Abstract
The telomerase is responsible for adding telomeric repeats to chromosomal ends and consists of the reverse transcriptase TERT and the RNA subunit TERC. The expression and activity of the telomerase are tightly regulated, and aberrant activation of the telomerase has been observed in >85% of human cancers. To better understand telomerase regulation, we performed immunoprecipitations coupled with mass spectrometry (IP-MS) and identified cold inducible RNA-binding protein (CIRP or hnRNP A18) as a telomerase-interacting factor. We have found that CIRP is necessary to maintain telomerase activities at both 32°C and 37°C. Furthermore, inhibition of CIRP by CRISPR-Cas9 or siRNA knockdown led to reduced telomerase activities and shortened telomere length, suggesting an important role of CIRP in telomere maintenance. We also provide evidence here that CIRP associates with the active telomerase complex through direct binding of TERC and regulates Cajal body localization of the telomerase. In addition, CIRP regulates the level of TERT mRNAs. At the lower temperature, TERT mRNA is upregulated in a CIRP-dependent manner to compensate for reduced telomerase activities. Taken together, these findings highlight the dual roles that CIRP plays in regulating TERT and TERC, and reveal a new class of telomerase modulators in response to hypothermia conditions.
INTRODUCTION
Mammalian telomeres consist of tandem repeats of 5′-TTAGGG-3′ and are essential terminal structures for maintaining genome integrity and stability (1,2). Telomeres are elongated and maintained by the telomerase. The mammalian telomerase core complex consists of the catalytic subunit TERT and the RNA component TERC/TR (3–6), where the TERC RNA serves as a template for TERT-mediated telomere elongation. Except in germ cells, stem cells, and certain highly proliferative cells, mammalian TERT expression and telomerase activity appear low and/or undetectable in most somatic cells (5,7–11). However, in the majority of human cancer cells (>85%) the telomerase is activated, underlining the significance in regulating TERT expression and telomerase activity to cell growth, survival and transformation (12–15). Dysfunctional telomere maintenance can lead to critically short telomeres and induce DNA damage and genomic instability, resulting in diseases including cancer and premature aging syndromes such as dyskeratosis congenita (11,16–23). Consequently, understanding the pathways that control TERT expression and telomerase activity is crucial to devising effective therapies against cancer and premature aging diseases. In addition to TERT and TERC, other accessory proteins have also proven important to TERC stability and telomerase assembly and trafficking in vivo. These factors include the core components of box H/ACA small nucleolar ribonucleoprotein particles (snoRNPs), DKC1 (dyskerin), GAR1, NHP2, NOP10, Pontin/Reptin, TCAB1, DAXX and telomeric proteins TIN2 and TPP1 (16,24–30). Indeed, mutations of these telomerase-associating factors have been documented in a variety of diseases including cancer and dyskeratosis congenita (17–19,31–35). In this study, we set out to identify additional telomerase-associated factors by carrying out large-scale immunoprecipitation coupled with mass spectrometry (IP-MS). We report the identification of cold-inducible RNA-binding protein CIRP (also called hnRNP A18) as a new telomerase regulator. Initially identified as a protein up-regulated when cells were exposed to UV irradiation, UV mimetic agents or mild cold stress (32°C or lower) (36–38), CIRP has since been implicated in DNA damage response (38–43), cell-cycle suppression (37), low temperature stress (44), cancer cell survival and transformation (45,46), proliferation of mouse immature male germ cells (47), mRNA stability (48) and circadian gene expression in cultured fibroblasts (49). Despite numerous studies, the exact mechanisms by which CIRP functions in these conditions remain poorly understood. Here, we show that CIRP regulates telomerase activity and is essential for telomere maintenance by regulating both TERT and TERC. This is the first study to demonstrate a role for CIRP in telomerase regulation and provide possible clues to CIRP function in response to damage and stress.
MATERIALS AND METHODS
Cell lines, cell culture and CIRP KO cells
HTC75, HeLa, U2OS and 293T cells were cultured with Dulbecco's modified Eagle's medium (DMEM, Gibco) containing 10% FBS and 1% penicillin/streptomycin at 32°C or 37°C and 5% CO2. HEK293T cells used for retrovirus production and transient transfection experiments. Full-length or mutant CIRP cDNAs were cloned into the pBabe based retroviral vector for mammalian expression (50). CIRP was tagged with S, FLAG, SBP at C terminus. Stable HTC75 cells were infected with appropriate retroviruses before puromycin selection (1 mg/ml for >7 days). For synchronization, cells were cultured in FBS-free DMEM medium for 24 h and then grown in fresh medium containing 10% FBS for >24 h and harvested at the indicated time points.
To generate CIRP KO HTC75 cells, we targeted exon 4. The sgRNA (5′-AACCGATCCCGTGGGTACCG) was cloned into the vector described by the Church Laboratory (Addgene) (51). Please see Supplementary Figure S1 for details about the clone. To generate CIRP KO HeLa cells, we targeted exon 2 (sgRNA, 5′-GCCATGGCATCAGATGA) and exon 7 (sgRNA, 5′-CATCGATGTTGTATTTGCAG), aiming to delete the entire region between the two sgRNA target sequences. The sgRNAs were cloned into the Lenti gRNA Blasticidin/Puromycin/Phage-Lenti-Inducible-Cas9-neo vector described by the Zhang Laboratory (Addgene) (52). KO clones were isolated as described (51,52) and their genomic DNA extracted for sequencing. Successful KO was also confirmed by immunoblotting.
shRNA sequences were cloned into pcl-mU6 retroviral vectors. The targeting sequences for various shRNAs and siRNAs are:
shGFP: 5′-CACAAGCTGGAGTACAACT-3′
shCIRP-1: 5′-GGAGGCTCCAGAGACTACTATAGCA-3′
shCIRP-2: 5′-GACAGATCTCTGAAGTGGTGGTTGT-3′
SiCIRP-1 (CIRBP/HSS101939) siRNA
Sense: 5′-GGAGGCUCCAGAGACUACUAUAGCA-3′
Anti-sense: 5′-UGCUCAUCCUCCUCGUUCUCCGCUG-3′
SiCIRP-2 (CIRBP/HSS174416) siRNA
Sense: 5′-GACAGAUCUCUGAAGUGGUGGUUGU-3′
Anti-sense: 5′-ACAACCACCACUUCAGAGAUCUGUC-3′
SiCIRP-3 (CIRBP/HSS174417) siRNA
Sense: 5′-GGGCGGGUCCUACAGAGACAGUUAU-3′
Anti-sense: 5′-AUAACUGUCUCUGUAGGACCCGCCC-3′
Immunoprecipitation-mass Spectrum (IP-MS)
293T cells stably expressing SFB-tagged full-length TERT were generated for tandem IP followed by mass spectrometry (IP-MS) as previously reported (53). The expression of TERT-SFB was confirmed by anti-Flag immunoblotting. For affinity purification, >2 × 108 cells were collected and lysed with NETN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) containing 1 μg/ml pepstatin A and aprotinin, 1 mM MgCl2 and 250 U/Ml Benzonase Nuclease (EMD Chemicals) for 25 min. Cleared lysates were incubated with 200 μl anti-Flag M2 affinity beads (A2220, Sigma) for 2 h at 4°C and eluted with 3 mg/ml biotin (Sigma) for 2 h at 4°C. The eluates were incubated with 100 μl S-protein agarose beads (Novagen) for 2 h at 4°C before SDS/PAGE and MS (Taplin Biological MS Facility, Harvard University). Proteins with <3 peptides or found in control IP samples were excluded (Supplementary Table S1).
Bimolecular fluorescence complementation (BiFC) assay, Co-immunoprecipitation (Co-IP) assay and western blotting
For BiFC, proteins respectively tagged by YFPn (residues 1–155 of Venus YFP) and YFPc (residues 156–239 of YFP) were stably co-expressed in HTC75 cells for microscopy and flow cytometry analysis as previously described (54). For Western blotting, cells were lysed in 1x NETN buffer (1 M Tris-HCl (pH 8.0), 1 mM EDTA, 100 mMNaCl, 0.5% NP-40, 1 mM DTT and proteinase inhibitor cocktail before SDS-PAGE and antibody probing.
The antibodies used are: rabbit polyclonal anti-CIRP (ab94999, Abcam), anti-FLAG M2 Affinity Gel (Sigma, A2220), rabbit polyclonal anti-flag (Sigma, F7425), mouse monoclonal anti-GST (Abmart, M20007M), rabbit polyclonal anti-actin (GeneTex, GTX109639) and mouse monoclonal anti-GAPDH (Abmart, M20006). GST-tagged proteins were pulled down by glutathione agarose beads (GE).
