Abstract
Differentiating erythroblasts execute a dynamic alternative splicing program shown here to include extensive and diverse intron retention (IR) events. Cluster analysis revealed hundreds of developmentally-dynamic introns that exhibit increased IR in mature erythroblasts, and are enriched in functions related to RNA processing such as SF3B1 spliceosomal factor. Distinct, developmentally-stable IR clusters are enriched in metal-ion binding functions and include mitoferrin genes SLC25A37 and SLC25A28 that are critical for iron homeostasis. Some IR transcripts are abundant, e.g. comprising ∼50% of highly-expressed SLC25A37 and SF3B1 transcripts in late erythroblasts, and thereby limiting functional mRNA levels. IR transcripts tested were predominantly nuclear-localized. Splice site strength correlated with IR among stable but not dynamic intron clusters, indicating distinct regulation of dynamically-increased IR in late erythroblasts. Retained introns were preferentially associated with alternative exons with premature termination codons (PTCs). High IR was observed in disease-causing genes including SF3B1 and the RNA binding protein FUS. Comparative studies demonstrated that the intron retention program in erythroblasts shares features with other tissues but ultimately is unique to erythropoiesis. We conclude that IR is a multi-dimensional set of processes that post-transcriptionally regulate diverse gene groups during normal erythropoiesis, misregulation of which could be responsible for human disease.
INTRODUCTION
Erythroid differentiation represents an excellent model system for exploring stage-specific post-transcriptional remodeling of gene expression during terminal differentiation. Fluorescence-activated cell sorting (FACS) makes possible isolation of discrete, highly purified populations of cells as they differentiate, enucleate to form reticulocytes and ultimately mature into red cells. Early progenitors known as burst-forming unit-erythroid (BFU-E) and colony-forming unit-erythroid (CFU-E) can be highly purified by this approach, as can proerythroblasts (proE) and several stages of terminally differentiating erythroblasts termed basophilic erythroblasts (basoE), polychromatophilic erythroblasts (polyE) and orthochromatophilic erythroblasts (orthoE). We and others have analyzed RNA-seq libraries prepared from these purified populations of human erythroid cells to gain new insights into the evolving erythroid transcriptome at the level of gene-level expression, alternative splicing, non-coding RNA expression, etc. (1–3). Moreover, similar analysis of mouse erythroblast populations allows for comparisons of gene expression patterns among mammalian species (1,3).
Proliferating mammalian erythroblasts exhibit a robust, dynamic alternative splicing program (2,4–5) enriched in genes involved in cell cycle, organelle organization, chromatin function and RNA processing (2). A prominent feature of the erythroblast splicing program is a number of alternative splicing ‘switches’ that increase PSI (percent spliced in) values predominantly in late erythroblasts at the polyE and orthoE stages, temporally correlated with major cellular remodeling as cells conclude their proliferation phase and prepare for enucleation. Splicing switches can alter protein function in physiologically important ways, e.g. upregulation of exon 16 splicing in protein 4.1R transcripts leads to synthesis of protein isoforms that bind spectrin and actin with high affinity, mechanically strengthening the red cell membrane prior to release into the circulation (6–8). In most cases, however, understanding the physiological functions of alternative protein isoforms generated via the erythroblast splicing program remains a challenge for future studies.
Intron retention (IR) is emerging as an unexpectedly rich contributor to transcriptome diversity, providing a mechanism for gene regulation during normal differentiation and development. Recent surveys have revealed extensive IR events with distinct tissue-, developmental- and stress-specific expression patterns (9–13), suggesting precise regulation by the splicing machinery. Widespread intron retention also characterizes many cancer transcriptomes (14). Global screens across many cell and tissue types from human and mouse show surprising abundance of IR, such that 35% of multi-exon genes contain intron(s) with ≥50% retention in at least one cell type (15). IR events are also particularly abundant in plants (16). Several functions have been proposed for IR, which could provide a post-transcriptional mechanism to downregulate gene expression by inducing degradation by nuclear surveillance machinery (13) or by nonsense-mediated decay (NMD) (12). Alternatively, IR could represent a conditional block to gene expression that might be relieved to facilitate intron removal in response to appropriate signaling events (17) or developmental cues (18).
Previous studies of the erythroid transcriptome entirely over-looked the IR component of the splicing program. To understand the role of IR in mammalian erythroblasts during terminal erythropoiesis, we developed custom software available at (https://github.com/pachterlab/kma) to analyze IR in RNA-seq data and applied these methods to study IR in populations of human erythroblasts from proE to orthoE. These new studies show that erythroblasts elaborate an extensive and diverse intron retention program encompassing numerous essential erythroid genes including those encoding splicing factors and proteins involved in iron homeostasis. Differentiation stage-specific changes in IR efficiency largely paralleled switches in splicing of cassette exons described earlier (2), reinforcing and expanding the concept that careful regulation of RNA processing plays a major role in terminal erythroid differentiation as cells mature along the path from proE to orthoE.
MATERIALS AND METHODS
Computational methods
RNA-seq reads were mapped using Bowtie v2.1.0 to an augmented transcriptome output by KeepMeAround (KMA) as described in our recently posted preprint (arXiv:1510.00696). Transcripts and introns were then quantified using eXpress v1.5.1 (19). We identified an unambiguous set of 186 838 quantifiable introns in the RefSeq transcriptome, but only 10 152 unique introns passed filters (below) in every condition. Intron retention values were calculated by taking the ratio of transcripts per million (TPM) (20) values of the intron (numerator) to the sum of TPM values of the overlapping isoforms and intron (denominator), resulting in IR levels on the [0, 1] scale. To reduce false positives, we removed introns with fewer than three uniquely mapped reads, denominator values of less than 1 TPM (excluding the intron expression) and introns with zero coverage regions longer than 20% of the intron length (Supplementary Information in (2)). KMA's hypothesis testing feature was then used on the filtered set of introns to test whether retention levels were higher than expected given the background retention levels in each experimental condition. This test also incorporates biological replicates to further reduce the chance of false positives.
Cluster analysis was performed using k-means clustering on a set of introns that passed the above filters in every sample and every condition. After clustering, GO analysis was performed using DAVID tools (21,22) on the genes from the introns in clusters C1-C6.
Code for analysis and plots can be found at https://github.com/pimentel/erythroid_ir_analysis.
Splice site strength was calculated using MaxEntScan with the maximum entropy score and default parameters (23).
RNA-seq data
RNA-seq data obtained from five highly purified human erythroblast populations—proerythroblasts (proE), early basophilic erythroblasts (e-basoE), late basophilic erythroblasts (l-basoE), polychromatophilic erythroblasts (polyE) and orthochromatophilic erythroblasts (orthoE) (24)—is available at GSE53635. The data include three biological replicates of each population. Human granulocyte RNA-seq data (12) was downloaded from GSE48307. For other tissues, we imported wiggle plots, showing RNA-seq coverage along the genome, that were generated from Illumina BodyMap 2.0 data available at http://www.ensembl.org/info/genome/genebuild/rnaseq_annotation.html.
Erythroblast cultures
To prepare RNA and protein for further analysis of IR, CD34+ cells were purified from cord blood and differentiated into erythroblasts over the course of 16 days as described (24).
Nuclear isolation
Nuclei were prepared from ∼20 million erythroblasts according to published methods (25), with minor modifications. In brief, the erythroblast plasma membrane was lysed using 0.05% NP40, and nuclei were separated from the reddish hemoglobin-rich cytoplasmic fraction by centrifugation through a sucrose cushion at ∼2000 rpm. The whitish nuclear pellet was rinsed with ice-cold phosphate buffered saline containing 1 mM ethylenediaminetetraacetic acid and was resuspended gently to generate a turbid suspension in which nuclei were microscopically verified. Purity of the nuclear fractions was further confirmed by immunoblotting with antibodies to U1–70K protein (a kind gift from D. Black, UCLA).
