Abstract
The complicated process of eukaryotic ribosome biogenesis involves about 200 assembly factors that transiently associate with the nascent pre-ribosome in a spatiotemporally ordered way. During the early steps of 60S subunit formation, several proteins, collectively called A3 cluster factors, participate in the removal of the internal transcribed spacer 1 (ITS1) from 27SA3 pre-rRNA. Among these factors is the conserved hetero-trimeric Nop7–Erb1–Ytm1 complex (or human Pes1–Bop1–Wdr12), which is removed from the evolving pre-60S particle by the AAA ATPase Rea1 to allow progression in the pathway. Here, we clarify how Ytm1 and Erb1 interact, which has implications for the release mechanism of both factors from the pre-ribosome. Biochemical studies show that Ytm1 and Erb1 bind each other via their ß-propeller domains. The crystal structure of the Erb1–Ytm1 heterodimer determined at 2.67Å resolution reveals an extended interaction surface between the propellers in a rarely observed binding mode. Structure-based mutations in the interface that impair the Erb1–Ytm1 interaction do not support growth, with specific defects in 60S subunit synthesis. Under these mutant conditions, it becomes clear that an intact Erb1–Ytm1 complex is required for 60S maturation and that loss of this stable interaction prevents ribosome production.
INTRODUCTION
Synthesis of new ribosomes is an ongoing process in the cell and absolutely essential for cell viability. In eukaryotes about 200 assembly factors are involved in the synthesis and formation of ribosomes (1–5). Biogenesis of the ribosomal subunits begins with the transcription of the rDNA by the RNA polymerase I. The transcript undergoes cleavages at the A0, A1 and A2 sites resulting in the 27SA2 product (6). During the A2 cleavage the large pre-rRNA is split into two major fragments, 20S and 27SA2, which after further processing leads to the small and large subunit, respectively. The 27SA2 undergoes additional maturation steps before leaving the nucleolus, including the removal of the internal transcribed spacer 1 (ITS1). One intermediate generated by the endonuclease mitochondrial RNA processing (MRP) (7,8), the 27 SA3, requires a number of factors (referred to as the A3 cluster) including the trimeric Nop7–Erb1–Ytm1 complex for further processing (4,9,10). None of the three proteins actually harbors nuclease activity but most likely play a role in recruitment of other factors or stabilization of the rRNA for further processing (11,12). The processing of the 27SA3 precursor requires the 5′-3′ exonucleases Rat1, Xrn1 and Rrp17 (13–15). The Nop7–Erb1–Ytm1 complex is held together by Erb1, which interacts with Nop7 via its N-terminal domain and with Ytm1 via a small region in the middle of the protein. Erb1 is composed of two large domains, a N-terminal helical domain and a C-terminal WD40 domain. An N-terminal truncation of Erb1 (aa265–807) was shown to be dominant negative when overexpressed and failed to support cell growth, while in contrast the C-terminal WD40 domain (aa420-807) has been shown to be dispensable for ribosome assembly and cell viability (10). Similar studies performed on the mammalian homologues, elucidated multiple roles of this conserved trimeric complex. In addition to their involvement in ribosome biogenesis (16,17), the mammalian homologues have also been linked to cell proliferation (18–21), chromosomal segregation (22) and colorectal tumorigenesis (23). Ytm1 is structurally homologous to the ribosome biogenesis factor Rsa4, as it also contains an N-terminal Ubiquitin-like (Ubl) domain and a C-terminal WD40 domain (Figure 1A). Both, Ytm1 and Rsa4 interact via a conserved region on their Ubl domains with the C-terminal MIDAS domain (metal ion dependent adhesion site) of the AAA ATPase Rea1 (24,25). Removal of Rsa4 from nucleoplasmic pre-ribosomal particles and their subsequent remodeling has been suggested to be an adenosine triphosphate (ATP)-consuming step carried out by the Rea1 ATPase (25). Our recent structural analyses of Rsa4 and Nsa2 revealed that a short linear motif of Nsa2 is sufficient for interaction with Rsa4, however during remodeling Nsa2 remains bound to the pre-ribosome, whereas Rsa4 is displaced (25,26). In contrast, interaction of the Rea1 MIDAS domain with the Ubl domain of Ytm1 causes removal of the Erb1–Ytm1 complex from the pre-ribosome (24). One possible explanation is that Nsa2 is deeply embedded in the pre-60S ribosome and only interacts with Rsa4 via a linear motif as observed in our recent structural study (24). Interaction of the Nop7–Erb1–Ytm1 complex at the pre-60S particle might be less intimate, though this is difficult to predict since early stages of ribosome biogenesis have not been structurally characterized.
Figure 1.
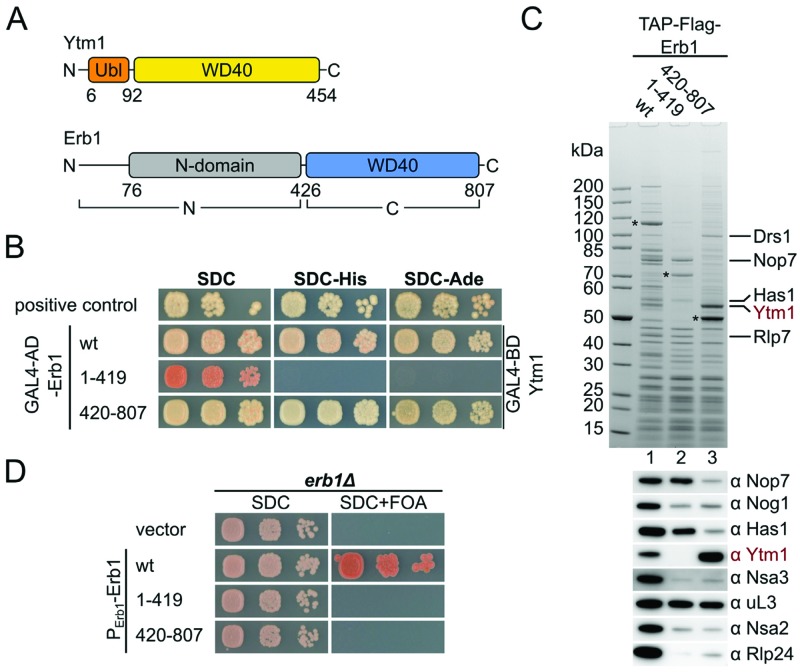
The β-propeller of Erb1 interacts with Ytm1 and is essential for cell growth. (A) Domain organization of Ytm1 and Erb1 from S. cerevisiae. Ytm1 contains an ubiquitin-like domain (Ubl; orange) and a β-propeller (WD40; yellow). Erb1 contains an N-terminal domain (gray) and a C-terminal β-propeller (blue). The domain boundaries are given in residue numbers. (B) The Erb1 β-propeller interacts with Ytm1 in yeast two-hybrid assays. Erb1 constructs were fused to an N-terminal GAL4-AD (activation domain) and Ytm1 full length was fused to an N-terminal GAL4-BD (binding domain). Constructs were co-transformed into the PJ69–4A strain and growth was monitored on SDC (SDC-Leu-Trp), SDC-His (SDC-Leu-Trp-His) and SDC-Ade (SDC-Leu-Trp-Ade) plates after 4 days at 30°C. (C) Affinity-purifications of Erb1 wt, the Erb1 N-terminus (aa1–419) and the Erb1 β-propeller (aa420–807). Final eluates were analysed by SDS-PAGE and Coomassie staining or western blotting using the indicated antibodies. (D) The Erb1 β-propeller is required for cell growth. An Erb1 shuffle strain was transformed with plasmids encoding for the indicated Erb1 alleles under control of the native Erb1 promoter. Growth on SDC (SDC-Leu) and SDC+FOA was monitored after 2 and 4 days, respectively.
We reasoned that the interaction between Erb1 and Ytm1, which has a similar domain architecture to Rsa4, might be homologous to the Rsa4–Nsa2 interaction, as it had been suggested that a short conserved region at the N-terminus (residues 383 to 419) of the WD40 domain of Erb1 was sufficient to bind to the WD40 domain of Ytm1 (10). In order to test this hypothesis and gain structural insight into the Erb1–Ytm1 complex, we analysed the interaction of these proteins in vivo and in vitro. We found that the interaction between Erb1 and Ytm1 requires the complete WD40 domain of Erb1, ruling out a short linear motif. Interface mutants that specifically perturb the interaction between Erb1 and Ytm1 decreased binding or completely abolished it, leading to specific defects in ribosome biogenesis.
MATERIALS AND METHODS
Yeast strains, genetic methods and plasmids
Saccharomyces cerevisiae (S. cerevisiae) strains used in this study are derived from W303 (27) and DS1–2b (5) and are listed in Supplemental Table S1. Gene disruption and tagging were performed according to standard procedures (28,29). For yeast two-hybrid analysis, the PJ69–4A strain was used (30). Recombinant DNA techniques were performed using standard procedures. E. coli DH5α was used for cloning and plasmid propagation. The CtYTM1 (CTHT_0061460) and CtERB1 (CTHT_0057570) genes were cloned from Chaetomium thermophilum (Ct) cDNA (31). Plasmids used in this study are listed in Supplemental Table S2.