Immunofluorescence (IF) and IF-fluorescent in situ hybridization (IF-FISH)
Cells grown on glass coverslips were fixed for 15 min on ice in 1x PBS (pH7.4) containing 4% paraformaldehyde, incubated in permeabilization solution (0.5% Trition-X 100, 20 mM HEPES, 50 mM NaCl, 3 mM MgCl2 and 300 mM Sucrose) for 10 min, followed by a second permeabilization for 30 min at RT after washes in 1x PBS. The coverslips were then blocked in 3% goat serum plus 0.1% BSA in 1x PBS followed by incubation with primary antibodies (overnight at 4°C) and secondary antibodies (1 h at room temperature). Primary antibodies include rabbit polyclonal anti-CIRP (ab94999, Abcam), mouse monoclonal anti-coilin [IH10] (ab87913, Abcam), mouse monoclonal anti-HA (H9658, Sigma), mouse monoclonal anti-TRF2 (OP129, Calbiochem). Secondary antibodies include fluorescein-conjugated goat ant-rabbit/mouse IgG (DyLight549, LK-GAR5492, Liankebio) and goat anti-rabbit/mouse IgG (DyLight488, LK-GAM4881, Liankebio). For IF-FISH, an additional incubation with PNA-TelC-FITC probe (Panagene) was conducted at 37°C for 2 h. Coverslips were mounted with Vectashield Mounting Medium containing 0.5 μg/ml DAPI and examined on a Nikon Ti fluorescence microscope.
Telomere repeat amplification protocol (TRAP) and IP-TRAP
Cells were cultured at indicated temperature (32°C/37°C, respectively) and harvested at various time points for straight TRAP assay or immunoprecipitation followed by TRAP (IP-TRAP) as previously described (17). The products were resolved on polyacrylamide gels (8%) and visualized with Gel Red (Biotium). Relative telomerase activity was calculated using the ImageQuant software (GE Healthcare).
Protein purification and electrophoretic mobility shift assay (EMSA)
Bacterially expressed GST-tagged CIRP and DKC1 were purified with glutathione-conjugated agarose beads, and eluted in 50 mM Tris (pH 8.0) buffer containing 50 mM reduced glutathione (GE). For EMSA, purified CIRP-GST proteins were incubated with 32P labeled T7 capped TERC probe (Ambion) (please see Supplementary Materials for details regarding probe preparation). Binding reactions were performed at 25°C for 30 min in binding buffer (100 mM NaCl, 10 mMTris (pH7.5), 5% (w/v) glycerol, 1 mM MgCl2, 1 U of RNasin Ribonuclease Inhibitor (Promega) and 0.3 pmol of 32P-labeled TERC probe). The products were resolved on 5% (wt/vol) native PAGE gel (100 V, 0.5x TBE, 180 min), visualized on a Typhoon PhosphorImager (General Electric Company) and quantified using ImageQuant (Amersham Biosciences).
Telomeric restriction fragment (TRF) analysis
Average length of telomeres was determined using the TRF assay analysis as described previously (12). Isolated genomic DNA was digested with Hinf I and Rsa I and resolved on 7% agarose gels (2 V/cm, 27 h). The denatured and dried gel was hybridized with 32P-labeled oligonucleotides [(TTAGGG)4], exposed to a PhosphorImager screen (55) and quantitated on a Typhoon PhosphorImager (General Electric Company).
RESULTS
CIRP is a telomerase-associated protein
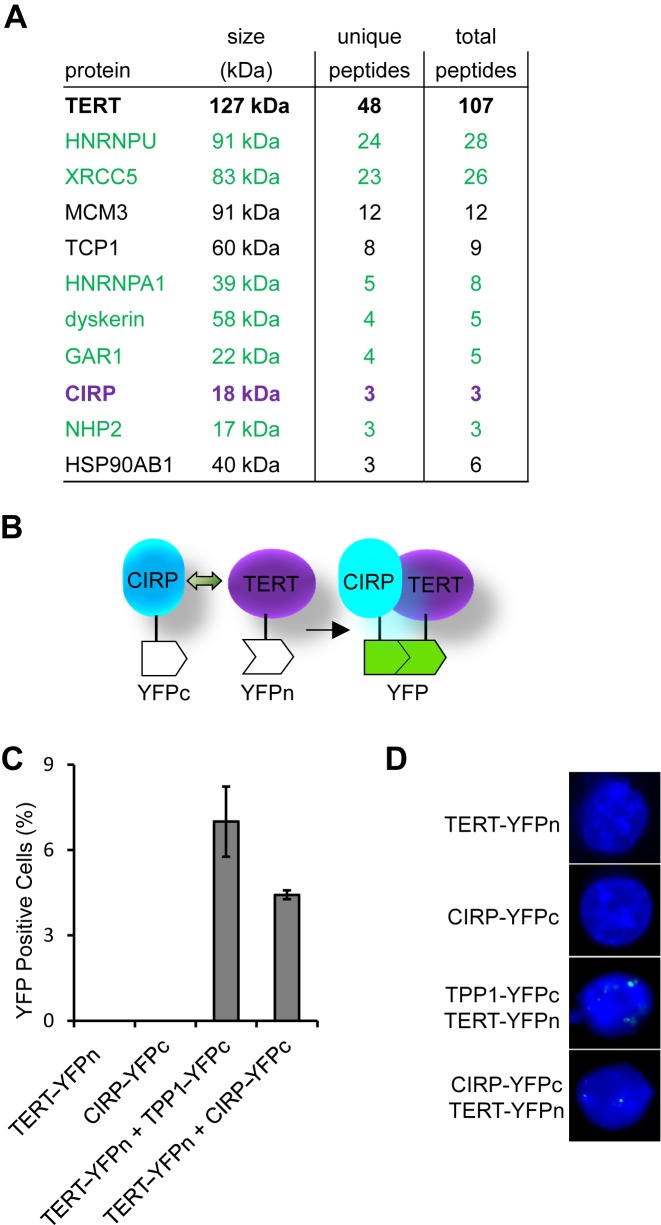
To isolate TERT-binding proteins, we first established 293T cells stably expressing TERT tagged with SFB (S tag, FLAG and Biotin-binding epitope tags) (TERT-SFB). TERT and its associated complexes were then isolated through a tandem immunoprecipitation strategy using anti-Flag antibodies and S-tag resins (56), and analyzed by SDS-PAGE and LC-MS/MS. As expected, known telomerase-associated proteins such as dyskerin (DKC1), Pontin/Reptin, glycine/arginine-rich domain containing protein 1 (GAR1), heterogeneous nuclear ribonucleoprotein (hnRNP) A1 and U, non-histone protein 2 (NHP2) and XRCC5/6 (Ku80/Ku70) were identified in our IP-MS (Figure 1A) (26,57–62). Interestingly, cold-inducible RNA-binding protein (CIRP) was also found in the TERT complex, suggesting that it may be a new telomerase-associated protein (Figure 1A).
Figure 1.
Cold inducible RNA-binding protein (CIRP) is a telomerase-associated protein. (A) 293T cells expressing SFB-tagged hTERT were first immunoprecipitated with anti-FLAG antibodies and then affinity purified with S-tag beads. The precipitates were then sequenced by mass spectrometry (MS). A select list of proteins identified is presented here. Previously reported telomerase-associated proteins are highlighted in green. Unique and total peptides are also listed. (B) Schematic diagram of the (Bimolecular fluorescence complementation) BiFC assay. (C) BiFC assays were carried out in HTC75 cells that stably co-expressed YFPn-tagged TERT together with YFPc-tagged CIRP or TPP1. Cells expressing TERT-YFPn or CIRP-YFPc alone served as negative controls. TPP1-YFPc served as a positive control. Cells were analyzed by FACS and the percentages of YFP+ cells were calculated and plotted. (D) Cells from (C) were analyzed by fluorescence microscopy. Green fluorescence indicate YFP signals in the nucleus.
CIRP is an 18-kD protein containing a well-characterized N-terminal RNA-binding domain (RRM) and a glycine-rich carboxyl-terminal motif (RGG) (63–68). To further confirm its interaction with TERT, we utilized the BiFC assay, which can detect protein–protein interactions in live cells (69). Here, CIRP and TERT were respectively tagged with the C- and N-terminal half of the yellow fluorescent protein (YFP) (Figure 1B). Interaction between CIRP and TERT would bring the two YFP fragments together for co-folding into a functional YFP molecule for fluorescence complementation. Indeed, we could detect fluorescence complementation by both FACS and fluorescence microscopy in cells co-expressing CIRP and TERT, at levels comparable to cells co-expressing TPP1 (a known TERT binding protein) and TERT (Figure 1C–D), indicating that CIRP is a telomerase-associated protein.
