Abstract
Mutations of the KRAS proto-oncogene are associated with several tumor types, which is why it is being considered as a target for anti-cancer drug development. The human KRAS promoter contains a nuclease hypersensitive element (NHE), which can bind to nuclear proteins and is believed to form G-quadruplex structures. Previous studies showed that a 32-nt oligonucleotide (32R-3n) mimicking the KRAS NHE can reduce gene transcription by sequestering MAZ, a crucial transcription factor. Here we show that 32R-3n has to dimerize in order to fold into a G-quadruplex structure. Individual 5′- and 3′-end G-quadruplex units are formed and both feature a symmetric head-to-head topology with edge-type loops. The MAZ binding sequence is located within the 3′-end unit. Nuclear magnetic resonance data complemented by CD and UV spectra show that nucleotides of the MAZ binding G-rich motif are dynamic and could be available for sequence or structure based recognition. Both stable G-quadruplex structures could protect 5′- and 3′-ends of 32R-3n and enhance its anti-cancer activity. Single stranded genomic KRAS NHE including nucleotides flanking the 32R-3n sequence could favor a different monomeric fold, which remains unknown.
INTRODUCTION
The ras family of proteins are ubiquitous small GTPases, which play an important role in cellular signal transduction (1). Point mutations in ras proto-oncogenes cause malignant transformations in human cells due to reduced capacity of the encoded protein to hydrolyze GTP to GDP, which leads to uncontrolled cell proliferation (2). Mammalian cells contain three proto-oncogenes of the ras family (HRAS, KRAS and NRAS) with different tissue expression levels. The KRAS protein is most abundant in the colon and thymus. Mutations of the KRAS gene are involved in various tumor types, especially lung, colorectal and pancreatic tumors (3). Due to its role in the pathogenesis of cancer, KRAS is a promising target for anti-cancer drugs (4–6).
The human KRAS proto-oncogene promoter contains a G-rich nuclease hypersensitive element (NHE) upstream of the major transcription initiation site, which is believed to form non-B-DNA structures, specifically G-quadruplexes (7–9). These four-stranded structures are comprised of stacks of G-quartets, which are planar arrangements of four guanine bases stabilized by monovalent cations (10,11). Continuous or interrupted guanine runs are interconnected by loops of which more common include edge-type (lateral), diagonal and propeller-type (12–15). The resulting strand orientation and subsequently G-quadruplex topology can therefore be of parallel, anti-parallel or mixed type. Accompanied by a plethora of less common structural elements G-quadruplex structures can reach tremendous diversity (16).
Initial interest in G-quadruplexes was sparked by their assumed presence in telomeric regions, however, focus has also shifted toward gene promoters. Bioinformatic data suggest that G-quadruplexes may act as cis-acting regulatory elements for a large portion (40%) of human genes (17). Recently, numerous attempts were made in order to elicit a therapeutic response by manipulating telomeric and promoter G-quadruplexes (18,19). One of the most popular approaches utilizes small molecule ligands that stabilize G-quadruplexes and/or interfere with potential protein-DNA interactions (20,21). G-quadruplex binding ligands are usually planar aromatic molecules, which interact with the outer G-quartets through π-π stacking. The large area of a G-quartet should offer such ligands enough discrimination against other DNA forms. However, the stabilization of a specific structure, despite the large degree of polymorphism exerted by G-quadruplexes, remains an open issue (22,23). Therefore, detailed information on the structure of potentially druggable G-quadruplexes lays the groundwork for successful drug design.
A Myc-associated zinc finger protein (MAZ) has been found to specifically bind to a GA-box with a consensus sequence GGG(A/C)GG, which is found in the KRAS promoter (24). Subsequently, it has been shown that MAZ is a transcription factor, which activates human KRAS transcription (25). A decoy strategy, using stable KRAS promoter analogues, has been devised in order to sequester the MAZ protein and prevent it from activating KRAS transcription. Xodo et al. found that a 32-nt fragment of the KRAS promoter, designated as 32R-3n, folds into a G-quadruplex structure (26). Furthermore, twisted intercalating nucleic acid (TINA) modified 32R-3n reduces the level of KRAS mRNA in Panc-1 cells to 20%, which in turn reduces tumor growth in mice carrying a Panc-1 xenograft (27). The TINA residues contain polycyclic aromatic hydrocarbon moieties, which are believed to stack on terminal G-quartets with a favorable effect on G-quadruplex stability (28,29).
Although several attempts have been made to determine structures of G-quadruplexes formed by various KRAS promoter oligonucleotides, utilization of low resolution techniques made unambiguous determination of folding topologies challenging (9,26,27,30,31). As a result the exact structural features of 32R-3n and its chemically modified analogues, which are important for KRAS attenuation, remain unknown. In the current study, nuclear magnetic resonance (NMR), CD and UV spectroscopy are used in order to determine folding topology of 32R-3n, d(GCG GTG TGG GAA GAG GGA AGA GGG GGA GGC AG). Our data show that two symmetric dimeric G-quadruplex units are formed in a solution containing K+ ions. The proposed topology offers rationale for anti-tumor activity of the 32R-3n oligonucleotide and is a basis for further development of decoy based KRAS suppression strategies in cancer cells.
MATERIALS AND METHODS
Sample preparation
DNA oligonucleotides were synthesized on solid support using a H-8 DNA synthesizer (K&A Laborgeraete). Oligonucleotides were removed from solid support using ultra fast treatment with 50% ammonium hydroxide/50% methyl amine (AMA). Incubation with AMA took place for 20 min at room temperature followed by 15 min at 65°C. The solution was evaporated and immediately redissolved in 10 mM TEAB buffer. Oligonucleotides were purified with reverse phase HPLC on a C18 column. The mobile phase was evaporated and the remaining DNA was treated for 20 min with 80% acetic acid to remove the DMT group. Subsequently, the DNA was precipitated with ethanol and again redissolved in TEAB buffer. A GE AktaPurifier with a HiPrep 26/10 column was used to desalt the DNA. After the final evaporation, the DNA was dissolved in the NMR buffer containing 10% 2H2O, 20 mM K phosphate buffer (pH 7) and 80 mM KCl. Final concentration of oligonucleotides was in the range between 0.5 and 1.0 mM.