RT-PCR analysis of IR transcripts
RNA was purified from cultured erythroblasts as described previously using RNeasy columns according to the manufacturer's instructions (Qiagen), but with the addition of a DNase step to eliminate potential contamination by genomic DNA. RNA from nuclear and cytoplasmic fractions was prepared using Trizol (Life Technologies). To provide additional assurance that intron-containing polymerase chain reaction (PCR) products were not derived from contaminating genomic DNA, we designed PCR assays to span at least one constitutively spliced intron as well as the candidate retained intron. PCR reaction conditions were adjusted to allow for amplification of IR products ≥3 kb in length (denaturation at 95°C for 20″, annealing at 60°C for 10″, extension at 70°C for 1′15″; 35 cycles) using KOD polymerase in the presence of betaine to enhance amplification. PCR products were analyzed on either 2% agarose gels (for products >1.5 kb) or 4.5% acrylamide gels. All PCR products discussed in the manuscript were confirmed by DNA sequencing.
RESULTS
IR is a major feature of gene expression in differentiating human erythroblasts
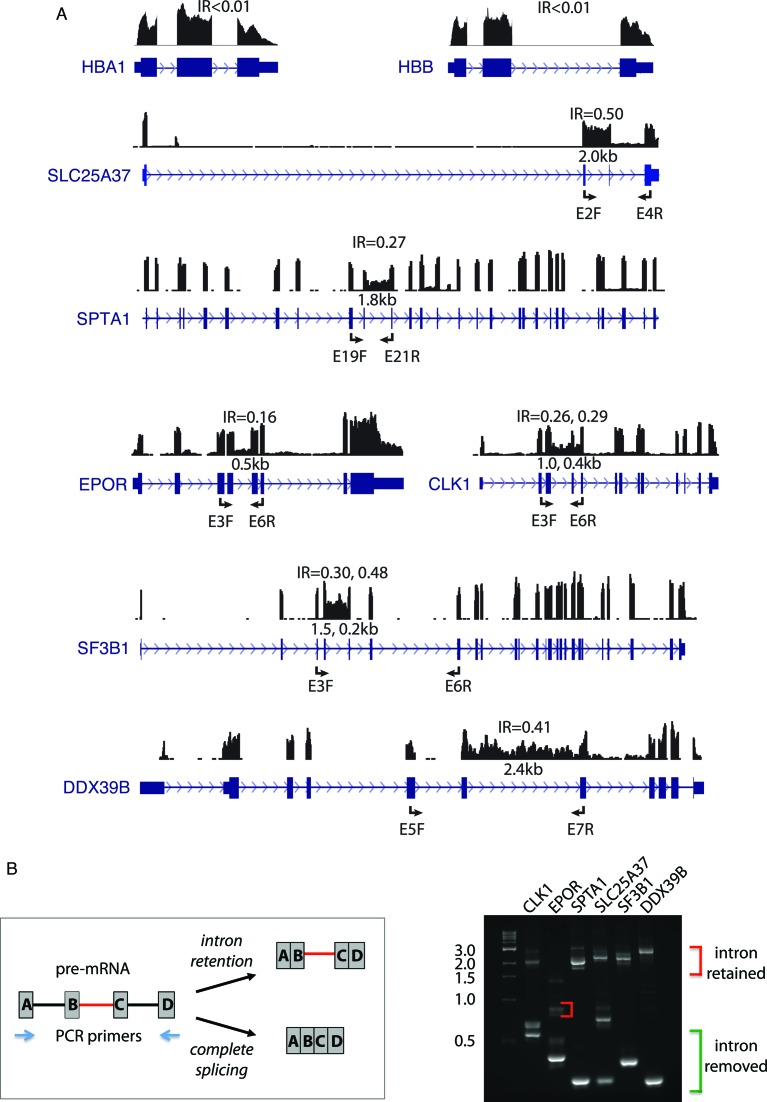
We mapped RNA-seq reads from highly purified human erythroblast populations to an augmented transcriptome, including introns, in order to detect all expressed regions independent of existing transcript annotations. Preliminary inspection of mapping data in the wiggle plot format, displaying RNA-seq read density along the genome, revealed that most introns were efficiently spliced in all erythroblast populations. For example, the α and β globin genes exhibited major peaks in read density over the exons and deep troughs in intronic regions due to highly efficient joining of exons and removal of introns during pre-mRNA splicing (Figure 1A, upper). Many housekeeping genes such as those encoding glycolytic enzymes also exhibited negligible IR (Supplementary Figure S1). In contrast, a number of important erythroid transcripts exhibited substantial IR (Figure 1A, lower). A very prominent IR event was found in the mitoferrin-1 gene (SLC25A37), which encodes a mitochondrial iron import protein that is critical for iron homeostasis and abundant heme biosynthesis in late erythroblasts. SLC25A37 intron 2, ∼2 kb in length, was highly retained in orthoE, while introns 1 and 3 were retained at much lower levels. Another major IR event occurs in the SPTA1 gene, encoding the structural protein α-spectrin best known for its essential role in promoting assembly of a mechanically stable erythroid membrane skeleton. Intron 20 (1.8 kb) exhibited substantial retention. We also observed moderate IR in EPOR (encoding the erythropoietin receptor), and in spliceosome-associated RNA binding proteins including UAP56 (encoded by DDX39B) and SAP155 (encoded by SF3B1). The latter is an important RNA splicing factor that is frequently mutated in the RARS (refractory anemia with ringed sideroblasts) subtype of myelodysplasia syndrome (MDS). As reported previously (17), IR also occurs in the CLK1 gene, encoding a tyrosine kinase that phosphorylates splicing factors of the SR protein family.
Figure 1.
Intron retention in important erythroid genes. (A) Wiggle plots showing RNA-seq reads from the orthoE stage mapped to genes with no IR (top panel, HBA1 and HBB) and genes with significant retention of one or more introns (SLC25A37, SPTA1, EPOR, CLK1, SF3B1 and DDX39B). 5′ and 3′ ends of the SPTA1 gene are not shown due to size constraints. Size of retained introns is indicated in kilobases and primer locations for PCR validations are shown. (B) RT-PCR confirmation of IR. The general PCR scheme is pictured at the left, while PCR products are shown at the right. Lane M, size standards.
Intron retention was validated by RT-PCR analysis of RNA isolated from multiple independent cultures of human erythroblasts. Amplicons spanned at least one retained intron and one constitutive intron, in order to demonstrate that intron-specific retention occurs within the context of a larger transcript, and to rule out artifacts that might be caused by DNA contamination. Figure 1B illustrates several examples in which both the fully spliced and the intron retention products were amplified. These results confirmed that RNA-seq reads mapping to these introns were expressed in the context of intact introns retained between flanking exons in stably expressed erythroblast transcripts.
Global analysis of IR in erythroblasts
Genome-wide study of IR in erythroblasts was performed using new computational tools that assign a retention value to every intron relative to expression of the flanking exons (see ‘Materials and Methods’ section). Applying these tools to RNA-seq data from the five erythroblast populations revealed wide variations in percent intron retention (IR), length of retained introns and number of introns retained per transcript. Hundreds of introns were retained at IR > 0.10 in at least one erythroblast population (see Supplementary Table S1 for a complete summary of calculated IR values). Some of these represented single IR events in otherwise efficiently-spliced transcripts; however, there were also many transcripts that retained multiple introns. The distribution of IR values across the erythroblast populations showed that overall IR increases as erythroblasts differentiate, with highest IR in cells at the orthoE stage (Supplementary Figure S2). These data demonstrate that a robust IR program affects the expression of many important erythroid genes.
