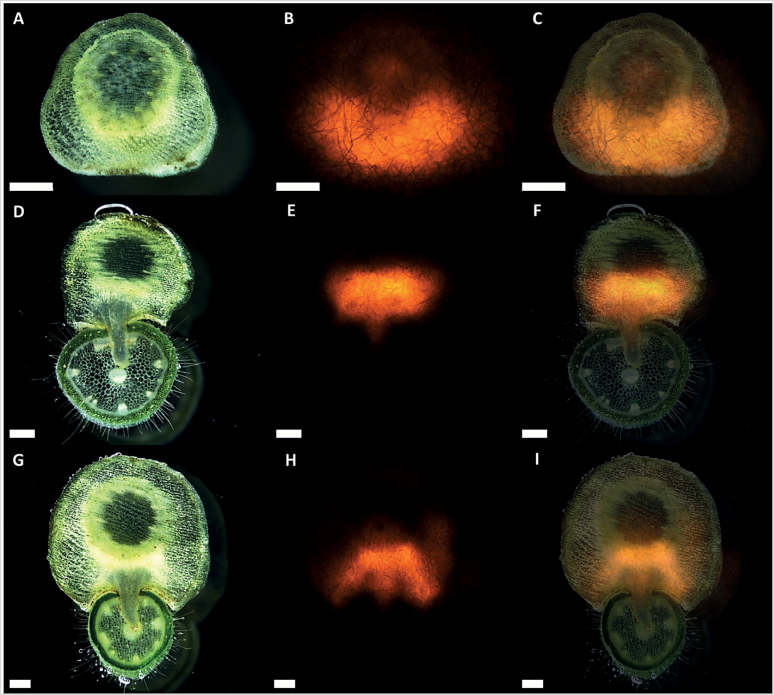
To approach the putative functionality of XTHs in Cuscuta haustorium development, an XTH-specific antibody (Jimenez et al., 2006) was used to label the proteins in cross-sections of FR light-induced haustoria. Fluorescence micrographs indicated that these xyloglucan-modifying enzymes are located in the cell walls of elongating cells in the areas flanking the haustorial initiation centre (Fig. 8); that is, in the part that is responsible for the swelling of the stem during attachment to a host plant. Furthermore, the xyloglucan endotransglucosylation action of XTHs was analysed in cross-sections of young FR light-induced haustoria as well as swelling, penetrating, and mature infection sites of C. reflexa infecting P. zonale by in situ XET action assays. Xyloglucan endotransglucosylation was detected in the parasite throughout haustorium development, with a clear bias towards the side facing the host plant (Fig. 9). Weaker XET action was also detected in the endophytic part of the haustorium. In contrast, the host plant did not display XTH-specific action. Tissue printing on control papers did not produce any fluorescent signal (data not shown). The detected XET action in the swelling areas of FR light-induced haustoria correlated with the immunolocalization patterns observed for these proteins (Supplementary Fig. S4 at JXB online). In contrast, enzyme extracts from Cuscuta haustoria had no detectable hydrolytic activity towards Tamarind xyloglucan (Supplementary Fig. S5).
Fig. 8.
Localization of XTHs in young FR light-induced haustoria. (A and C) Bright field and (B and D) fluorescence micrographs of toluidine blue O-stained and XTH-labelled cross-sections of haustoria 3 d after treatment with FR light. (E) Bright field and (F) fluorescence micrographs of control with pre-immune IgGs. Scale bars are 500 µm.
Fig. 9.
XET action during infection of P. zonale by C. reflexa. Cross-sections of (A) swelling, (D) penetrating, and (G) mature infection stages tissue printed on XET test paper. (B, E, and H) Fluorescence micrographs showing XET action in the printed tissues in (A), (D), and (G), respectively. Merged pictures of (A–B), (D–E), and (G–H) are presented in (C), (F), and (I), respectively. Scale bars are 500 µm. Visible fibres are integral to test papers.
