Highlight
Improved salinity stress tolerance in wheat Nax lines is achieved by transcriptional and post-translational down-regulation of Na+ loading into the xylem by a SOS1 transporter.
Key words: HKT transporter, potassium, salinity stress, sequestration, sodium, xylem loading.
Abstract
Salinity stress tolerance in durum wheat is strongly associated with a plant’s ability to control Na+ delivery to the shoot. Two loci, termed Nax1 and Nax2, were recently identified as being critical for this process and the sodium transporters HKT1;4 and HKT1;5 were identified as the respective candidate genes. These transporters retrieve Na+ from the xylem, thus limiting the rates of Na+ transport from the root to the shoot. In this work, we show that the Nax loci also affect activity and expression levels of the SOS1-like Na+/H+ exchanger in both root cortical and stelar tissues. Net Na+ efflux measured in isolated steles from salt-treated plants, using the non-invasive ion flux measuring MIFE technique, decreased in the sequence: Tamaroi (parental line)>Nax1=Nax2>Nax1:Nax2 lines. This efflux was sensitive to amiloride (a known inhibitor of the Na+/H+ exchanger) and was mirrored by net H+ flux changes. TdSOS1 relative transcript levels were 6–10-fold lower in Nax lines compared with Tamaroi. Thus, it appears that Nax loci confer two highly complementary mechanisms, both of which contribute towards reducing the xylem Na+ content. One enhances the retrieval of Na+ back into the root stele via HKT1;4 or HKT1;5, whilst the other reduces the rate of Na+ loading into the xylem via SOS1. It is suggested that such duality plays an important adaptive role with greater versatility for responding to a changing environment and controlling Na+ delivery to the shoot.
Introduction
Soil salinity severely affects plant growth and limits agricultural crop production (Qadir et al., 2014; Shabala et al., 2014). Approximately 20% of the world’s cultivated land, which accounts for over 6% of the world total area, is currently threatened by salinity (Rengasamy, 2010). An elevated salt concentration in the soil leads to the accumulation of toxic concentrations of Na+ in the leaves (Munns and Tester, 2008; Rahnama et al., 2010). Consequently, control of Na+ long-distance transport and the ability to retrieve Na+ from the xylem are considered amongst the most essential traits conferring salinity tolerance (Munns and Tester, 2008; Shabala et al., 2013).
Wheat is one of the most important cereal crops worldwide, providing approximately one-fifth of the total calorific input of the world’s population (Shewry, 2009). Among wheat species, durum (pasta) wheat (Triticum turgidum ssp. durum) is generally less tolerant to salt stress than bread wheat (Triticum aestivum) (Cuin et al., 2009), mainly due to the high rates of Na+ accumulation and poor K+/Na+ discrimination (Gorham et al., 1990; Munns and James, 2003).
Two loci that reduced Na+ accumulation in the shoot, named Nax1 and Nax2, were discovered in an unusual durum wheat (James et al., 2006). These loci had been transferred there from the ancestral wheat species einkorn (Triticum monococcum) in order to introduce rust resistance genes into modern wheat (James et al., 2006). These loci are not present in modern durum or bread wheat, so they were crossed into the current durum cultivar Tamaroi, and near-isogenic lines were developed containing either the Nax1 or Nax2 loci or both. These lines had lower rates of Na+ transport from the roots to the shoots, the result of a lower rate of net Na+ loading into the xylem (James et al., 2006). Both loci could unload Na+ from the xylem in the root, while Nax1 could also unload Na+ from the xylem at the leaf base (the sheath) so leading to a high Na+ ratio between the sheath and the blade (James et al., 2006). Nax1 and Nax2 lines also had higher rates of K+ transport from the root to the shoot, resulting in an enhanced discrimination of K+ over Na+ (James et al., 2006). Using fine-mapping, the candidate gene for Nax1 was identified on chromosome 2A as a Na+ transporter of the HKT gene family TmHKT1;4 (Huang et al., 2006). The candidate gene for Nax2 was localized on chromosome 5A and identified as TmHKT1;5 (Byrt et al., 2007). It was localized on the plasma membrane of cells surrounding the xylem and, when crossed into an elite Australian durum cultivar, was found to confer a yield benefit of 25% on saline soil in a farmer’s field (Munns et al., 2012). A recent study found that a closely related gene in bread wheat, TaHKT1;5-D, is also localized on the plasma membrane in the root stele and operates in retrieving Na+ from the xylem vessels thus restricting the transport of Na+ from the root to the leaves in bread wheat (Byrt et al., 2014).
While Na+ retrieval from the xylem before it reaches the sensitive photosynthetic tissues is indeed essential for plant performance under saline conditions, it is also important to reduce the amounts of Na+ initially loaded into the xylem. It was argued (Shabala et al., 2010, 2013) that the ideal scenario for a plant would be quickly to send the amount of Na+ to the shoot that is required in order to achieve full osmotic adjustment rapidly and to maintain a normal growth rate (hence, no yield penalties). Once this is achieved, it would be advantageous for a plant to reduce the rate of xylem Na+ loading to the absolute minimum for maintaining cell turgor in growing tissues while, at the same time, preventing excessive Na+ from being accumulated in photosynthetically-active and fully-grown leaf tissues. This poses a question: what is the molecular nature of the mechanisms mediating xylem Na+ loading and the modes of their control? Recent thermodynamic analysis has suggested that channel-mediated xylem Na+ loading dominates at the early stages of salt stress (minutes to hours), while longer exposure to salinity (hours and days) will require thermodynamically-active xylem Na+ loading (Shabala, 2013). Two possible candidates were proposed, one a SOS1 Na+/H+ exchanger (Shi et al., 2002) and another, a cation–Cl (CCC) co-transporter (Colmenero-Flores et al., 1999).
The salt-overly-sensitive (SOS) signal transduction pathway is regarded as a key mechanism for maintaining intracellular ion homeostasis under saline conditions (Zhu et al., 1998; Hasegawa et al., 2000; Sanders et al., 2000; Zhu, 2001). According to the current view, elevated Na+ causes increases in cytosolic free Ca2+ (Knight et al., 1997), which can be sensed by SOS3, a myristoylated Ca2+-binding protein (Liu and Zhu, 1998; Ishitani et al., 2000). SOS3 interacts with and activates SOS2 (a serine/threonine protein kinase), thus forming a SOS2/SOS3 complex (Halfter et al., 2000; Liu et al., 2000). AtSOS1 is identified as a Na+/H+ antiporter, localized in epidermal cells at the root tip and also in parenchyma cells at the xylem/symplast boundary of roots, stems, and leaves where it controls long-distance Na+ transport (Shi et al., 2002). Overexpression of SOS1 in transgenic Arabidopsis has been shown to improve salt tolerance (Shi et al., 2003). Furthermore, Feki et al. identified the TdSOS1 orthologue from durum wheat. The heterologous expression of TdSOS1 in a yeast strain lacking endogenous Na+ efflux proteins showed complementation involving cation efflux (Feki et al., 2011). Importantly, expression of a truncated form of wheat TdSOS1 in the Arabidopsis sos1-1 mutant exhibited improved salt tolerance (Feki et al., 2014).