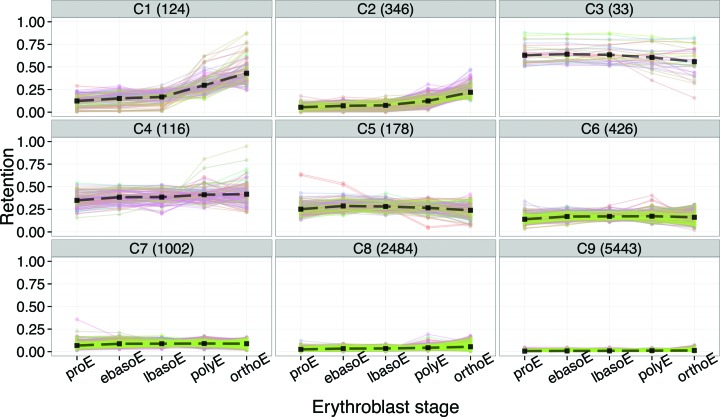
We reasoned that dynamic regulation of IR events might be an important gene regulatory mechanism during terminal erythropoiesis, similar to stage-specific exon splicing switches executed in late erythroblasts (2). Cluster analysis of IR values for each intron at all five maturational stages revealed nine groups of introns (Figure 2). Clusters C1 and C2, comprising ∼470 introns, represent developmentally dynamic events that substantially increase IR in the last two differentiation stages. In contrast, clusters C3–C9 constitute a graded series of developmentally stable intron groups with differentiation-independent IR values, i.e. relatively little change from proE to orthoE. IR is relatively high in C3 but much lower in C9.
Figure 2.
Cluster analysis of IR during erythroblast differentiation. The number of introns in each cluster C1–C9 is indicated in parentheses.
Analysis of intragenic IR patterns revealed that C3, and to a lesser extent in C4, differed qualitatively from the other clusters in that many of the highly retained introns mapped to the first or last intron of a transcript (Supplementary Figure S3). Some of these events might therefore represent alternative initiation or termination of transcription, rather than intron retention per se. However, a few high-level IR events did localize to internal introns (e.g. in SLC25A37).
Computational predictions of developmentally dynamic and developmentally stable IR classes were validated by examination of RNA-seq read-mapping patterns for individual genes (Supplementary Figure S4A), and by experimental analysis of selected introns using via RT-PCR (Supplementary Figure S4B). Both approaches confirmed that some introns exhibited increased IR in late erythroblasts, while other introns maintained a more constant IR during terminal erythropoiesis. These findings mirror earlier observations of exon splicing patterns: erythroblasts alternatively splice hundreds of cassette exons, many of which maintain stable PSI values throughout terminal erythropoiesis, while a select subset undergo significant increases in PSI in late-stage polyE and orthoE (2).
To evaluate the relative abundance of IR transcripts in erythroblasts, we compared IR values with gene expression levels. SLC25A37, one of the most highly expressed genes in orthoE, exhibits ∼50% retention of intron 2, so that the IR isoform is estimated to rank as the 10th most abundant polyadenylated non-globin transcript. Other genes expressing abundant IR isoforms include SPTA1 (182nd in abundance with IR ≈ 0.27) and splicing factor SF3B1 (315th in abundance with IR ≈ 0.50). IR transcripts can thus be highly expressed in late erythroblasts, potentially acting as a major post-transcriptional control point to limit functional mRNA levels.
Dynamic and stable IR events modulate expression of functionally distinct gene classes
Gene ontology (GO) analysis was performed to test whether different IR clusters with might be enriched for different biological functions. We found that the dynamically increased IR events in clusters C1 and C2 were greatly enriched for terms related to RNA processing (Table 1). As shown in Table 2, prominent among these were spliceosome-associated factors including U1 snRNP components U1–70K and U1A (encoded by SNRNP70 and SNRPA1), the U2 snRNP subunit SAP155 (encoded by SF3B1), the U2-associated factor UAP56 (DDX39B) and its paralog DDX39A. Other related genes with introns clustering in C1 or C2 include several hnRNP proteins (HNRNPA2B1, HNRNPD, HNRNPH1, HNRNPH3, HNRNPL), RBM proteins (RBM17, RBM3, RBM39, RBM6), SR proteins (SRSF6, SRSF10) and other proteins with important functions in RNA metabolism (FUS, ACIN1, EWSR, NXF1). Importantly, maturation-associated IR events shown in Table 2 were confirmed by inspection of gene-specific wiggle plots, a few of which are shown in Supplementary Figure S4.
Table 1. Biological processes enriched in IR clusters.
| GO terms for Clusters 1 and 2 | Raw P-value | Corrected P-value |
|---|---|---|
| GO:0008380∼RNA splicing | 9.11E-17 | 1.67E-13 |
| GO:0006397∼mRNA processing | 2.84E-13 | 2.14E-10 |
| GO:0016071∼mRNA metabolic process | 4.72E-13 | 2.37E-10 |
| GO:0000375∼RNA splicing, via transesterification reactions | 4.48E-12 | 1.35E-09 |
| GO:0000377∼RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 3.88E-12 | 1.46E-09 |
| GO:0000398∼nuclear mRNA splicing, via spliceosome | 3.88E-12 | 1.46E-09 |
| GO:0006396∼RNA processing | 1.68E-11 | 4.23E-09 |
| GO:0006350∼transcription | 1.19E-05 | 0.003 |
| GO:0003723∼RNA binding | 7.98E-06 | 0.004 |
| GO:0016044∼membrane organization | 9.07E-05 | 0.017 |
| GO:0016265∼death | 1.45E-04 | 0.024 |
| GO:0051168∼nuclear export | 1.79E-04 | 0.024 |
| GO:0045449∼regulation of transcription | 1.76E-04 | 0.026 |
| GO:0051276∼chromosome organization | 2.43E-04 | 0.030 |
| GO:0009057∼macromolecule catabolic process | 2.95E-04 | 0.034 |
| GO:0012502∼induction of programmed cell death | 4.22E-04 | 0.037 |
| GO:0008219∼cell death | 3.54E-04 | 0.037 |
| GO:0043122∼regulation of I-kappaB kinase/NF-kappaB cascade | 4.09E-04 | 0.038 |
| GO:0043122∼regulation of I-kappaB kinase/NF-kappaB cascade | 4.09E-04 | 0.038 |
| GO:0006917∼induction of apoptosis | 4.09E-04 | 0.040 |
| GO:0007049∼cell cycle | 5.36E-04 | 0.044 |
| GO:0008270∼zinc ion binding | 0.02 | 0.63 |
| GO:0043167∼ion binding | 0.04 | 0.68 |
| GO:0043169∼cation binding | 0.12 | 0.85 |
| GO:0046872∼metal ion binding | 0.16 | 0.90 |
| GO:0046914∼transition metal ion binding | 0.21 | 0.94 |
| GO Terms for Clusters 4 and 5 | Raw P-value | Corrected P-value |
| GO:0046914∼transition metal ion bindinga | 1.63E-04 | 0.028 |
| GO:0008270∼zinc ion binding | 6.17E-04 | 0.036 |
| GO:0046872∼metal ion binding | 5.16E-04 | 0.036 |
| GO:0019899∼enzyme binding | 1.05E-04 | 0.036 |
| GO:0043167∼ion binding | 4.57E-04 | 0.039 |
| GO:0045449∼regulation of transcription | 3.78E-05 | 0.040 |
| GO:0043169∼cation binding | 4.04E-04 | 0.046 |
| GO:0016563∼transcription activator activity | 9.46E-04 | 0.046 |
| GO:0008380∼RNA splicing | 0.01 | 0.83 |
aIncludes the mitoferrins SLC25A37 and SLC25A28.