UV melting
UV melting experiment was carried out on a Varian CARY-100 BIO Spectrophotometer. The low concentration experiment was carried out at 295 nm using a 1 cm cell. The concentration of DNA was adjusted to the absorbance at 295 nm in the range between 0.4 and 0.8. In the high concentration experiment, NMR samples were used directly in a 1 mm cell and the wavelengths were adjusted between 300 and 305 nm to obtain an absorbance of less than 1.0. The temperature was increased/decreased from 10 to 80°C with the rate of 0.5°C/min. Melting temperatures were determined from the first order derivatives of melting profiles.
Circular dichroism spectroscopy
Circular dichroism (CD) spectra were recorded on an Applied Photophysics Chirascan CD spectrometer at 25°C using 1.0 mm and 0.1 mm path length quartz cells for samples with oligonucleotide concentrations of 50 and 500 μM, respectively. All samples were prepared with the NMR buffer (20 mM K phosphate buffer, pH 7 and 80 mM KCl). The wavelength range was from 220 to 320 nm.
NMR spectroscopy
NMR spectra were collected on Agilent VNMRS 800 and 600 MHz NMR spectrometers at 25°C. 1D and 2D NMR spectra were acquired using DPFGSE water suppression sequence. NOESY spectra were acquired with mixing times (τm) from 80 to 250 ms. Spectra were processed with VNMRJ (Agilent Technologies) and NMRPipe 8.2 software packages. Assignment was carried out with CcpNmr's Analysis module v2.4.
Electrophoretic mobility shift assay
Non-denaturing 12% polyacrylamide gel was prepared with TBE buffer containing 100 mM KCl. Same buffer was also used in both reservoirs. Gel plates were cooled to 10°C for the 6 h run at 90 V. Thermo Scientific GeneRuler Ultra Low Range DNA Ladder was applied to the central lane. Stains-All was used for staining.
RESULTS
KRAS promoter oligonucleotide forms two G-quadruplex units at 5′- and 3′-ends
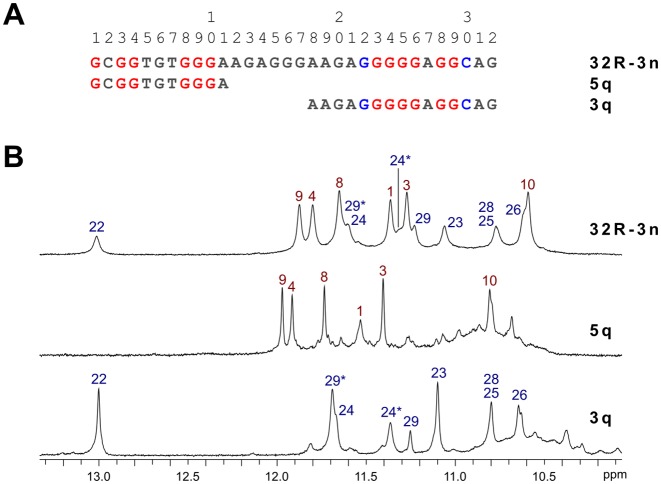
An aqueous K+ ion containing solution of 32R-3n was prepared and its 1D 1H NMR spectrum gave a quick insight into G-quadruplex formation. The imino region of 32R-3n exhibited two sets of signals with different characteristics. Six signals with chemical shifts ranging from δ 10.6 to 11.9 ppm were relatively sharp, while remaining eight signals exhibited broader linewidths in a wider chemical shift range between δ 10.6 and 13.0 ppm (Figure 1). All guanine sites in the sequence of 32R-3n were probed for hydrogen bonding with a series of residue specific partially 15N isotopically enriched oligonucleotides. 1D 15N-edited HSQC spectra revealed that guanine tracts, which are hydrogen bonded and possibly involved in G-quartet formation, can be split formally into two regions (Supplementary Figures S1 and S2). The first region near the 5′-end of the 32R-3n sequence includes guanine residues 1, 3, 4 and 8–10, which correspond to the six sharper imino resonances in the 1D NMR spectrum. The second region near the 3′-end includes the long G-tract from residue 22 to 26 with the addition of G28 and G29. It is noteworthy that G24 and G29 each give rise to two imino resonances, which suggests some sort of conformational switching, which is slow on the NMR chemical shift timescale (vide infra). Oligonucleotides with 15N labeled guanines in the center of the sequence (residues 13–20) did not produce observable imino resonances in 15N-edited HSQCs, which suggested that they were not hydrogen bonded.
Figure 1.
Sequences and NMR characterization of G-quadruplex forming KRAS oligonucleotides. (A) Sequences of the wild type 32R-3n and its truncated 5q and 3q analogues corresponding to 5′- and 3′-end quadruplex forming region, respectively. Red guanines are involved in Hoogsteen hydrogen bonding, while blue guanines and cytosines form Watson–Crick base pairs. (B) Imino regions of 1D 1H NMR spectra of 32R-3n, 5q and 3q oligonucleotides with assignments. Spectra were acquired at 25°C with oligonucleotide concentrations in the range between 0.5 and 2.0 mM in 10% 2H2O, 80 mM KCl and 20 mM K phosphate buffer (pH 7).
