Abstract
G-quadruplexes are non-canonical structures of nucleic acids, in which guanine bases form planar G-tetrads (G·G·G·G) that stack on each other in the core of the structure. G-quadruplexes generally contain multiple times of four (4n) guanines in the core. Here, we study the structure of G-quadruplexes with only (4n - 1) guanines in the core. The solution structure of a DNA sequence containing 11 guanines showed the formation of a parallel G-quadruplex involving two G-tetrads and one G-triad with a vacant site. Molecular dynamics simulation established the formation of a stable G-triad·water complex, where water molecules mimic the position of the missing guanine in the vacant site. The concept of forming G-quadruplexes with missing guanines in the core broadens the current definition of G-quadruplex-forming sequences. The potential ability of such structures to bind different metabolites, including guanine, guanosine and GTP, in the vacant site, could have biological implications in regulatory functions. Formation of this unique binding pocket in the G-triad could be used as a specific target in drug design.
INTRODUCTION
G-quadruplex is a non-canonical structure of nucleic acids, in which guanine bases are associated by Hoogsteen hydrogen bonds to form planar G-tetrads that stack on each other in the core of the structure (1,2). G-quadruplexes have been detected in cells (3,4). They are considered functionally important in numerous cellular processes, such as DNA replication (5,6) recombination (7), transcription (8), translation (9) and telomere maintenance (10–12) and shown to cause genomic instability (5,13).
Aside from the simple sequence pattern G3+N1–7G3+N1–7G3+N1–7G3+ predicting G-quadruplex formation (14,15), several new motifs (16–23) were found to form stable intramolecular G-quadruplexes in vitro, including motifs with bulges (24) or duplex-stems (18) in the core of G-quadruplex structures. Recently, high-resolution sequencing data suggested the formation of over 700 000 intramolecular G-quadruplexes in the human genome, including those with bulges or long loops (25).
Overall, these predicted intramolecular G-quadruplexes contain multiple times of four (4n) guanines in the G-tetrad core. G-quadruplexes with less than (4n) guanines in the core have been investigated by computational (26) and experimental techniques (22,27–32), showing the formation of intermediate structures in the G-quadruplex folding pathway. Nuclear magnetic resonance (NMR) studies on a DNA sequence containing three G-tracts derived from the thrombin aptamer showed the formation of a G-triplex structure (22,23). Such intermediate structures were shown to be stabilized by G-quadruplex ligands (33), opening an avenue for the development of new drugs binding to intermediate structures. However, G-quadruplex with (4n - 1) guanines in the core has not been studied. These sequences could have a new mode of interaction with small molecules such as metabolites that are present in cells at relatively high concentration (34) and involved in numerous biological processes (35). Such binding abilities were observed for RNA riboswitches (36) and aptamers (37–40).
In this study, we investigate the G-quadruplex structures formed by sequences containing 11 guanines in the core. The solution structure of one such sequence revealed a G-quadruplex containing a G-triad·water complex with a vacant site. The elucidation of this novel G-quadruplex topology broadens the description of G-quadruplex forming sequences. The potential ability of the structure to bind metabolites could have important biological implications such as metabolite sensing and regulation. Formation of the vacant site in G-quadruplexes could be used as a platform for drug design.
MATERIALS AND METHODS
Sample preparation
Unlabeled, 4% 15N-labeled and 2H-labeled DNA oligonucleotides were chemically synthesized using an ABI 394 DNA/RNA synthesizer. Necessary reagents were purchased from Glen Research. Samples were purified with a Poly-Pak II cartridge (Glen Research) and dialyzed successively against water, 25 mM KCl solution and water. Prior to all experiments, DNA (except for T95–2T and 93del) samples were heated at ∼95°C for 10 min and quickly cooled down to room temperature using cold water (termed as ‘quenching’ procedure). The samples typically contained 35 mM KCl and 10 mM potassium phosphate (KPi) (pH 7.0), supplemented with 1 mM EDTA and 0.1 mM NaN3. Guanosine was purchased from Sigma-Aldrich and preapred at 150-200 mM in 1 M KOH solution.
Circular dichroism (CD)
CD spectra were recorded at 20°C on a JASCO-815 spectropolarimeter using a 1-cm path-length quartz cuvette. The DNA concentration was 3–10 μM. For each experiment, an average of three scans was recorded between 220 and 320 nm. Baseline correction was applied for all spectra and the data were zero-corrected at 320 nm.