Table 2. Transcripts for RNA binding proteins that exhibit increased IR in late erythroblasts.
| Gene | Intron coordinates | retention (proE to orthoE)1 | Cluster | Comments |
|---|---|---|---|---|
| SNRNP70 | chr19:49606845–49607890 | 0.12–0.32 | 1 | U1 snRNP protein U1–70K |
| SF3B1 | chr2:198283676–198285151 | 0.09–0.30 | 2 | U2 snRNP protein SAP155 |
| chr2:198283313–198283520 | 0.23–0.52 | 1 | ||
| DDX39B | chr6:31499183–31500556 | 0.08–0.18 | 2 | U2 snRNP-associated protein UAP56 |
| chr6:31500689–31503143 | 0.20–0.41 | 1 | ||
| SRRM2 | chr16:2819285–2820352 | 0.12–0.48 | 1 | |
| DDX39A | chr19:14520260–14520347 | 0.06–0.15 | 2 | Paralog of UAP56 |
| chr19:14520685–14521027 | 0.16–0.49 | 1 | ||
| chr19:14521147–14521800 | 0.04–0.22 | 2 | ||
| chr19:14521985–14522317 | 0.10–0.28 | 2 | ||
| chr19:14523491–14523824 | 0.07–0.25 | 2 | ||
| SNRPA1 | chr15:101826007–101826418 | 0.12–0.39 | 1 | U2 snRNP protein |
| SRSF3 | chr6:36568054–36568928 | 0.03–0.29 | 2 | SR protein family |
| SRSF5 | chr14:70235614–70235898 | 0.04–0.25 | 2 | SR protein family |
| chr14:70235969–70237183 | 0.07–0.31 | 2 | ||
| SRSF6 | chr20:42088061–42088410 | 0.14–0.37 | 1 | SR protein family |
| SRSF7 | chr2:38975796–38976039 | 0.30–0.60 | SR protein family | |
| chr2:38976489–38976670 | 0.26–0.40 | 2 | ||
| SRSF9 | chr12:120901926–120903429 | 0.03–0.17 | 2 | SR protein family |
| SRSF10 | chr1:24301566–24304400 | 0.08–0.33 | 2 | SR protein family |
| SRSF11 | chr1:70694238–70697950 | 0.12–0.29 | 6 | SR protein family |
| HNRNPD | chr4:83275308–83275907 | 0.13–0.34 | 1 | hnRNP family |
| HNRNPDL | chr4:83346037–83346715 | 0.31–0.50 | 4 | hnRNP family |
| chr4:83346821–83347189 | 0.36–0.53 | 4 | ||
| HNRNPH1 | chr5:179043220–179043869 | 0.06–0.17 | 2 | hnRNP family |
| chr5:179048037–179048242 | 0.35–0.49 | 4 | ||
| chr5:179048400–179048843 | 0.15–0.40 | 1 | ||
| chr5:179048979–179050037 | 0.06–0.19 | 2 | ||
| HNRNPL | chr19:39331200–39334481 | 0.03–0.16 | 2 | hnRNP family |
| DDX5 | chr17:62496892–62498127 | 0.11–0.26 | 6 | RNA helicase |
| chr17:62498342–62498556 | 0.10–0.24 | 6 | RNA helicase | |
| RBM3 | chrX:48434056–48434701 | 0.07–0.49 | 1 | RNA binding motif (RBM) family |
| RBM6 | chr3:50098981–50099394 | 0.13–0.42 | 1 | RBM family |
| RBM15 | chr1:110884891–110888160 | 0.54–0.30 | 4 | RBM family |
| RBM17 | chr10:6154325–6155470 | 0.05–0.16 | 2 | RBM family |
| RBM39 | chr20:34326941–34327314 | 0.15–0.34 | 1 | RBM family |
| chr20:34327471–34328446 | 0.11–0.22 | 2 | ||
| NXF1 | chr11:62566048–62567848 | <0.01–0.16 | NA | Nuclear RNA export protein |
| ACIN1 | chr14:23537881–23538684 | 0.08–0.24 | 2 | Exon junction complex |
| FUS | chr16:31196501–31198122 | 0.15–0.45 | 1 | RNA and DNA binding protein |
| chr16:31198158–31199645 | 0.15–0.43 | 1 | ||
| chr16:31199679–31200443 | 0.06–0.36 | 2 | ||
| TRA2A | chr7:23561460–23561750 | 0.11–0.50 | Alternative splicing regulator | |
| EIF4A2 | chr3:186505672–186506913 | 0.11–0.22 | Translation factor |
A very different picture emerged from GO analysis of stable clusters C4 and C5, which encompass 294 IR events in 248 genes, and which maintain moderate to high IR throughout terminal erythropoiesis. RNA processing functions were not significantly enriched in C4 and C5. Instead, GO terms related to metal ion binding were over-represented in these clusters (Table 1). Genes in these functional categories include several that are important for erythroid iron homeostasis, such as the mitochondrial iron importers mitoferrin-1 (SLC25A37) and mitoferrin-2 (SLC25A28) and two heme biosynthetic enzymes (PPOX and HMBS). Notably, metal ion binding functions were not significantly enriched in clusters C1 and C2.
We also observed high IR in several genes with functions related to cell division (Supplementary Figure S5), although they were not as consistently enriched in any one cluster. REEP4 helps clear the ER from metaphase chromatin, thereby ensuring correct progression through mitosis and proper nuclear envelope architecture (26); three REEP4 introns that were upregulated in late erythroblasts map to C1 and C2. MICALL2 is a paralog of MICALL1, a protein that influences microtubule dynamics during early and late mitosis (27); MICALL2 transcripts exhibit widespread high levels of retention in introns distributed among clusters C1, C2, C3 and C4. In transcripts for pericentrin, a key component of the centrosome that plays a role in centriole disengagement (28), two introns that map to C4 and C5 were moderately retained across all five erythroblast populations.
Developmentally dynamic IR is not due to weak splice sites
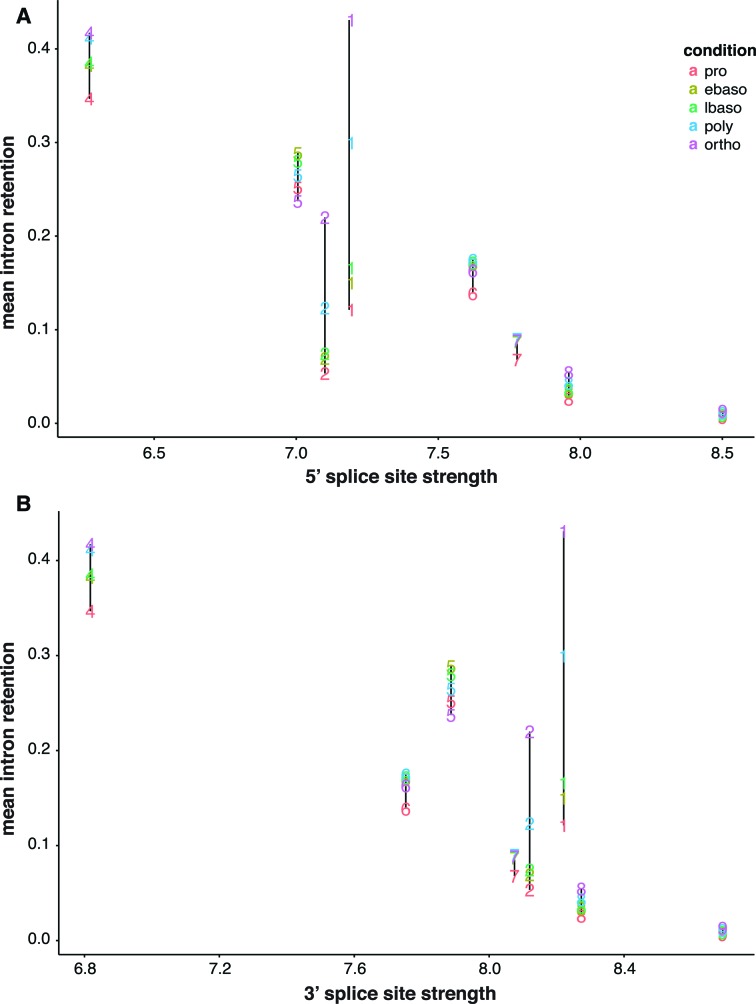
Retained introns on average are flanked by weaker splice sites than constitutively spliced introns (9,15). Here we explored this relationship in greater detail and asked whether differences in splice site strength might distinguish developmentally dynamic IR from developmentally stable IR. Cluster C3 was excluded from analysis due to its small size. For stable clusters C4–C9, IR was inversely related to both 5′ and 3′ splice site strength (Figure 3A and B), implicating splice sites in determination of IR events. This relationship appeared mostly independent of differentiation status. For C1 introns at the early proE to basoE stages, a similar relationship between splice site strength and IR was observed. In contrast, however, C1 introns at the polyE and orthoE stages increased IR values far beyond the expected range. A similar but more modest effect was observed for C2 introns. Looked at another way, introns in C1 and C2 exhibited a much greater dynamic range in IR values during erythroblast differentiation than introns in C4 and C5 that had much weaker splice sites. Together these results support a model in which splice site strength, while correlated with base line IR values in proE cells, cannot explain dynamic increases in IR in late erythroblasts .Splice site strength is thus a determinant of IR for a large group of developmentally stable introns; however, dynamic clusters C1 and C2 represent a distinct class for which features other than splice site strength must be responsible for upregulating IR during terminal erythropoiesis.