In this study, we used Nax1 and Nax2 durum wheat lines to provide supporting evidence of a role for SOS1-mediated Na+ loading into the xylem in these species. We tested the hypothesis that reduced Na+ accumulation in the shoot of Nax lines could be conferred not only by higher Na+ retrieval from the xylem, but also by reduced Na+ loading into the xylem. Our electrophysiological and molecular data fully support this hypothesis and suggest that the Nax loci regulate activity and expression levels of a SOS1-like Na+/H+ exchanger in the xylem tissue of wheat and that down-regulation of this transporter in Nax lines improves plant performance under saline conditions. This mechanism operates in addition to, or instead of, the reported increased Na+ retrieval from the xylem by the HKT transporter. This reduces the overall net xylem Na+ loading and accumulation in the shoot, thus increasing salinity tolerance.
Materials and methods
Plant material and growth conditions
Durum wheat (Triticum turgidum L. ssp. durum Desf.) seeds of cv. Tamaroi and BC5F2 Nax lines were a kind gift from Dr Richard James (CSIRO Plant Industry, Canberra). Tamaroi, like all durum and bread wheat cultivars, lacks the Nax loci which originated from the diploid ancestral wheat, Triticum monococcum (James et al., 2006). These loci were back-crossed into the current durum cultivar Tamaroi, and near-isogenic BC5F2 lines were developed using specific molecular markers (James et al., 2006). Nax lines were homozygous for Nax1, Nax2, or both Nax1 and Nax2. The candidate gene for Nax1 is TmHKT1;4-A2 on chromosome 2A (Huang et al., 2006) and for Nax2 is TmHKT1;5-A on chromosome 5A (Byrt et al., 2007). SOS1 is located on chromosome 3 (Mullan et al., 2007), and there was no selection for this gene during back-crossing, so all lines have the same SOS1 gene allele as the parental cultivar Tamaroi.
Seeds were surface-sterilized with 10% bleach (King White, Victoria, Australia) for 10min, rinsed thoroughly with deionized water, then grown hydroponically for 6 d in the dark in an aerated Basic Salt Medium (BSM) containing 0.1mM CaCl2 and 0.5mM KCl (pH=5.6; non-buffered).
Non-invasive ion flux measurements
Net ion fluxes were measured using non-invasive ion-selective vibrating microelectrodes (the MIFE technique; University of Tasmania, Hobart, Australia). The principles of MIFE ion flux measurements are described in full elsewhere (Shabala et al., 1997) and all the details of microelectrode fabrication and calibration are available in our previous publications (Shabala and Shabala, 2002; Shabala et al., 2006). Liquid ionic exchangers used in this work are the commercially available ionophore cocktails (60031 for K+; 71176 for Na+; 95297 for H+; all from Fluka, Busch, Switzerland).
Root K+ and H+ flux measurements
One hour prior to measurement, 6-d-old wheat seedlings were immobilized, with their roots placed horizontally in a 10ml Perspex measuring chamber containing the bathing medium as described elsewhere (Chen et al., 2005; Bose et al., 2014). Two ion-selective microelectrodes, one for K+, the other for H+, were used simultaneously, with the electrode tips aligned and positioned 50 μm above the root surface. Once steady-state fluxes were reached (40–60min after immobilization), measurements commenced. Net fluxes of ions were measured for 5–10min from the mature (~15–20mm from the tip) root zone. The salinity treatment (80mM NaCl) was then administered and net K+ and H+ fluxes measured for a further 60min.
Root Na+ flux measurements
A so-called ‘recovery protocol’ (Cuin et al., 2011) was used to quantify the activity of a Na+ efflux system in epidermal and stelar root tissues. Six-day-old wheat seedlings were treated with 150mM NaCl for a further 24h in the dark. A seedling was then transferred to a 10ml Perspex measuring chamber containing the bathing medium and 150mM NaCl. After 1h adaption, roots were quickly but thoroughly rinsed with a 10mM CaCl2 solution to remove surface and apoplastic NaCl before being transferred to a clean chamber containing Na+-free BSM solution. Net ion fluxes were measured for approximately 60min from either the mature root epidermis of the seminal root (~15–20mm from the tip), or root elongation zone. The first 2min of recording were ignored during analysis to eliminate any confounding effect of the Donnan exchange in the cell wall (see Cuin et al., 2011, for justification and details).
For measurements from the xylem parenchyma, the root stele was mechanically isolated as described earlier (Shabala et al., 2010). An apical stelar segment was cut (the first 5–7mm of the stele), immobilized in a Perspex chamber in BSM and left to recover for 4–6h in the presence of 50mM NaCl. The recovery protocol (see above) was then applied to quantify the activity of a Na+ efflux system at the xylem parenchyma interface.
RNA extraction and RT-qPCR experiments
Total RNA was extracted from ~100mg of roots from Tamaroi and Nax lines using the RNeasy Plant Mini Kit (Qiagen). First-strand cDNA was synthesized using the QuantiTect Reverse Tanscription Kit (Qiagen), which includes the genomic DNA removal step. Relative transcript levels were assayed by real-time PCR analysis using the Qiagen Rotor-gene Real-Time PCR system. The TdSOS1 gene was amplified using two specific primers TdSOS1; 5′SOS (5′-ATTCCCTCAGGTGCTTCGTG-3′) and 3′SOS (5′-TTTCCTCGAGCAACCCAGTC-3′). The wheat actin gene (Genebank Accession No. AB181991.1) was used as an internal control for gene expression. The actin primers were AF (5′-TACACGAAGCGACATACAA-3′) and AR (5′-AATAGAGCCACCGATCCA-3′). RT-qPCR conditions were as follows: 95 °C for 5min, 94 °C for 30s (50 cycles), 58 °C for 30s, and 72 °C for 1min. Amplified products were detected using QuantiNova SYBR Green PCR Kit (Qiagen). Each data point represents six biological replicates in each sample, presented as the mean ±SE. The experiment was repeated three times, with consistent results.