Ultraviolet (UV) absorption
UV spectra were recorded on a JASCO V-650 spectrophotometer using a 1-cm path-length quartz cuvette. The DNA concentration was 3–10 μM. UV absorbance at 295 nm was recorded as a function of temperature (from 20 to 95°C) and used to define the thermal stability of S4 and T4. The heating rate was 0.5°C per minute.
Gel electrophoresis
Polyacrylamide gels were prepared at 20% of acrylamide (acrylamide/bis-acrylamide ratio of 1:37.5) with Tris-Borate-EDTA as a running buffer supplemented by 10 mM KCl. All gels were resolved at 120 volts for ∼1 h and visualized using UV shadowing.
Nuclear magnetic resonance (NMR) spectroscopy
Experiments were performed on 600 and 700 MHz Bruker spectrometers at 10 and 25°C. DNA concentrations were 0.1–1.2 mM. Various salt concentrations (from 10 to ∼100 mM K+) were tested and similar NMR spectra were observed, indicating the formation of the same G-quadruplex structure. At every 20–30 min interval, NOESY experiment was suspended, and the sample was quenched before the experiment was resumed. All spectra were processed and analyzed using the TopSpin 2.1 (Bruker), Sparky 3.1 (41) and Spinworks 4.0 (http://home.cc.umanitoba.ca/∼wolowiec/spinworks) programs.
Structure calculations
Inter-proton distances for T4 were deduced from NOESY experiments performed in H2O and D2O (mixing time, 200 ms). Structure computations were performed using the XPLOR-NIH program (42) in two general steps: (i) distance geometry simulated annealing, (ii) distance-restrained molecular dynamics refinement. During structure calculations hydrogen bond restraints, inter-proton distance restraints, dihedral restraints, repulsive restraints as well as planarity restraints were imposed. The detailed procedure is described in our earlier work (18). These calculations led to ten lowest-energy structures in vacuo.
These structures were subsequently used for molecular dynamics refinements in water using the AMBER program (43). For the initial DNA structure, 17 K+ counter ions (two of them in the center of the G-tetrad core) were added to obtain an electroneutral system. The system was then placed in a truncated octahedral water-box with 4146 TIP3P water molecules (44). During the refinement in explicit water, planarity and repulsive restraints were removed. The K+ ion coordinating the G-triad was restricted in the G-core using seven distance constraints from the K+ ion to the O6 atom of G5, G8, G9, G12, G13, G16 and G17 residues. The final structures were then subjected to short minimization in vacuo. Analysis of the structures and the MD trajectories were carried out with VMD (45) and the ‘ptraj’ module of AMBER. Structures were displayed using PYMOL (46) or VMD (45).
Molecular dynamics (MD) simulation
MD simulation was run for 52 ns using the lowest-energy structure of T4 as a starting point. AMBER 10 with the Parmbsc0 force field was used (43,47). The detailed protocol is similar to that described in structure calculations section.
Data deposition
The coordinates for the T4 G-quadruplex structure have been deposited in the Protein Data Bank (accession code 2N60).
RESULTS
G-quadruplex formation by sequences containing 11 guanines
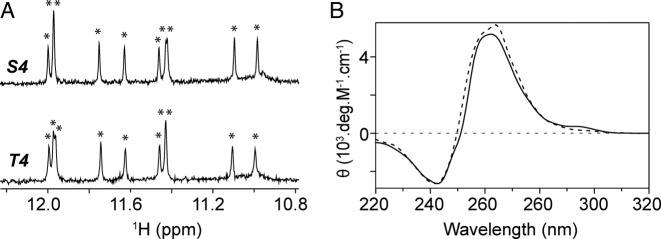
We examined the G-quadruplex formation by the S4 and T4 sequences containing 11 guanines (Table 1). These sequences were derived from T95–2T (Table 1), which adopts an intramolecular three-layer parallel G-quadruplex (Supplementary Figure S1) (48), following an abasic site or a G-to-T substitution in the middle of the first G-tract. NMR imino proton spectra of S4 and T4 were nearly identical, displaying ten sharp peaks between 10.8 and 12.0 ppm (Figure 1A), indicating the formation of the same G-quadruplex fold. The CD spectra of S4 and T4 exhibit a positive peak at ∼260 nm and a negative peak at ∼240 nm (Figure 1B), characteristic of parallel-stranded G-quadruplexes. In a native gel electrophoresis (Supplementary Figure S2), the migration of S4 and T4 was similar to that of T95–2T (48) and faster than that of the 93del dimeric G-quadruplex (49), suggesting the formation of a monomeric G-quadruplex. In a melting experiment, S4 and T4 unfolded at 51.2 and 45.6°C, respectively (Supplementary Figure S3). These values are much lower than the melting temperature of the parent sequence T95–2T (24).