CIRP is required for maintaining telomerase activity at both 32°C and 37°C
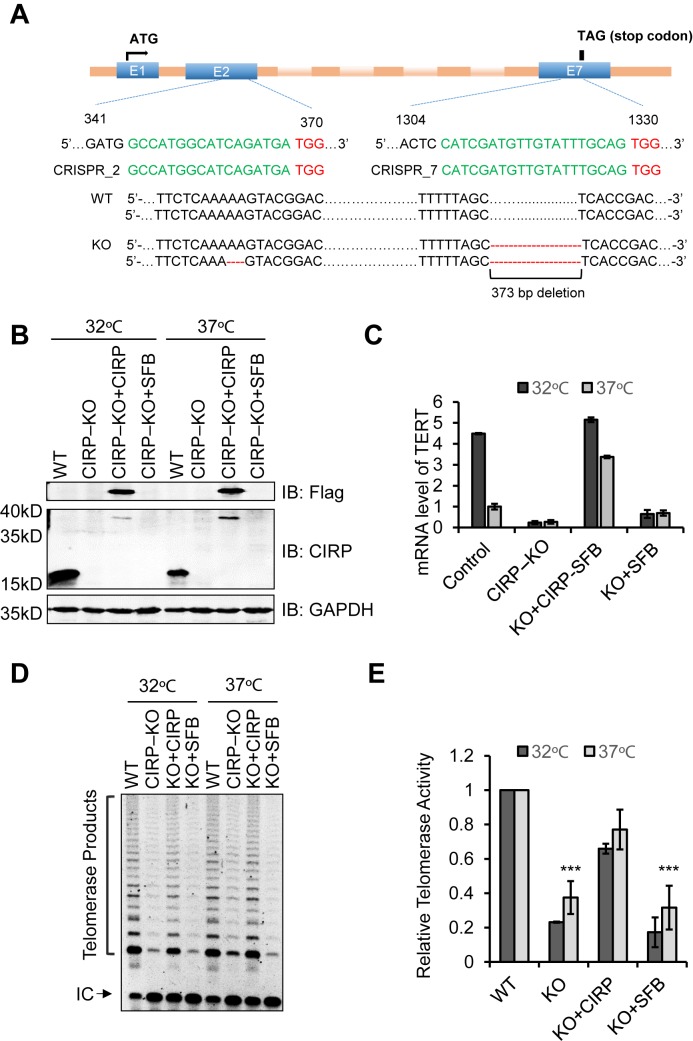
Next, we decided to probe the function of CIRP using CIRP knockout (KO) HeLa cells generated by the CRISPR-Cas9 technology (51,70). We designed two sgRNAs respectively targeting the sequences in exon 2 and 7 of the CIRP gene, located in the RRM and SY domain. Successful targeting could lead to truncation of the CIRP protein and disrupt telomerase binding. Genomic sequencing of the CIRP KO clone revealed that both alleles contained frame-shift indels in the sgRNA target region (Figure 2A). CIRP KO was also confirmed by Western blotting of cells maintained at different temperatures with anti-CIRP antibodies, given a role of CIRP in regulating cellular responses to temperature stress (Figure 2B). Interestingly, both TERT mRNA levels and the total telomerase activities decreased in the absence of CIRP at both temperatures (Figure 2C–D). Importantly, when SFB-tagged sgRNA-resistant CIRP (CIRP-SFB) was ectopically expressed in the KO cell line (Figure 2B), the reduction in TERT mRNA expression and telomerase activity at different temperatures was rescued (Figure 2C–E). These findings point to CIRP as an important regulator of the telomerase at both 37°C as well as lower temperatures.
Figure 2.
CIRP is required for maintaining telomerase activities at both 32°C and 37°C. (A) Two sgRNAs respectively targeting exon 2 and exon7 of the CIRP locus were used to generate the CIRP KO HeLa cells. Genomic DNA was extracted from the KO clone for Sanger sequencing. Top, organization of the first seven exons of CIRP, with the translation start site (ATG in exon 1) and stop codon (TAG in exon 7) indicated. In expanded view is the sequence for the target region, with PAM sequences in red and sgRNA sequences in green. Based on sequencing results, one allele has a 2 bp deletion (red dashes) in exon 2 and both alleles have a 373 bp deletion between exon 5 and exon 7 (red dashes). Both mutations resulted in frame-shifts. (B) Cells from (A) were probed by Western blotting with anti-CIRP antibodies. An anti-GAPDH antibody served as a loading control. Cells from (A) that also stably expressed SFB-tagged sgRNA-resistant CIRP were similarly examined using anti-CIRP and FLAG antibodies. Cells were cultured at either 32°C or 37°C. (C) Quantification of the TERT mRNA level in CIRP KO and rescue cells. Cells were cultured at 32°C or 37°C for 48 h. (D) TRAP assays were carried out with extracts from control HeLa cells and CIRP KO and rescue cells. Cells were cultured at 32°C or 37°C for 48 h. A representative gel image of the TRAP assay is shown here. (E) Quantification of the relative telomerase activity of cells from (D) using the ImageQuant software. Error bars represent the s.d. of three independent experiments. P values were determined by Student's t-test. *P < 0.05; **P <0.01; ***P <0.001.
CIRP also has a role in circadian clock gene expression (71), and a recent study reported the regulation of telomerase activities by circadian rhythms in cultured fibroblasts (72). We therefore speculated that CIRP might modulate telomerase activities through the circadian clock. To test this, we first generated HTC75 cells, human fibrosarcoma cell line, that were knocked out for CIRP by CRISPR-Cas9 (Supplementary Figure S1A–B). The cells were cultured at either 32°C or 37°C and synchronized by serum shock. At different time points following cell cycle release, we measured total telomerase activities of the cells (Supplementary Figure S1C–D). Again, CIRP KO led to decreased telomerase activities at both temperatures. Notably, no apparent circadian rhythms of telomerase activities were observed in either control or CIRP KO cells (Supplementary Figure S1E, F), indicating that the telomerase activity was likely not regulated by the circadian clock in these cells.
CIRP knockdown reduces telomere length
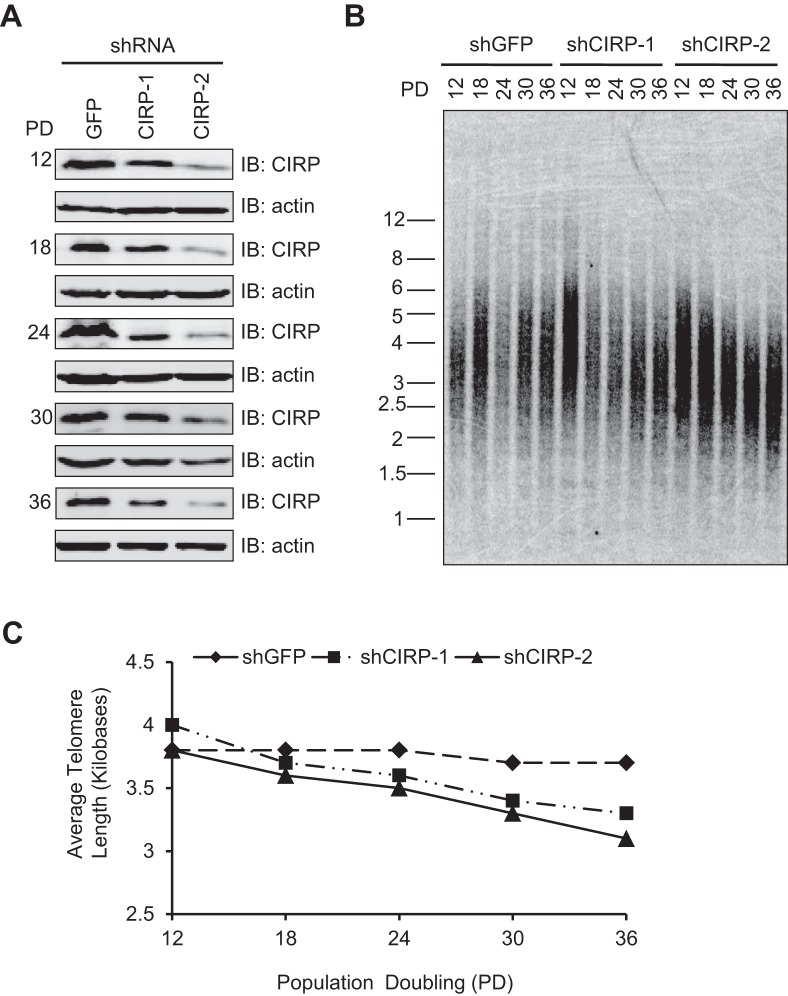
The association of CIRP with the telomerase and its likely participation in the regulation of telomerase activity implicate CIRP in telomere maintenance. To test this hypothesis, HTC75 cells stably expressing control or CIRP shRNAs were generated. Efficient long-term inhibition of CIRP expression was confirmed by Western blotting (Figure 3A). The CIRP knockdown cells were passaged over time and their average telomere length compared to control cells (Figure 3B–C). In support of our hypothesis, inhibition of CIRP indeed led to shortened telomeres compared to controls. Of the two shRNAs tested, shRNA2 appeared more efficient at knocking down CIRP. Consistent with its higher knockdown efficiency, cells stably expressing shRNA2 exhibited more accelerated telomere shortening than those expressing shRNA1, providing support to the idea that CIRP participates in maintaining telomere homeostasis.