Discussion
Most existing studies on the specific gene expression changes unfolding during the interaction between species of Cuscuta and their hosts have focused on genes expressed by the host upon parasitization. Differentially expressed host genes representing various functions, including general plant pathogen defence responses and cell wall metabolism, were identified by differential display or SSH techniques (Borsics and Lados, 2002; Albert et al., 2004; Li et al., 2009). A study on differential gene expression in Cuscuta has identified a cysteine protease, Cuscutain (Bleischwitz et al., 2010), but with the exception of this, corresponding studies are very scarce. Two aspects potentially aggravate the identification of bona fide differentially expressed genes during Cuscuta haustorium development: the uptake of host RNAs (Kim and Westwood, 2015) and the lack of a genome sequence from any Cuscuta species. In this study, these risks were mitigated by inducing haustoriogenesis in the absence of a host plant and by generating reference transcriptomes from two Cuscuta species using tissue that was not in contact with the host.
FR light in the range of 740nm together with physical contact between the parasite and a surface (normally the host) were described to be effective inducers of haustorium development in the absence of a host plant (Tada et al., 1996). The reversal of the inductive effect by red light (660nm) further suggested the involvement of phytochrome in haustorium differentiation (Furuhashi et al., 1997). FR light also triggers positive phototropism in seedlings of Cuscuta, prompting speculations that this fraction of the visible light that passes the green foliage provides an indication to the parasite that hosts are present (Orr et al., 1996). Morphologically, the early stages of haustorium development on a host plant and after FR light induction are similar (Figs 1, 2). Moreover, the present study demonstrated that all differentially expressed candidate genes, whose expression levels were quantified by RT-qPCR, displayed the same tendencies in the first stage of FR light- and host-induced haustoriogenesis (Fig. 6). This suggests that the physical stimuli sufficed to induce the transcriptional reprogramming and the ensuing tissue differentiation. The initial swelling stage during which the parasite attaches itself to the surface is followed by the protrusion of the discernible haustorium at ~6 d after FR light treatment (Fig. 2). Also at this advanced stage, gene-specific transcript abundances were comparable with those in penetrating stages of host plant infection (Supplementary Table S3 at JXB online), substantiating the conclusion that in these stages of haustorium development, changes in the transcriptional pattern do not depend on any host-derived signals. Ultimately, however, the plastic Petri dish hinders further growth of the haustorium, while in a host the differentiation of feeding hyphae commences. This last step that leads to an establishment of a physiological connection appears to be dependent on specific signals from the tissue that is invaded (Christensen et al., 2003; Vaughn, 2006) and can therefore not be easily mimicked in a host-free system.
An analysis of the 394 SSH clones whose inserts were sequenced (198 and 196, respectively, for each library), already showed a clear bias towards different functional categories represented by the transcripts in each of the two SSH libraries. The most conspicuous bias was that towards cell wall-related functions in the haustorium-specific library (Figs 3, 4). This finding conforms well with recent reports of changes to the cell wall composition during host-invasive growth of Cuscuta (Johnsen et al., 2015; Striberny and Krause, 2015). We therefore did not conduct more in-depth sequencing of the differentially expressed cDNA libraries and instead proceeded to profile the expression of cell wall-related genes during haustorium development and in comparison with other Cuscuta tissues exhibiting rapid growth. In haustoria, cell division and cell elongation are most pronounced in the shoot tips and a few centimetres below the tips, respectively (Supplementary Figure S3 at JXB online). Quantitative expression analysis in these parts of the parasite can thus reveal whether the identified genes are associated with growth in general or whether they are specific to haustoriogenesis.
Cr-GH17 and Cr-GH31 are glycoside hydrolases that displayed expression levels in elongating stem regions and/or shoot tips of C. reflexa that were comparable with the expression levels in the young haustorial tissues (Fig. 6). The products of these genes are therefore not unique to the haustorium, but rather are characteristic for growing tissues of the parasite. The GHs are a large and widespread group of enzymes characterized by their ability to hydrolyse glycosidic bonds, thus facilitating breakdown of carbohydrates. The majority of GHs in plants are involved in cell wall metabolism (Frankova and Fry, 2013).