Table 1. DNA sequences used in this study.
| Name | Sequence (5′ ─ 3′) |
|---|---|
| T95–2T | TT GGG T GGG T GGG T GGG T |
| S4 | TT GSG T GGG T GGG T GGG T |
| T4 | TT GTG T GGG T GGG T GGG T |
| T4-I17 | TT GTG T GGG T GGG T GGI T |
S and I refer to a dspacer (deoxyribose lacking a base) and an Inosine, respectively.
Figure 1.
(A) Nuclear magnetic resonance (NMR) imino proton spectra of S4 (top) and T4 (bottom) and (B) CD spectra of S4 (dotted line) and T4 (continuous line). θ represents the molar ellipticity. Imino proton peaks are labeled with asterisks.
T4 forms an intramolecular parallel-stranded G-quadruplex containing a G-triad
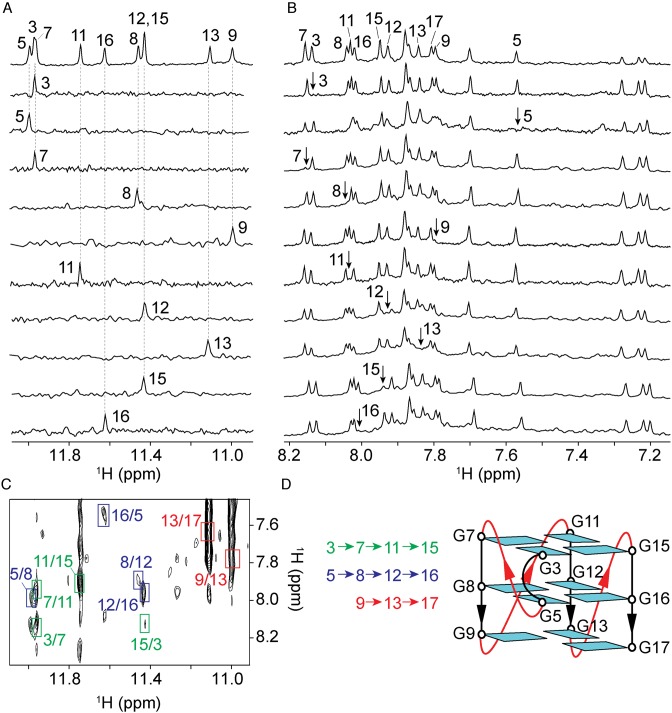
Given the high spectral similarity between S4 and T4, we only focused our structural study on T4. Guanine imino (H1) and aromatic (H8) protons were unambiguously assigned using site-specific 15N-labeling and site-specific deuteration approaches (50,51) (Figure 2A and B; Supplementary Table S1). Spectral assignments were supported by using through-bond [1H-13C]-JRHMBC (52) and through-space (NOESY) experiments (Supplementary Figures S4 and S5). The H6/8-H1′ NOE sequential connectivities could be traced from T1 to T18 (Supplementary Figure S5). The arrangement of guanines in the G-tetrad core was deduced from the characteristic H1-H8 NOE connectivity (Figure 2C), showing the formation of a parallel-stranded G-quadruplex with two G-tetrads (G3·G7·G11·G15 and G5·G8·G12·G16) and one G-triad (G9·G13·G17) (Figure 2D). All guanines adopted anti glycosidic conformations, consistent with the intensities of the observed H1’-H8 NOEs (Supplementary Figure S5).
Figure 2.
NMR spectral assignment and folding topology of T4. (A) Assignment of guanine imino (H1) protons by using site-specific15N-labeled samples and (B) H8 protons by using 2H-labeled samples (Supplementary Table S1) at the indicated positions. Reference spectrum is on top. (C) NOESY spectrum (mixing time, 200 ms) used for the folding topology determination. The characteristic H1-H8 cross-peaks are framed and labeled with residue number of H1 and H8 proton in the first and second position, respectively. (D) Folding topology of T4, all guanines are colored in cyan.
The positioning of G17 within the G9·G13·G17 triad was consistent with the lower solvent protection of its imino proton as compared to other ones in the G-tetrad core. At room temperature, among 11 guanines only G17 did not show imino proton peak (Figure 2A). At low temperature (10°C) an additional peak appeared in the imino proton region, which was assigned to H1 of G17 (Supplementary Figure S6). These results were supported by the data on an analog sequence T4-I17 (Table 1), in which G17 was replaced by an inosine, displaying a highly similar spectrum to that of T4 (Supplementary Figure S7) with no characteristic downfield inosine imino proton peak (53).