Figure 3.
Analysis of splice site strength in IR clusters. Average 5′ splice site strength (panel A) and 3′ splice site strength (panel B) is indicated for each cluster at each stage of terminal erythropoiesis, color coded according to differentiation stage (“condition"). In clusters C4–C9, IR is relatively stable across erythroblast populations and is inversely correlated with splice site strength. Clusters C1 and C2 display a much greater dynamic range of IR that is not correlated with splice site strength.
IR is associated with PTC exons
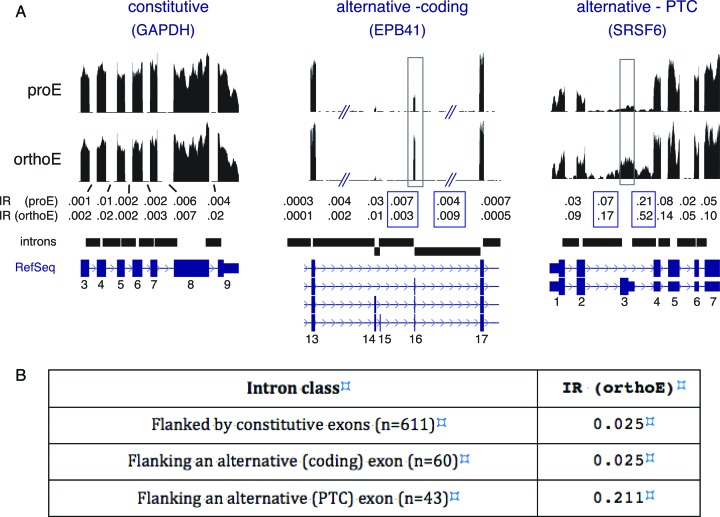
Most introns are spliced co-transcriptionally; however, introns that flank alternative exons are sometimes excised post-transcriptionally (29,30). To investigate whether IR events in erythroblast transcripts might be associated with properties of the adjacent exons, we examined three sets of introns derived from our previous studies of erythroblast splicing (2): (i) introns flanking alternative ‘coding’ exons; (ii) introns flanking alternative ‘PTC’ exons that introduce premature termination codons (PTC); and (iii) a control set of introns, taken from the same gene sets, filtered to contain only introns flanked on both sides by constitutive exons. Figure 4A illustrates examples of each group. Introns of the GAPDH gene are all members of the constitutive class; EPB41 exon 16 is a well-studied alternative exon that is upregulated in late erythropoiesis and encodes a peptide functionally implicated in membrane stability (7); and SRSF6 exon 3 is a PTC exon that is upregulated in late erythroblasts (2) and is known to induce NMD (31,32). IR values for these three transcripts were low for introns flanking constitutive exons and the alternative coding exon in EPB41, but much higher for the PTC-flanking introns of SRSF6. Extending the analysis to many additional introns yielded similar results (Figure 4B). Introns adjacent to constitutive exons (IRavg = 0.025) or coding alternative exons (IRavg = 0.025) exhibited low retention values, while introns flanking PTC exons were retained to a much higher extent (IRavg = 0.21).
Figure 4.
IR characteristics of three intron classes. (A) RNA-seq read mapping data for Refseq annotated gene regions with no alternative splicing (GAPDH), alternative splicing of a coding exon (exon 16 in EPB41) and alternative splicing of a PTC exon (exon 3 in SRSF6). Boxed regions indicate the alternative exons of interest and the IR values of their flanking introns. (B) Summary of IR results for introns adjacent to alternative exons studied in (2), Figures 3, 4 and 6. Constitutive exons are from the same gene sets.
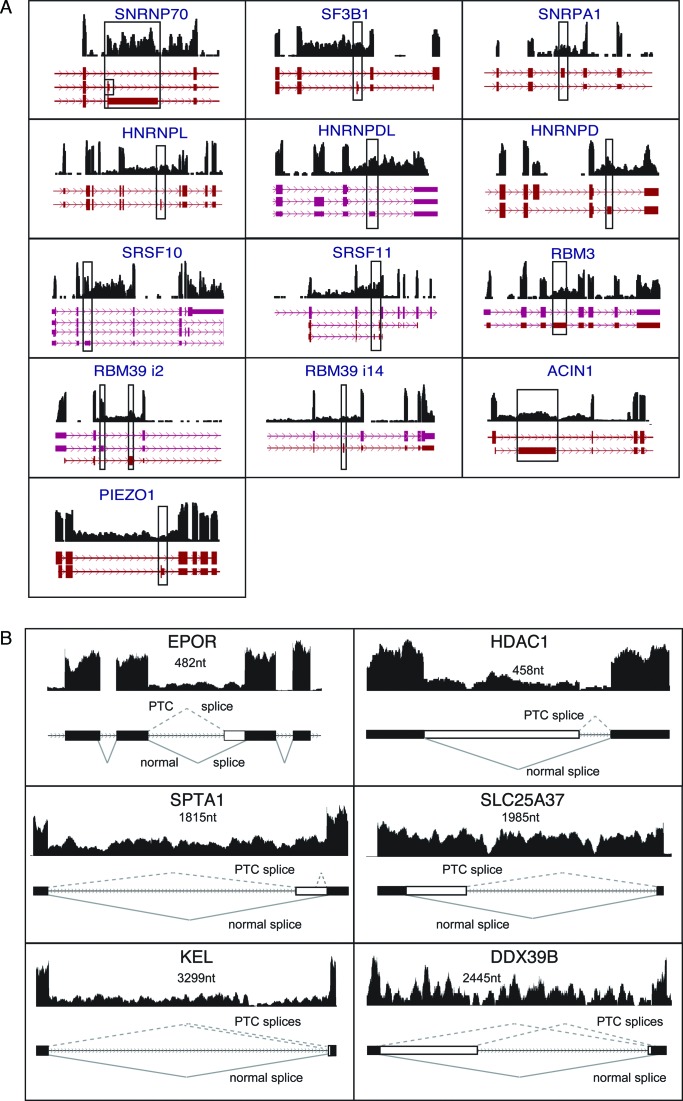
The association between PTC exons and IR was evident in a number of transcripts for RNA processing factors. Figure 5A shows several examples of Ensembl-annotated PTC exons (boxed) that are flanked by retained introns, while other nearby introns generally exhibit much less retention. Included in this group are major spliceosome-associated factors, SR and hnRNP proteins frequently implicated in splicing regulation, and RBM-containing proteins. However, the PTC exon-IR link was not limited to RNA processing functions since it was also an important feature of the PIEZO1 gene encoding a mechanosensitive ion channel.