A third gene, Cr-RGP, which encodes a reversibly glycosylated polypeptide, also showed elevated expression levels in non-haustorial growing tissues of the parasite. RGPs are a family of proteins suggested to be involved in the synthesis of plant cell wall polysaccharides (Dhugga et al., 1997), but whose exact function remains elusive. Based on sequence similarity, Cr-RGP belongs to class 1 RGPs (Langeveld et al., 2002), which are tentatively associated with plasmodesmata (Sagi et al., 2005).
Several studies connect pectins and its modifiers to host plant infection by Cuscuta (Nagar et al., 1984; Srivastava et al., 1994; Bar Nun and Mayer, 1999; Vaughn, 2002, 2003, 2006; Johnsen et al., 2015). In the present study, the transcript abundance of a pectin acetylesterase, Cr-PAE, was found to be higher in young haustoria than in all other tested tissues of Cuscuta (Fig. 6), making it a potential marker gene for haustorium initiation and corroborating the role of pectin modification during the infection process.
Cr-PX-2 and Cr-PX-3 transcripts were also more abundant in young haustoria than in any of the other tissues (Fig. 6). Peroxidases are a large group of enzymes that catalyse the oxidation of a number of substrates (Battistuzzi et al., 2010). In plants, PXs are associated with cell wall metabolism (Duroux and Welinder, 2003; Veitch, 2004) and with the defence against plant pathogens (Lamb and Dixon, 1997; Lopez-Curto et al., 2006) so their relative abundance may indicate a ‘state of alert’ in the haustorial cells of Cuscuta.
Expression differences that were similarly large or even larger than those of the peroxidase genes and likewise specific to the young haustoria of C. reflexa were observed for Cr-XTH-1 and Cr-XTH-2. XTHs are assigned roles in loosening of the plant cell wall by restructuring xyloglucans, allowing turgor-driven expansive cell growth (Rose et al., 2002; Van Sandt et al., 2007). Xyloglucans are the most abundant hemicelluloses in primary cell walls of dicotyledonous plants (Scheller and Ulvskov, 2010) so that all parasitic plants can be expected to display xyloglucan-modifying activities. The differential expression of closely related XTH genes in FR light-induced haustoria of C. gronovii, which belongs to a subgenus of Cuscuta different than C. reflexa, substantiates the assumption (Fig. 6). Also, Ranjan et al. (2014) found seven XTH genes in C. pentagona that were more highly expressed at the pre-haustorial stage compared with seedlings and stems, suggesting that the early expression of these genes is in fact part of a common developmental pattern in most, if not all, Cuscuta species.
Recent epitope deletion assays (Vidal-Melgosa et al., 2015) showed that specifically the group of xyloglucans containing the XXXG-motif is a target of enzymatic activity in haustoria and in the infected host (Johnsen et al., 2015). In the present study, these previous investigations on xyloglucan-modifying enzymes were extended by immunolocalization studies and XET action assays using tissue prints on XET test papers. Anti-XTH labelling of 3-day-old FR light-induced haustoria confined the xyloglucan-modifying enzymes to the cell walls of elongating cells in the swelling areas of young haustoria (Fig. 8), indicating that restructuring of this hemicellulose is taking place during the initial stage of haustoriogenesis. In vitro activity assays using crude extracts suggested that the xyloglucan restructuring activity was not predominantly hydrolytic (see Supplementary Fig. S5 at JXB online). Correspondingly, in situ XET action assays clearly revealed xyloglucan endotransglucosylation in the swelling area of FR light-induced haustoria (Supplementary Fig. S4) and in the comparable area of haustoria that faces the host plant during infection (Fig. 9). Vaughn (2006) reported reduced cellulose and xyloglucan levels in the cell walls of phloic hyphae during the parasitization of Impatiens by C. pentagona and proposed that this loosening of the cell wall facilitates apoplastic transfer of sugars into the parasite. A reduction in xyloglucan in the neighbouring phloem cells of the host was also observed in the same study. Some XET action was detected in the endophytic part of the haustorium in the present study (Fig. 9). Since Cr-XTH-1 and Cr-XTH-2 expression levels drop in the more advanced stages of haustoriogenesis, a full investigation of the expression patterns of all other XTH gene candidates that have been identified in the reference transcriptomes of both Cuscuta species will be necessary to reveal which of the genes take over the hypothetical task of modifying host xyloglucans.