Solution structure of T4 reveals a G-quadruplex with an accessible G-triad
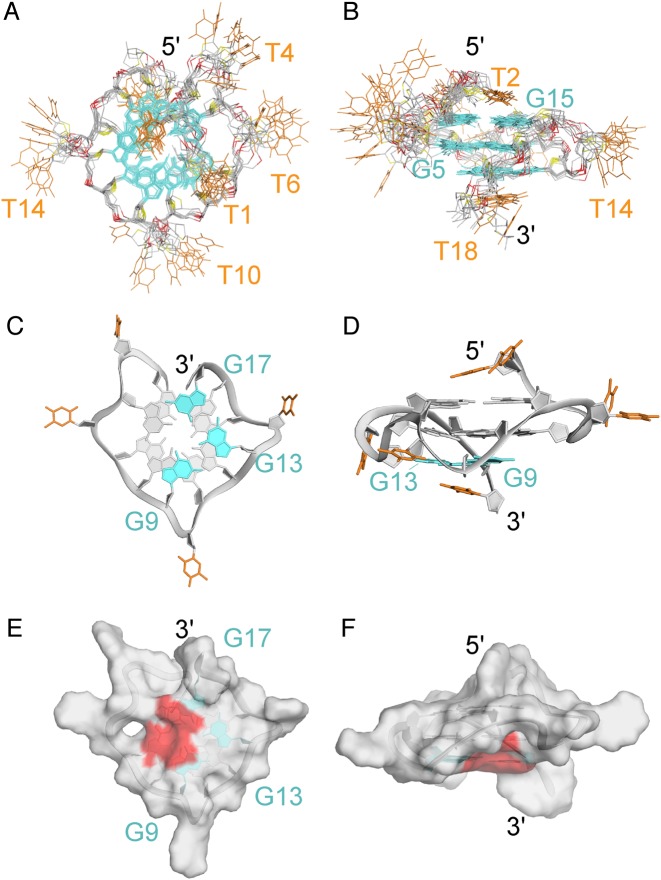
The solution structure of T4 was calculated on the basis of NMR restraints, extracted from NOE cross-peaks in NOESY experiments. The ten lowest energy structures (Figure 3) are well converged (see Table 2 for statistics). The structure contains three single-residue propeller loops (T6, T10 and T14) and a bulge (T4). Residues T1, T2 stack on the 5′-end, while T18 stacks on the 3′-end (Figure 3A and B) (54). Our structure reassembles to a previously reported parallel G-quadruplex with a bulge (24) (Supplementary Figure S8). Similar effects on bulge length and residue type (Supplementary Table S2 and Figures S9 and S10) were also observed (24).
Figure 3.
NMR solution structure of T4. (A) top and (B) side view of the ten superimposed lowest-energy structures. (C) Bottom and (D) side view of a representative structure. Surface representation of the (E) bottom and (F) side view of a representative structure. Guanines are colored in cyan and grey, thymines in orange. The accessible surface of the vacant site is colored in red.
Table 2. Statistics of the computed structures of T4 G-quadruplex.
| A. NMR restraints | ||
| Distance restraints | D2O | H2O |
| Intraresidue | 91 | 0 |
| Sequential (i, i + 1) | 40 | 5 |
| Long-range (i, ≥i + 2) | 5 | 7 |
| Other restraints | ||
| Hydrogen bond | 40 | |
| Dihedral angle | 11 | |
| Repulsive | 16 | |
| B. Structure statistics | ||
| NOE violations | ||
| Number (>0.2 Å) | 0.2 ± 0.4 | |
| Maximum violation (Å) | 0.169 ± 0.050 | |
| RMSD of violations (Å) | 0.021 ± 0.004 | |
| Deviations from the ideal covalent geometry | ||
| Bond lengths (Å) | 0.005 ± 0.002 | |
| Bond angles (°) | 0.758 ± 0.089 | |
| Impropers (°) | 0.467 ± 0.144 | |
| Pairwise all heavy atom RMSD values (Å) | ||
| All heavy atoms of G-tetrad core | 1.11 ± 0.18 | |
| All heavy atoms | 2.72 ± 0.41 | |
The G9·G13·G17 triad is equivalent to a G-tetrad with a missing (vacant) guanine (with a R.M.S.D. <1Å). The guanines of the G-triad show regular stacking observed in parallel G-quadruplex structures (55). Interestingly, the vacant site present in the G-triad is highly accessible to solvent (Figure 3E and F).