Figure 3.
CIRP knockdown reduces telomere length. (A) HTC75 cells stably expressing different shRNA sequences were generated and passaged over time. Cells were harvest at the indicated time points for Western blots. PD, population doubling. Control, shRNA-GFP. Anti-actin antibodies were used for loading controls. (B) Telomere restriction fragment (TRF) analysis of cells from (A) that were passaged for the indicated PDs to determine average telomere length. PD, population doubling. (C) Quantification of data from (B) using the Total Lab software.
CIRP binds directly to the telomerase RNA template TERC
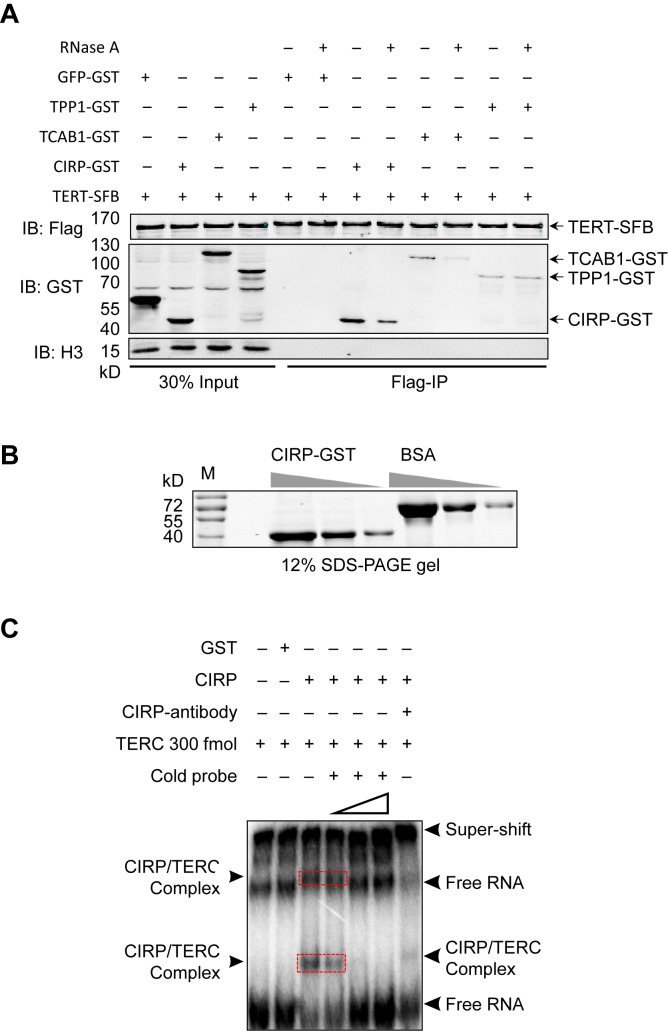
The two RNA-binding domains of CIRP, the N-terminal RRM and C-terminal RGG motifs have both been demonstrated to bind RNA in vitro and in vivo in mammalian cells (39,73). It has also been reported that under low temperature stress (cold-shock), CIRP is induced to high levels and preferentially binds to poly(U)/polypyrimidine tracks, which are located at the 3′-end of introns of mRNAs (39). We therefore speculated that CIRP might associate with TERC, the RNA component of telomerase. To explore this possibility, we first determined whether the interaction between CIRP and TERT depended on TERC. To this end, we carried out the RNase A sensitivity assay using 293T cells co-expressing TERT-SFB with CIRP-GST. Cell extracts treated with or without RNase A were then analyzed by IP-western (Figure 4A). The telomeric protein TPP1 can directly bind to TERT (74), an interaction that should not be affected by RNase A treatment. In comparison, TCAB1-TERT binding has been shown to be sensitive to RNase A treatment (75). We found that treating samples with RNase A resulted in decreased co-precipitation of TERT and CIRP, indicating decreased binding between CIRP and TERT-SFB (Figure 4A and Supplementary Figure S4A), suggesting that CIRP and TERT association might be RNA-dependent.
Figure 4.
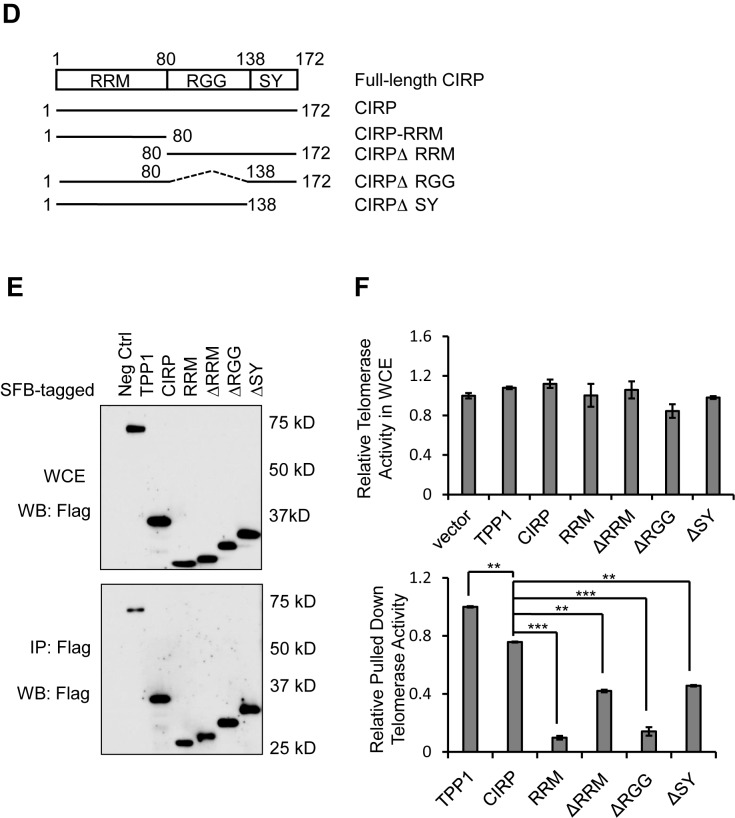
CIRP binds directly to the telomerase RNA template TERC. (A) Whole cell extracts from 293T cells transiently co-expressing SFB-tagged TERT and various GST-fusion proteins were treated with RNase A (100 μg/ml) and then incubated with anti-Flag M2 beads for pull-down assays. The precipitates were analyzed by Western blotting with the indicated antibodies. GST-tagged TCAB1 and TPP1 were used as positive and negative controls respectively. IB, immunoblotting. (B) Recombinant CIRP-GST proteins were purified from Escherichia coli and examined by SDS-PAGE and Commassie Blue staining. Different amounts of CIRP (3-fold difference between different lanes) and BSA (1, 3 and 9 μg) were loaded for comparison. (C) CIRP EMSA assays were carried out where recombinant CIRP-GST proteins (0.45 μg) were incubated with the radiolabeled TERC probe. The reaction mixtures were then resolved by non-denaturing PAGE. Increasing amounts (0.3, 3 and 6 pmol) of unlabeled TERC probes were added to reactions that contained CIRP-GST (0.9 μg) and 32P-labeled FL-TERC (300 fmol). GST alone (4 μg) served as a negative control. For supershift experiments, an anti-CIRP antibody (0.25 mg/ml) was added during the binding reaction. (D) Schematic representation of CIRP domain structure and deletion mutants. RRM, RNA binding domain. RGG, glycine-rich domain. SY, serine and tyrosine-rich domain. (E, F) SFB-tagged full-length and deletion mutants of CIRP were transiently expressed in 293T cells and immunoprecipitated using anti-FLAG antibodies. Whole cell extracts (WCE) and the immunoprecipitates were eluted for Western blotting (E) or TRAP assays (F). TPP1 and vector alone served as positive and negative controls respectively. TRAP assay results were normalized to protein amount, where error bars represent s.d. (n = 3). P values were determined by Student's t -test. *P < 0.05; **P < 0.01; ***P < 0.001.
Next, we investigated whether CIRP could bind directly to TERC in vitro. For this purpose, bacterially purified CIRP-GST proteins (Figure 4B) were used in the EMSA (76). The TERC RNA probe was in vitro transcribed and radio-labeled before being incubated with the recombinant proteins. The TERC-interacting protein DKC1 was included as a positive control (77,78). Even at very low protein concentrations (e.g. 0.45 μg), CIRP was able to shift the TERC RNA probe, resulting in slower migration forms on gels (Supplementary Figure S4B). Compared to DKC1, the CIRP-TERC complexes appeared much more heterogeneous, possibly the results of multiple binding sites. In addition, the binding of CIRP to TERC could be competed away by unlabeled TERC RNA probe in a concentration-dependent manner, and the CIRP-TERC complexes could be super-shifted with the addition of anti-CIRP antibodies (Figure 4C). These results underline the TERC-dependent association of CIRP with the telomerase in vivo and support a direct interaction between CIRP and TERC.