Previous studies on 32R-3n (27) included PAGE assays, which suggested it adopts a dimeric structure in the presence of K+ ions. This led us to believe that two G-quadruplexes are formed by two molecules of 32R-3n. Truncated oligonucleotides comprised of one of the two G-quartet forming regions (G1-G10 and G22-C30) of 32R-3n with different overhang lengths were screened for G-quadruplex forming ability. All oligonucleotides exhibited NMR signals characteristic of G-quadruplex formation albeit with chemical shift perturbations. 5q and 3q were found to exhibit 1H NMR resonance patterns, which corresponded to individual G-quadruplex units within full-length 32R-3n and were further characterized (Figure 1). An attempt to induce unimolecular folding of 32R-3n included preparation of a diluted NMR sample (100 μM), which was annealed for 5 min at 95°C followed by snap cooling on ice. The resulting 1D 1H NMR spectrum was identical to that of a concentrated sample and indicated persistence of the dimeric structure.
Additionally, an oligonucleotide of the same length has been prepared where the central segment (A11-A21) of the sequence of 32R-3n (between the two G-quadruplex forming regions) was replaced with a T11 element. Nine observable imino 1H resonances (including the downfield G22 resonance) support the formation of both G-quadruplex units (Supplementary Figure S3). However, due to considerable chemical shift perturbations assignment of the Hoogsteen hydrogen bonded guanine imino resonances could not be carried over from 32R-3n. Compared to 32R-3n, three imino resonances are not observed in 1D NMR spectra of the T11 mutant indicating increased dynamics of some guanine residues at the 3′-end. Interestingly, a shorter oligonucleotide with a T5 element replacing the central segment in 32R-3n formed a complex mixture of G-quadruplex structures (Supplementary Figure S3). This shows that a shorter central segment reduces flexibility and increases structure heterogeneity.
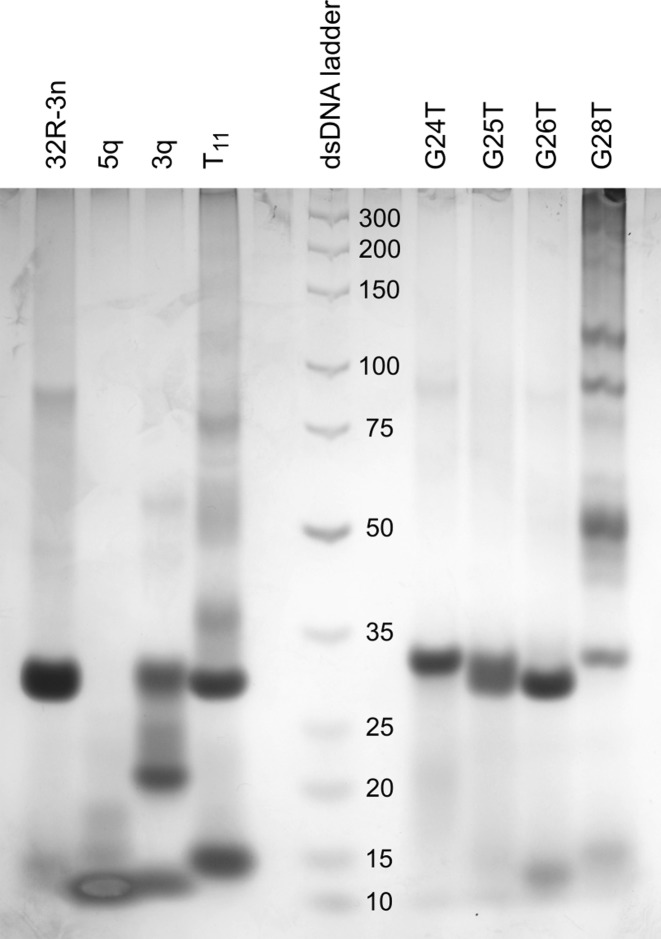
While NMR spectra of 32R-3n suggest a single structure is present in solution containing K+ ions, spectra of both truncated oligonucleotides (5q and 3q) show the presence of a major species and one or more minor species (Figure 1). This is also evident from PAGE (Figure 2). 32R-3n migrates as a single band. As expected, the major component of 5q migrates faster, but several minor species with lower electrophoretic mobility, possibly due to different loop orientations, are also present. On the other hand, 3q also has a fast migrating component, however a substantial portion of the oligonucleotide migrates as slowly as 32R-3n (with two G-quadruplex units), which suggest oligomerization of 3q. The T11 mutant migrates as two distinct bands with electrophoretic mobilities corresponding to a dimeric (32R-3n like) and monomeric form.
Figure 2.
PAGE of 32R-3n, 5q, 3q, T11 and oligonucleotides with G to T point mutations. The DNA ladder with corresponding dsDNA lengths is in the central lane.
3′- and 5′-end G-quadruplex units feature symmetric head-to-head topologies with edge-type loops
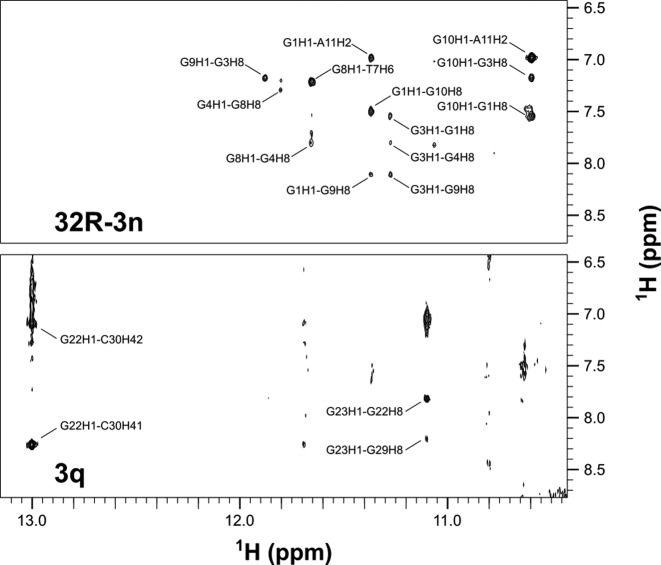
A series of 2D COSY and NOESY spectra were acquired and used for assignment of anomeric, aromatic, amino and imino 1H resonances of 32R-3n, 5q and 3q. On the basis of NOE connectivities folding topologies of both G-quadruplex units were determined (Figure 3). Signals widths in 1D and 2D spectra are favorable in the case of truncated oligonucleotides compared to the full length 32R-3n. Importantly, identical sequential walks of both G-quadruplex units can be traced in NOESY spectra of the full-length and truncated oligonucleotides.