Yeast two-hybrid analysis
Plasmids expressing YTM1 alleles fused to an N-terminal GAL4-BD (binding domain) and N-terminal GAL4-AD (activation domain) fused to the different Erb1 alleles were co-transformed into the PJ69–4A yeast two-hybrid reporter strain (30). Representative transformants were spotted in 10-fold serial dilution on SDC (SDC-Leu-Trp), SDC-His (SDC-Leu-Trp-His) and SDC-Ade (SDC-Leu-Trp-Ade) plates and growth at 30°C was analysed after the indicated days. Growth on SDC-His indicates weak interactions whereas growth on SDC-Ade plates suggests strong interactions.
Yeast affinity purification
All TAP-Flag (ProteinA-TEVsite-CBP-Flag) constructs were expressed from plasmids under the control of the respective endogenous promoter. Affinity purifications from yeast extracts were performed as previously described (32). All purification steps were performed in buffer containing 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 1.5 mM MgCl2, 5% glycerol, 0.1% NP-40 and 1 mM DTT. Yeast cells were lysed in a planetary mill (Fritsch) and the lysate was cleared by centrifugation. For the first purification step, IgG sepharose (GE Healthcare) was added to the supernatant and incubated for 1 h at 4°C. Beads were extensively washed and the bound material was released from the beads by tobacco etch virus (TEV) protease cleavage for 1.5 h at 16°C. In a second purification step, Flag agarose (Sigma-Aldrich) was added to the TEV eluate and incubated for 1 h at 4°C. The beads were washed with buffer and samples were released from the beads by incubation with buffer supplemented with 1.5× Flag peptide for 30 min at 4°C. The eluates were precipitated with TCA (10% final concentration) and resuspended in SDS loading buffer.
Sucrose gradient analysis
Yeast cultures were grown in YPD medium. When cultures reached an OD600 of 0.8 cycloheximide was added to a final concentration of 100 μg/ml and cultures were incubated for 10 min on ice. Cells were lysed in buffer containing 50 mM Tris-HCl pH 7.5, 100 mM KCl, 12 mM MgCl2 and 100 μg/ml cycloheximide (vortexing with glass beads, 5 × 30s). The lysate was cleared by centrifugation and loaded on a 10–50% sucrose gradient (50 mM Tris-HCl pH 7.5, 100 mM KCl, 12 mM MgCl2). Sucrose gradients were centrifuged for 2 h 45 min with 39 000 rpm at 4°C (SW40, Beckman Coulter) and analysed on a Foxy junior (Teledyne ISCO) at 254 nm.
In vitro pull-down assay
Binding assays were performed in buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 10 mM MgCl2, 10 mM KCl, 5% glycerol, 0.01% NP-40 and 1 mM DTT. Samples were incubated for 30 min at 4°C. Flag agarose (ANTI-Flag M2 Affinity Gel, Sigma Aldrich) was added and samples were incubated for additional 90 min at 4°C. The wash step was performed in 1 ml Mobicol columns (MoBiTec) in a table-top centrifuge at 4°C. Flag beads were washed once with 800 μl buffer and twice with 500 μl buffer. Bound material was eluted with buffer supplemented with 1.5× Flag peptide. Eluates were analysed by SDS-PAGE and Coomassie staining.
Expression and purification of the CtErb1WD–CtYtm1 complex
Expression of (His)6-CtErb1WD40 (aa423–801) was carried out in E. coli BL21-CodonPLUS(DE3). Cells were grown in LB medium at 37°C until an OD600 of 0.6 was reached, then shifted to 23°C and protein expression was induced by addition of 0.25 mM IPTG (isopropyl-β-D-thiogalactoside). Cells were harvested after 3 h and cell pellets were stored at - 80°C.
CtYtm1 was insoluble when expressed in E. coli. Therefore we generated a modified YEplac112 (TRP1) plasmid containing a LEU2d marker to enhance the plasmid copy number in the yeast cell. The plasmid contained a GAL1–10 promoter and an N-terminal pA-TEV (protA-TEVsite) tag for purification. A W303 strain was transformed with YEplac112Leu2d-GAL1–10-pA-TEV-CtYtm1 and cells were selected on SDC-Trp medium. Colonies were restreaked on SDC-Leu medium to enhance the plasmid copy number. A SRC-Leu overnight culture was used to inoculate the YPG main culture (start OD600 0.1) and cells were grown for 20–24 h at 30°C and 110 rpm until they reached an OD600 of about 6–8. Lysis was performed in a buffer containing 20 mM HEPES, 200 mM NaCl, 10 mM MgCl2, 10 mM KCl, 5% glycerol and 1 mM DTT supplemented with 0.01% NP-40 and protease inhibitor (Sigma-Aldrich) using a planetary mill (Fritsch). The lysate was centrifuged for 12 min at 4000 rpm, 4°C and again at 17 500 rpm for 25 min using a JA25.50 rotor (Beckmann-Coulter). IgG sepharose beads (GE Healthcare) were added to the supernatant and incubated for 90 min at 4°C. Beads were extensively washed with buffer supplemented with 0.01% NP-40. Cleared lysate of E. coli cells overexpressing (His)6-CtErb1WD40 was added to the beads and incubated for 1 h at 4°C under gentle agitation. IgG beads were extensively washed with a buffer containing 20 mM HEPES, 100 mM NaCl, 10 mM MgCl2, 10 mM KCl, 5% glycerol and 1 mM DTT supplemented with 0.01% NP-40 and TEV protease cleavage was carried out at 4°C, overnight. The cleaved protein was incubated with SP Sepharose beads for 1 h at 4°C and the proteins were eluted by stepwise addition of buffer (without NP-40) with increasing salt concentrations (100 mM–1 M NaCl). Fractions containing the CtErb1WD40–CtYtm1 heterodimer were pooled and the buffer was exchanged via a PD-10 column to 20 mM HEPES pH 7.5, 200 mM NaCl, 10 mM MgCl2, 10 mM KCl, 5% glycerol and 1 mM DTT.
SEC-MALS
The purified CtErb1WD40–CtYtm1 complex was analysed by size exclusion chromatography (SEC) with online multi-angle light scattering (MALS). The analysis was performed in a buffer containing 20 mM HEPES pH 7.5, 200 mM NaCl, 10 mM MgCl2, 10 mM KCl, 5% glycerol and 1 mM DTT. The sample was loaded onto a Superdex 200 Increase 10/300 GL column for sample separation. The ÄKTA basic system (GE Healthcare) was connected to an eight-angle light scattering detector (DAWN HELEOS, Wyatt Technology) and a refractometer (SEC-3010, WGE Dr Bures). The analysis was carried out at 4°C with 0.5 ml/min flow rate and the data was analysed using the ASTRA 6.1 software. Peak fractions were analysed by SDS-PAGE and Coomassie staining.
Structure determination of the CtErb1–CtYtm1 complex
The purified complex was concentrated to 10–20 mg/ml and crystallized in 28% ethylene-glycol using the sitting drop vapor diffusion method at 293 K. For data-collection crystals were harvested into reservoir solution and flash cooled in liquid nitrogen. Diffraction data were collected at ID23–2 (ESRF, Grenoble, France) (33). X-ray diffraction data were integrated and scaled with XDS (34) and AIMLESS (35) from the CCP4-package (36). The crystal belongs to the space group P21 with cell dimensions of a = 91.66 Å, b = 81.35 Å, c = 141.69 Å and β = 100.23°.
The structure was solved by molecular replacement as implemented in MOLREP (37). Based on cell content analysis (77% and 55% solvent for one and two heterodimers, respectively) the initial search consisted of 4 β-propellers and 2 Ubl domains. Inclusion of the Ubl domains failed to give any meaningful result and was therefore omitted from further trials. The best scoring solution was obtained by searching with four copies of the WDR5-coordinates (PDB-ID: 2h9p, 21% sequence identity to CtYtm1) (38), giving a contrast of 4.8 in MOLREP, indicating a clear solution. The initial map was of very low quality and required manual rebuilding, which was done in Coot (39). After several rounds of manual building in Coot and refinement with Refmac5 (40), the two molecules could be un-ambiguously identified and the majority of the residues could be placed automatically with Buccaneer (41). Remaining residues were built manually with Coot. Further refinement was carried out in PHENIX (42). Figures were prepared with PyMOL (43). Superpositions were calculated with GESAMT (36,44) from the CCP4-package. The final model contains two molecules of CtErb1WD40 and CtYtm1 in the ASU. The data collection and refinement statistics are summarized in Table 1.