The RRG domain is essential for CIRP-telomerase interaction
To gain better insight into the mechanism by which CIRP binds the telomerase, we examined the regions on CIRP that could mediate such interaction. SFB-tagged full-length or truncated CIRP was transiently expressed in 293T cells to compare their telomerase binding abilities (Figure 4E). Here, anti-FLAG immunoprecipitates, which should contain CIRP and the associated telomerase, were used in TRAP assays to determine the amount of telomerase activities that came down with SFB-CIRP (30,79). As shown in Figure 4F–G, full-length CIRP was able to bring down telomerase activities at levels comparable to TPP1, a key telomerase-associating protein (80–83). Removing any of the motifs, especially the RGG domain, reduced the amount of co-precipitated telomerase activity. These data suggest that all three domains may be needed for CIRP in vivo binding to the telomerase, with the RGG domain being the most crucial (Figure 4G).
CIRP regulates telomerase activity through two different mechanisms
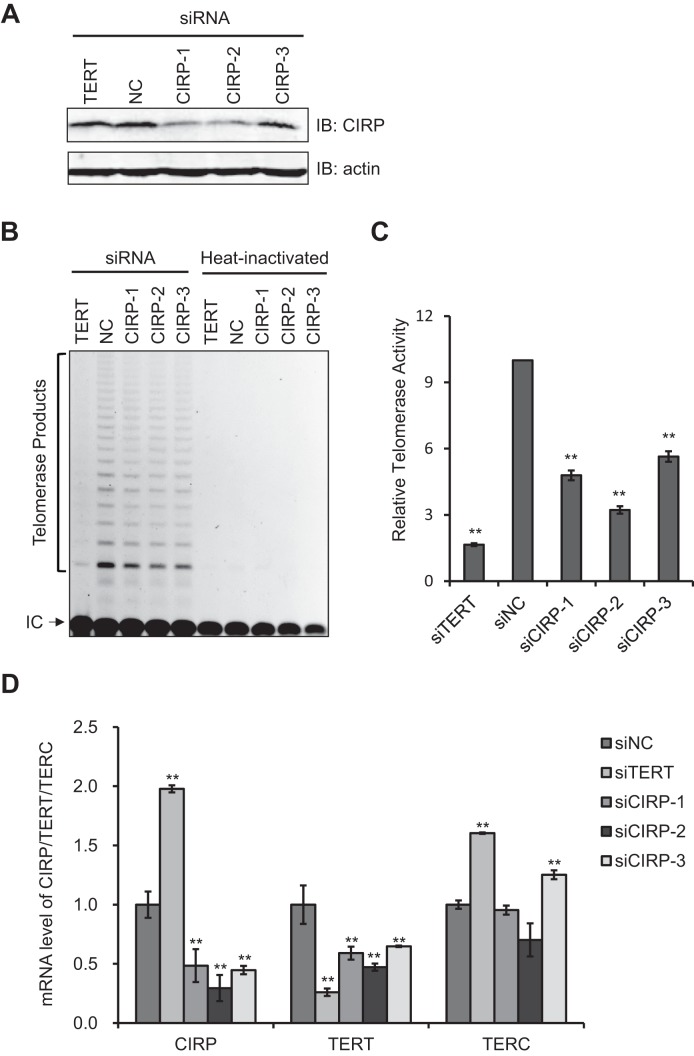
Binding of CIRP to the 3′ untranslated region of specific mRNA transcripts has been reported to stabilize the mRNAs and facilitate their transport to ribosomes (48,84). Changes in telomerase transcription, post-translational modification, assembly and telomeres recruitment can all have an impact on telomerase activity (75,85–90). We therefore first determined whether CIRP could modulate the expression of TERT and/or TERC using HTC75 cells transiently depleted of CIRP by siRNAs. All three siRNAs used were able to reduce CIRP expression, two of which achieved a knockdown efficiency of ≥70% (Figure 5A and Supplementary Figure S2A). As a control, cells were also transfected with a siRNA oligo against TERT. As expected, these cells displayed vastly reduced TERT mRNA expression and telomerase activity (Figure 5B–D). Consistent with our findings thus far, siRNA depletion of CIRP led to reduced telomerase activities in the cells as measured by TRAP assays (Figure 5B–C). In fact, the level of telomerase activity inhibition was consistent with the knockdown efficiency of the siRNA oligos. Interestingly, CIRP knockdown also significantly decreased the mRNA level of TERT without affecting overall TERC RNA levels (Figure 5D), a finding that is consistent with the reduction of total telomerase activity in CIRP knockdown cells (Figure 5C). Taken together, our findings suggest that CIRP-mediated telomerase regulation occurs at least in part through modulating TERT mRNA expression.
Figure 5.
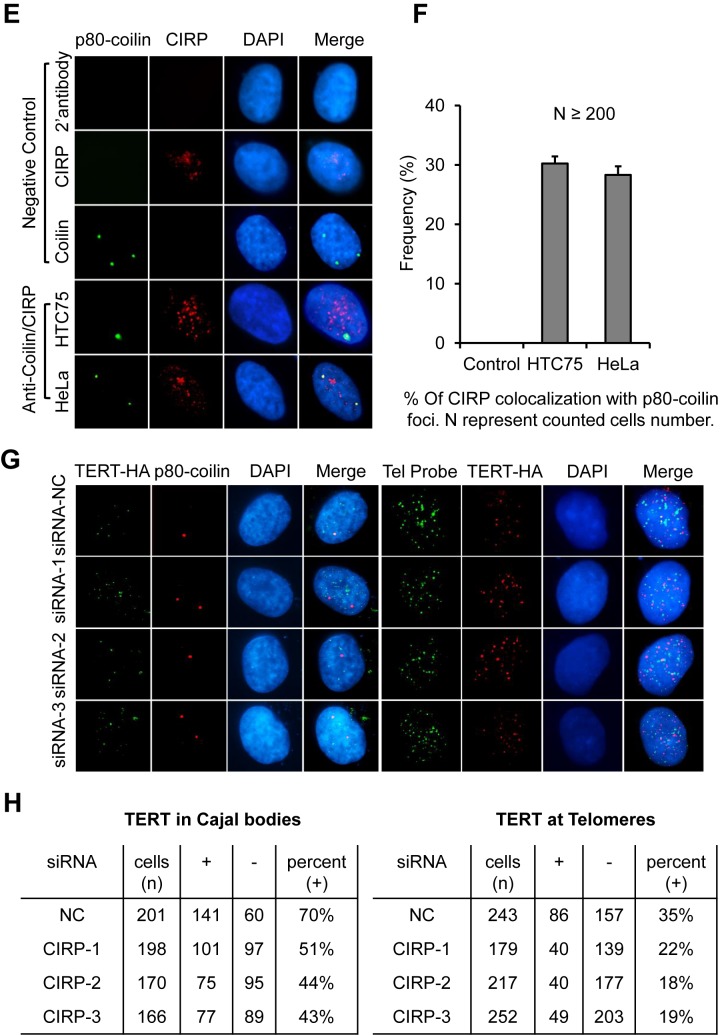
CIRP regulates telomerase activity through two different mechanisms. (A) HTC75 cells transiently transfected with siRNA oligos against CIRP were examined by WB at 48 h after transfection. A siRNA oligo against TERT served as a positive control. NC, negative control. Anti-actin blotting serves as a loading control. (B) TRAP assays were performed using cells from (A) (∼100 cells/reaction). Heat-inactivated samples served as negative controls. IC, internal control. (C) Quantification of relative telomerase activity in the whole cell extracts of cells from (B) using the ImageQuant software. Error bars represent the s.d. of three independent experiments. P values were determined by Student's t-test. *P < 0.05; **P < 0.01; ***P < 0.001. (D) Real-time qPCR assays were carried out to quantify mRNA levels of TERT, TERC and CIRP in cells from (A). Error bars represent S.E. from three independent experiments. P-values were determined by the Student t-test. **P < 0.005. (E) Immunofluorescence analysis was performed using HTC75 and HeLa cells and antibodies against endogenous CIRP (red) and p80-coilin (green). DAPI (blue) was used to stain the nuclei. (F) Data from (E) were quantified and graphed. Error bars represent S.E. from three independent experiments. At least 200 cells were examined for each sample. (G) HTC75 cells stably expressing HA-TERT were examined by immunostaining. Left, cells were stained with antibodies HA (green) and p80-coilin (red). Right, the same cells were stained with anti-HA antibodies (red) a telomere FISH probe (green). (H) Quantification of data from (G). Left, percentages of cells in which TERT co-stained with p80-coilin (+) versus those that did not (-). Right, percentages of cells in which TERT co-stained with the telomere probe (+) versus those that did not (-).