Statistical analysis
Data were analysed using one-way of variance, and treatment mean separations were performed using Duncan’s multiple range tests at the 5% level of significance in IBM Statistics 21.
Results
NaCl-induced ion flux response in root
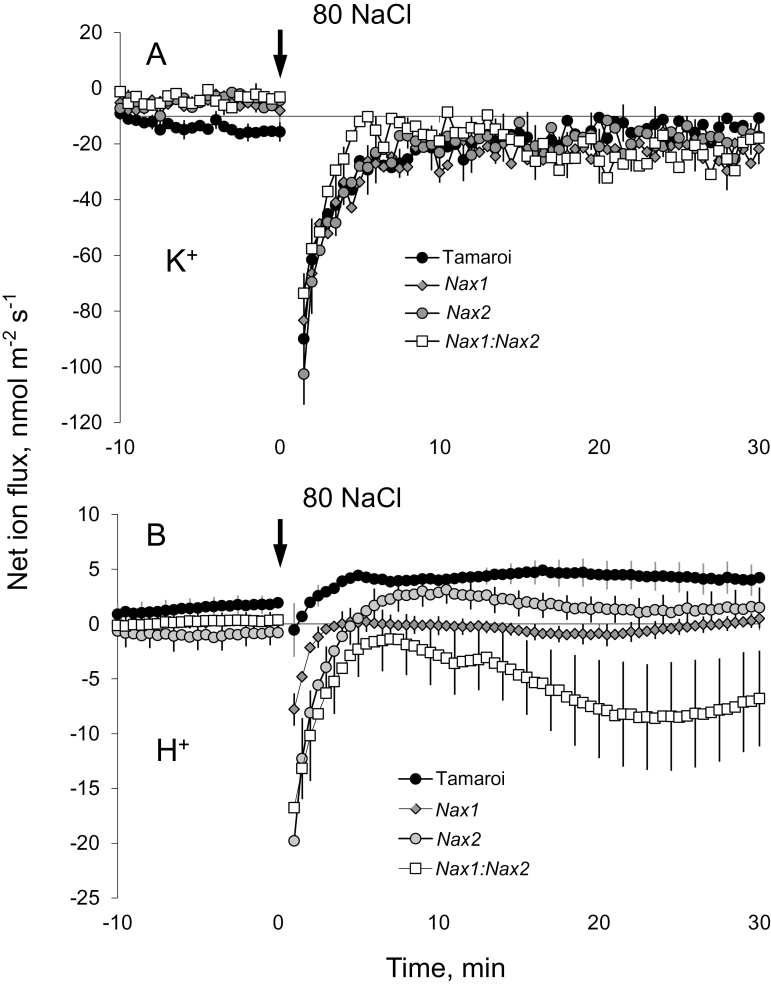
The addition of 80mM NaCl caused significant changes in net ion (K+ and H+) fluxes from the mature root zone of the durum wheat cultivar Tamaroi, which lacks the Nax loci, as well as near-isogenic lines that contained the Nax loci; Fig. 1). Peak K+ efflux was reached within a few minutes of stress onset (Fig. 1A), followed by a gradual recovery of K+ flux (although it always remained negative, i.e. net efflux). No significant (P <0.05) differences in NaCl-induced K+ efflux kinetics were found between Tamaroi and any of the Nax lines. H+ fluxes measured in response to salt treatment were lower for the Nax lines compared with Tamaroi (Fig. 1B), with steady-state H+ flux values (measured 30min after stress onset) being Tamaroi>Nax1=Nax 2>Nax1:Nax2 lines (Fig. 1B). Interestingly, an H+ efflux was observed in the Nax1:Nax2 line, in contrast to the slight H+ influx found in Tamaroi and the other two Nax lines.
Fig. 1.
Transient K+ (A) and H+ (B) flux responses measured from the mature zone of the root epidermis from Nax lines and the parental Tamaroi (lacking Nax loci) in response to acute 80mM NaCl treatment. Mean ±SE (n=6). For all MIFE measurements, the sign convention is ‘efflux negative’.
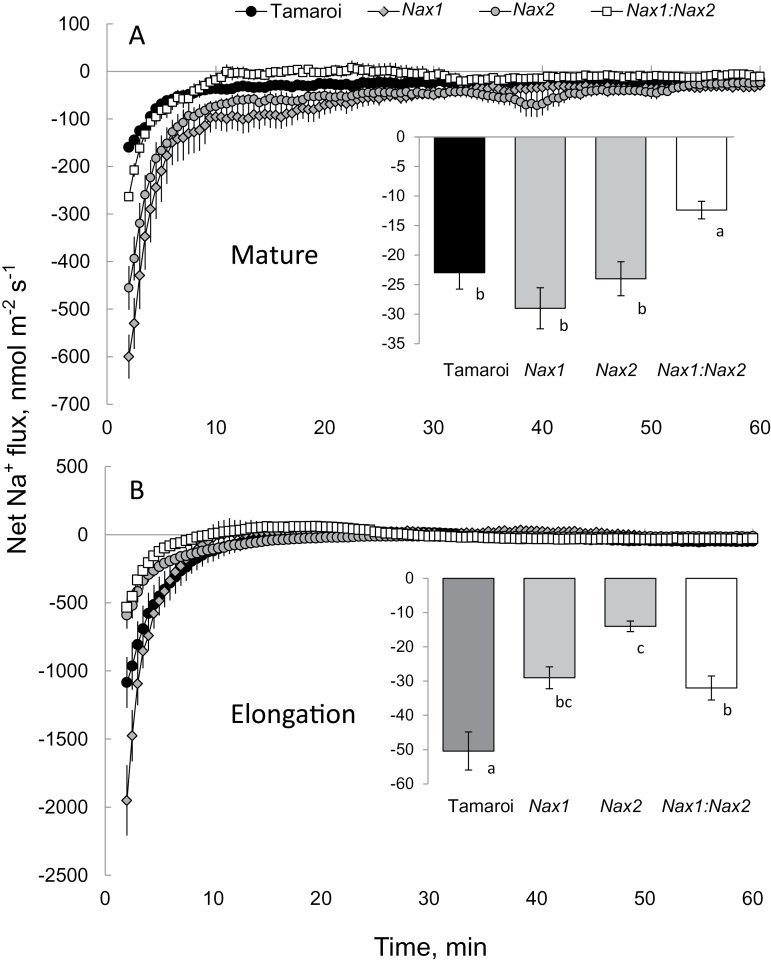
Transfer of salt-treated roots (150mM NaCl for 24h) to Na+-free solution resulted in a significant Na+ efflux from the root epidermis in both the mature and elongation zones (Fig. 2). Inserts in each panel denote steady-state Na+ efflux during the final 30min of the measurement. No clear trends emerged for the mature zone (see insert in Fig. 2A), but the Na+ efflux in the root elongation zone (where SOS1 is predominantly expressed; Shi et al., 2002) in all Nax lines was significantly lower than in Tamaroi (significant at P <0.05).