Molecular dynamics simulation reveals the formation of a G-triad·water complex
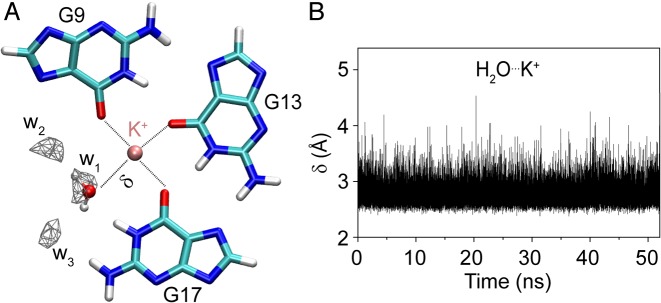
In order to study the water distribution around the G-triad, we performed restrained molecular dynamics (MD) simulation (52 ns) in water, using the AMBER program (43). In our initial simulation, the departure of K+ ions from the G-tetrad core led to a distorted structure, similar to what observed in long (μs timescale) MD simulation of G-triplex and G-quadruplex intermediates (26). Thus, the K+ ions in the core were restricted throughout the simulation (see ‘Materials and methods’ section).
Our simulation revealed the presence of three well-defined water molecules in the G-triad plane (Figure 4 and Supplementary Figure S11), occupying the vacant site with a density about four times higher than that of bulk water. The first water molecule (w1) is located close to the K+ ion, so that its oxygen atom is coordinated to the ion (with an average distance between K+ and water oxygen of 2.77 ± 0.18 Å) and hydrogen-bonded to H1 of G17 (Figure 4 and Supplementary Figure S12). The bonding pattern observed for w1 is strikingly similar to that of a guanine C=O group in a G-tetrad. The two other water molecules (w2 and w3) form hydrogen bonds with N7 of G9 and amino hydrogen of G17 respectively. These three water molecules mimic the position of the missing (ghost) guanine in the vacant site, with oxygen of w1, w2 and w3 being equivalent to O6, N1 and N7 atoms of the ghost guanine (Supplementary Figure S13). The interaction between the G-triad and water (formation of a G-triad-water complex) might be important for its stabilization in the G-quadruplex context.
Figure 4.
Formation of a G-triad-water complex (A) G9·G13·G17 base triad extracted from a MD simulation snapshot. The density of water oxygen is calculated using 0.5 Å3 grid box. Only densities of the water molecules with more than four times the water bulk are shown. Water molecule coordinating the cation is represented. (B) Distance (δ) between water oxygen and K+ as a function of time. K+ is colored in pink, carbon in cyan, hydrogen in white, oxygen in red and nitrogen in blue.
DISCUSSION
Triad arrangements in G-quadruplexes
We have determined the structure of a G-quadruplex containing two G-tetrads and one G-triad. The arrangement of guanines in this G-triad is similar to that in a G-tetrad, creating a vacant site. The same G-triad was observed in a structure of a truncated thrombin aptamer (22) and detected in a G-quadruplex formed by a human minisatellite sequence (19).
A different G-triad arrangement (56) and other base triads, including G·C·A, G·G·A, G·G·T, T·T·A (57), have been reported in the context of G-quadruplexes. The arrangement of bases in the G-triad studied here is different from other base triads in term of ion coordination and a vacant site (pocket) formation.
Sequences that can form G-quadruplexes with vacant sites
NMR data of other sequences containing 11 guanines derived from T95–2T (Supplementary Table S3 and Figure S14) showed ten major imino proton peaks in the 10.8–12.2 ppm region suggesting the formation of different G-quadruplexes containing a G-triad. Single abasic substitutions in human telomeric sequences, resulting in sequences with 11 guanines, were shown to form G-quadruplexes (58,59), possibly containing a G-triad. In this study, we show the formation of three-layer (n = 3) G-quadruplexes with a vacant site. We expect that this concept could be extended to n layers with (4n - 1) guanines in the core. Formation of n-layer G-quadruplexes containing more than one vacant site (e.g. (4n - 2) or (4n - 3) guanines in the core), which can occur in the same or different layers, is also possible. Indeed, the formation of two consecutive G-triads was observed in the structure of a G-triplex (22). It is likely that the concept of forming G-quadruplexes with missing guanines could also be applied to RNA sequences.
Fast folding of G-quadruplexes with a vacant site
In our experiments, the formation of a single G-quadruplex structure with a vacant site was obtained using a fast cooling procedure (see ‘Materials and Methods’ section). In principle, such structures could be kinetically trapped in any G-rich sequences, including sequences with multiple times of 4 guanines (4n). The folding kinetics of G-quadruplexes were shown to be slower or equivalent to many cellular processes (32), therefore the rapid formation of G-quadruplexes with G-triads could be favored in vivo. Hence, these structures might be a valuable target for drug design.