Figure 3.
Aromatic-anomeric regions of 2D NOESY (τm = 250 ms) NMR spectra of 32R-3n, 5q and 3q oligonucleotides. The sequential walk is shown for the truncated oligonucleotides.
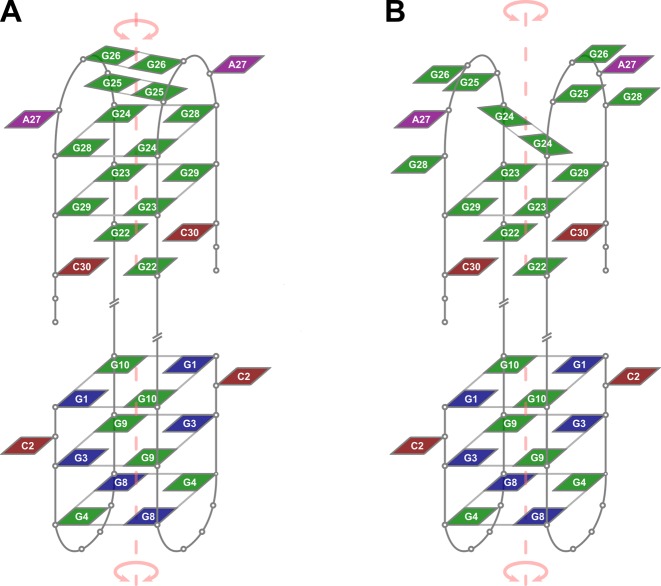
Assignment of the 5′-end structured region (G1-A11) was performed with the full length 32R-3n and its truncated oligonucleotide 5q. High intensity intranucleotide cross-peaks and characteristic internucleotide aromatic-anomeric NOESY correlations indicate that G1, G3 and G8 are in syn conformation. Imino to aromatic 1H NOE contacts revealed that individual G-quartets in the 5′-end region are comprised of G1/G10, G3/G9 and G4/G8 pairs (Figure 4). Each guanine is both hydrogen-bond acceptor and donor to its G-quartet partner. NOE connectivities support alternating G1:G10:G1:G10, G3:G9:G3:G9 and G4:G8:G4:G8 configurations of the three G-quartets of the 5′-end unit. The order of G-quartets in the structure was determined through examination of inter-quartet guanine imino-imino and imino-aromatic cross-peaks (Supplementary Figure S4). Based on the available NMR data we propose that the 5′-end G-quadruplex folds into an anti-parallel dimeric fold-back head-to-head topology with edge-type loops (Figure 5). The first G-tract (G1-G3-G4) is interrupted by C2. A limited number of weak NOE contacts with C2 suggest that it is flexible and protrudes away from the G-quadruplex core (Supplementary Figure S4). The trinucleotide TGT loop is followed by the second continuous G-tract (G8-G10). T7's methyl group exhibits intense cross-peaks with imino protons of G4 and G8, which suggests efficient stacking of T7 on the adjacent G-quartet. Similarly, A11's H2 exhibits cross-peaks with imino protons of G1 and G10 comprising the G-quartet on the opposite side of the G-quadruplex unit. The resulting fold-back topology exhibits an axis of symmetry running through the center of G-quartets, perpendicular to G-quartet planes. Resonance signal overlap precludes determination of clockwise or anticlockwise orientation of nucleotides within G-quartets. The proposed topology is however consistent with a structure exhibiting C2 symmetry.
Figure 4.
Imino-aromatic regions of 2D NOESY (τm = 250 ms) NMR spectra of 32R-3n and 3q oligonucleotides.
Figure 5.
Proposed topologies of low- (A) and high-temperature (B) forms of dimeric G-quadruplexes formed by 32R-3n in the presence of K+ ions. Guanines in anti and syn conformation are depicted in green and blue, respectively. Adenines and cytosines are in purple and red, respectively. Some loop and overhang residues are omitted for clarity. An axis of symmetry traverses central cavities of both G-quadruplex units.
The complete sequential walk found for the 5′-end structured region can be traced in NOESY spectra of 32R-3n. On the other hand, some sequential steps of the 3′-end structured region are obscured in NOESY spectra of 32R-3n due to broader cross-peaks and resonance overlap (Figure 3). Since equivalent imino 1H chemical shift patterns of the 3′-end structured region are observed in spectra of 32R-3n and 3q (Supplementary Figures S1 and S2), identical topology of this region is implied. Therefore, the sample of 3q was used for assignment of the 3′-end structured region in 32R-3n (G22-C30). 1D NMR spectra of 3q exhibits several imino resonances in the Hoogsteen chemical shift range and a single downfield resonance at δ 13.0 ppm, which has been assigned to G22. On the basis of G22 imino to C30 amino 1H NOE contacts the formation of G22:C30 Watson–Crick base pairs could be unequivocally determined (Figure 4). 1H chemical shifts of the C30 amino group show that one proton is hydrogen bonded (δ 8.26 ppm) and the other is not (δ 7.10 ppm). The non-hydrogen bonded proton rules out the possibility of cross-strand hydrogen bonds between two G:C base pairs in the dimeric structure and suggests their slipped arrangement. A G23 H1 to G29 H8 NOE contact suggests formation of a G23:G29:G23:G29 quartet adjacent to the two G:C base pairs. Interestingly, G24, G25 and G26 also give rise to observable, albeit weaker imino resonances (Figure 1). The G28 imino resonance was identified through 15N-edited HSQC spectra and was found to be broader and overlapping with G25 (Supplementary Figure S2). Despite hydrogen bonding observed NOE contacts do not support the formation of a single set of well defined G-quartets in the G24-G28 region.