Table 1. Data collection and refinement statistics.
| CtErb1WD40/CtYtm1 | |
|---|---|
| Data collection | |
| Beamline | ESRF ID23–2 |
| Wavelength (Å) | 0.87260 |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 91.66, 81.35, 141.69 |
| αβγ (°) | 90, 100.23, 90 |
| Resolution (Å) | 48.61–2.67 (2.76–2.67)* |
| Rmerge | 0.1621 (1.135) |
| I/σ(I) | 10.63 (1.56) |
| Reflections total | 306006 (30257) |
| Reflections unique | 58434 (5794) |
| Completeness (%) | 99.73 (99.95) |
| Multiplicity | 5.2 (5.2) |
| Refinement | |
| Rwork | 0.2210 (0.3325) |
| Rfree | 0.2509 (0.3676) |
| Number of atoms | |
| Protein | 12526 |
| Water | 183 |
| Ligands | 45 |
| B-factors | |
| Protein | 51.10 |
| Water | 39.90 |
| Ligands | 47.80 |
| RMS deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (º) | 0.92 |
| Ramachandran plot | |
| Most favoured (%) | 96.10 |
| Disallowed | 0.31 |
*Values in parenthesis refer to the highest resolution shell.
Multiple sequence alignment
Alignments were generated with Clustal Omega (45) and visualized with ESPRIPT (46). Surface representations of conserved residues were generated using the ConSurf server (47).
Miscellaneous
Additional methods used in this study are fluorescence microscopy and western blot analysis. The following antibodies were used: anti-Arc1 (1:5,000), anti-Nog1 (1:30,000), anti-Nsa2 (1:10,000), anti-Rlp24 (1:15,000), anti-Has1 (1:10,000), anti-Nop7 (1:5000), anti-Nsa3 (1:5,000), anti-Rpl3 (1:5,000), anti-Noc1 (1:500), anti-Ytm1 (1:100), anti-Flag at 1:2,000 (Sigma-Aldrich, HRP-conjugated, A8592), anti-HA at 1:1,000 (Covance Research Products, MMS-101R), anti-myc at 1:400 (9E10, ThermoFisher Scientific) and HRP conjugated secondary antibodies: goat anti-rabbit 1:2,000 (BioRad, #170–6515) and goat anti-mouse 1:2,000 (BioRad, #170–6516). NuPAGE (4–12%) Bis-Tris gels and NuPAGE MOPS SDS running buffer (Invitrogen) were used for SDS-PAGE and gels were stained with Roti-Blue (ROTH).
RESULTS
Erb1 interacts with Ytm1 via its essential WD40 domain
In order to gain detailed insights into the interaction between Ytm1 and Erb1, we cloned the corresponding orthologous genes from Chaetomium thermophilum (Ct), a eukaryotic thermophile, with the goal to better exploit them for biochemical and structural studies (31,48). Utilizing in vitro reconstitution and yeast two-hybrid (Y2H) assays, we could identify a robust interaction between CtYtm1 and CtErb1 when the C-terminal WD40 domain of CtErb1 was present (aa423–801). The complete N-terminal part (aa1–454) of CtErb1 failed to interact with CtYtm1 full-length protein (Supplementary Figure S1). To test whether the S. cerevisiae orthologs interact via the same mechanism, we performed Y2H analysis with the yeast constructs (Figure 1A and B and Supplementary Figure S2A). This confirmed that Erb1 full length and the Erb1-WD40 domain (aa420–807; Erb1WD40) strongly interact with full-length Ytm1, whereas the N-terminal part (Erb1N; aa1–419) that includes the previously suggested binding motif (aa383 to 419) revealed no interaction (10). One likely explanation for this discrepancy is that the previous study employed a C-terminal tag on Erb1, whereas we use an N-terminal tag. In our structure (see below), the C-terminus of Erb1 is part of the WD40 fold and is located directly in the interface to Ytm1. Introduction of additional residues at the C-terminus most likely interfere with the fold and interaction. Furthermore, purification of TAP-Flag-Erb1 or TAP-Flag-Erb1WD40 robustly enriched Ytm1, whereas TAP-Flag-Erb1N did not co-purify Ytm1 (Figure 1C and Supplementary Figure S2B). However, consistent with previous findings, the N-terminal truncation of Erb1 (aa1–419) enriched Nop7 during affinity-purification (10). In order to show that the interaction between Erb1 and Ytm1 is of functional importance, we generated an Erb1 shuffle strain (erb1Δ), and transformed it with plasmids encoding full-length Erb1, N-terminal or C-terminal truncations (aa1–419 and aa420–807). Only full-length Erb1 was able to complement the erb1Δ strain (Figure 1D). Taken together, these data show that the Erb1WD40 domain is essential for interaction with Ytm1 and cell growth.
Structure of the CtErb1–CtYtm1 complex
To gain insight into the structural basis of the Erb1–Ytm1 interaction, we also focused on the CtErb1–CtYtm1 complex assembled with the subunits from Chaetomium thermophilum (31,48). We recombinantly expressed full-length CtYtm1 and the CtErb1WD40 domain in S. cerevisiae and E. coli, respectively, reconstituted the complex and determined its crystal structure to a resolution of 2.67 Å. Data collection and refinement statistics are summarized in Table 1. The asymmetric unit contains two CtErb1–CtYtm1 heterodimers, which are very similar, indicated by low RMSD values of 0.13 Å over 357 residues and 0.39 Å over 457 residues for CtErb1 and CtYtm1, respectively (Figure 2A and Supplementary Figure S3A). Although the heterodimers form a quasi-dimer in the crystal structure, such interaction could not be observed in solution by SEC-MALS analysis (Supplementary Figure S3B and C).
Figure 2.
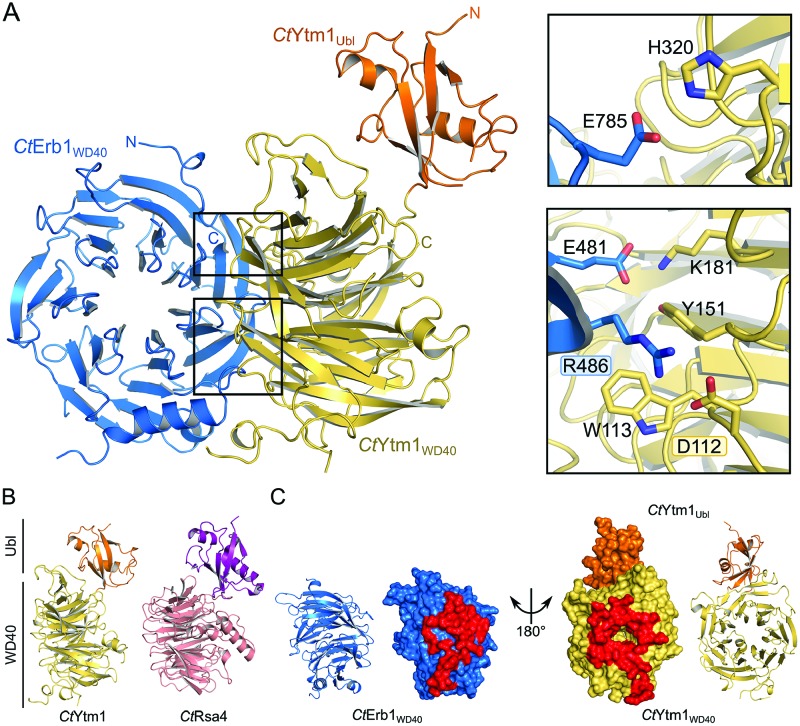
Crystal structure of the Erb1WD40–Ytm1 complex from Chaetomium thermophilum. (A) Overall structure of the C. thermophilum Erb1WD40–Ytm1 complex. CtYtm1WD40 (yellow) packs with its top surface against the side of CtErb1WD40 (blue), CtYtm1Ubl (orange) is not involved in binding. The interface is stabilized by several salt-bridges (E785-H320, E481-K181, R486-D112, between Erb1WD40 and Ytm1, respectively). In addition Erb1 R486 is stabilized by CtYtm1 W113 and Y151 by cation-π-stacking. (B) Side-by-side comparison of the crystal structures of CtYtm1 and CtRsa4 (Ubl domains in orange and purple; WD40 domains in yellow and salmon, respectively). (C) Interaction between both molecules involves a large surface area. CtYtm1 and CtErb1 are shown in cartoon and surface representation, with interacting residues highlighted (red) in the surface representation.
The crystal structure of the CtErb1 WD40 domain (CtErb1WD40) reveals a seven bladed β-propeller with an insertion in blade 2 and is very similar to a recently published structure of the yeast homologue (RMSD of 0.97 Å over 346 residues) (49), suggesting very little flexibility in the overall fold. The insertion in blade 2, also observed in the yeast homologue, consists of a fifth beta strand (β10) and two alpha helices (α1 and 2) (Supplementary Figure S4A). Although conserved among different organisms, the sequence identity in this region is lower than in the rest of the domain, especially in helix α2 and β-strand β10 (Supplementary Figures S4B and S5). CtErb1WD40 contains a large number of conserved arginine and lysine residues opposite to the CtYtm1 interface, creating a positively charged patch that could potentially interact with rRNA (Supplementary Figure S4C), as proposed recently (49).