Although the exact function of neither Cr-XTH-2 nor XTH1_SLY is known, their intriguing similarity (Fig. 7) gives rise to a bold train of thought where the infective mechanisms in parasites might not have evolved de novo but, rather, through re-purposing of defence pathways that existed already in their non-parasitic ancestors. A similar scenario was suggested as an explanation for similarities in the mutual recognition systems by pathogens and hosts (Vasta, 2012). XTH1_SLY is expressed very strongly in expanding tomato fruits (Matas et al., 2011) (see also Tomato eFP Browser, http://bar.utoronto.ca/efp_tomato/cgi-bin/efpWeb.cgi), suggesting a role during fruit ripening. While this can be best reconciled with a molecular function in cell wall loosening, XTHs were also shown to have the opposite effect and can tighten cell walls (Maris et al., 2009). In fact, the latter process would be expected to exist in the parasite to protect its walls from its own hydrolytic enzyme cocktail, but such details of the complex array of actions and reactions between Cuscuta and its hosts need to be addressed in future work.
Our observation that the expression of cell wall-related genes in Cuscuta was up-regulated at the onset of haustorium development is in agreement with other recent reports on gene expression in parasitic plants (Ranjan et al., 2014; Yang et al., 2014), which indicates that changes to cell walls are essential to the formation of the infection organ. Furthermore, the site-specific action of XTHs during host plant infection points towards the relevance of xyloglucan modification in host–Cuscuta interactions. Consequently, cell wall genes and XTH genes in particular might prove effective gene silencing targets for the control of Cuscuta in agriculture, and, as such, efforts to unravel the role of xyloglucan and its modifiers in haustorium development should be continued.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Sequences of gene-specific primers with respective amplicon sizes and PCR efficiencies.
Supplementary Table S2. Gene expression levels in single biological replicates.
Supplementary Table S3. Mean gene expression levels of biological triplicates.
Supplementary Fig. S1. Melt peaks of qPCR amplicons.
Supplementary Fig. S2. Size separation of qPCR amplicons on agarose gel.
Supplementary Fig.S3. Stem elongation in the region just below the apical shoot tip.
Supplementary Fig. S4. XET action in FR light-induced haustoria.
Supplementary Fig. S5. Hydrolytic activity of a haustorial enzyme extract from C. reflexa towards xyloglucan.
Acknowledgements
The generous gift of IgGs from pre-immune serum and immune serum against XTHs by Dr Emilia Labrador, Dr Berta Dopico, and Dr Ignacio Martín (University of Salamanca, Spain) is gratefully acknowledged. Ave Tooming-Klunderud is thanked for 454 pyro-sequencing. The sequencing service was provided by the Norwegian High-Throughput Sequencing Centre, a national technology platform supported by the ‘Functional Genomics’ and ‘Infrastructure’ programmes of the Research Council of Norway and the Southeastern Regional Health Authorities (http://www.sequencing.uio.no). We are indebted to the Phytotron staff at Holt (University of Tromsø, Norway), in particular Leidulf Lund, for the care and maintenance of our plants. Eli Robertsen is thanked for technical assistance in plasmid isolation. Dr Hanne R. Johnsen, Dr Karsten Fischer (University of Tromsø, Norway), and Anja Striberny are thanked for fruitful discussions and/or comments to the manuscript. The manuscript is part of the doctoral thesis of SO.
Glossary
Abbreviations:
- Cg
Cuscuta gronovii
- Cq
quantification cycle
- Cr
Cuscuta reflexa
- FR
far-red
- GH
glycoside hydrolase
- GT
glycosyltransferase
- PAE
pectin acetylesterase
- PX
peroxidase
- RGP
reversibly glycosylated polypeptide
- RT-qPCR
reverse transcription quantitative real-time PCR
- SF2
pre-mRNA splicing factor 2
- SSH
suppression subtractive hybridization
- XET
xyloglucan endotransglucosylation
- XTH
xyloglucan endotransglucosylase/hydrolase.