The importance of G24, G25, G26 and G28 for folding of the whole structure has been tested by their substitution with thymines (in the full length 32R-3n). 1H NMR signal patterns of the mutated oligonucleotides contain all imino 1H NMR resonances of the 5′-end G-quadruplex unit, which shows that the 5′-end region is not affected by mutations near the 3′-end (Supplementary Figure S5). On the other hand, the effect on 3′-end G-quadruplex unit varies with mutation site. G24T suppresses the formation of the 3′-end G-quadruplex unit as none of the imino 1H resonances can be observed for the sequence at the 3′-end. For G25T very weak imino 1H NMR resonances of G22, G23 and G29 can be observed. On the other hand, the G26T mutation seems to have less effect as G22, G23 and G29 imino 1H resonance intensities of the 3′-end region are comparable to 32R-3n.
Differences in the structure of the 3′-end are reflected in PAGE (Figure 2). G26T migrates as a single band at the same rate as 32R-3n. G24T also migrates as a single band, however, slightly, but noticeably slower. The loss of 3′-end G-quadruplex structure results in a minimal decrease in electrophoretic mobility. G25T produces a wide band corresponding to an equilibrium between a structure with only the 5′-end G-quadruplex and a structure with both G-quadruplex units formed. NMR spectrum of the G28T oligonucleotide exhibits only imino 1H resonances assigned to the 5′-end G-quadruplex (Supplementary Figure S5), which shows that the mutation restricts formation of the 3′-end G-quadruplex. The importance of G28 for folding of the 3′-end G-quadruplex forming region is also evident from PAGE as the G28T mutant forms only a minor fraction of the dimeric structure in favor of several higher order structures (Figure 2). The electrophoretic mobility of the G28T dimer corresponds to a structure with only the 5′-end G-quadruplex formed, which is in agreement with the NMR spectrum. A very broad background signal in the imino region of the NMR spectrum corresponds to the higher order structures.
Importance of residues G24 and G28 for folding of the 3′-end G-quadruplex unit suggests that a G24:G28:G24:G28 quartet is formed, but due to dynamic properties does not give observable characteristic NMR cross-peaks. Additionally, formation of G25:G25 and G26:G26 base pairs is in accordance with observable G25 (δ 10.8 ppm) and G26 (δ 10.6 ppm) imino 1H NMR resonances. The major form of the 3′-end G-quadruplex is therefore comprised of two G:C base pairs and G23:G29:G23:G29 quartet, which form the core of an anti-parallel dimeric head-to-head fold-back topology (Figure 5A). The proposed formation of a G24:G28:G24:G28 quartet and two G:G base pairs agrees with available NMR data although is not directly supported by clear NOE interactions among resolved and assigned imino and/or aromatic resonances in a structure involved in dynamic conformational exchange.
Observation of a second pair of imino resonances for G24 and G29 (designated as G24* and G29* in Figure 1) implies an alternate structure of the 3′-end G-quadruplex unit. We have acquired 1D 1H NMR spectra at temperatures ranging from 25 to 40°C (Supplementary Figure S6). Due to overlap of imino resonances additional 15N-edited HSQCs were acquired for G24-selectively 15N-labeled 3q at 25 and 40°C. The ratios of G24/G24* and G29/G29* signal intensities are highly temperature dependent. 1D 1H NMR spectra acquired at 40°C are dominated by imino resonances of G22, G23, G24* and G29*. The simultaneous decrease of imino proton signals of G24, G25, G26 and G29 upon temperature increase suggest melting of G25:G25 and G26:G26 base pairs and rearrangement of the G24:G29:G24:G29 quartet in favor of a G24:G24 base pair (Figure 5B). It is noteworthy that aromatic and anomeric resonances of G24 and G29 of both forms were not resolved on the proton chemical shift time-scale. Interestingly, G23 exhibits a single imino proton resonance, which is possibly due to efficient stacking of the adjacent G24 in both forms. G29 on the other hand loses stacking of adjacent G28 at higher temperatures (cf. structures in Figure 5).
1H signals of the region connecting 3′- and 5′-end G-quadruplex units (A11-A21) in spectra of 32R-3n get progressively broader from both 3′- and 5′-ends. A11 and G21 exhibit slight broadening of cross-peaks in aromatic-anomeric region of NOESY spectra, while no peaks could be assigned to residues A12-G20 in 1D or 2D spectra (Figure 3 and Supplementary Figure S7). This suggests that this region is flexible and allows for relative movement of the two G-quadruplex units.
G:C base pairs are critical for stability of the 3′-end G-quadruplex
Melting temperature experiment by UV absorption of the full length 32R-3n (Table 1) is in agreement with previous studies (27). 5q melts at a twelve degree lower temperature. Interestingly, the temperature at half transition (T1/2) of 3q was higher than that of 5q, which could be attributed to the stable Watson–Crick base pairs. First derivative of the melting curve of 3q shows a very weak premelting transition at around 33°C (Supplementary Figure S8), which could correspond to the melting of G25:G25 and G26:G26 base pairs and rearrangement of the G24:G29:G24:G29 quartet into a G24:G24 base pair as suggested by NMR. T11 is destabilized by the change in sequence of the segment connecting the two G-quadruplex units, which results in a 9°C lower T1/2 versus 32R-3n. Interestingly, no hysteresis can be observed for the T11 mutant. 32R-3n and 5q exhibit only slight hysteresis of a few degrees, while the hysteresis in the case of 3q is 19°C (considering the major transition at 42°C).