Fig. 2.
Net Na+ fluxes measured in ‘recovery protocols’ from (A) mature and (B) apical (elongation zone; ~2mm from root tip) zones of epidermal root cells of Nax lines and Tamaroi after transfer from 150mM NaCl solution (24h treatment) to sodium-free BSM. Mean ±SE (n=6). Inserts in each panel denote the steady-state Na+ efflux 60min after the removal of the salt treatment.
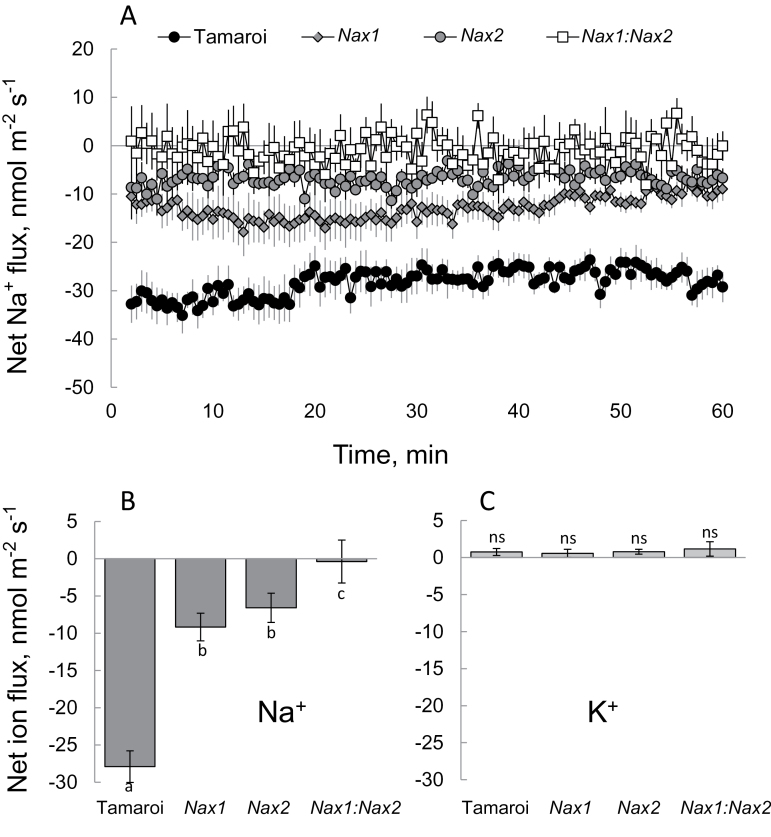
To quantify the activity of the Na+ efflux system at the xylem parenchyma interface, the root stele was mechanically isolated as described in the Materials and Methods, and net ion fluxes were measured after transferring the stele from 50mM NaCl (pre-treated for 6h) to a Na+-free BSM. The net Na+ efflux in Tamaroi was significantly (P <0.05) higher than in the other three genotypes, while the Nax1:Nax2 line had the lowest Na+ efflux. Net K+ fluxes were not significantly different from zero (Fig. 3C), indicating that the observed difference in measured Na+ flux was not an artefact originating from the possible poor discrimination of the Na+ microelectrode LIX between Na+ and K+ (see Chen et al., 2005, for details).
Fig. 3.
(A) Net Na+ fluxes measured in ‘recovery protocols’ from the root stele after 6h exposure to 50mM NaCl. (B, C) Mean Na+ (B) and K+(C) values measured from stelar tissue over the 60min interval after transferring the stele to a Na+-free solution. Mean ±SE (n=6).
Ion flux profiles in leaf sheath and blade tissues
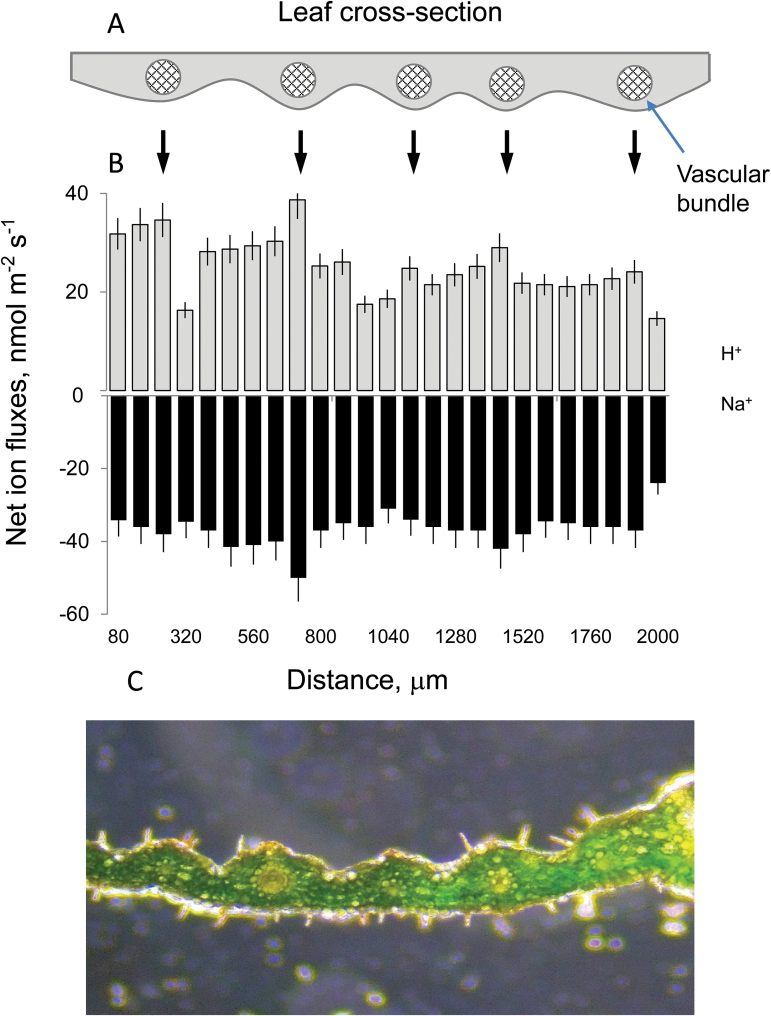
Previous studies have pointed to the leaf sheath, as well as the roots, as the most likely locations of HKT1;4 gene expression conferred by the Nax1 loci (Huang et al., 2006; James et al. 2006). Accordingly, the difference in net ion fluxes between the vascular bundles and leaf mesophyll in Tamaroi and Nax lines was compared. As the HKT genes involved in Na+ retrieval from the xylem are considered to be expressed in tissues surrounding the vascular bundle (Horie et al., 2009), it was critical that the electrodes were positioned exactly above this tissue. To ensure this, methodological experiments were conducted, mapping cross-sectional ion flux profiles in wheat leaves (Fig. 4). Similar to our previous reports for bean mesophyll (Shabala et al., 2002), the ion flux profiles in wheat leaves showed a strong correlation with leaf anatomy, with both the highest H+ influx and the highest Na+ efflux occurring from vascular bundles (Fig. 4B). Consequently, locations with the highest H+ influx and Na+ efflux activity were used to compare parental and Nax lines (depicted in Fig. 5).