The structure of CtYtm1 contains an N-terminal ubiquitin-like domain (CtYtm1Ubl) followed by a β-propeller composed of seven blades (CtYtm1WD40, Figure 2A and Supplementary Figure S6A). Similar to CtErb1WD40 the CtYtm1WD40 domain contains a number of conserved and organism-specific extensions throughout the blades (Supplementary Figures S6A and S7). CtYtm1WD40 contains one helix in blade 1 and one between blade 6 and 7 (α3 and α4 respectively, Supplementary Figures S6A and S7). Blade 1 has a slightly shortened fourth beta strand (β9), which is ‘compensated’ by the extended β-strands in blade 2 (β12 and 13). Previously, a putative NLS (residues 274 to 278 in CtYtm1) was predicted in blade 3 (50). In the structure, this region is part of an extended loop, which is surface-exposed and partially disordered (Supplementary Figure S6A). Interestingly, CtYtm1 is structurally similar to Rsa4 (26) with a similar overall domain arrangement (Figure 2B). However, the Rsa4 β-propeller comprises eight blades instead of seven and a α-helical insertion in blade 5 (Supplementary Figure S6B). In addition, the Rsa4 Ubl domain shows a different orientation with respect to the β-propeller in the different crystal structures, indicative of flexibility (26). The orientation of the CtYtm1 Ubl domain with respect to the β-propeller is essentially the same in both molecules of the asymmetric unit, although they are stabilized by different crystal contacts (Supplementary Figure S6C). It is likely that in solution the CtYtm1 Ubl domain can adopt different orientations, as no specific interactions can be observed between the two domains.
The Erb1–Ytm1 complex employs a rare binding mode
Multiple interfaces have been proposed for the interaction between two WD40 domains using either the top/bottom or even a specific side to bind the other protein (51). The arrangement of CtErb1WD40 and CtYtm1 in the complex follows one of the possible interaction sites proposed between WD40 domains (side-to-bottom). Although the binding of CtErb1WD40 is along the central pore at the bottom of CtYtm1, it does not employ a linear motif but instead makes extensive contacts to CtYtm1WD40 (Figure 2A and C). The interaction involves two neighboring blades of CtErb1WD40 and covers 1278 Å2 and 1154 Å2 surface of CtErb1WD40 and CtYtm1, respectively. This corresponds to 8.4% (CtErb1WD40) and 5.7% (CtYtm1) of each molecules total accessible surface area, indicating a very stable dimer interface (Figure 2C). The interface is made up of CtErb1WD40 blades 7 and 1 and involves multiple salt bridges (CtErb1–CtYtm1, E481-K181, R486-D112, E785-H320) and hydrogen bonds, but also more rare interactions such as cation-π-stacking (W113/Y151-R486) (Figure 2A). In conclusion, the interaction between CtErb1WD40 and CtYtm1 is mediated by specific, mutual recognition of their three-dimensional protein surface. This is conceptually very different from the common interaction mode of WD40 domains with linear motifs.
Role of Erb1-R470 and Ytm1-D104 in ribosome biogenesis
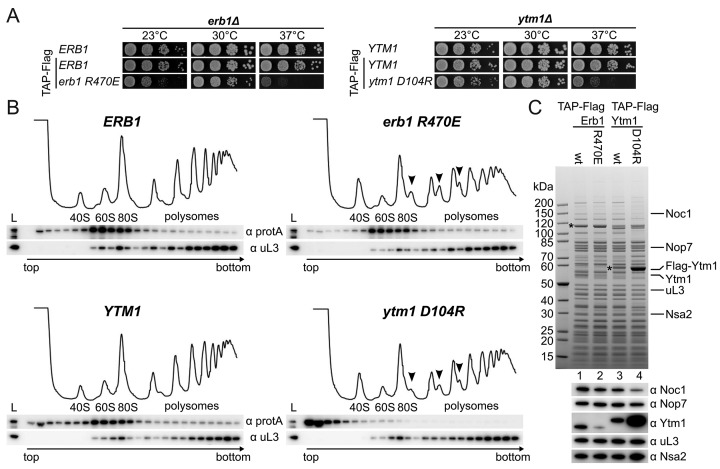
Based on the crystal structure, we sought to introduce reverse charge point mutations in Erb1 and Ytm1 to abolish interaction between the highly conserved Erb1-R470 and Ytm1-D104 pair (R486 and D112 in the Ct orthologs, respectively) and analyse their growth at different temperatures in an erb1Δ or ytm1Δ shuffle strain, respectively (Figure 3A and Supplementary Figure S2E). Although Erb1-R470E could complement the erb1Δ strain at 30°C, growth was impaired slightly at 23°C and strongly at 37°C. The Ytm1-D104R mutant showed strongly reduced growth at 37°C, suggesting that in both cases the interaction has been compromised but not fully abolished. To find out whether these growth defects are caused by an altered ribosome assembly pathway, we performed polysome gradient analysis. This revealed that wild-type Ytm1 and Erb1 co-sediment with pre-60S particles (Figure 3B), but the viable Ytm1-D104R and Erb1-R470E mutants exhibited different profiles, as we observed the occurrence of half-mers in both strains (indicated by arrows in Figure 3B) indicative of defects in 60S maturation. Although Erb1-R470E remained stably ribosome associated as in the wild-type, Ytm1-D104R was almost entirely shifted to the soluble fraction. While the gradient analysis shows a shift to the soluble fraction of the Ytm1-D104R mutant, affinity purification still showed co-purification of pre-ribosomes with a large excess of free Ytm1-D104R. However compared to the wild-type and Erb1-R470E, these particles had a slightly different composition. These data suggest that Erb1 itself is strongly associated with the pre-ribosome and provides the primary, but not the sole, recruiting point of Ytm1. In addition, affinity purification of Erb1-R470E showed strongly reduced levels of Ytm1 (Figure 3C). Together, this indicates that the interaction to Erb1 has been impaired, and highlights the importance of Erb1 and Ytm1 in ribosome biogenesis.
Figure 3.
Characterization of the conserved Erb1-R470/Ytm1-D104 salt bridge. (A) Disruption of the conserved salt bridge between Erb1 and Ytm1 leads to strong temperature sensitive (ts) phenotypes at 37°C. Growth analysis of ERB1 wt and erb1-R470E as well as YTM1 wt and ytm1-D104R. Constructs were transformed into Erb1 and Ytm1 shuffle strains, respectively, and selected on SDC+FOA plates. Strains were spotted in 10-fold serial dilution on YPD plates and incubated for 2 days at the indicated temperatures. (B) Ribosome profile analysis of the N-terminal TAP-Flag tagged (ProteinA-TEVsite-CBP-Flag) erb1-R470E and the ytm1-D104R mutants and comparison with the respective wt alleles. Half-mers in the 80S and polysome fractions are indicated with arrows. (C) Tandem affinity purification of the N-terminal TAP-Flag tagged Erb1 wt and R470E mutant in comparison with Ytm1 wt and the D104R mutant. Final eluates were analysed by SDS-PAGE and Coomassie staining or western blotting with the indicated antibodies. Bait proteins are marked with an asterisk.