Table 1. Melting temperatures of the full length and truncated oligonucleotides.
| Oligonucleotide | T1/2 (°C) | |
|---|---|---|
| Low concentration | High concentration | |
| 32R-3n | 48 | 52 |
| 5q | 36 | 55 |
| 3q | 42 (33) | 47 |
| T11 | 39 | 40 |
*Concentration of oligonucleotides was in the range between 5 and 10 μM for the low concentration experiment (measured at 295 nm) and between 0.5 and 1.0 mM for the high concentration experiment (measured at 300–305 nm). The temperature was increased/decreased from 10 to 80°C with the rate of 0.5°C/min.
We have also determined the T1/2 for 32R-3n, 5q, 3q and T11 at NMR concentrations. T1/2 of 32R-3n, 3q and T11 are higher for up to 5°C, which is caused by a crowding effect due to increased oligonucleotide concentration. On the other hand, a dramatic increase in T1/2 of 19°C is observed with 5q (Table 1), which can only be explained by a considerable change in the structure of 5q at different concentrations.
Dimeric structures are resolved at lower concentrations
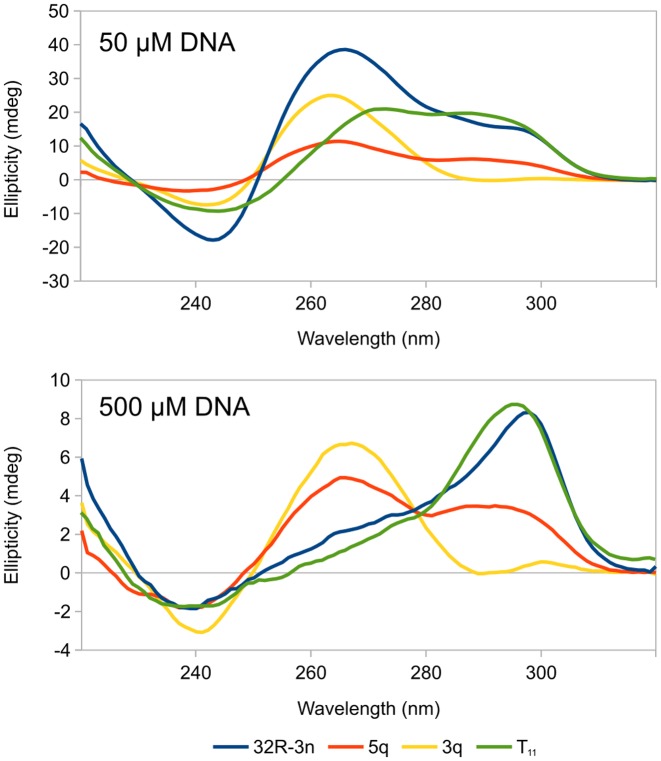
CD spectra of 32R-3n, 5q, 3q and T11 exhibit positive peaks at 265 and 295 nm and negative peaks at 240 nm (Figure 6). Spectra were recorded with two oligonucleotide concentrations (50 and 500 μM). Relative intensities of signals in CD spectra vary for each oligonucleotide and concentration. None of the spectra are characteristic of pure parallel or anti-parallel G-quadruplexes. NMR data suggest that at high concentrations major G-quadruplexes species of all oligonucleotides are anti-parallel, which is reflected in CD spectra as a maximum at around 295 nm. The positive and negative signals at 265 and 240 nm, respectively, are typically attributed to parallel G-quadruplex species. Interestingly, the positive 295 nm signals rapidly decrease with oligonucleotide dilution suggesting unfolding of the anti-parallel G-quadruplex structures and a transition to parallel ones at low concentrations.
Figure 6.
CD spectra of 32R-3n (blue), 5q (red), 3q (yellow) and T11 (green) oligonucleotides at 50 and 500 μM oligonucleotide concentrations. Spectra were acquired at 25°C in an 80 mM KCl and 20 mM K phosphate buffer (pH 7) solution with optical paths of 0.1 mm (500 μM DNA) and 1.0 mm (50 μM DNA).
32R-3n and T11 exhibit almost identical CD spectra corresponding to mostly anti-parallel dimeric G-quadruplexes at high concentrations. Upon dilution both exhibit a shift of equilibria toward parallel (possibly monomeric) structures with 32R-3n being more affected of the two. CD spectra of 5q show a comparable degree of parallel and anti-parallel species at both high and low concentrations, while 3q is mostly parallel. However, NMR data show that the major species of 5q and 3q at high concentrations are anti-parallel. Broad background signals in NMR spectra and multiple bands on PAGE suggest that both oligonucleotides form ensembles of minor species, which could be parallel G-quadruplexes and give rise to characteristic CD signals while not being explicitly identified in NMR spectra.
DISCUSSION
The complete KRAS NHE contains four runs of at least three guanines. Despite high guanine frequency features like G-tracts of various lengths, single guanines in loops, interrupted G-tracts etc. allow for a number of folding options. Due to the notoriously polymorphic nature of G-quadruplexes, a complex array of structures and interconversion between them is expected and observed (30). Consequently, previous studies on KRAS NHE oligonucleotides of various lengths resulted in different folding topologies.