Fig. 4.
Ion flux profiles along the cross-section of wheat leaf. (A) Schematic model depicting the position of major veins in a wheat leaf. (B) Net H+ (grey bars) and Na+ (black bars) fluxes measured at different parts of the leaf (see panel A) from control plants to assess the leaf profile. Mean ±SE (n=4 individual leaves). (C) A microscopic image depicting a cross-section of a wheat leaf.
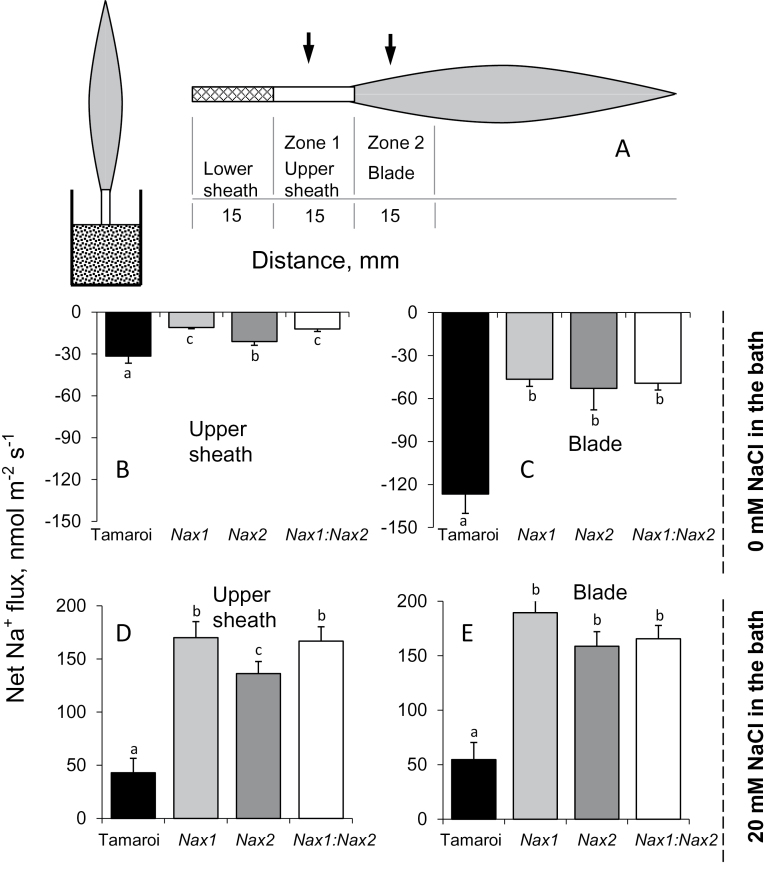
Fig. 5.
Activity of Na+ efflux systems in the leaf blade and upper sheath tissues of wheat lines. (A) A schematic diagram of the experimental protocol. Excised leaves were placed in a beaker containing 20mM NaCl solution with the sheath immersed in the solution to a depth of 15mm and treated for 2 d to accumulate salt. Particular attention was paid to ensure that the depth of insertion and the total surface area of the leaf exposed to salinity was the same in all treatments. (B, C) Steady-state Na+ fluxes measured from leaf segments isolated from the upper sheath (B, zone 1 in panel A) and leaf blade tissue (C, zone 2 in panel A) of leaves treated with 20mM NaCl as described above and then transferred into Na+-free BSM medium for MIFE measurements. (D, E) Net Na+ fluxes from the upper sheath (D) and leaf blade tissue (E) exposed to 20mM NaCl for 2 d, and measured in the presence of 20mM NaCl in the bathing medium. Mean ±SE (n=5).
When detached leaves were placed in 20mM NaCl, Tamaroi leaves showed significantly lower net Na+ uptake compared with all the Nax lines (a 3-fold difference; Fig. 5D, E; significant at P <0.05) in both the upper sheath and blade tissue. As the measured net flux is a sum of channel-mediated Na+ uptake and transporter-mediated Na+ efflux, one possible explanation for this observation may be higher Na+ leaf exclusion ability in Tamaroi. To check this, leaf segments were transferred to a Na+-free medium to reveal the specific contribution of Na+-efflux systems. Consistent with these data, the net Na+ efflux in the leaf blade tissue was much more pronounced in Tamaroi than in any of the Nax lines (Fig. 5B, C; significant at P <0.05). The same pattern was observed both in the upper sheath (Fig. 5D) and leaf blade (Fig. 5E) regions, although net Na+ efflux was about 4-fold stronger from the leaf blade.
Effect of Nax loci on SOS1 transcript level in roots
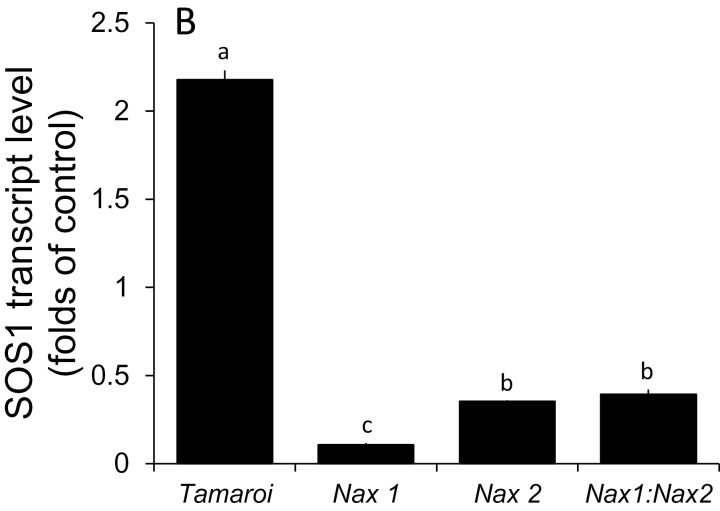
The relative SOS1 transcript level was up-regulated (compared with the control) in salt-treated (150mM for 3 weeks) Tamaroi roots (2.18-fold; Fig. 6), while it was down-regulated in all Nax lines (0.11-, 0.35-, and 0.39-fold for Nax1, Nax2, Nax1:Nax2 lines, respectively, all significant at P <0.001).