Vacant site in G-quadruplexes can act as a platform for small-molecule binding
G-quadruplexes with (4n - 1) guanines in the core can have a vacant site, creating a binding pocket for small-molecule recognition through hydrogen bonds with two guanines in the G-triad and metal ion coordination (Figure 5). Such structures are potentially natural candidates for binding to metabolites. For example, guanosine can interact with the vacant site and complete the G-triad to form a G-tetrad (Supplementary Figure S15) (60).
Figure 5.
G-triad with a vacant site for small-molecule recognition. Hydrogen-bond donors and acceptors are indicated with arrows; possible metal coordination is shown with a dotted arrow. Metal ion M+ is colored in pink, carbon in cyan, hydrogen in white, oxygen in red and nitrogen in blue.
The G-triad can be engineered to obtain a different acceptor/donor pattern in the vacant site. For example, G-triads containing modified bases, such as 8-oxoguanine (O) and xanthine (X), could be formed (O·G·G and G·G·X triads, derived from a G·G·X·O tetrad (61)) creating a specific vacant site for pairing with matching ligands (xanthine and 8-oxoguanine, respectively). On the other hand, ligands with desirable properties could also be optimized to fit the vacant site of a G-triad.
Several RNA riboswitches are known to bind metabolites and regulate gene expression (62). Riboswitches were found to bind guanine and guanine derivatives with high affinity and specificity (63). These structures contain hairpin loops with specific binding pockets for metabolite recognition (36). In principle, G-quadruplexes with a vacant site can also bind to several metabolites, including mono- and dinucleotides (64), and play regulatory roles.
CONCLUSION
We have shown the formation of G-quadruplexes containing (4n - 1) guanines in the core. The structure of a parallel-stranded G-quadruplex involving two G-tetrads and one G-triad with a vacant site was solved by NMR. Molecular dynamics simulation showed the formation of a G-triad·water complex mimicking a ghost guanine. The vacant site in the G-triad could be used as a recognition spot for metabolite and drug binding.
NOTE
Most of this work was presented at the 5th International Meeting on Quadruplex Nucleic Acids: G4thering in Bordeaux; May 26–28, 2015, Bordeaux, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore Ministry of Education Academic Research Fund Tier 3 [MOE2012-T3-1-001] and grants from Nanyang Technological University (to A.T.P). Funding for open access charge: Singapore Ministry of Education Academic Research Fund Tier 3 [MOE2012-T3-1-001].
Conflict of interest statement. None declared.
REFERENCES
- 1.Gellert M., Lipsett M.N., Davies D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U.S.A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 3.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M., Jr, Sen D., Lansdorp P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopes J., Piazza A., Bermejo R., Kriegsman B., Colosio A., Teulade-Fichou M.P., Foiani M., Nicolas A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paeschke K., Capra J.A., Zakian V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cahoon L.A., Seifert H.S. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumari S., Bugaut A., Huppert J.L., Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackburn E.H. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 11.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 12.Lei M., Podell E.R., Cech T.R. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 13.Piazza A., Adrian M., Samazan F., Heddi B., Hamon F., Serero A., Lopes J., Teulade-Fichou M.P., Phan A.T., Nicolas A. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015;34:1718–1734. doi: 10.15252/embj.201490702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phan A.T., Kuryavyi V., Ma J.-B., Faure A., Andréola M.-L., Patel D.J. An interlocked dimeric parallel-stranded DNA quadruplex: A potent inhibitor of HIV-1 integrase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amrane S., Adrian M., Heddi B., Serero A., Nicolas A., Mergny J.L., Phan A.T. Formation of pearl-necklace monomorphic G-quadruplexes in the human CEB25 minisatellite. J. Am. Chem. Soc. 2012;134:5807–5816. doi: 10.1021/ja208993r. [DOI] [PubMed] [Google Scholar]