In mammalian cells, telomerase assembly and TERC maturation take place in Cajal bodies where RNP processing and DKC1 recruitment of TERC occur (75). Our findings that CIRP could bind directly to TERC and form a complex with TERT raised the possibility that CIRP might also regulate non-coding RNAs and participate in telomerase assembly and/or trafficking. When we examined endogenous CIRP subcellular localization by immunofluorescence, we found it to distribute uniformly throughout the nucleoplasm (Figure 5E). Interestingly, a percentage of CIRP foci overlapped with staining for p80-coilin, a marker of Cajal bodies (Figure 5E). Co-localization of CIRP and p80-coilin was observed in both HTC75 and HeLa cells (30% and 28%, respectively) (Figure 5F), pointing to a more general function of CIRP. We suspected that CIRP might co-localize with TERT in these cells as well. To test this idea, we generated a HTC75 cell line stably overexpressing HA-tagged TERT and assessed the localization of endogenous CIRP and HA-TERT in these cells by immunostaining. As we predicted, endogenous CIRP could indeed co-localize with HA-TERT (≥40% co-localized foci) (Supplementary Figure S2B, C). These results implicate CIRP in specifically regulating the telomerase in Cajal bodies, where the telomerase is assembled and subsequently targeted to telomeres (86,87,91).
To probe the potential function of CIRP in telomerase assembly, we examined HA-TERT localization in cells transiently depleted of CIRP by siRNAs. Immunofluorescence analysis showed that CIRP knockdown significantly reduced the percentage of cells in which TERT co-stained with the Cajal body marker coilin (Figure 5G–H). Because Cajal bodies have been implicated in targeting the telomerase to telomeres during S phase, we also determined the effect of CIRP knockdown on the telomeric localization of HA-TERT by immuno-FISH using a fluorescent telomere DNA probe and anti-HA antibodies. Again, depleting CIRP significantly reduced the presence of TERT at telomeres as well (Figure 5G–H), indicating that CIRP is important for TERT localization in Cajal bodies as well as its subsequent telomeric targeting. Together, these data reveal two different mechanisms by which CIRP regulates telomerase activities, through modulating TERT mRNA expression and telomerase assembly.
CIRP maintains telomerase activity at lower temperatures by controlling TERT mRNA
Moderately low temperatures are essential for normal spermatogenesis and fertility in both mice and humans. The telomerase is highly active in the testis during spermatogenesis (92–94). However, low temperature may pose problems for the telomerase, affecting TERT mRNA stability and telomerase assembly and function (95). Interestingly, the telomerase activity appeared very similar at 32°C versus 37°C (Supplementary Figure S1C), pointing to potentially unknown mechanisms for maintaining telomerase activities at low temperatures. Previous studies have shown that maintaining cells at 32°C can induce the expression of CIRP both in vivo and in vitro (96–98). We therefore hypothesized that CIRP might be critical for maintaining telomerase activities at hypothermia conditions.
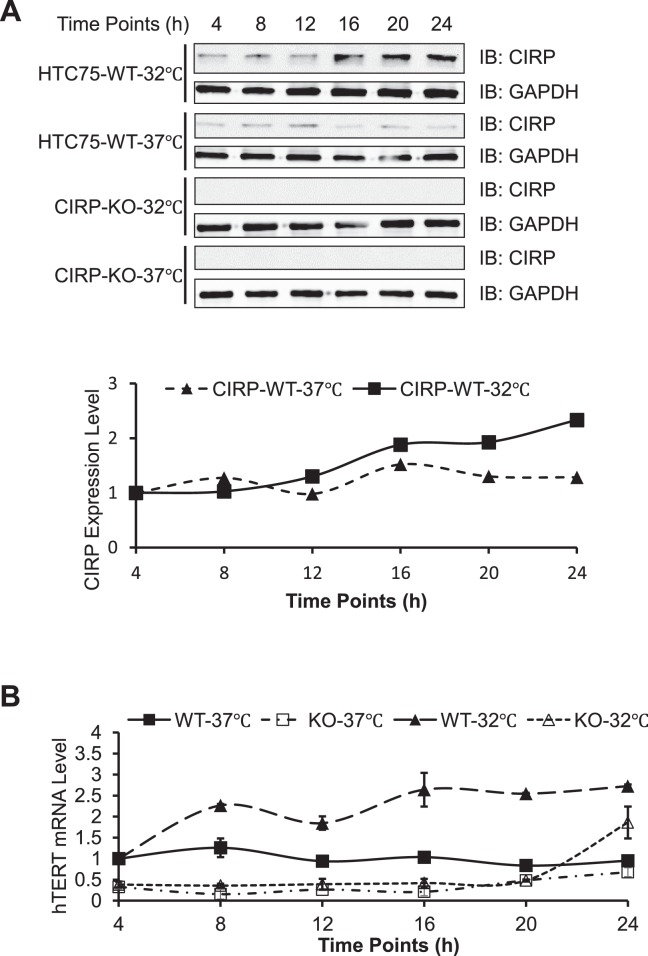
In line with previous findings, CIRP protein levels gradually increased after cells were shifted to lower temperatures (Figure 6A), where a 2-fold increase of CIRP proteins was evident by 24 h after temperature shift. Notably, the expression of TERT mRNA appeared sensitive to temperature stress and correlated with the expression of CIRP (Figure 6B). While the mRNA level of TERT was up-regulated (2- to 3-fold) in cells cultured at 32°C, the total telomerase activity remained relatively stable at 32°C versus 37°C (Supplementary Figure S1C, D). Instead of exhibiting reduced telomerase activities in response to physiological temperature decreases (95), cells appeared to up-regulate TERT expression to maintain telomerase activities at lower temperatures. Because CIRP expression directly correlated with TERT mRNA levels, we predicted that maintenance of telomerase activity at lower temperature might be regulated by CIRP. Consistent with this model, the increase in TERT mRNA level at 32°C was abolished in CIRP-KO cells, and total telomerase activities were also reduced at 32°C (Supplementary Figure S1). These results suggest a crucial role for CIRP in the regulation of TERT expression and telomerase activity, particularly at lower temperatures.
Figure 6.
CIRP is required for maintaining TERT mRNA levels at low temperatures. (A) Parental (WT) and CIRP KO (CIRP-KO) HTC75 cells were synchronized by serum withdrawal for 24 h. At different time points following release, cells were exposed to mild cold shock and maintained at 32°C. Cells that were continuously maintained at 37°C served as controls. Western blot assays (top) were subsequently performed using whole cell extracts from these cells and the indicated antibodies. The expression levels of CIRP were quantified, normalized to the level GAPDH, and plotted (bottom). (B) q-RT-PCR assays were performed using cell extracts from (A) to determine the level of TERT mRNAs. The values were then normalized to the internal control first and then to the results from 4 h after release. Error bars represent the s.d. of three independent experiments.
DISCUSSION
Telomerase regulation is critical to telomere homeostasis (1). In this study, we have identified CIRP as a telomerase- and telomere-associating factor. Our immunofluorescence experiments indicate that only a small percentage of CIRP localized to telomeres in asynchronous cells (Supplementary Figure S3), underlining the dynamic and/or transient nature of CIRP association with telomeres. We have also uncovered important clues to CIRP function in telomere maintenance. Through its interaction with the telomerase, CIRP contributes to proper telomerase assembly and telomere targeting. Disrupting CIRP-mediated telomerase control in telomerase-positive cells, either by CRISPR-Cas9 mediated KO or RNAi-mediated KD, led to compromised telomerase activities and shortened telomere. And depletion of CIRP also resulted in pronounced reduction of TERT at telomeres and in Cajal bodies. These findings have shed new light on the molecular mechanisms of telomerase and telomere regulation. In addition to directly participating in modulating telomerase assembly and targeting, we also found CIRP to regulate TERT mRNA expression. Taken together with CIRP's role in target mRNA stabilization in mouse testis (48), our data support the notion that CIRP may bind TERT mRNA through its RGG domain and stabilize TERT mRNA in a similar fashion.
It is likely that the coordinated trafficking of both TERT and TERC is required to bring the telomerase to telomeres. It is also possible that CIRP may work together with other telomerase and telomere-associated factors (e.g. TPP1). How CIRP is able to achieve its regulatory role and how CIRP establishes crosstalk with other telomere maintenance pathways warrant further investigation.
Our knockout and rescue experiments highlight the critical role of CIRP in the maintenance of TERT mRNA stability and telomerase activity, particularly at low temperatures. CIRP binds directly to TERC and CIRP KO reduced telomerase activity to 20%, suggesting CIRP is a key component of the telomerase complex. It is possible that reduced telomerase activity in the absence of CIRP might contribute to genomic instability and cell proliferation defects observed with low temperature shock (99). Moderately low temperatures in the testis are essential for normal spermatogenesis and fertility in humans and rodents. Our data suggest that CIRP may be crucial to maintain high telomerase activities in testis stem cells (100,101), counteracting potential problems in TERT mRNA stability, and telomerase assembly and function at low temperatures.