Fig. 6.
The relative transcript level of TdSOS1 in roots of Tamaroi and Nax lines (6-d-old seedlings exposed to 150mM NaCl for 24h). Each data point represents six biological replicates in each sample, presented as mean ±SE.
Discussion
The Nax loci restrict Na+ transport from the roots to the leaves. Their effect on net xylem loading of Na+ (James et al., 2006), subsequent Na+ exclusion from leaves, and leaf longevity under saline conditions (James et al., 2011), was, until now, presumed to be due entirely to the action of the Na+ transporters HKT1:4 and HKT1:5, respectively contained within them (Huang et al., 2006; Byrt et al., 2007). We show here that the Nax loci also affect SOS1-like Na+/H+ exchanger expression and activity in durum wheat. While interpreting our data we assume a similarity in SOS1 function and expression pattern between Arabidopsis and wheat.
Activity of a SOS1-like Na + /H + exchanger in root epidermis is suppressed in Nax lines
As shown in Fig. 1B and Fig. 2B, net H+ influx and Na+ efflux in root epidermal cells in Nax lines were much lower than those in Tamaroi. These observations are fully consistent with the notion that the Nax loci also affect SOS1-like Na+/H+ exchanger activity in the root epidermis. First, the observed patterns were only found at the root apex (where SOS1 transporters are most strongly expressed; Shi et al., 2002) but not in the mature root zone (Fig. 2). Second, the measured Na+ flux was sensitive to amiloride, a known inhibitor of the Na+/H+ exchanger (data not shown; but see Cuin et al., 2011, for supporting evidence). In contrast to animals, higher plants lack ATP-driven Na+-pumps, so rely on Na+/H+ exchangers to efflux Na+ back to the apoplast. It is estimated that a typical glycophyte plant effluxes about 90% of the Na+ that enters a root cell (Davenport et al., 2005); the operation of a SOS1 plasma membrane Na+/H+ exchanger seems to be essential to achieve this goal (Shi et al., 2002, 2003). Overexpression of AtSOS1 has been shown to improve salt tolerance in transgenic Arabidopsis (Shi et al., 2003). In addition, OsSOS1 has been characterized in rice and it demonstrates a capacity for Na+/H+ exchange in plasma membrane vesicles of yeast (Saccharomyces cerevisiae) cells, reducing their net cellular Na+ content (Martinez-Atienza et al., 2007). When the activity of the SOS1 exchanger is suppressed under saline conditions, Na+ exclusion and H+ uptake in the root epidermis is reduced. This is what is observed here for all Nax lines.
The SOS1-like exchanger plays a substantial role in xylem Na + loading in wheat and its activity is reduced in stelar tissues of Nax lines
In Arabidopsis, SOS1 genes are preferentially expressed in stelar root tissues (Shi et al., 2002) and are considered to function in xylem Na+ loading (Shi et al., 2002). Our finding that net Na+ efflux is significantly higher in Tamaroi compared with any Nax lines (Fig. 3) is consistent with this proposal and also provides strong supportive evidence for the inhibition of a SOS1-like exchanger by the Nax loci. Qualitatively similar patterns were observed in both the upper sheath (Fig. 5B) and blade of the leaf (Fig. 5C). Inevitably, a certain amount of Na+ will penetrate ‘the first line of defence’ (Na+ exclusion from the root epidermis) and enter the xylem. Here, it will either be (i) retrieved back into the stele by HKT transporters or (ii) transported to the shoot to be compartmentalized by leaf vacuoles where it can contribute to osmotic adjustment. Tamaroi was observed to have significantly more xylem Na+ loading than any of the Nax lines, due to the normal function of SOS1. Thus, it appears that the Nax loci confer two highly complementary mechanisms: an enhanced retrieval of Na+ back into the root stele (as reported elsewhere: Blumwald et al., 2000; Shi et al., 2002), and a reduced rate of Na+ loading into the xylem in the first instance (reported here). Both contribute to the same aim: reducing the xylem Na+ content. It can be speculated that such duality plays an important adaptive role and provides more flexibility to plants. Indeed, as shown in this work, the Nax loci may suppress the activity of a SOS1-like Na+/H+ exchanger in both epidermal (Fig. 2) and stelar (Fig. 3) tissues. This suppression reduces a plant’s ability to exclude Na+ from uptake but, at the same time, the rate of Na+ entering the xylem is also reduced. This should result in more Na+ staying in the roots, to be used either for osmotic adjustment (Shabala and Lew, 2002), or salt stress-signalling (Wu et al., 2015) purposes. Also, from general point of view, it may be beneficial for plants to have another ‘back-up’ mechanism when challenged by salinity stress, in case one of the systems fails to operate.
The reduced SOS1-like activity in the Nax lines could be explained (at least partially) by the reduced level of the SOS1-transcript, as revealed by RT-qPCR experiments (Fig. 6). The down-regulation of TdSOS1 in the root of the Nax line under saline condition could explain why the function of the SOS1 exchanger was altered in both the root epidermis and stele compared with the parental line Tamaroi. It could be speculated that the Nax loci, which consist of a short chromosome segment originating from Triticum monococcum as well as the HKT genes, contain some ‘regulating genes’, which have a negative feedback on the TdSOS1 expression at the transcription level. This issue will be addressed in a future study.