Earlier study of Xodo et al. found that 32R-3n, a truncated portion of the KRAS NHE lacking the 5′ GGG run, adopts both monomeric and dimeric G-quadruplex forms in a KCl solution (27). At low concentrations shortly after annealing a monomeric form is preferred, while an overnight incubation shifts the equilibrium toward the dimeric form. We have shown that dimers of 32R-3n actually form two individual G-quadruplex units at 5′- and 3′-ends. The central (A11-A21) segment of 32R-3n, despite being G-rich, only connects the two G-quadruplex units. Despite our efforts using various annealing protocols and different oligonucleotide concentrations we were unable to induce unimolecular folding of 32R-3n. Therefore we cannot propose a model for the secondary structure of 32R-3n region within genomic DNA. It is noteworthy that the concentrations in our experiments were at least an order of magnitude higher than those used by Xodo et al. due to sensitivity limitations of NMR spectroscopy. Interestingly, T11 PAGE migration pattern shows a monomer/dimer equilibrium even after prolonged incubation of the oligonucleotide in a KCl solution. This suggests that the central (A11-A21) segment of 32R-3n is responsible for the formation of a dimeric structure in diluted solutions.
The two G-quadruplex units at 5′- and 3′-ends of 32R-3n have been extensively characterized. While both G-quadruplex folds can be obtained by 5q or 3q oligonucleotides, a correct length of overhangs is required to achieve similar stacking interactions as in 32R-3n. According to imino 1H NMR chemical shift pattern in the case of 5q this requires at least one A after the last G-tract, while 3q clearly requires additional stabilization and the first G-tract has to be preceded by four nucleotides (AAGA) in order to adopt the same structure as in 32R-3n.
The 5′-end G-quadruplex unit exhibits three G-quartets with 3-nt edge-type loops. On the other hand, NOE connectivities support the formation of only a single G-quartet flanked by two G:C base pairs in the 3′-end G-quadruplex unit. The G24-G28 segment could form 5-nt loops as stable G-quadruplex structures with long loops do exist (32,33). However, due to observable imino 1H NMR resonances we have assigned an additional G-quartet and two G:G base pairs to the structure of the 3′-end G-quadruplex unit. Broad NMR resonances suggest increased dynamics of the G24-G28 segment. Structural elements within the G24-G28 segment of the 3′-end G-quadruplex unit are melted at higher temperatures and replaced by a single G:G base pair, while the core G:C base pairs and the adjacent G-quartet remain structured.
Although UV melting shows that 3q melts at a higher temperature compared to 5q, this can be attributed to a pair of stable G:C base pairs in 3q. Compared to the full length 32R-3n and 5q, which feature simpler two-state melting, 3q melts over a wide temperature range (Supplementary Figure S8). Although only a single broad premelt can be identified in the melting profile of 3q, we hypothesize on the basis of NMR data that some structural elements melt first, followed by complete melting of the 3′-end G-quadruplex unit. It is noteworthy that the melting profile of 3q also exhibits considerable hysteresis, which suggests slow folding kinetics. On the other hand, only minimal hysteresis is present with 5q and 32R-3n. The two-state melting of 32R-3n indicates that the folding of the 3′-end G-quadruplex unit within the full length oligonucleotide is cooperative and heavily dependent on the formation of the 5′-end G-quadruplex.
It has been shown that 32R-3n is taken up by MAZ with a KD in the nanomolar range and reduces cell growth of pancreatic cancer cells (27). Furthermore, an oligonucleotide modified with two polycyclic aromatic hydrocarbon TINA insertions (compound 637_3 in (26)) exhibits a substantial increase in G-quadruplex stability. Two additional LNA modifications at the 3′-end (compound 2998 in (27)) increase nuclease stability and cause a large increase in anti-cancer activity. Polycyclic aromatic hydrocarbons supposedly interact with G-quadruplex units via stacking on the outer G-quartets. According to topology proposed in this study ideal positioning of such moieties within 32R-3n is between sites 10–11 and 21–22, which are the exact sites where the oligonucleotides (637_3 and 2998) have been modified. However, a dimer of modified oligonucleotides would position pairs of TINA residues in close proximity and most likely destabilize the structure. This is in agreement with previous work of Xodo et al., which shows that oligonucleotide 2998 is a monomer in K+ solutions (27). On the other hand, an equimolar mixture of modified and unmodified 32R-3n could produce hybrids with enhanced stability. The possibility of oligonucleotide 2998 invading the NHE and forming a dimer with the genomic G-rich strand is unlikely, due to the G:C base pair rich dsNHE.
Dimeric 32R-3n and, even more so, monomeric 2998 were found to exhibit anti-proliferative effects in pancreatic cancer cells via sequestering MAZ and downregulating KRAS transcription. Which structural elements of dimeric 32R-3n are recognized by MAZ remains unknown. The 3′-end G-quadruplex unit includes the GGGAGG (G24-G29) MAZ binding sequence. Furthermore, the G24-G29 region within 32R-3n is dynamic and involved in a conformational equilibrium, which could be important for its availability for binding to MAZ. This could allow for a sequence based recognition of the G-rich motif. Additionally, the two stable G-quadruplex structures could serve as a stability enhancer and protect both ends of 32R-3n. While the dimeric G-quadruplex structures are not possible within a single G-rich strand of genomic KRAS NHE, molecular crowding conditions could favor a monomeric topology including additional nucleotides flanking the 5′- or 3′-ends of the 32R-3n sequence.
Supplementary Material
Acknowledgments
We would like to thank Professors LE Xodo and EB Pedersen for their fruitful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Slovenian research agency (ARRS) [P1-0242, J1-6733]. Funding for open access charge: ARRS [P1-242].
Conflict of interest statement. None declared.