References
- Alakonya A, Kumar R, Koenig D, et al. 2012. Interspecific RNA interference of SHOOT MERISTEMLESS-like disrupts Cuscuta pentagona plant parasitism. The Plant Cell 24, 3153–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M, Werner M, Proksch P, Fry SC, Kaldenhoff R. 2004. The cell wall-modifying xyloglucan endotransglycosylase/hydrolase LeXTH1 is expressed during the defence reaction of tomato against the plant parasite Cuscuta reflexa . Plant Biology 6, 402–407. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Aly R. 2007. Conventional and biotechnological approaches for control of parasitic weeds. In Vitro Cellular and Developmental Biology-Plant 43, 304–317. [Google Scholar]
- Aly R. 2013. Trafficking of molecules between parasitic plants and their hosts. Weed Research 53, 231–241. [Google Scholar]
- Bar Nun N, Mayer AM. 1999. Culture of pectin methylesterase and polyphenoloxidase in Cuscuta campestris . Phytochemistry 50, 719–727. [Google Scholar]
- Battistuzzi G, Bellei M, Bortolotti CA, Sola M. 2010. Redox properties of heme peroxidases. Archives of Biochemistry and Biophysics 500, 21–36. [DOI] [PubMed] [Google Scholar]
- Birnboim HC, Doly J. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Research 7, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanca JM, Pascual L, Ziarsolo P, Nuez F, Canizares J. 2011. ngs_backbone: a pipeline for read cleaning, mapping and SNP calling using next generation sequence. BMC Genomics 12, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleischwitz M, Albert M, Fuchsbauer HL, Kaldenhoff R. 2010. Significance of Cuscutain, a cysteine protease from Cuscuta reflexa, in host–parasite interactions. BMC Plant Biology 10, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsics T, Lados M. 2002. Dodder infection induces the expression of a pathogenesis-related gene of the family PR-10 in alfalfa. Journal of Experimental Botany 53, 1831–1832. [DOI] [PubMed] [Google Scholar]
- Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GBC) 99, 45–56. [Google Scholar]
- Christensen NM, Dorr I, Hansen M, van der Kooij TA, Schulz A. 2003. Development of Cuscuta species on a partially incompatible host: induction of xylem transfer cells. Protoplasma 220, 131–142. [DOI] [PubMed] [Google Scholar]
- Costea M, Garcia MA, Stefanovic S. 2015. A phylogenetically based infrageneric classification of the parasitic plant genus Cuscuta (dodders, Convolvulaceae). Systematic Botany 40, 269–285. [Google Scholar]
- Dawson JH, Musselman LJ, Wolswinkel P, Dorr I. 1994. Biology and control of Cuscuta . Reviews of Weed Science 6, 265–317. [Google Scholar]
- Dhugga KS, Tiwari SC, Ray PM. 1997. A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: purification, gene cloning, and trans-Golgi localization. Proceedings of the National Academy of Sciences, USA 94, 7679–7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, et al. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Sciences, USA 93, 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroux L, Welinder KG. 2003. The peroxidase gene family in plants: a phylogenetic overview. Journal of Molecular Evolution 57, 397–407. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, et al. 2014. Pfam: the protein families database. Nucleic Acids Research 42, D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankova L, Fry SC. 2013. Biochemistry and physiological roles of enzymes that ‘cut and paste’ plant cell-wall polysaccharides. Journal of Experimental Botany 64, 3519–3550. [DOI] [PubMed] [Google Scholar]
- Fry SC. 1997. Novel ‘dot-blot’ assays for glycosyltransferases and glycosylhydrolases: optimization for xyloglucan endotransglycosylase (XET) activity. The Plant Journal 11, 1141–1150. [Google Scholar]
- Furuhashi K, Tada Y, Okamoto K, Sugai M, Kubota M, Watanabe M. 1997. Phytochrome participation in induction of haustoria in Cuscuta japonica, a holoparasitic flowering plant. Plant and Cell Physiology 38, 935–940. [Google Scholar]