Figure 5.
Intron retention flanking PTC exons in RNA processing genes. (A) Wiggle plots showing RNA-seq reads from orthoE cells are aligned with Ensembl-annotated gene regions spanning PTC exons. Boxes indicate PTC exons. (B) Wiggle plots showing retained introns that are associated with unproductive ‘PTC’ splice sites supported by RNA-seq reads, either Ensembl-annotated (SLC25A37, DDX39B, HDAC1, KEL, EPOR) or novel (SPTA1, KEL). Size of the retained intron in nucleotides is indicated.
A number of prominent IR events in erythroblasts were not associated with annotated PTC exons, e.g. introns in transcripts for erythropoietin receptor (EPOR), α-spectrin (SPTA1), mitoferrin1 (SLC25A37) and membrane protein KEL. Careful analysis of such cases revealed that even when PTC exons are not evident, unproductive splicing events can be mediated via cryptic 5′ or 3′ splice site(s) (Figure 5B). For example, the cryptic sites in KEL transcripts represented novel splices that were not annotated in the UCSC genome browser but were supported by erythroblast RNA-seq reads and/or by RT-PCR analysis of erythroblast RNA. Even after careful analysis, however, there remained introns for which no cryptic splice sites could be identified. This latter group might represent a functionally different IR class, or there could be unannotated ‘decoy’ sites that are difficult to detect because they splice with very low efficiency.
Transcripts with highly retained introns are localized to the nucleus and are resistant to nonsense-mediated decay
Retained introns generally introduce PTCs that can induce NMD of cytoplasmic IR transcripts (12), but stable expression of NMD-resistant nuclear IR transcripts has also been reported (9,15). We isolated nuclear and cytoplasmic fractions from cultured human erythroblasts and assayed relative abundance of IR transcripts using RT-PCR. Figure 6A shows that IR transcripts from six different highly expressed erythroblast genes were predominantly localized to the nuclear fraction. In each case small PCR products representing spliced RNAs were detected in both nuclear and cytoplasmic fractions, but the much larger IR bands were predominantly in the nuclear fraction. Interestingly, RNA-seq data from K562 erythroleukemia cells at the UCSC genome browser (ENCODE RNA-seq Tracks) supports the preferential nuclear localization of many retained introns; data for SPTA1 and SF3B1 are shown in Supplementary Figure S6. These results show that nuclear restriction of incompletely spliced transcripts is a post-transcriptional mechanism for limiting expression of translatable mRNAs in the cytoplasm.
Figure 6.
(A) Nuclear localization of IR transcripts. Nuclear (N) and cytoplasmic (C) fractions of human erythroblasts were assayed for intron retention by RT-PCR. Migration of IR isoforms is indicated by filled arrowheads, unproductive splicing by open arrowheads and productive splicing by open circles. IR isoforms are greatly enriched in the nucleus relative to spliced transcripts. (B) IR isoform is not degraded by NMD. Erythroblast RNA from cells cultured without (−) or with (+) cycloheximide plus emetine were amplified by RT-PCR. Enhanced detection of the PTC isoform of the SNRNP70 transcript indicates successful inhibition of NMD, but the IR isoform of SLC25A28 did not increase under the same conditions. A novel PTC isoform of SLC25A28, which was NMD sensitive, was revealed in this experiment. * indicates a PCR artifact. CHI, cycloheximide.
Nuclear-localized IR transcripts are expected to be NMD-resistant. We tested this prediction by inhibiting NMD with cycloheximide plus emetine, a treatment that enhances steady state levels of PTC-containing erythroblast transcripts from genes such as SNRNP70 (ref. (2) and Figure 6B). For SLC25A28, in contrast, NMD inhibition actually decreased the amount of IR product relative to the fully spliced product. Similar results were obtained when nuclear and cytoplasmic fractions were examined separately (Supplementary Figure S7) and when other IR transcripts were assayed. Interestingly, this experiment also revealed novel isoforms of SLC25A28 (Figure 6B) and SPTA1 (not shown) that were characterized by inclusion of previously unannotated PTC exons.
IR events upregulated in late stage erythroblasts are expressed at lower levels in other tissues
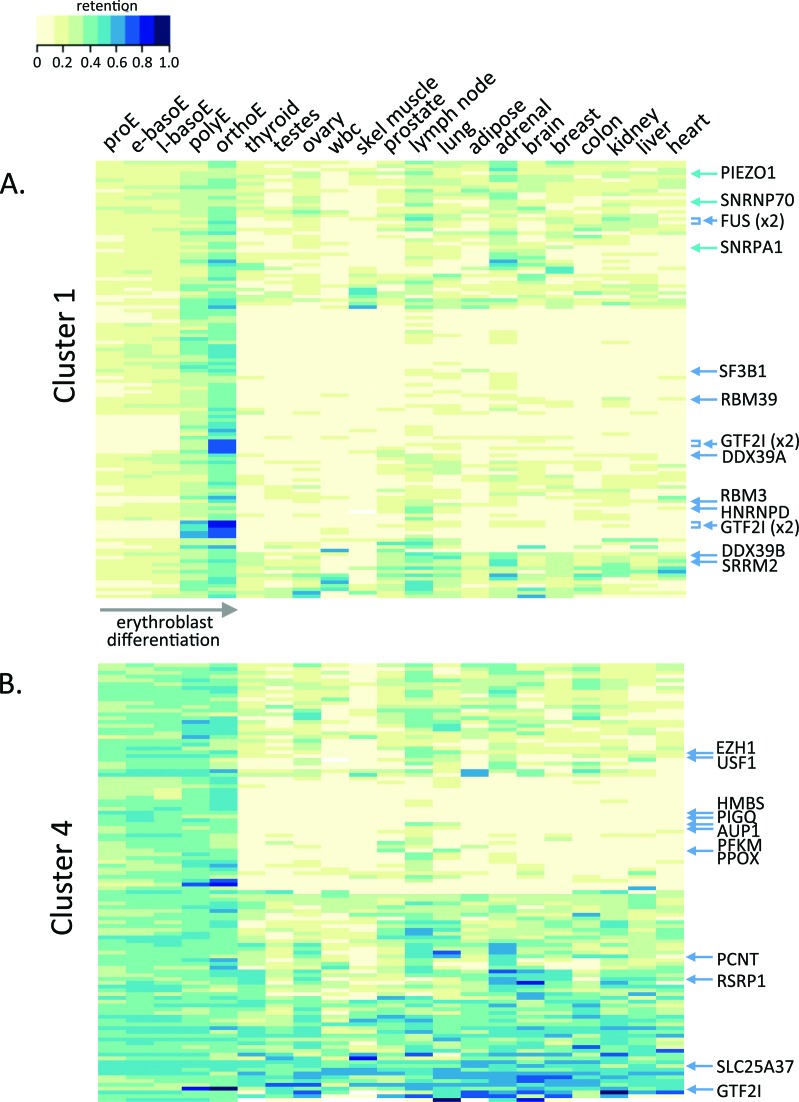
To explore whether prominent erythroblast IR events are unique to erythroblasts or are more broadly relevant in human biology, we examined IR in a variety of tissues. Retention of introns from each erythroblast IR cluster was assessed using KMA to analyze RNA-seq data from 16 other tissues in the llumina Human BodyMap 2.0 project. Many of the upregulated introns in clusters C1 and C2 also exhibited widespread retention in other tissues, sometime comparable to levels in late erythroblasts, but more often at lower levels. These data are shown in heat map format in Figure 7A for C1 and in Supplementary Figure S8 for C2; retention values are summarized in Supplementary Table S2). For example, the spliceosomal components SF3B1, DDX39B, SNRNP70 and RNPA1 all showed highest retention in orthoE but also widespread retention in other tissues (Figure 7A; for wiggle plots see Supplementary Figure S9). In some cases, e.g. SPTA1, IR is quite erythroid specific because the gene itself is not expressed in non-erythroid tissues.