Root ion homeostasis under saline conditions: an improved model
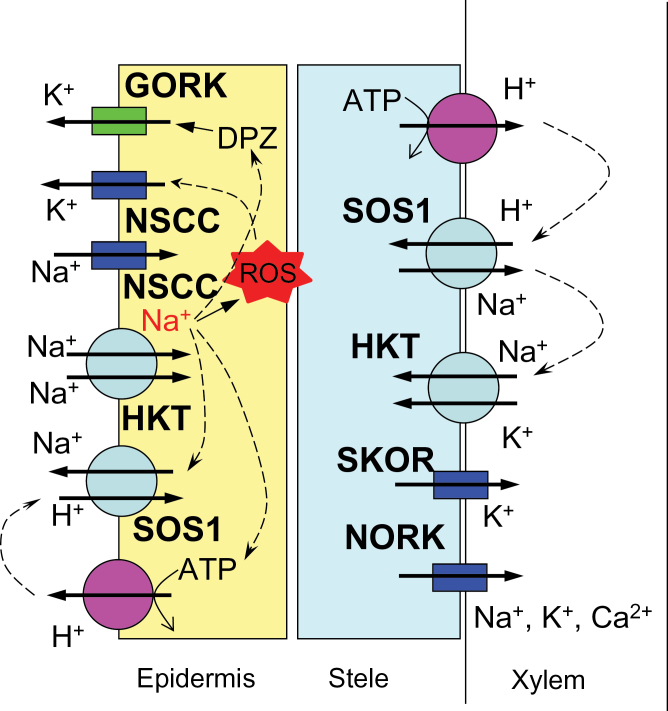
Based on our reported results, the following model can be suggested (Fig. 7). Na+ enters the cell via non-selective cation channels (NSCC, Demidchik and Maathuis, 2007) and/or HKT transporters (Laurie et al., 2002; Garciadeblas et al., 2003; Haro et al., 2005; Horie et al., 2001, 2007), depolarizing the plasma membrane and resulting in a substantial K+ leak from the root epidermis (Fig. 1), mediated by GORK channels (Anschutz et al., 2014; Pottosin and Shabala, 2014). Increased cytosolic Na+ can lead to the accumulation of ROS (Vass et al., 1992; Allakhverdiev et al., 2002), further exacerbating K+ efflux from cytosol via ROS-activated K+-permeable NSCC (Pottosin and Shabala, 2014). No difference in any of above mechanisms exists between Tamaroi and the Nax lines. The major bulk of Na+ will be excluded by plasma membrane-located SOS1 Na+/H+ exchangers, fuelled by the H+-ATPase. Nax loci suppress (directly or indirectly) the transcript level of the SOS1 gene (Fig. 6) and its activity (Fig. 2). Some of the Na+ accumulated in the root cortex is loaded into the xylem, mediated by both passive (at the early stages of salt stress; Shabala et al., 2013) and active (SOS1-mediated) transport systems. In Nax lines, the rate of Na+ loading is suppressed, at either the transcriptional or functional level (or both). Some of the loaded Na+ is removed by HKT transporters located at the xylem parenchyma interface (Munns and Tester, 2008; Horie et al., 2009); an ability much more pronounced in the Nax lines. Hence, xylem Na+ loading is controlled by two highly complementary uptake and release systems, providing the plant with a greater versatility to respond to a changing environment and to control Na+ delivery to the shoot.
Fig. 7.
A model depicting the mechanisms contributing to root ion homeostasis under saline conditions. See the text for details.
Acknowledgements
This work was supported by the Australian Research Council and Grain Research and Development Corporation grants to Sergey Shabala.
References
- Allakhverdiev SI, Nishiyama Y, Miyairi S, Yamamoto H, Inagaki N, Kanesaki Y, Murata N. 2002. Salt stress inhibits the repair of photodamaged photosystem II by suppressing the transcription and translation of psbA genes in Synechocystis . Plant Physiology 130, 1443–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anschutz U, Becker D, Shabala S. 2014. Going beyond nutrition: regulation of potassium homoeostasis as a common denominator of plant adaptive responses to environment. Journal of Plant Physiology 171, 670–687. [DOI] [PubMed] [Google Scholar]
- Blumwald E, Aharon GS, Apse MP. 2000. Sodium transport in plant cells. Biochimica et Biophysica Acta–Biomembranes 1465, 140–151. [DOI] [PubMed] [Google Scholar]
- Bose J, Rodrigo-Moreno A, Shabala S. 2014. ROS homeostasis in halophytes in the context of salinity stress tolerance. Journal of Experimental Botany 65, 1241–1257. [DOI] [PubMed] [Google Scholar]
- Byrt CS, Platten JD, Spielmeyer W, James RA, Lagudah ES, Dennis ES, Tester M, Munns R. 2007. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1 . Plant Physiology 143, 1918–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrt CS, Xu B, Krishnan M, et al. 2014. The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat. The Plant Journal 80, 516–526. [DOI] [PubMed] [Google Scholar]
- Chen Z, Newman I, Zhou M, Mendham N, Zhang G, Shabala S. 2005. Screening plants for salt tolerance by measuring K+ flux: a case study for barley. Plant, Cell and Environment 28, 1230–1246. [Google Scholar]
- Colmenero-Flores JM, Moreno LP, Smith CE, Covarrubias AA. 1999. Pvlea-18, a member of a new late-embryogenesis-abundant protein family that accumulates during water stress and in the growing regions of well-irrigated bean seedlings. Plant Physiology 120, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuin TA, Bose J, Stefano G, Jha D, Tester M, Mancuso S, Shabala S. 2011. Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: in planta quantification methods. Plant, Cell and Environment 34, 947–961. [DOI] [PubMed] [Google Scholar]
- Cuin TA, Tian Y, Betts SA, Chalmandrier R, Shabala S. 2009. Ionic relations and osmotic adjustment in durum and bread wheat under saline conditions. Functional Plant Biology 36, 1110–1119. [DOI] [PubMed] [Google Scholar]
- Davenport RJ, Munoz-Mayor A, Jha D, Essah PA, Rus A, Tester M. 2007. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis . Plant, Cell and Environment 30, 497–507. [DOI] [PubMed] [Google Scholar]
- Demidchik V, Maathuis FJM. 2007. Physiological roles of nonselective cation channels in plants: from salt stress to signalling and development. New Phytologist 175, 387–404. [DOI] [PubMed] [Google Scholar]
- Feki K, Quintero FJ, Khoudi H, Leidi EO, Masmoudi K, Pardo JM, Brini F. 2014. A constitutively active form of a durum wheat Na+/H+ antiporter SOS1 confers high salt tolerance to transgenic Arabidopsis. Plant Cell Reports 33, 277–288. [DOI] [PubMed] [Google Scholar]
- Feki K, Quintero FJ, Pardo JM, Masmoudi K. 2011. Regulation of durum wheat Na+/H+ exchanger TdSOS1 by phosphorylation. Plant Molecular Biology 76, 545–556. [DOI] [PubMed] [Google Scholar]
- Garciadeblas B, Senn ME, Banuelos MA, Rodriguez-Navarro A. 2003. Sodium transport and HKT transporters: the rice model. The Plant Journal 34, 788–801. [DOI] [PubMed] [Google Scholar]
- Gorham J, Wyn Jones RG, Bristol A. 1990. Partial characterization of the trait for enhanced K+–Na+ discrimination in the D genome of wheat. Planta 180, 590–597. [DOI] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK. 2000. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proceedings of the National Academy of Sciences, USA 97, 3735–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro R, Banuelos MA, Senn MAE, Barrero-Gil J, Rodriguez-Navarro A. 2005. HKT1 mediates sodium uniport in roots. Pitfalls in the expression of HKT1 in yeast. Plant Physiology 139, 1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. 2000. Plant cellular and molecular responses to high salinity. Annual Review of Plant Physiology and Plant Molecular Biology 51, 463–499. [DOI] [PubMed] [Google Scholar]
- Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI. 2007. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO Journal 26, 3003–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Hauser F, Schroeder JI. 2009. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends in Plant Science 14, 660–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A. 2001. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa . The Plant Journal 27, 129–138. [DOI] [PubMed] [Google Scholar]
- Huang S, Spielmeyer W, Lagudah ES, James RA, Platten JD, Dennis ES, Munns R. 2006. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiology 142, 1718–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Liu JP, Halfter U, Kim CS, Shi WM, Zhu JK. 2000. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. The Plant Cell 12, 1667–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James RA, Blake C, Byrt CS, Munns R. 2011. Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. Journal of Experimental Botany 62, 2939–2947. [DOI] [PubMed] [Google Scholar]
- James RA, Davenport RJ, Munns R. 2006. Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2 . Plant Physiology 142, 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR. 1997. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. The Plant Journal 12, 1067–1078. [DOI] [PubMed] [Google Scholar]
- Laurie S, Feeney KA, Maathuis FJM, Heard PJ, Brown SJ, Leigh RA. 2002. A role for HKT1 in sodium uptake by wheat roots. The Plant Journal 32, 139–149. [DOI] [PubMed] [Google Scholar]
- Liu JP, Ishitani M, Halfter U, Kim CS, Zhu JK. 2000. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proceedings of the National Academy of Sciences, USA 97, 3730–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Zhu JK. 1998. A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945. [DOI] [PubMed] [Google Scholar]
- Martinez-Atienza J, Jiang XY, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, Quintero FJ. 2007. Conservation of the salt overly sensitive pathway in rice. Plant Physiology 143, 1001–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullan DJ, Colmer TD, Francki MG. 2007. Arabidopsis–rice–wheat gene orthologues for Na+ transport and transcript analysis in wheat–L. elongatum aneuploids under salt stress. Molecular Genetics and Genomics 277, 199–212. [DOI] [PubMed] [Google Scholar]
- Munns R, James RA. 2003. Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant and Soil 253, 201–218. [Google Scholar]
- Munns R, James RA, Xu B, et al. 2012. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nature Biotechnology 30, 360–364. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annual Review of Plant Biology 59, 651–681. [DOI] [PubMed] [Google Scholar]
- Pottosin I, Shabala S. 2014. Polyamines control of cation transport across plant membranes: implications for ion homeostasis and abiotic stress signaling. Frontiers in Plant Science 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadir M, Quillerou E, Nangia V, Murtaza G, Singh M, Thomas RJ, Drechsel P, Noble AD. 2014. Economics of salt-induced land degradation and restoration. Natural Resources Forum 38, 282–295. [Google Scholar]
- Rahnama A, James RA, Poustini K, Munns R. 2010. Stomatal conductance as a screen for osmotic stress tolerance in durum wheat growing in saline soil. Functional Plant Biology 37, 255–263. [Google Scholar]
- Rengasamy P. 2010. Soil progress affecting crop production in salt-affected soils. Functional Plant Biology 37, 613–620. [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB. 2000. The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. The Plant Cell 12, 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S. 2013. Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Annals of Botany 112, 1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S, Bose J, Hedrich R. 2014. Salt bladders: do they matter? Trends in Plant Science 19, 687–691. [DOI] [PubMed] [Google Scholar]
- Shabala S, Hariadi Y, Jacobsen S-E. 2013. Genotypic difference in salinity tolerance in quinoa is determined by differential control of xylem Na+ loading and stomatal density. Journal of Plant Physiology 170, 906–914. [DOI] [PubMed] [Google Scholar]
- Shabala S, Shabala L. 2002. Kinetics of net H+, Ca2+, K+, Na+ , NH4+, and Cl– fluxes associated with post-chilling recovery of plasma membrane transporters in Zea mays leaf and root tissues. Physiologia Plantarum 114, 47–56. [DOI] [PubMed] [Google Scholar]
- Shabala S, Shabala L, Gradmann D, Chen ZH, Newman I, Mancuso S. 2006. Oscillations in plant membrane transport: model predictions, experimental validation, and physiological implications. Journal of Experimental Botany 57, 171–184. [DOI] [PubMed] [Google Scholar]
- Shabala S, Shabala S, Cuin TA, Pang J, Percey W, Chen Z, Conn S, Eing C, Wegner LH. 2010. Xylem ionic relations and salinity tolerance in barley. The Plant Journal 61, 839–853. [DOI] [PubMed] [Google Scholar]
- Shabala SN, Lew RR. 2002. Turgor regulation in osmotically stressed Arabidopsis epidermal root cells. Direct support for the role of inorganic ion uptake as revealed by concurrent flux and cell turgor measurements. Plant Physiology 129, 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala SN, Newman IA, Morris J. 1997. Oscillations in H+ and Ca2+ ion fluxes around the elongation region of corn roots and effects of external pH. Plant Physiology 113, 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR. 2009. Wheat. Journal of Experimental Botany 60, 1537–1553. [DOI] [PubMed] [Google Scholar]
- Shi HZ, Lee BH, Wu SJ, Zhu JK. 2003. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana . Nature Biotechnology 21, 81–85. [DOI] [PubMed] [Google Scholar]
- Shi HZ, Quintero FJ, Pardo JM, Zhu JK. 2002. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. The Plant Cell 14, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass I, Styring S, Hundal T, Koivuniemi A, Aro EM, Andersson B. 1992. Reversible and irreversible intermediates during photoinhibition of photosystem .2. Stable reduced QA species promote chlorophyll triplet formation. Proceedings of the National Academy of Sciences, USA 89, 1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HH, Shabala L, Liu XH, Azzarello E, Zhou M, Pandolfi C, Chen ZH, Bose J, Mancuso S, Shabala S. 2015. Linking salinity stress tolerance tissue-specific Na+ sequestration in wheat roots. Frontiers in Plant Science 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. 2001. Plant salt tolerance. Trends in Plant Science 6, 66–71. [DOI] [PubMed] [Google Scholar]
- Zhu JK, Liu JP, Xiong LM. 1998. Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. The Plant Cell 10, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]