- 18.Lim K.W., Phan A.T. Structural basis of DNA quadruplex-duplex junction formation. Angew. Chem. Int. Ed. Engl. 2013;52:8566–8569. doi: 10.1002/anie.201302995. [DOI] [PubMed] [Google Scholar]
- 19.Adrian M., Ang D.J., Lech C.J., Heddi B., Nicolas A., Phan A.T. Structure and conformational dynamics of a stacked dimeric G-quadruplex formed by the human CEB1 minisatellite. J. Am. Chem. Soc. 2014;136:6297–6305. doi: 10.1021/ja4125274. [DOI] [PubMed] [Google Scholar]
- 20.Lim K.W., Nguyen T.Q., Phan A.T. Joining of multiple duplex stems at a single quadruplex loop. J. Am. Chem. Soc. 2014;136:17969–17973. doi: 10.1021/ja5078816. [DOI] [PubMed] [Google Scholar]
- 21.Kocman V., Plavec J. A tetrahelical DNA fold adopted by tandem repeats of alternating GGG and GCG tracts. Nat. Commun. 2014;5:5831. doi: 10.1038/ncomms6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerofolini L., Amato J., Giachetti A., Limongelli V., Novellino E., Parrinello M., Fragai M., Randazzo A., Luchinat C. G-triplex structure and formation propensity. Nucleic Acids Res. 2014;42:13393–13404. doi: 10.1093/nar/gku1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Limongelli V., De Tito S., Cerofolini L., Fragai M., Pagano B., Trotta R., Cosconati S., Marinelli L., Novellino E., Bertini I., et al. The G-Triplex DNA. Angew. Chem. Int. Ed. 2013;52:2269–2273. doi: 10.1002/anie.201206522. [DOI] [PubMed] [Google Scholar]
- 24.Mukundan V.T., Phan A.T. Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 2013;135:5017–5028. doi: 10.1021/ja310251r. [DOI] [PubMed] [Google Scholar]
- 25.Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 26.Stadlbauer P., Trantírek L., Cheatham T.E. III, Koča J., Šponer J. Triplex intermediates in folding of human telomeric quadruplexes probed by microsecond-scale molecular dynamics simulations. Biochimie. 2014;105:22–35. doi: 10.1016/j.biochi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Mashimo T., Yagi H., Sannohe Y., Rajendran A., Sugiyama H. Folding pathways of human telomeric type-1 and type-2 G-quadruplex structures. J. Am. Chem. Soc. 2010;132:14910–14918. doi: 10.1021/ja105806u. [DOI] [PubMed] [Google Scholar]
- 28.Rosu F., Gabelica V., Poncelet H., De Pauw E. Tetramolecular G-quadruplex formation pathways studied by electrospray mass spectrometry. Nucleic Acids Res. 2010;38:5217–5225. doi: 10.1093/nar/gkq208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boncina M., Lah J., Prislan I., Vesnaver G. Energetic basis of human telomeric DNA folding into G-quadruplex structures. J. Am. Chem. Soc. 2012;134:9657–9663. doi: 10.1021/ja300605n. [DOI] [PubMed] [Google Scholar]
- 30.Gray R.D., Trent J.O., Chaires J.B. Folding and unfolding pathways of the human telomeric G-quadruplex. J. Mol. Biol. 2014;426:1629–1650. doi: 10.1016/j.jmb.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W., Hou X.M., Wang P.Y., Xi X.G., Li M. Direct measurement of sequential folding pathway and energy landscape of human telomeric G-quadruplex structures. J. Am. Chem. Soc. 2013;135:6423–6426. doi: 10.1021/ja4019176. [DOI] [PubMed] [Google Scholar]
- 32.Zhang A.Y., Balasubramanian S. The kinetics and folding pathways of intramolecular G-quadruplex nucleic acids. J. Am. Chem. Soc. 2012;134:19297–19308. doi: 10.1021/ja309851t. [DOI] [PubMed] [Google Scholar]
- 33.Rajendran A., Endo M., Hidaka K., Teulade-Fichou M.-P., Mergny J.-L., Sugiyama H. Small molecule binding to a G-hairpin and a G-triplex: a new insight into anticancer drug design targeting G-rich regions. Chem. Commun. 2015;51:9181–9184. doi: 10.1039/c5cc01678a. [DOI] [PubMed] [Google Scholar]
- 34.Bennett B.D., Kimball E.H., Gao M., Osterhout R., Van Dien S.J., Rabinowitz J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patti G.J., Yanes O., Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serganov A., Patel D.J. Molecular recognition and function of riboswitches. Curr. Opin. Struct. Biol. 2012;22:279–286. doi: 10.1016/j.sbi.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hermann T., Patel D.J. Biochemistry — adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 38.Liu J., Cao Z., Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lauhon C.T., Szostak J.W. RNA aptamers that bind flavin and nicotinamide redox cofactors. J. Am. Chem. Soc. 1995;117:1246–1257. doi: 10.1021/ja00109a008. [DOI] [PubMed] [Google Scholar]
- 40.Li Y., Geyer C.R., Sen D. Recognition of anionic porphyrins by DNA aptamers. Biochemistry. 1996;35:6911–6922. doi: 10.1021/bi960038h. [DOI] [PubMed] [Google Scholar]