The Erb1–Ytm1 interaction is essential for cell growth
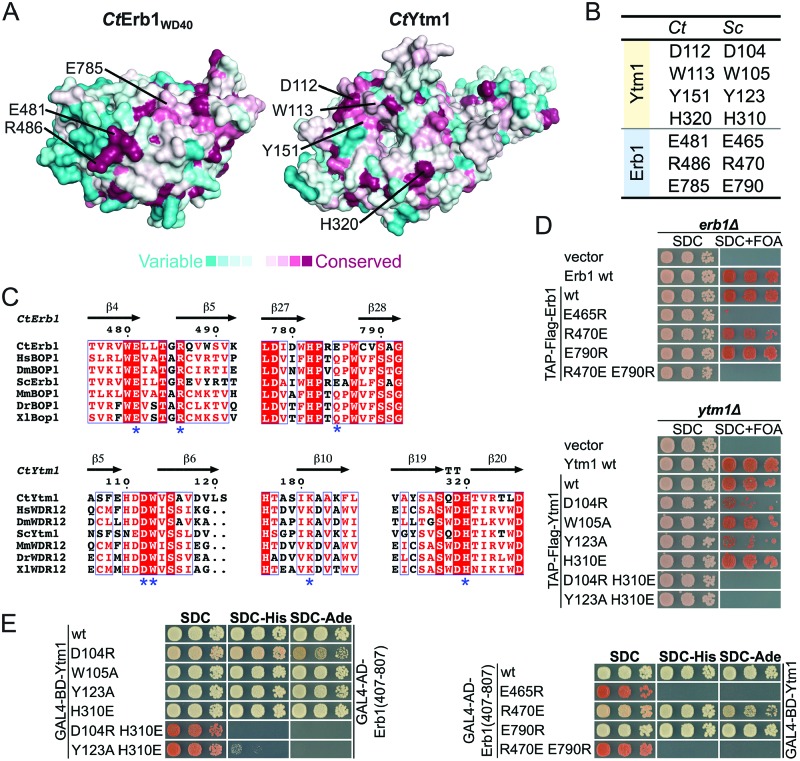
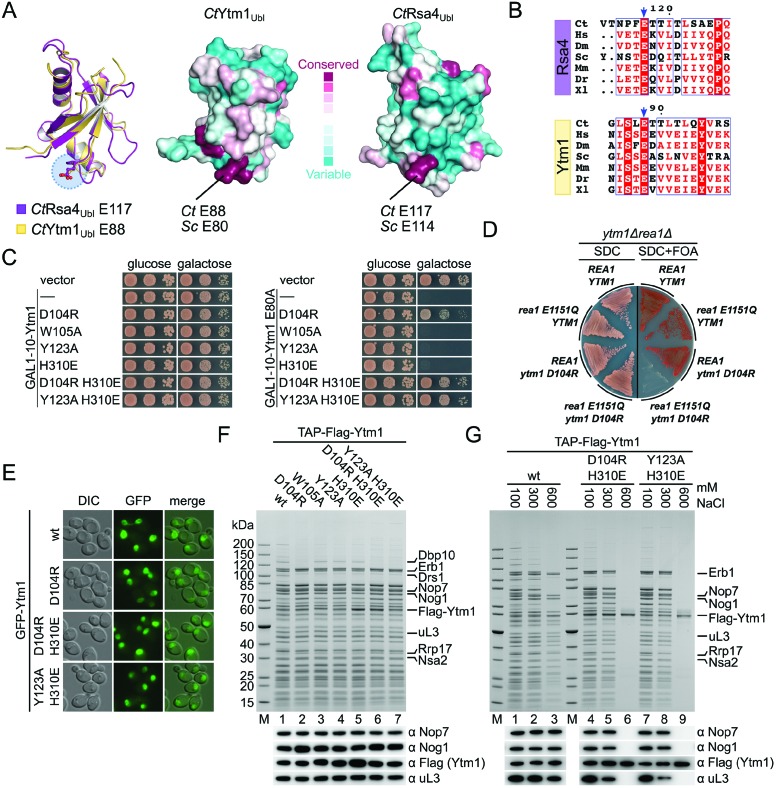
As the Erb1-R470 and Ytm1-D104 mutants exhibit defects in ribosome assembly, we hypothesized that the Erb1–Ytm1 interaction might have been weakened in vivo, but still allow partial interaction during biochemical purifications. Therefore, we mutated other conserved residues (Figure 4A–C) to further de-stabilize this interaction, and analysed them by yeast two-hybrid and yeast complementation assays. In an otherwise lethal erb1Δ background, growth could be restored with a plasmid-borne copy of Erb1-R470E (Figure 4D and Supplementary Figure S2C) and Erb1-E790R (R486 and E785 in Ct, respectively), but neither the single Erb1 mutant E465R nor the double mutant Erb1 R470E/E790R (E481 and R486/E785 in Ct, respectively) allowed cell growth. This effect was weaker when we altered the interface residues on Ytm1 and introduced these into a ytm1Δ shuffle strain (Figure 4D and Supplementary Figure S2C). Growth could be complemented with the addition of Ytm1 mutants D104R, Y123A, H310E and W105A (D112, Y151, H320 and W113 in Ct, respectively). However, the combination of two mutations, Ytm1 D104R/H310E and Y123A/H310E, lead to a loss of complementation, suggesting that the interface might no longer be intact in vivo.
Figure 4.
The Erb1–Ytm1 interaction is required for cell viability. (A) ConSurf analysis of CtErb1WD40 and CtYtm1. Amino acids are colored according to their conservation. Variable amino acids are shown in turquoise and highly conserved amino acids in maroon (see below). Amino acids chosen for the mutational analysis are indicated. (B) Table showing the equivalent residues in C. thermophilum (Ct) and S. cerevisiae (Sc). (C) Multiple sequence alignments of Erb1 and Ytm1 (in higher eukaryotes named Bop1 and WDR12, respectively). The sequences of C. thermophilum (Ct), Homo sapiens (Hs), Drosophila melanogaster (Dm), S. cerevisiae (Sc), Mus musculus (Ms), Danio rerio (Dr) and Xenopus laevis (Xl) were aligned using Clustal Omega and visualized with ESPRIPT. Highly conserved residues are highlighted (red boxes) and amino acids mutated in this study are indicated with an asterisk. (D) Complementation analysis of the indicated Erb1 and Ytm1 alleles. An Erb1 shuffle strain (upper panel) and a Ytm1 shuffle strain (lower panel) were transformed with an empty vector control, the respective untagged wt allele on a plasmid under control of the respective native promoter or the wt allele and the indicated interface mutants with an N-terminal TAP-Flag tag. Cells were spotted in 10-fold serial dilutions on SDC (SDC-Leu) and SDC+FOA plates. Cell growth at 30°C was monitored after 2 and 5 days respectively. (E) Yeast two-hybrid analysis of Ytm1 interface mutants (left panel) and Erb1 interface mutants (right panel). Ytm1 alleles were fused to an N-terminal GAL4-BD (binding domain) and the different Erb1WD40 variants (aa407–807, corresponding to the construct boundaries of the CtErb1 WD40 domain used for crystallization) were fused to an N-terminal GAL4-AD (activation domain). Constructs were co-transformed into the PJ69–4A Y2H strain and growth was monitored on SDC (SDC-Leu-Trp), SDC-His (SDC-Leu-Trp-His) and SDC-Ade (SDC-Leu-Trp-Ade) plates after 3 days at 30°C.
To assess whether the interaction between Erb1 and Ytm1 has been abolished in these mutants, we performed Y2H and in vitro binding assays. Indeed results from the Y2H analysis (Figure 4E and Supplementary Figure S2D) correlate with in vivo growth complementation analysis, where mutants which were inviable also failed to show an interaction. The binding of these mutants in vitro were more compromised compared to the in vivo experiments, where all Erb1 mutants, except the E790R allele, failed to bind to Flag-Ytm1 wt (Supplementary Figure S8A). Mutations in Ytm1 weakened the binding (W105R, Y123A and H310E) or failed to interact (D104R, D104R/H310E and Y123A/H310E) (Supplementary Figure S8B).
Based on our in vivo and in vitro analyses, we conclude that the Erb1–Ytm1 interaction is conserved, and essential for cell viability and ribosome assembly. Mutations in the interface can be designed, which either weaken or abolish this interaction.
Generation of structure-based dominant-negative Ytm1 mutants
Previous studies showed that the Nop7–Erb1–Ytm1 complex is removed from the pre-60S particles through the action of the AAA ATPase Rea1 (24). This step involves interaction between a conserved glutamate residue (E80) on the Ytm1Ubl domain with the Rea1MIDAS domain. Analogously, removal of Rsa4 by Rea1 at a later biogenesis stage also proceeds via a similarly positioned and conserved glutamate residue (E114) on the Rsa4Ubl (Figure 5A and B). To dissect the role of the Rea1MIDAS–Ytm1Ubl interaction in the context of the Erb1–Ytm1 complex, we overexpressed Ytm1 variants in a wild-type background with the goal to identify dominant-negative mutants.
Figure 5.
Ytm1 interface mutations abolish the dominant lethal effect of the E80A mutation. (A) Comparison of the Ubl domains of Rsa4 (PDB-ID: 4WJS) and Ytm1. Left panel: overlay of the Rsa4 (maroon) and Ytm1 (yellow) Ubl domains. The conserved glutamic acid residues required for interaction with the MIDAS domain of Rea1 are highlighted (blue circle). Right panel: ConSurf analysis of Ytm1 and Rsa4 Ubl domains (PDB-ID: 4WJS). Variable amino acids are colored in turquoise and conserved ones in maroon. The color-coding is shown. (B) Multiple sequence alignments of Rsa4 and Ytm1. The sequences of Chaetomium thermophilum (Ct), Homo sapiens (Hs), Drosophila melanogaster (Dm), Saccharomyces cerevisiae (Sc), Mus musculus (Ms), Danio rerio (Dr) and Xenopus laevis (Xl) were aligned using Clustal Omega and visualized with ESPRIPT. The regions containing the conserved glutamic acid residues are shown and the conserved glutamate residue is marked with a blue arrow. (C) Dominant negative analysis of the indicated Ytm1 interface mutants (left panel) and in combination with the dominant lethal E80A mutation (right panel). Plasmids containing the different Ytm1 alleles under control of the GAL1–10 promoter were transformed in a wt (W303) strain. Cells were spotted in 10-fold serial dilutions on glucose (SDC-Leu) or galactose (SGC-Leu) containing medium and growth was monitored after 2 days at 30°C. (D) Synthetic lethal interaction analysis between TAP-Flag-ytm1-D104R and rea1-E1151Q. The ytm1Δ rea1Δ double shuffle strain was transformed with the indicated Rea1 and TAP-Flag-Erb1 alleles. Cells were streaked on SDC (SDC-Leu-Trp) and SDC+FOA plates and growth at 30°C was monitored after 3 and 5 days, respectively. (E) Fluorescence microscopy of N-terminal GFP tagged Ytm1 alleles. Plasmids were transformed into a GAL1–3×HA-YTM1 strain. Cells were grown for 6 h in YPD (glucose) before microscopy to deplete the endogenous protein. (F) Affinity purifications of the different Ytm1 alleles. Plasmids containing the N-terminal TAP-Flag tagged Ytm1 alleles under control of the native Ytm1 promoter were transformed into a GAL1–3×HA-YTM1 strain. Cells were grown for 6 h in glucose medium (YPD) to deplete endogenous Ytm1. Final eluates were analysed with SDS-PAGE and Coomassie staining or western blotting with the indicated antibodies. (G) Salt treatment releases Ytm1 mutants from the pre-ribosome. Affinity purification of strains described in (F). Particles bound to IgG sepharose beads were treated with buffer containing different salt concentration. Subsequent steps were carried out in buffer containing 100 mM NaCl. Eluates were analysed through SDS-PAGE and Coomassie staining or western blot analysis.