Figure 7.
Comparison of IR in erythroblasts and other tissues. (A) Heat map displaying IR values for introns in cluster 1, with the five erythroblast populations at the left. Individual genes of interest are indicated at the right. (B) Heat map displaying IR values for introns in cluster 4.
In contrast, many C4 and C5 introns were retained at similar levels across erythroid differentiation and in non-erythroid tissues (Figure 7B and Supplementary Figure S7B), e.g. SLC25A37 in C4 and SLC25A28 in C5. Notable exceptions to this generalization included introns in HMBS, PPOX and PIGQ, which had high IR in erythroblasts but lower IR in non-erythroid tissues. These unique IR properties might be related to their much higher expression level in erythroblasts than in other tissues (data not shown). Together these results demonstrate that erythroblasts execute a complex intron retention program that shares many aspects with other tissues but ultimately is unique to terminal erythropoiesis.
DISCUSSION
Erythroblast differentiation is an excellent model system for studying the role of RNA processing in shaping the transcriptome during development. During the final four cell divisions that comprise terminal erythropoiesis, erythroblasts execute a highly dynamic, stage-specific RNA processing program that encompasses not only a robust network of alternative exon splicing (2), but also a broad array of intron retention events (this paper). Cluster analysis revealed many developmentally-dynamic IR events and splice site analysis supported active regulation of these events rather than a general decline in splicing activity in late erythroblasts. We hypothesize that IR is a mechanistically diverse, regulated set of processes with a complexity that could parallel alternative exon splicing and that it plays an important role in human erythroblasts as they undergo extensive remodeling prior to enucleation and maturation into red cells. Further studies will be needed to investigate the various pathways and RNA binding proteins that regulate IR in erythroblasts.
Consistent with studies in other cells (9), IR in erythroblast transcripts is enriched in introns flanking alternative exons. Our results further indicate that much of the enrichment is associated with the subset of cassette exons that have PTCs, and that retained introns lacking PTC exons often exhibit other unproductive splicing events. Whether these unproductive sites are causally related to IR by promoting assembly into immature spliceosomal complexes (33–36) is unknown. Another possibility is that IR and unproductive splicing are both consequences of other features that inhibit use of the normal splice sites and allow for regulation by accessory factors. Interestingly, retention of introns flanking PTC exons in the SR protein genes was noticed previously and has been shown to be conserved (9,31–32).
More broadly, a pool of nuclear localized IR transcripts might have several alternative fates: degradation by nuclear RNA surveillance machinery (13), non-productive splicing of the retained intron followed by degradation in the cytoplasm via NMD (12,32,37–38) or completion of intron excision to produce mature translatable mRNA (18). These choices might offer a flexible post-transcriptional regulatory mechanism to modulate important erythroblast pathways such as iron homeostasis, e.g. by controlling output of the mitoferrin-1 gene. Future studies will be aimed at exploring these and other potential functions for IR during terminal erythropoiesis.
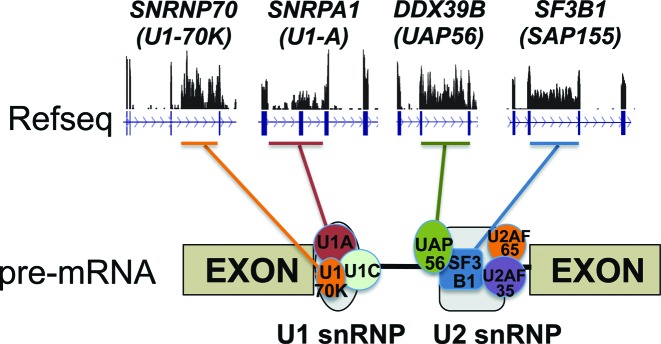
What might be the function of regulated IR during normal erythropoiesis? One major function might be to post-transcriptionally modulate protein output from selected genes. This interpretation is consistent with the high expression levels of intron-retaining transcripts from SLC25A37, SPTA1 and SF3B1 genes that cannot encode full length proteins due to their localization to the nucleus and the presence of multiple PTCs in the long retained introns. Given the prevalence of IR in splicing factor genes—especially in several spliceosome-associated factors—one function of IR might be to down-modulate splicing capacity of the cells in a quantitative sense as they prepare for enucleation and express less RNA per cell in late erythroblasts (Figure 8). In fact, IR levels for SF3B1, SNRNP70 and DDX39B co-vary not only temporally during terminal erythropoiesis, but also more or less spatially across various tissues (see BodyMap data, Supplementary Figure S8). Another possible role for IR might be to act as a post-transcriptional regulator of iron homeostasis genes, e.g. by modulating the expression of selected heme biosynthetic enzymes (PPOX and HMBS) and iron transporters (SLC25A37 and SLC25A28) in response to the iron/heme demands of erythroblasts producing huge quantities of hemoglobin. Coordinate regulation of IR in splicing factor genes or iron homeostasis genes could thus be an important cellular control mechanism (Figure 7).
Figure 8.
Model showing that major spliceosomal genes can be regulated by IR. Shown above are the wiggle plots in mature erythroblasts (orthoE); much reduced IR is evident in earlier stages.
Finally, given the capability of IR to modulate gene expression, we speculate that inappropriate regulation of IR may be an under-appreciated cause of human disease. There is precedence for mutations that induce inappropriate intron retention, for example, in the case of BRCA2 (39), and aberrant intron retention events are prominent in breast cancer (40). Recently it was shown that ZRSR2-mutated MDS patients exhibit IR specifically for introns spliced by the minor spliceosome (41), and it would not be surprising if IR is a feature of other MDS subtypes. Although intron retention in the mitoferrin gene SLC25A37 was proposed to contribute to abnormal iron accumulation in MDS erythroblasts (42), the observation that considerable IR occurs normally in erythroblast SLC25A37 transcripts suggests that this should be interpreted with caution.