- 41.Goddard T.D., Kneller D.G. SPARKY 3. San Francisco: University of California; [Google Scholar]
- 42.Schwieters C.D., Kuszewski J.J., Tjandra N., Marius Clore G. The XPLOR-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 43.Case D.A., Berryman J.T., Betz R.M., Cerutti D.S., Cheatham I.T.E., Darden T.A., Duke R.E., Giese T.J., Gohlke H., Goetz A.W., et al. San Francisco: University of California; 2015. [Google Scholar]
- 44.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 45.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 46.DeLano W.L. Palo Alto: DeLano Scientific; 2002. [Google Scholar]
- 47.Pérez A., Marchán I., Svozil D., Sponer J., Cheatham T.E., Laughton C.A., Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of α/γ conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Do N.Q., Phan A.T. Monomer-dimer equilibrium for the 5′-5′ stacking of propeller-type parallel-stranded G-quadruplexes: NMR structural study. Chemistry. 2012;18:14752–14759. doi: 10.1002/chem.201103295. [DOI] [PubMed] [Google Scholar]
- 49.Phan A.T., Kuryavyi V., Ma J.B., Faure A., Andreola M.L., Patel D.J. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phan A.T. A site-specific low-enrichment 15N,13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002;124:1160–1161. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]
- 51.Huang X., Yu P., LeProust E., Gao X. An efficient and economic site-specific deuteration strategy for NMR studies of homologous oligonucleotide repeat sequences. Nucleic Acids Res. 1997;25:4758–4763. doi: 10.1093/nar/25.23.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phan A.T. Long-range imino proton-13C J-couplings and the through-bond correlation of imino and non-exchangeable protons in unlabeled DNA. J. Biomol. NMR. 2000;16:175–178. doi: 10.1023/a:1008355231085. [DOI] [PubMed] [Google Scholar]
- 53.Smith F.W., Feigon J. Strand orientation in the DNA quadruplex formed from the Oxytricha telomere repeat oligonucleotide d(G4T4G4) in solution. Biochemistry. 1993;32:8682–8692. doi: 10.1021/bi00084a040. [DOI] [PubMed] [Google Scholar]
- 54.Martin-Pintado N., Heddi B., Teuku A.K., Serimbetov Z., Phan A.T. 5th International Meeting on Quadruplex Nucleic Acids: G4thering in Bordeaux, May 26–28. Bordeaux, France: 2015. G-quadrulpexes with (4n-1) guanines in the G-tetrad core: formation of a G-triad·H2O complex [Abstract No Thu-P03] [Google Scholar]
- 55.Lech C.J., Heddi B., Phan A.T. Guanine base stacking in G-quadruplex nucleic acids. Nucleic Acids Res. 2013;41:2034–2046. doi: 10.1093/nar/gks1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lim K.W., Amrane S., Bouaziz S., Xu W., Mu Y., Patel D.J., Luu K.N., Phan A.T. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patel D.J., Phan A.T., Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujimoto T., Nakano S., Miyoshi D., Sugimoto N. The effects of molecular crowding on the structure and stability of G-quadruplexes with an abasic site. J. Nucleic Acids. 2011;2011:857149. doi: 10.4061/2011/857149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Virgilio A., Petraccone L., Esposito V., Citarella G., Giancola C., Galeone A. The abasic site lesions in the human telomeric sequence d[TA(G3T2A)3G3]: a thermodynamic point of view. Biochim. Biophys. Acta. 2012;1820:2037–2043. doi: 10.1016/j.bbagen.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 60.Pang S.M., Heddi B., Martin-pintado N., Serimbetov Z., Phan A.T. 5th International Meeting on Quadruplex Nucleic Acids: G4thering in Bordeaux, May 26–28. Bordeaux, France: 2015. Interaction between guanosine and a G-quadruplex with a vacant site [Abstract No Wed-P30] [Google Scholar]
- 61.Cheong V.V., Heddi B., Lech C.J., Phan A.T. Xanthine and 8-oxoguanine in G-quadruplexes: formation of a G·G·X·O tetrad. Nucleic Acids Res. 2015;43:10506–10514. doi: 10.1093/nar/gkv826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nudler E., Mironov A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 63.Serganov A., Yuan Y.R., Pikovskaya O., Polonskaia A., Malinina L., Phan A.T., Hobartner C., Micura R., Breaker R.R., Patel D.J. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Danilchanka O., Mekalanos J.J. Cyclic dinucleotides and the innate immune response. Cell. 2013;154:962–970. doi: 10.1016/j.cell.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.