REFERENCES
- 1.Malumbres M., Barbacid M. RAS oncogenes: the first 30 years. Nat. Rev. Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 2.Macaluso M., Russo G., Cinti C., Bazan V., Gebbia N., Russo A. Ras family genes: an interesting link between cell cycle and cancer. J. Cell. Physiol. 2002;192:125–130. doi: 10.1002/jcp.10109. [DOI] [PubMed] [Google Scholar]
- 3.Stephen A.G., Esposito D., Bagni R.K., McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 4.Friday B.B., Adjei A.A. K-ras as a target for cancer therapy. Biochim. Biophys. Acta. 2005;1756:127–144. doi: 10.1016/j.bbcan.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Spiegel J., Cromm P.M., Zimmermann G., Grossmann T.N., Waldmann H. Small-molecule modulation of Ras signaling. Nat. Chem. Biol. 2014;10:613–622. doi: 10.1038/nchembio.1560. [DOI] [PubMed] [Google Scholar]
- 6.Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J. Drugging the undruggable RAS: mission possible? Nat. Rev. Drug. Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordano J., Perucho M. Chromatin structure of the promoter region of the human c-K-ras gene. Nucleic Acids Res. 1986;14:7361–7378. doi: 10.1093/nar/14.18.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman E.K., Trusko S.P., Murphy M., George D.L. An S1 nuclease-sensitive homopurine/homopyrimidine domain in the c-Ki-ras promoter interacts with a nuclear factor. Proc. Natl. Acad. Sci. USA. 1990;87:2705–2709. doi: 10.1073/pnas.87.7.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cogoi S., Xodo L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neidle S., Balasubramanian S. Quadruplex Nucleic Acids. Cambridge: Royal Society of Chemistry; 2006. [Google Scholar]
- 11.Monchaud D. Biological Relevance & Therapeutic Applications of DNA- & RNA-Quadruplexes. London: Future Science Ltd; 2015. [Google Scholar]
- 12.Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webba da Silva M. Geometric formalism for DNA quadruplex folding. Chem. Eur. J. 2007;13:9738–9745. doi: 10.1002/chem.200701255. [DOI] [PubMed] [Google Scholar]
- 14.Trajkovski M., Webba da Silva M., Plavec J. Unique structural features of interconverting monomeric and dimeric G-quadruplexes adopted by a sequence from the intron of the N-myc gene. J. Am. Chem. Soc. 2012;134:4132–4141. doi: 10.1021/ja208483v. [DOI] [PubMed] [Google Scholar]
- 15.Marušič M., Šket P., Bauer L., Viglasky V., Plavec J. Solution-state structure of an intramolecular G-quadruplex with propeller, diagonal and edgewise loops. Nucleic Acids Res. 2012;40:6946–6956. doi: 10.1093/nar/gks329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kocman V., Plavec J. A tetrahelical DNA fold adopted by tandem repeats of alternating GGG and GCG tracts. Nat. Commun. 2014;5:5831. doi: 10.1038/ncomms6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huppert J.L., Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Cian A., Lacroix L., Douarre C., Temime-Smaali N., Trentesaux C., Riou J.F., Mergny J.L. Targeting telomeres and telomerase. Biochimie. 2008;90:131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Balasubramanian S., Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monchaud D., Teulade-Fichou M.P. A hitchhiker's guide to G-quadruplex ligands. Org. Biomol. Chem. 2008;6:627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S., Wu Y., Zhang W. G-quadruplex structures and their interaction diversity with ligands. ChemMedChem. 2014;9:899–911. doi: 10.1002/cmdc.201300566. [DOI] [PubMed] [Google Scholar]
- 22.Patel D.J., Phan A.T., Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discovery. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bossone S.A., Asselin C., Patel A.J., Marcu K.B. MAZ, a zinc finger protein, binds to c-MYC and C2 gene sequences regulating transcriptional initiation and termination. Proc. Natl. Acad. Sci. USA. 1992;89:7452–7456. doi: 10.1073/pnas.89.16.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cogoi S., Paramasivam M., Membrino A., Yokoyama K.K., Xodo L.E. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 2010;285:22003–22016. doi: 10.1074/jbc.M110.101923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cogoi S., Paramasivam M., Filichev V., Géci I., Pedersen E.B., Xodo L.E. Identification of a new G-quadruplex motif in the KRAS promoter and design of pyrene-modified G4-decoys with antiproliferative activity in pancreatic cancer cells. J. Med. Chem. 2009;52:564–568. doi: 10.1021/jm800874t. [DOI] [PubMed] [Google Scholar]
- 27.Cogoi S., Zorzet S., Rapozzi V., Géci I., Pedersen E.B., Xodo L.E. MAZ-binding G4-decoy with locked nucleic acid and twisted intercalating nucleic acid modifications suppresses KRAS in pancreatic cancer cells and delays tumor growth in mice. Nucleic Acids Res. 2013;41:4049–4064. doi: 10.1093/nar/gkt127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filichev V.V., Gaber H., Olsen T.R., Jørgensen P.T., Jessen C.H., Pedersen E.B. Twisted intercalating nucleic acids - intercalator influence on parallel triplex stabilities. Eur. J. Org. Chem. 2006;2006:3960–3968. [Google Scholar]
- 29.Paramasivam M., Cogoi S., Filichev V.V., Bomholt N., Pedersen E.B., Xodo L.E. Purine twisted-intercalating nucleic acids: a new class of anti-gene molecules resistant to potassium-induced aggregation. Nucleic Acids Res. 2008;36:3494–3507. doi: 10.1093/nar/gkn242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paramasivam M., Cogoi S., Xodo L.E. Primer extension reactions as a tool to uncover folding motifs within complex G-rich sequences: analysis of the human KRAS NHE. Chem. Commun. 2011;47:4965–4967. doi: 10.1039/c1cc10269a. [DOI] [PubMed] [Google Scholar]
- 31.Cogoi S., Paramasivam M., Spolaore B., Xodo L.E. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 2008;36:3765–3780. doi: 10.1093/nar/gkn120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yue D.J.E., Lim K.W., Phan A.T. Formation of (3+1) G-quadruplexes with a long loop by human telomeric DNA spanning five or more repeats. J. Am. Chem. Soc. 2011;133:11462–11465. doi: 10.1021/ja204197d. [DOI] [PubMed] [Google Scholar]
- 33.Phan A.T., Modi Y.S., Patel D.J. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.