TERT expression and telomerase activities are low or undetectable in most human somatic cells (9,10). However, telomerase activation is evident in ≥85% of cancers (102,103), suggesting that upregulation of TERT expression and telomerase activation is a key step in cancer development. Telomere dysfunction has been intimately linked to cell proliferation, survival and tumorigenesis (12), and studies on the control of telomerase expression and activity are crucial to our understanding of the pathways that lead to cellular transformation and carcinogenesis. Recent evidence suggests that CIRP may act as an oncogene (104). Our identification of CIRP as a telomerase and telomere regulator provides new avenues to explore previously unknown mechanisms of telomerase control and telomere maintenance.
Supplementary Material
Acknowledgments
We would like to thank Zhen Zhang for excellent technical assistance. And we would like to thank Yujing Li, Yanyan Miao, Yan Huang and Guang Shi for their critical reading of the manuscript and helpful suggestions on experimental design and data analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Program [973 Program 2012CB911201]; National Natural Science Foundation of China [NSFC 81330055, 31171397, 31271533, 31570827,91213302 and 31371508]; GM095599, Welch Foundation Q-1673; C-BASS shared resource of the Dan L. Duncan Cancer Center [P30CA125123]. Funding for open access charge: National Natural Science Foundation of China [NSFC 31271533 and MOST 2012CB911201].
Conflict of interest statement. None declared.
REFERENCES
- 1.Blackburn E.H. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. How Telomeres Solve the End-Protection Problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrington L., Zhou W., McPhail T., Oulton R., Yeung D.S.K., Mar V., Bass M.B., Robinson M.O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kilian A., Bowtell D.D.L., Abud H.L., Hime G.R., Venter D.J., Keese P.K., Duncan E.L., Reddel R.R., Jefferson R.A. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura T.M., Morin G.B., Chapman K.B., Weinrich S.L., Andrews W.H., Lingner J., Harley C.B., Cech T.R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 6.Nandakumar J., Cech T.R. Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 2013;14:69–82. doi: 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryan T.M., Reddel R.R. Telomere dynamics and telomerase activity in vitro immortalised cells. Eur. J. Cancer. 1997;33:767–773. doi: 10.1016/S0959-8049(97)00065-8. [DOI] [PubMed] [Google Scholar]
- 8.Hiyama E., Hiyama K. Telomere and telomerase in stem cells. Br. J. Cancer. 2007;96:1020–1024. doi: 10.1038/sj.bjc.6603671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avilion A., Piatyszek M.A., Gupta J., Shay J.W., Bacchetti S., Gredider C.W. Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res. 1996;56:645–650. [PubMed] [Google Scholar]
- 10.Takakura M., Kyo S., Kanaya T. Expression of human telomerase subunits and correlation with telomerase activity in cervical cancer. Cancer Res. 1998;58:1558–1561. [PubMed] [Google Scholar]
- 11.Urquidi V., Tarin D., Goodison S. Role of telomerase in cell senescence and oncogenesis. Annu. Rev. Med. 2000;51:65–79. doi: 10.1146/annurev.med.51.1.65. [DOI] [PubMed] [Google Scholar]
- 12.Ouellette M.M. Subsenescent telomere lengths in fibroblasts immortalized by limiting amounts of telomerase. J. Biol. Chem. 2000;275:10072–10076. doi: 10.1074/jbc.275.14.10072. [DOI] [PubMed] [Google Scholar]
- 13.Bodnar A.G., Ouellette M., Frolkis M., Holt S.E., Chiu C.P., Morin G.B., Harley C.B., Shay J.W., Lichtsteiner S., Wright W.E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 14.Halvorsen T.L., Leibowitz G., Levine F. Telomerase activity is sufficient to allow transformed cells to escape from crisis. Mol. Cell. Biol. 1999;19:1864–1870. doi: 10.1128/mcb.19.3.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaziri H., Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr. Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell J.R., Wood E., Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 17.Yang D., He Q., Kim H., Ma W., Songyang Z. TIN2 protein dyskeratosis congenita missense mutants are defective in association with telomerase. J. Biol. Chem. 2011;286:23022–23030. doi: 10.1074/jbc.M111.225870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong F., Savage S.A., Shkreli M., Giri N., Jessop L., Myers T., Chen R., Alter B.P., Artandi S.E. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev. 2011;25:11–16. doi: 10.1101/gad.2006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vulliamy T., Marrone A., Szydlo R., Walne A., Mason P.J., Dokal I. Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat. Genet. 2004;36:447–449. doi: 10.1038/ng1346. [DOI] [PubMed] [Google Scholar]
- 20.Murnane J.P. Telomere loss as a mechanism for chromosome instability in human cancer. Cancer Res. 2010;70:4255–4259. doi: 10.1158/0008-5472.CAN-09-4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batista L.F., Pech M.F., Zhong F.L., Nguyen H.N., Xie K.T., Zaug A.J., Crary S.M., Choi J., Sebastiano V., Cherry A., et al. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature. 2011;474:399–402. doi: 10.1038/nature10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alter B.P., Rosenberg P.S., Giri N., Baerlocher G.M., Lansdorp P.M., Savage S.A. Telomere length is associated with disease severity and declines with age in dyskeratosis congenita. Haematologica. 2012;97:353–359. doi: 10.3324/haematol.2011.055269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartwig F.P., Collares T. Telomere dysfunction and tumor suppression responses in dyskeratosis congenita: balancing cancer and tissue renewal impairment. Ageing Res. Rev. 2013;12:642–652. doi: 10.1016/j.arr.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Pogacic E., Dragon C., Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 2000;20:9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C., Meier U.T. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23:1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu D., Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell. 2007;28:773–785. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang M., Li Y., Zhang Y., Chen Y., Huang W., Wang D., Zaug A.J., Liu D., Zhao Y., Cech T.R., et al. Disease mutant analysis identifies a novel function of DAXX in telomerase regulation and telomere maintenance. J. Cell Sci. 2014;128:331–341. doi: 10.1242/jcs.159467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D., Safari A., O'Connor M.S., Chan D.W., Laegeler A., Qin J., Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 29.O'Connor M.S., Safari A., Xin H., Liu D., Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11874–11879. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O'Connor M.S., Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 31.Marrone A., Walne A., Tamary H., Masunari Y., Kirwan M., Beswick R., Vulliamy T., Dokal I. Telomerase reverse-transcriptase homozygous mutations in autosomal recessive dyskeratosis congenita and Hoyeraal-Hreidarsson syndrome. Blood. 2007;110:4198–4205. doi: 10.1182/blood-2006-12-062851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rostamiani K., Klauck S.M., Heiss N., Poustka A., Khaleghi M., Rosales R., Metzenberg A.B. Novel mutations of the DKC1 gene in individuals affected with dyskeratosis congenita. Blood Cells Mol. Dis. 2010;44:88. doi: 10.1016/j.bcmd.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Sharma A., Myers K., Ye Z., D'Orazio J. Dyskeratosis congenita caused by a novel TERT point mutation in siblings with pancytopenia and exudative retinopathy. Pediatr. Blood Cancer. 2014;61:2302–2304. doi: 10.1002/pbc.25161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trahan C., Martel C., Dragon F. Effects of dyskeratosis congenita mutations in dyskerin, NHP2 and NOP10 on assembly of H/ACA pre-RNPs. Hum. Mol. Genet. 2010;19:825–836. doi: 10.1093/hmg/ddp551. [DOI] [PubMed] [Google Scholar]
- 35.Vulliamy T., Beswick R., Kirwan M., Marrone A., Digweed M., Walne A., Dokal I. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8073–8078. doi: 10.1073/pnas.0800042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.JR A., JR I.A., Hollander M.C. DNA damage-inducible transcripts in mammalian cells. Biochemistry. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishiyama H., Itoh K., Kaneko Y., Kishishita M., Yoshida O., Fujita J. A Glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J. Cell Biol. 1997;137:899–908. doi: 10.1083/jcb.137.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheikh M., Carrier F., Papathanasiou M.A., Hollander M.C., Zhan Q.M., Yu K., Fornace A.J., Jr Identification of several human homologs of hamster DNA damage-inducible transcripts. J. Biol. Chem. 1997;272:26720–26726. doi: 10.1074/jbc.272.42.26720. [DOI] [PubMed] [Google Scholar]