Of all the Ytm1 variants tested, none showed any growth defect upon overexpression (D104R, W105A, Y123A, H310E, D104R/H310E, Y123A/H310E). However when the corresponding Erb1 interface mutants were overexpressed (E465R, R470E, E790R, R470E/E790R) we observed a dominant negative phenotype, which was especially pronounced at a low temperature (23°C) for the Erb1-E465R and Erb1-R470E/E790R alleles, which inhibit binding to Ytm1 (Figure 5C and Supplementary Figure S9A). These data suggest that under mutant conditions a block in 60S maturation is caused by a failure of mutant Erb1 to be released from the particle. In contrast, the ribosome association of the Ytm1 mutants appears to be more labile.
These results prompted us to combine the Ytm1 interface mutants with the dominant lethal Ytm1 E80A mutation, that shows impaired interaction with the Rea1-Midas (Figure 5C, right panel). Overexpression of Ytm1 mutants harboring the E80A mutation together with W105A, Y123A or H310E had a dominant negative effect comparable to the E80A-single mutation. These three mutants had shown no significant defects in Erb1 binding or cell growth (Figure 4D and E). These findings suggest that the E80A double mutants had efficiently replaced endogenous wild-type Ytm1 comparable to the E80A single mutant, causing a similar block of ribosome biogenesis through an impaired Rea1 interaction. On the other hand, mutants impaired in Erb1 binding (D104R, D104R/H310E, Y123A H310E) (see also Figure 4E and Supplementary Figure S8) abolished the dominant negative growth defect of the E80A mutation, suggesting a failure to efficiently associate with the pre-60S ribosome via Erb1. To further explore the link between Rea1 and the Erb1–Ytm1 interface, we performed genetic interaction studies. The combinations of TAP-Flag-ytm1-D104R or TAP-Flag-erb1-R470E with rea1-E1151Q lead to synthetic lethality (Figure 5D and Supplementary Figure S9B). This Rea1 mutant harbors a mutation in the Walker B motif of AAA domain 3, highlighting the role of ATP hydrolysis and removal of the Ytm1–Erb1 complex.
Since we had observed that the Ytm1-D104R variant was still able to associate with pre-60S particles (Figure 3C), we next asked whether the mutants deficient in Erb1 binding (Ytm1 D104R/H310E and Y123A/H310E) were incompetent of associating with pre-ribosomes. We generated a strain expressing 3×HA tagged Ytm1 under the control of a repressible GAL1 promoter (Supplementary Figure S10). Fluorescence microscopy, to analyse the cellular localization of the different GFP-Ytm1 alleles, as well as affinity purifications of TAP-Flag tagged Ytm1 constructs were performed after depletion of the endogenous Ytm1 through shifting cells from galactose containing medium (SGC-Leu) to glucose medium (YPD) for 6 h. The GFP-tagged Ytm1-mutants impaired in Erb1 binding (D104R, D104R/H310E and Y123A/H310E) showed a normal nucleolar distribution when compared to the Ytm1 wild-type suggesting correct localization of the mutants (Figure 5E). To analyse this in more detail, we performed affinity purification of all Ytm1 mutants generated based on the crystal structure. We observed that all Ytm1 mutants were still able to co-purify pre-ribosomal particles (Figure 5F). However, compared to the wild-type purification, all mutants were depleted in Drs1, an ATP-dependent helicase involved in 60S ribosome biogenesis (52,53). The human ortholog DDX27 was recently described to interact with Bop1 (Erb1) and Pes1 (Nop7) via a conserved FxF-motif in its N-terminus (54). This interaction most likely involves the β-propeller of Erb1 (see also Figure 1C). In addition, only the Ytm1 W105A, Y123A and H310E mutants co-purified the helicase Dbp10 (55,56) to wild-type levels whereas particles from Ytm1 mutants impaired in Erb1 interaction showed no Dbp10 association (Figure 5F). Most surprisingly, all mutants enriched similar amounts of Erb1 although previous experiments probing the Erb1–Ytm1 interface showed reduced or abolished interaction (see also Figure 4E). One possible explanation for this observation could be that Ytm1 may interact with the pre-ribosome via (an) additional factor(s) and/or rRNA. To address this, we performed affinity-purifications of wild-type Ytm1 and the lethal double mutants Ytm1 D104R/H310E and Y123A/H310E, but included washing steps with increasing salt concentrations to discriminate between a potentially weak Erb1–Ytm1 and additional interactions of Ytm1 (Figure 5G). Based on our observations during the in vitro reconstitution of the Erb1–Ytm1 complex, we expected the wild-type complex to be salt stable. With increasing salt concentrations, pre-60S particles purified via wild-type Ytm1 gradually decreased in complexity but still retained multiple ribosome biogenesis factors including Erb1 and Nop7, even at the highest salt concentrations. However in the case of the double mutants, both biogenesis factors and ribosomal proteins were no longer associated with purified Ytm1 at high salt concentration. These data indicate that Ytm1 might have additional binding sites on the pre-ribosome, which contribute to the recruitment to the pre-60S surface. Even though our results strongly indicate an abolished Erb1–Ytm1 interaction with the described mutants, a residual binding sufficient to recruit Ytm1 to the pre-ribosome cannot be excluded.
DISCUSSION
Ribosome biogenesis in eukaryotes is tightly regulated and a highly hierarchical process that depends on a large number of non-ribosomal proteins. Recent advances in the structural characterization of pre-ribosomal particles have provided first insights into the structural and mechanistic details of ribosome biogenesis, but are still restricted to late assembly intermediates. Here, we provide functional and structural data highlighting the role of the Erb1–Ytm1 complex during early stages of ribosome biogenesis. We define the essential interface of the heterodimer and analyse its impact on pre-60S assembly.
Erb1 exclusively interacts via its WD40 domain with Ytm1
A common mode of interaction between WD40 domains and their ligands is the binding of short linear motifs near the central pore or occasionally on the sides of the blades (57). Previously it was suggested that the interaction of Erb1 to Ytm1 requires only a small region (residues 383 to 419) of its N-terminal domain and not the C-terminal WD40 domain, which was suggested to be non-essential (10). However we were unable to observe this interaction in vivo and complementation of an erb1Δ strain could only be achieved with a plasmid encoding full-length Erb1. Based on our crystal structure and in vivo analysis, we show that the main interaction, between Erb1 and Ytm1, is mediated by their respective WD40 domains.
Although there are reports of heterodimeric WD40 complexes, their binding mode often involves either complementation (58,59) of the β−propeller or interaction of a linear motif along the central pore, reviewed in (57). To our knowledge the interaction between two different proteins via their WD40 domains, which does not involve a linear motif, has only been observed once before—in the structure of yeast transcription factor IIIC τ60/Δτ91-subcomplex (60). This complex, involved in RNA transcription, has a highly similar arrangement of the WD40 domains as the Erb1–Ytm1 complex presented here (Supplementary Figure S11). In both cases one molecule uses the top surface for interaction, whereas the other one employs a distinct side of the WD40 domain. Given the strong representation of WD40 proteins in various cellular processes including ribosome biogenesis, nuclear transport or membrane fusion, it seems surprising that the mode of interaction described in the current study is underrepresented in the available literature.