Supplementary Material
Acknowledgments
J.G.C. and L.P. designed the research; H.P., M.P., S.L.G. performed research and analyzed data; and J.G.C., H.P., N.M., and L.P. wrote the article.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [DK094699 to J.G.C., L.P.; DK32094 and DK26263 to N.M.]; Director, Office of Science and Office of Biological & Environmental Research of the US Department of Energy [DE-AC02-05CH1123]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.An X., Schulz V.P., Li J., Wu K., Liu J., Xue F., Hu J., Mohandas N., Gallagher P.G. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood. 2014;123:3466–3477. doi: 10.1182/blood-2014-01-548305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pimentel H., Parra M., Gee S., Ghanem D., An X., Li J., Mohandas N., Pachter L., Conboy J.G. A dynamic alternative splicing program regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2014;42:4031–4042. doi: 10.1093/nar/gkt1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paralkar V.R., Mishra T., Luan J., Yao Y., Kossenkov A.V., Anderson S.M., Dunagin M., Pimkin M., Gore M., Sun D., et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123:1927–1937. doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng A.W., Shi J., Wong P., Luo K.L., Trepman P., Wang E.T., Choi H., Burge C.B., Lodish H.F. Muscleblind-like 1 (Mbnl1) regulates pre-mRNA alternative splicing during terminal erythropoiesis. Blood. 2014;124:598–610. doi: 10.1182/blood-2013-12-542209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi L., Lin Y.H., Sierant M.C., Zhu F., Cui S., Guan Y., Sartor M.A., Tanabe O., Lim K.C., Engel J.D. Developmental transcriptome analysis of human erythropoiesis. Hum. Mol. Genet. 2014;23:4528–4542. doi: 10.1093/hmg/ddu167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chasis J.A., Coulombel L., Conboy J., McGee S., Andrews K., Kan Y.W., Mohandas N. Differentiation-associated switches in protein 4.1 expression. Synthesis of multiple structural isoforms during normal human erythropoiesis. J. Clin. Invest. 1993;91:329–338. doi: 10.1172/JCI116189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Discher D., Parra M., Conboy J.G., Mohandas N. Mechanochemistry of the alternatively spliced spectrin-actin binding domain in membrane skeletal protein 4.1. J. Biol. Chem. 1993;268:7186–7195. [PubMed] [Google Scholar]
- 8.Horne W.C., Huang S.C., Becker P.S., Tang T.K., Benz E.J., Jr Tissue-specific alternative splicing of protein 4.1 inserts an exon necessary for formation of the ternary complex with erythrocyte spectrin and F-actin. Blood. 1993;82:2558–2563. [PubMed] [Google Scholar]
- 9.Boutz P.L., Bhutkar A., Sharp P.A. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015;29:63–80. doi: 10.1101/gad.247361.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho V., Mei Y., Sanny A., Chan S., Enders A., Bertram E.M., Tan A., Goodnow C.C., Andrews T.D. The RNA-binding protein hnRNPLL induces a T cell alternative splicing program delineated by differential intron retention in polyadenylated RNA. Genome Biol. 2014;15:R26. doi: 10.1186/gb-2014-15-1-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shalgi R., Hurt J.A., Krykbaeva I., Taipale M., Lindquist S., Burge C.B. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell. 2013;49:439–452. doi: 10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong J.J., Ritchie W., Ebner O.A., Selbach M., Wong J.W., Huang Y., Gao D., Pinello N., Gonzalez M., Baidya K., et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 13.Yap K., Lim Z.Q., Khandelia P., Friedman B., Makeyev E.V. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012;26:1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dvinge H., Bradley R.K. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7:45. doi: 10.1186/s13073-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braunschweig U., Barbosa-Morais N.L., Pan Q., Nachman E.N., Alipanahi B., Gonatopoulos-Pournatzis T., Frey B., Irimia M., Blencowe B.J. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24:1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filichkin S.A., Priest H.D., Givan S.A., Shen R., Bryant D.W., Fox S.E., Wong W.K., Mockler T.C. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ninomiya K., Kataoka N., Hagiwara M. Stress-responsive maturation of Clk1/4 pre-mRNAs promotes phosphorylation of SR splicing factor. J. Cell Biol. 2011;195:27–40. doi: 10.1083/jcb.201107093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boothby T.C., Zipper R.S., van der Weele C.M., Wolniak S.M. Removal of retained introns regulates translation in the rapidly developing gametophyte of Marsilea vestita. Dev. Cell. 2013;24:517–529. doi: 10.1016/j.devcel.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Roberts A., Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B., Ruotti V., Stewart R.M., Thomson J.A., Dewey C.N. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26:493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 24.Hu J., Liu J., Xue F., Halverson G., Reid M., Guo A., Chen L., Raza A., Galili N., Jaffray J., et al. Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood. 2013;121:3246–3253. doi: 10.1182/blood-2013-01-476390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandya-Jones A., Black D.L. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlaitz A.L., Thompson J., Wong C.C., Yates J.R., 3rd, Heald R. REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev. Cell. 2013;26:315–323. doi: 10.1016/j.devcel.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinecke J.B., Katafiasz D., Naslavsky N., Caplan S. Novel functions for the endocytic regulatory proteins MICAL-L1 and EHD1 in mitosis. Traffic. 2015;16:48–67. doi: 10.1111/tra.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee K., Rhee K. Separase-dependent cleavage of pericentrin B is necessary and sufficient for centriole disengagement during mitosis. Cell Cycle. 2012;11:2476–2485. doi: 10.4161/cc.20878. [DOI] [PubMed] [Google Scholar]
- 29.Pandya-Jones A., Bhatt D.M., Lin C.H., Tong A.J., Smale S.T., Black D.L. Splicing kinetics and transcript release from the chromatin compartment limit the rate of Lipid A-induced gene expression. RNA. 2013;19:811–827. doi: 10.1261/rna.039081.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khodor Y.L., Menet J.S., Tolan M., Rosbash M. Cotranscriptional splicing efficiency differs dramatically between Drosophila and mouse. RNA. 2012;18:2174–2186. doi: 10.1261/rna.034090.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lareau L.F., Brenner S.E. Regulation of splicing factors by alternative splicing and NMD is conserved between kingdoms yet evolutionarily flexible. Mol. Biol. Evol. 2015;32:1072–1079. doi: 10.1093/molbev/msv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 33.Chiou N.T., Shankarling G., Lynch K.W. hnRNP L and hnRNP A1 induce extended U1 snRNA interactions with an exon to repress spliceosome assembly. Mol. Cell. 2013;49:972–982. doi: 10.1016/j.molcel.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma S., Maris C., Allain F.H., Black D.L. U1 snRNA directly interacts with polypyrimidine tract-binding protein during splicing repression. Mol. Cell. 2011;41:579–588. doi: 10.1016/j.molcel.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonnal S., Martinez C., Forch P., Bachi A., Wilm M., Valcarcel J. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol. Cell. 2008;32:81–95. doi: 10.1016/j.molcel.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Cote J., Dupuis S., Jiang Z., Wu J.Y. Caspase-2 pre-mRNA alternative splicing: identification of an intronic element containing a decoy 3′ acceptor site. Proc. Natl. Acad. Sci. U.S.A. 2001;98:938–943. doi: 10.1073/pnas.031564098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eom T., Zhang C., Wang H., Lay K., Fak J., Noebels J.L., Darnell R.B. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. eLife. 2013;2:e00178. doi: 10.7554/eLife.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng S., Gray E.E., Chawla G., Porse B.T., O'Dell T.J., Black D.L. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat. Neurosci. 2012;15:381–388. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Acedo A., Sanz D.J., Duran M., Infante M., Perez-Cabornero L., Miner C., Velasco E.A. Comprehensive splicing functional analysis of DNA variants of the BRCA2 gene by hybrid minigenes. Breast Cancer Res. 2012;14:R87. doi: 10.1186/bcr3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eswaran J., Horvath A., Godbole S., Reddy S.D., Mudvari P., Ohshiro K., Cyanam D., Nair S., Fuqua S.A., Polyak K., et al. RNA sequencing of cancer reveals novel splicing alterations. Sci. Rep. 2013;3:1689. doi: 10.1038/srep01689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madan V., Kanojia D., Li J., Okamoto R., Sato-Otsubo A., Kohlmann A., Sanada M., Grossmann V., Sundaresan J., Shiraishi Y., et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun. 2015;6:6042. doi: 10.1038/ncomms7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visconte V., Avishai N., Mahfouz R., Tabarroki A., Cowen J., Sharghi-Moshtaghin R., Hitomi M., Rogers H.J., Hasrouni E., Phillips J., et al. Distinct iron architecture in SF3B1-mutant myelodysplastic syndrome patients is linked to an SLC25A37 splice variant with a retained intron. Leukemia. 2015;29:188–195. doi: 10.1038/leu.2014.170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.