- Garcia MA, Costea M, Kuzmina M, Stefanovic S. 2014. Phylogeny, character evolution, and biogeography of Cuscuta (dodders; Convolvulaceae) inferred from coding plastid and nuclear sequences. American Journal of Botany 101, 670–690. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Research 33, D121–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y, Mutuku JM, Yoshida S, Shirasu K. 2015. Transcriptomics exposes the uniqueness of parasitic plants. Briefings in Functional Genomics 14, 275–282. [DOI] [PubMed] [Google Scholar]
- Jiang L, Wijeratne AJ, Wijeratne S, Fraga M, Meulia T, Doohan D, Li Z, Qu F. 2013. Profiling mRNAs of two Cuscuta species reveals possible candidate transcripts shared by parasitic plants. PLoS One 8, e81389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez T, Martin I, Labrador E, Dopico B. 2006. The immunolocation of a xyloglucan endotransglucosylase/hydrolase specific to elongating tissues in Cicer arietinum suggests a role in the elongation of vascular cells. Journal of Experimental Botany 57, 3979–3988. [DOI] [PubMed] [Google Scholar]
- Johnsen HR, Striberny B, Olsen S, Vidal-Melgosa S, Fangel JU, Willats WG, Rose JK, Krause K. 2015. Cell wall composition profiling of parasitic giant dodder (Cuscuta reflexa) and its hosts: a priori differences and induced changes. New Phytologist 207, 805–816. [DOI] [PubMed] [Google Scholar]
- Kaiser B, Vogg G, Furst UB, Albert M. 2015. Parasitic plants of the genus Cuscuta and their interaction with susceptible and resistant host plants. Frontiers in Plant Science 6, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G, LeBlanc ML, Wafula EK, dePamphilis CW, Westwood JH. 2014. Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science 345, 808–811. [DOI] [PubMed] [Google Scholar]
- Kim G, Westwood JH. 2015. Macromolecule exchange in Cuscuta–host plant interactions. Current Opinion in Plant Biology 26, 20–25. [DOI] [PubMed] [Google Scholar]
- Kosik O, Farkas V. 2008. One-pot fluorescent labeling of xyloglucan oligosaccharides with sulforhodamine. Analytical Biochemistry 375, 232–236. [DOI] [PubMed] [Google Scholar]
- Lamb C, Dixon RA. 1997. The oxidative burst in plant disease resistance. Annual Review of Plant Physiology and Plant Molecular Biology 48, 251–275. [DOI] [PubMed] [Google Scholar]
- Langeveld SMJ, Vennik M, Kottenhagen M, van Wijk R, Buijk A, Kijne JW, de Pater S. 2002. Glucosylation activity and complex formation of two classes of reversibly glycosylated polypeptides. Plant Physiology 129, 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc M, Kim G, Patel B, Stromberg V, Westwood J. 2013. Quantification of tomato and Arabidopsis mobile RNAs trafficking into the parasitic plant Cuscuta pentagona . New Phytologist 200, 1225–1233. [DOI] [PubMed] [Google Scholar]
- Leblanc M, Kim G, Westwood JH. 2012. RNA trafficking in parasitic plant systems. Frontiers in Plant Science 3, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DM, Staehelin C, Zhang YS, Peng SL. 2009. Identification of genes differentially expressed in Mikania micrantha during Cuscuta campestris infection by suppression subtractive hybridization. Journal of Plant Physiology 166, 1423–1435. [DOI] [PubMed] [Google Scholar]
- Li KB. 2003. ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics 19, 1585–1586. [DOI] [PubMed] [Google Scholar]
- Lohse M, Nagel A, Herter T, May P, Schroda M, Zrenner R, Tohge T, Fernie AR, Stitt M, Usadel B. 2014. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant, Cell and Environment 37, 1250–1258. [DOI] [PubMed] [Google Scholar]
- Lopez-Curto L, Marquez-Guzman J, Diaz-Pontones DM. 2006. Invasion of Coffea arabica (Linn.) by Cuscuta jalapensis (Schlecht): in situ activity of peroxidase. Environmental and Experimental Botany 56, 127–135. [Google Scholar]
- Maris A, Suslov D, Fry SC, Verbelen JP, Vissenberg K. 2009. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. Journal of Experimental Botany 60, 3959–3972. [DOI] [PubMed] [Google Scholar]