The Erb1–Ytm1 interface has to withstand the Rea1 powerstroke
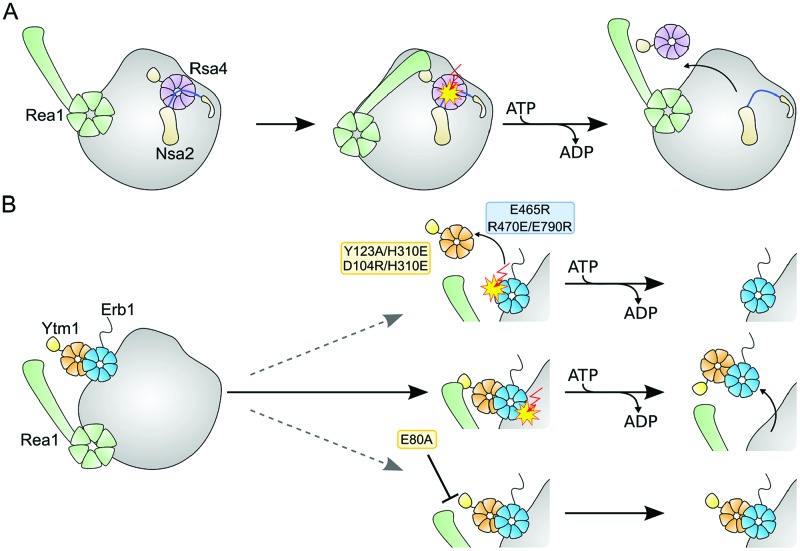
In contrast to the complex, three-dimensional surface recognition employed in the Erb1–Ytm1 interface, Rsa4 is recruited to the pre-60S particle by a small linear motif of Nsa2, which itself is tightly anchored to the pre-ribosome (26). The affinity between Rsa4 and Nsa2 is in the low-nano molar range, yet the addition of the AAA ATPase Rea1 and ATP only succeeds in removal of Rsa4, leaving Nsa2 bound to the pre-ribosome (25). A subtly distinct mechanism seemed likely for the Erb1–Ytm1 complex, since addition of Rea1 and ATP caused removal of Ytm1 and Erb1 from pre-ribosomal assemblies (24). Indeed we identified some subtle yet significant differences. Erb1 recruits Ytm1 exclusively via its intact WD40 domain to the pre-60S particle. The Erb1–Ytm1 interaction is stabilized by extensive interactions and involves a large surface on both proteins rather than a short linear motif. This interface seems to hold both proteins together so tightly that the addition of Rea1 results in the dissociation of the entire Nop7–Erb1–Ytm1 complex or the Erb1–Ytm1 complex from the pre-ribosome (24). Weakening the Erb1–Ytm1 interaction leads to defects in ribosome biogenesis probably because removal of Ytm1 alone blocks subsequent maturation steps as rRNA remodeling and/or recruitment and exchange of assembly factors is compromised. Similar to the intricate network of interactions observed for Rsa4 at later stages in pre-60S maturation (26,32,61,62), the Erb1–Ytm1 complex may also be involved in multiple interactions with rRNA and assembly factors. While structural data on Ytm1/pre-60S interaction are not available, a potential binding site of Erb1 on the pre-60S particle has been mapped by CRAC (11). It seems to contact the 25S rRNA including helices H16, H21 and H22. However, the structure of this region with respect to mature 60S is not known or whether it has to undergo remodeling steps to reach its final structure. At a later stage, for the formation of the central protuberance extensive remodeling of 25S rRNA helices H82 to H89 is necessary (26,32,61,62). Since the AAA ATPase Rea1 is required for the removal of both factors (Erb1–Ytm1 and Rsa4) (24,25) at spatially and temporally different points, it is difficult to envisage that Rea1 does so from a single binding site. Rea1 has an elongated shape and has been shown to exhibit structural flexibility (25). Whether the position of Rea1 for both steps is fixed or has to change is not known. Removal of a remodeling factor in an early stage in ribosome biogenesis and repositioning at a later stage would offer attractive possibilities for the ordered assembly and reuse of existing factors.
Structure-based model for Erb1–Ytm1 in pre-60S maturation
The formation of the central protuberance requires the correct positioning of the 5S RNP, which first occupies a position about 180° rotated with respect to its mature orientation (32). The 25S rRNA including helices H82 to H89 has to undergo quite dramatic rearrangements in pre-60S maturation. Rearrangement appears to be blocked by the Rpf2–Rrs1 complex, which establishes a network of interactions reaching from the 5S RNP to Rsa4 (26,61–63). Removal of Rsa4 by Rea1 (probably after the removal of Rpf2-Rrs1) seems to allow relocation of rRNA elements during ribosome maturation (Figure 6A), necessary for formation of the central protuberance. While this part of 60S biogenesis has been structurally well characterized, the early stages where the Erb1–Ytm1 complex is involved remain largely unexplored. Based on our data, we propose the following speculative model. The Nop7, Erb1 and Ytm1 are recruited to early pre-60S particles (12). Following recruitment of Rea1, Erb1–Ytm1 and partially also Nop7 are removed from the pre-ribosome. Loss of these factors may allow rearrangement of rRNA, concomitant with binding of new factors, which could prevent re-binding of Erb1–Ytm1. Failure to remove Erb1 or Erb1–Ytm1, as result of a weakened Erb1–Ytm1 or Rea1MIDAS–Ytm1Ubl interface, might lead to an intermediate that retains Erb1 or Erb1–Ytm1 at its initial position on the pre-ribosome (helices 16/21/22, Figure 6B). Remodeling of rRNA, binding of downstream factors and further biogenesis steps could be impaired for this intermediate.
Figure 6.
Removal of various factors by the AAA ATPase Rea1 during ribosome biogenesis. (A) Rsa4 (purple) interacts at the pre-ribosome (gray) with Nsa2 (light brown, Rsa4 binding motif in blue), which is deeply embedded into the pre-ribosome. Interaction of Rsa4Ubl with Rea1MIDAS (green) causes removal of Rsa4 but not Nsa2 from the pre-ribosome. (B) In the case of Erb1/Ytm1 under normal conditions Rea1 removes both proteins from the pre-ribosome. An impaired Erb1/Ytm1 interface (Ytm1 Y123A/H310E or D104R/H310E, Erb1 E465R or R470E/E790R) might cause removal of Ytm1 but not Erb1 from the pre-ribosome. In the Ytm1 E80A-background both proteins remain bound to the pre-ribosome. Further steps in ribosome biogenesis do not proceed correctly.
Our study provides insights into an underrepresented, stable interaction between two WD40 domains and its role in ribosome biogenesis. While the Erb1–Ytm1 interface needs to withstand the powerstroke of Rea1 ensuring concerted removal of the complex, the Rsa4–Nsa2 interface does not. Rsa4 has been proposed to transmit remodeling energy from Rea1 into the developing ribosome (26). The Erb1–Ytm1 complex seems to be linked to Rea1 in a similar manner, but functions in a mechanistically distinct manner. Structural insights into this early stage of ribosome biogenesis are instrumental in resolving this question.
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited at the RCSB Protein Data Bank (PDB) with the accession number 5EM2.
Supplementary Material
Acknowledgments
We thank J. Kopp, C. Siegmann and G. Müller from the BZH/Cluster of Excellence: CellNetworks crystallization platform for protein crystallization, A. Gumiero for data collection, M. Gnädig for technical assistance, E. Thomson for suggestions on the manuscript and ESRF for support and access to beamlines. I.S. and E.H. are investigators of the Cluster of Excellence: CellNetworks and acknowledge support through EcTop1. We are grateful to M. Fromont-Racine (anti-Nog1, anti-Nsa2 and anti-Rlp24), P. Linder (anti-Has1), B. Stillman (anti-Nop7), H. Tschochner (anti-Noc1), J.R. Warner (anti-Rpl3), D.H. Wolf (anti-Nsa3) and J. Woolford (anti-Ytm1) for their kind gift of antibodies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [HU363/10-5, HU363/12-1 to E.H.]; [SFB 638 to I.S.]; DFG [GRK1188 to M.T.]. Funding for open access charge: DFG/Uni Heidelberg EcTop1.
Conflict of interest statement. None declared.
REFERENCES
- 1.Woolford J.L., Jr, Baserga S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tschochner H., Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 3.Henras A.K., Soudet J., Gerus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harnpicharnchai P., Jakovljevic J., Horsey E., Miles T., Roman J., Rout M., Meagher D., Imai B., Guo Y., Brame C.J., et al. Composition and functional characterization of yeast 66s ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 5.Nissan T.A., Bassler J., Petfalski E., Tollervey D., Hurt E.C. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Udem S.A., Warner J.R. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J. Mol. Biol. 1972;65:227–242. doi: 10.1016/0022-2836(72)90279-3. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt M.E., Clayton D.A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu S., Archer R.H., Zengel J.M., Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc. Natl. Acad. Sci. U.S. A. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miles T.D., Jakovljevic J., Horsey E.W., Harnpicharnchai P., Tang L., Woolford J.L., Jr Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell Biol. 2005;25:10419–10432. doi: 10.1128/MCB.25.23.10419-10432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang L., Sahasranaman A., Jakovljevic J., Schleifman E., Woolford J.L., Jr Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-subcomplex in yeast preribosomes. Mol. Biol. Cell. 2008;19:2844–2856. doi: 10.1091/mbc.E07-12-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Granneman S., Petfalski E., Tollervey D. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J. 2011;30:4006–4019. doi: 10.1038/emboj.2011.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahasranaman A., Dembowski J., Strahler J., Andrews P., Maddock J., Woolford J.L., Jr Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J. 2011;30:4020–4032. doi: 10.1038/emboj.2011.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oeffinger M., Zenklusen D., Ferguson A., Wei K.E., El Hage A., Tollervey D., Chait B.T., Singer R.H., Rout M.P. Rrp17p is a eukaryotic exonuclease required for 5′ end processing of Pre-60S ribosomal RNA. Mol. Cell. 2009;36:768–781. doi: 10.1016/j.molcel.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Hage A., Koper M., Kufel J., Tollervey D. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev. 2008;22:1069–1081. doi: 10.1101/gad.463708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry Y., Wood H., Morrissey J.P., Petfalski E., Kearsey S., Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pestov D.G., Strezoska Z., Lau L.F. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol. Cell Biol. 2001;21:4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohrmoser M., Hölzel M., Grimm T., Malamoussi A., Harasim T., Orban M., Pfisterer I., Gruber-Eber A., Kremmer E., Eick D. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol. Cell Biol. 2007;27:3682–3694. doi: 10.1128/MCB.00172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strezoska Z., Pestov D.G., Lau L.F. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J. Biol. Chem. 2002;277:29617–29625. doi: 10.1074/jbc.M204381200. [DOI] [PubMed] [Google Scholar]
- 19.Lerch-Gaggl A., Haque J., Li J., Ning G., Traktman P., Duncan S.A. Pescadillo is essential for nucleolar assembly, ribosome biogenesis, and mammalian cell proliferation. J. Biol. Chem. 2002;277:45347–45355. doi: 10.1074/jbc.M208338200. [DOI] [PubMed] [Google Scholar]
- 20.Holzel M., Rohrmoser M., Schlee M., Grimm T., Harasim T., Malamoussi A., Gruber-Eber A., Kremmer E., Hiddemann W., Bornkamm G.W., et al. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 2005;170:367–378. doi: 10.1083/jcb.200501141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimm T., Holzel M., Rohrmoser M., Harasim T., Malamoussi A., Gruber-Eber A., Kremmer E., Eick D. Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res. 2006;34:3030–3043. doi: 10.1093/nar/gkl378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Killian A., Le Meur N., Sesboue R., Bourguignon J., Bougeard G., Gautherot J., Bastard C., Frebourg T., Flaman J.M. Inactivation of the RRB1-Pescadillo pathway involved in ribosome biogenesis induces chromosomal instability. Oncogene. 2004;23:8597–8602. doi: 10.1038/sj.onc.1207845. [DOI] [PubMed] [Google Scholar]
- 23.Killian A., Sarafan-Vasseur N., Sesboue R., Le Pessot F., Blanchard F., Lamy A., Laurent M., Flaman J.M., Frebourg T. Contribution of the BOP1 gene, located on 8q24, to colorectal tumorigenesis. Genes Chromosomes Cancer. 2006;45:874–881. doi: 10.1002/gcc.20351. [DOI] [PubMed] [Google Scholar]
- 24.Bassler J., Kallas M., Ulbrich C., Thoms M., Pertschy B., Hurt E. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol. Cell. 2010;38:712–721. doi: 10.1016/j.molcel.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulbrich C., Diepholz M., Bassler J., Kressler D., Pertschy B., Galani K., Böttcher B., Hurt E. Mechanochemical removal of ribosome biogenesis factors from Nascent 60S ribosomal subunit. Cell. 2009;138:911–922. doi: 10.1016/j.cell.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 26.Bassler J., Paternoga H., Holdermann I., Thoms M., Granneman S., Barrio-Garcia C., Nyarko A., Stier G., Clark S.A., Schraivogel D., et al. A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation. J. Cell Biol. 2014;207:481–498. doi: 10.1083/jcb.201408111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas B.J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 28.Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 29.Longtine M.S., McKenzie A. 3rd, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.James P., Halladay J., Craig E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amlacher S., Sarges P., Flemming D., van Noort V., Kunze R., Devos D.P., Arumugam M., Bork P., Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 32.Leidig C., Thoms M., Holdermann I., Bradatsch B., Berninghausen O., Bange G., Sinning I., Hurt E., Beckmann R. 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat. Commun. 2014;5:3491. doi: 10.1038/ncomms4491. [DOI] [PubMed] [Google Scholar]
- 33.Flot D., Mairs T., Giraud T., Guijarro M., Lesourd M., Rey V., van Brussel D., Morawe C., Borel C., Hignette O., et al. The ID23–2 structural biology microfocus beamline at the ESRF. J. Synchrotron Radiat. 2010;17:107–118. doi: 10.1107/S0909049509041168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans P.R., Murshudov G.N. How good are my data and what is the resolution. Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vagin A., Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 38.Schuetz A., Allali-Hassani A., Martin F., Loppnau P., Vedadi M., Bochkarev A., Plotnikov A.N., Arrowsmith C.H., Min J. Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 2006;25:4245–4252. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 42.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The PyMOL Molecular Graphics System, Schrödinger, LLC. 2010. https://www.pymol.org/citing https://www.pymol.org/citing
- 44.Krissinel E. Enhanced fold recognition using efficient short fragment clustering. J. Mol. Biochem. 2012;1:76–85. [PMC free article] [PubMed] [Google Scholar]
- 45.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Noort V., Bradatsch B., Arumugam M., Amlacher S., Bange G., Creevey C., Falk S., Mende D.R., Sinning I., Hurt E., et al. Consistent mutational paths predict eukaryotic thermostability. BMC Evol. Biol. 2013;13:7. doi: 10.1186/1471-2148-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wegrecki M., Neira J.L., Bravo J. The carboxy-terminal domain of Erb1 is a seven-bladed ss-propeller that binds RNA. PLoS One. 2015;10:e0123463. doi: 10.1371/journal.pone.0123463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nal B., Mohr E., Silva M.I., Tagett R., Navarro C., Carroll P., Depetris D., Verthuy C., Jordan B.R., Ferrier P. Wdr12, a mouse gene encoding a novel WD-Repeat Protein with a notchless-like amino-terminal domain. Genomics. 2002;79:77–86. doi: 10.1006/geno.2001.6682. [DOI] [PubMed] [Google Scholar]
- 51.Chen S., Spiegelberg B.D., Lin F., Dell E.J., Hamm H.E. Interaction of Gbetagamma with RACK1 and other WD40 repeat proteins. J. Mol. Cell. Cardiol. 2004;37:399–406. doi: 10.1016/j.yjmcc.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 52.Merl J., Jakob S., Ridinger K., Hierlmeier T., Deutzmann R., Milkereit P., Tschochner H. Analysis of ribosome biogenesis factor-modules in yeast cells depleted from pre-ribosomes. Nucleic Acids Res. 2010;38:3068–3080. doi: 10.1093/nar/gkp1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams C.C., Jakovljevic J., Roman J., Harnpicharnchai P., Woolford J.L., Jr Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA. 2002;8:150–165. doi: 10.1017/s1355838202010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kellner M., Rohrmoser M., Forne I., Voss K., Burger K., Muhl B., Gruber-Eber A., Kremmer E., Imhof A., Eick D. DEAD-box helicase DDX27 regulates 3′ end formation of ribosomal 47S RNA and stably associates with the PeBoW-complex. Exp. Cell Res. 2015;334:146–159. doi: 10.1016/j.yexcr.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 55.Burger F., Daugeron M.C., Linder P. Dbp10p, a putative RNA helicase from Saccharomyces cerevisiae, is required for ribosome biogenesis. Nucleic Acids Res. 2000;28:2315–2323. doi: 10.1093/nar/28.12.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Talkish J., Zhang J., Jakovljevic J., Horsey E.W., Woolford J.L., Jr Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:8646–8661. doi: 10.1093/nar/gks609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stirnimann C.U., Petsalaki E., Russell R.B., Muller C.W. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Jennings B.H., Pickles L.M., Wainwright S.M., Roe S.M., Pearl L.H., Ish-Horowicz D. Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol. Cell. 2006;22:645–655. doi: 10.1016/j.molcel.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 59.Fath S., Mancias J.D., Bi X., Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 60.Mylona A., Fernandez-Tornero C., Legrand P., Haupt M., Sentenac A., Acker J., Muller C.W. Structure of the tau60/Delta tau91 subcomplex of yeast transcription factor IIIC: insights into preinitiation complex assembly. Mol. Cell. 2006;24:221–232. doi: 10.1016/j.molcel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 61.Kharde S., Calvino F.R., Gumiero A., Wild K., Sinning I. The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis. Nucleic Acids Res. 2015;43:7083–7095. doi: 10.1093/nar/gkv640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Madru C., Lebaron S., Blaud M., Delbos L., Pipoli J., Pasmant E., Rety S., Leulliot N. Chaperoning 5S RNA assembly. Genes Dev. 2015;29:1432–1446. doi: 10.1101/gad.260349.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asano N., Kato K., Nakamura A., Komoda K., Tanaka I., Yao M. Structural and functional analysis of the Rpf2-Rrs1 complex in ribosome biogenesis. Nucleic Acids Res. 2015;43:4746–4757. doi: 10.1093/nar/gkv305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.