- Matas AJ, Yeats TH, Buda GJ, et al. 2011. Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. The Plant Cell 23, 3893–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar R, Singh M, Sanwal GG. 1984. Cell-wall degrading enzymes in Cuscuta reflexa and its hosts. Journal of Experimental Botany 35, 1104–1112. [Google Scholar]
- Orr GL, Haidar MA, Orr DA. 1996. Smallseed dodder (Cuscuta planiflora) phototropism toward far-red when in white light. Weed Science 44, 233–240. [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research 41, D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Ichihashi Y, Farhi M, Zumstein K, Townsley B, David-Schwartz R, Sinha NR. 2014. De novo assembly and characterization of the transcriptome of the parasitic weed Cuscuta pentagona identifies genes associated with plant parasitism. Plant Physiology 166, 1186–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. 2002. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant and Cell Physiology 43, 1421–1435. [DOI] [PubMed] [Google Scholar]
- Sagi G, Katz A, Guenoune-Gelbart D, Epel BL. 2005. Class 1 reversibly glycosylated polypeptides are plasmodesmal-associated proteins delivered to plasmodesmata via the Golgi apparatus. The Plant Cell 17, 1788–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. 2010. Hemicelluloses. Annual Review of Plant Biology 61, 263–289. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Nighojkar A, Kumar A. 1994. Multiple forms of pectin methylesterase from Cuscuta reflexa filaments. Phytochemistry 37, 1233–1236. [Google Scholar]
- Striberny B, Krause K. 2015. Cell wall glycoproteins at interaction sites between parasitic giant dodder (Cuscuta reflexa) and its host Pelargonium zonale . Plant Signaling and Behavior (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Sugai M, Furuhashi K. 1996. Haustoria of Cuscuta japonica, a holoparasitic flowering plant, are induced by the cooperative effects of far-red light and tactile stimuli. Plant and Cell Physiology 37, 1049–1053. [Google Scholar]
- UniProt-Consortium. . 2015. UniProt: a hub for protein information. Nucleic Acids Research 43, D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, et al. 2005. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiology 138, 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sandt VST, Suslov D, Verbelen JP, Vissenberg K. 2007. Xyloglucan endotransglucosylase activity loosens a plant cell wall. Annals of Botany 100, 1467–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasta GR. 2012. Galectins as pattern recognition receptors: structure, function, and evolution. Advances in Experimental Medicine and Biology 946, 21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn KC. 2002. Attachment of the parasitic weed dodder to the host. Protoplasma 219, 227–237. [DOI] [PubMed] [Google Scholar]
- Vaughn KC. 2003. Dodder hyphae invade the host: a structural and immunocytochemical characterization. Protoplasma 220, 189–200. [DOI] [PubMed] [Google Scholar]
- Vaughn KC. 2006. Conversion of the searching hyphae of dodder into xylic and phloic hyphae: a cytochemical and immunocytochemical investigation. International Journal of Plant Sciences 167, 1099–1114. [Google Scholar]
- Veitch NC. 2004. Horseradish peroxidase: a modern view of a classic enzyme. Phytochemistry 65, 249–259. [DOI] [PubMed] [Google Scholar]
- Vidal-Melgosa S, Pedersen HL, Schuckel J, Arnal G, Dumon C, Amby DB, Monrad RN, Westereng B, Willats WG. 2015. A new versatile microarray-based method for high throughput screening of carbohydrate-active enzymes. Journal of Biological Chemistry 290, 9020–9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. 1994. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Analytical Biochemistry 223, 7–12. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wafula EK, Honaas LA, et al. 2014. Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Molecular Biology and Evolution 32, 767–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.