- 39.Yang C., Carrier F. The UV-inducible RNA-binding protein A18 (A18 hnRNP) plays a protective role in the genotoxic stress response. J. Biol. Chem. 2001;276:47277–47284. doi: 10.1074/jbc.M105396200. [DOI] [PubMed] [Google Scholar]
- 40.Zhou K.Z., Zheng X.M., Yang Z.W., Zhang L., Chen H.D. Overexpression of CIRP may reduce testicular damage induced by cryptorchidism. Clin. Invest. Med. 2009;32:E103–E111. doi: 10.25011/cim.v32i2.6027. [DOI] [PubMed] [Google Scholar]
- 41.Li S., Zhang Z., Xue J., Liu A., Zhang H. Cold-inducible RNA binding protein inhibits H(2)O(2)-induced apoptosis in rat cortical neurons. Brain Res. 2012;1441:47–52. doi: 10.1016/j.brainres.2011.12.053. [DOI] [PubMed] [Google Scholar]
- 42.Xue J.H., Nonoguchi K., Fukumoto M., Sato T., Nishiyama H., Higashitsuji H., Itoh K., Fujita J. Effects of ischemia and H2O2 on the cold stress protein CIRP expression in rat neuronal cells. Free Radic. Biol. Med. 1999;27:1238–1244. doi: 10.1016/s0891-5849(99)00158-6. [DOI] [PubMed] [Google Scholar]
- 43.Lee H.N., Ahn S.M., Jang H.H. Cold-inducible RNA-binding protein, CIRP, inhibits DNA damage-induced apoptosis by regulating p53. Biochem. Biophys. Res. Commun. 2015;464:916–921. doi: 10.1016/j.bbrc.2015.07.066. [DOI] [PubMed] [Google Scholar]
- 44.Liu A., Zhang Z., Li A., Xue J. Effects of hypothermia and cerebral ischemia on cold-inducible RNA-binding protein mRNA expression in rat brain. Brain Res. 2010;1347:104–110. doi: 10.1016/j.brainres.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 45.Guo X., Wu Y., Hartley R.S. Cold-inducible RNA-binding protein contributes to human antigen R and cyclin E1 deregulation in breast cancer. Mol. Carcinog. 2009;49:130–140. doi: 10.1002/mc.20582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng Y., Kulkarni P., Inoue T., Getzenberg R.H. Down-regulating cold shock protein genes impairs cancer cell survival and enhances chemosensitivity. J. Cell. Biochem. 2009;107:179–188. doi: 10.1002/jcb.22114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masuda T., Itoh K., Higashitsuji H., Higashitsuji H., Nakazawa N., Sakurai T., Liu Y., Tokuchi H., Fujita T., Zhao Y., et al. Cold-inducible RNA-binding protein (Cirp) interacts with Dyrk1b/Mirk and promotes proliferation of immature male germ cells in mice. Proc. Natl. Acad. Sci. U.S.A. 2012;109:10885–10890. doi: 10.1073/pnas.1121524109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia Z., Zheng X., Zheng H., Liu X., Yang Z., Wang X. Cold-inducible RNA-binding protein (CIRP) regulates target mRNA stabilization in the mouse testis. FEBS Lett. 2012;586:3299–3308. doi: 10.1016/j.febslet.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 49.Morf J., Rey G., Schneider K., Stratmann M., Fujita J., Naef F., Schibler U. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012;338:379–383. doi: 10.1126/science.1217726. [DOI] [PubMed] [Google Scholar]
- 50.Wan M., Qin J., Songyang Z., Liu D. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J. Biol. Chem. 2009;284:26725–26731. doi: 10.1074/jbc.M109.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mali P., Yang L.H., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan J., Ghosal G., Chen J. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol. cell. 2012;47:410–421. doi: 10.1016/j.molcel.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee O.H., Kim H., He Q., Baek H.J., Yang D., Chen L.Y., Liang J., Chae H.K., Safari A., Liu D., et al. Genome-wide YFP fluorescence complementation screen identifies new regulators for telomere signaling in human cells. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.001628. M110 001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Y., Sfeir A.J., Zou Y., Buseman C.M., Chow T.T., Shay J.W., Wright W.E. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138:463–475. doi: 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim H., Lee O.H., Xin H.W., Chen L.Y., Qin J., Chae H.K., Lin S.Y., Safari A., Liu D., Zhou S.Y. TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat. Struct. Mol. Biol. 2009;16:372–379. doi: 10.1038/nsmb.1575. [DOI] [PubMed] [Google Scholar]
- 57.Thacker J., Zdzienicka M.Z. The XRCC genes: expanding roles in DNA double-strand break repair. DNA Rep. 2004;3:1081–1090. doi: 10.1016/j.dnarep.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 58.Fujii Y., Zhao F.X., Fu S.C., Nakai N., Lai C.Y. Stable preparation of aldose reductase isoenzymes from human placenta. Protein Expr. Purif. 1991;2:420–425. doi: 10.1016/1046-5928(91)90103-p. [DOI] [PubMed] [Google Scholar]
- 59.LaBranche H., Dupuis S., Ben-David Y., Bani M.R., Wellinger R.J., Chabot B. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- 60.Dragon F., Pogacic V., Filipowicz W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 2000;20:3037–3048. doi: 10.1128/mcb.20.9.3037-3048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pogacic V., Dragon F., Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 2000;20:9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ting N.S.Y., Yu Y.P., Pohorelic B., Lees-Miller S.P., Beattie T.L. Human Ku70/80 interacts directly with hTR, the RNA component of human telomerase. Nucleic Acids Res. 2005;33:2090–2098. doi: 10.1093/nar/gki342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kiledjian M., Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ding J., Hayashi M.K., Zhang Y., Manche L., Krainer A.R., Xu R.M. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nichols R.C., Wang X.W., Tang J., Hamilton B.J., High F.A., Herschman H.R., Rigby W.F. The RGG domain in hnRNP A2 affects subcellular localization. Exp. Cell Res. 2000;256:522–532. doi: 10.1006/excr.2000.4827. [DOI] [PubMed] [Google Scholar]
- 66.Alfano C., Sanfelice D., Babon J., Kelly G., Jacks A., Curry S., Conte M.R. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 2004;11:323–329. doi: 10.1038/nsmb747. [DOI] [PubMed] [Google Scholar]
- 67.Nagata T., Suzuki S., Endo R., Shirouzu M., Terada T., Inoue M., Kigawa T., Kobayashi N., Guntert P., Tanaka A., et al. The RRM domain of poly(A)-specific ribonuclease has a noncanonical binding site for mRNA cap analog recognition. Nucleic Acids Res. 2008;36:4754–4767. doi: 10.1093/nar/gkn458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thandapani P., O'Connor T.R., Bailey T.L., Richard S. Defining the RGG/RG motif. Mol. Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 69.Ma W., Kim H., Songyang Z. Studying of telomeric protein-protein interactions by Bi-molecular fluorescence complementation (BiFC) and peptide array-based assays. Methods Mol. Biol. 2011;735:161–171. doi: 10.1007/978-1-61779-092-8_16. [DOI] [PubMed] [Google Scholar]
- 70.Cong L., Ran F.A., Cox D., Lin S.L., Barretto R., Habib N., Hsu P.D., Wu X.B., Jiang W.Y., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morf J., Rey G., Schneider K., Stratmann M., Fujita J., Naef F., Schibler U. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012;338:379–383. doi: 10.1126/science.1217726. [DOI] [PubMed] [Google Scholar]
- 72.Chen W.D., Wen M.S., Shie S.S., Lo Y.L., Wo H.T., Wang C.C., Hsieh I.C., Lee T.H., Wang C.Y. The circadian rhythm controls telomeres and telomerase activity. Biochem. Biophys. Res. Comm. 2014;451:408–414. doi: 10.1016/j.bbrc.2014.07.138. [DOI] [PubMed] [Google Scholar]
- 73.Yang R., Zhan M., Nalabothula N.R., Yang Q., Indig F.E., Carrier F. Functional significance for a heterogenous ribonucleoprotein A18 signature RNA motif in the 3′-untranslated region of ataxia telangiectasia mutated and Rad3-related (ATR) transcript. J. Biol. Chem. 2010;285:8887–8893. doi: 10.1074/jbc.M109.013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zaug A.J., Podell E.R., Nandakumar J., Cech T.R. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Venteicher A.S., Abreu E.B., Meng Z., McCann K.E., Terns R.M., Veenstra T.D., Terns M.P., Artandi S.E. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feng X., Luo Z., Jiang S., Li F., Han X., Hu Y., Wang D., Zhao Y., Ma W., Liu D., et al. The telomere-associated homeobox-containing protein TAH1/HMBOX1 participates in telomere maintenance in ALT cells. J. Cell Sci. 2013;126:3982–3989. doi: 10.1242/jcs.128512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mitchell J.R., Wood E., Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 78.Wang C., Meier U.T. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23:1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chai W.H., Ford L.P., Lenertz L., Wright W.E., Shay J.W. Human Ku70/80 associates physically with telomerase through interaction with hTERT. J. Biol. Chem. 2002;277:47242–47247. doi: 10.1074/jbc.M208542200. [DOI] [PubMed] [Google Scholar]
- 80.Nandakumar J., Bell C.F., Weidenfeld I., Zaug A.J., Leinwand L.A., Cech T.R. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012;492:285–289. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhong F.L., Batista L.F., Freund A., Pech M.F., Venteicher A.S., Artandi S.E. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012;150:481–494. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y., Chen L.Y., Han X., Xie W., Kim H., Yang D., Liu D., Songyang Z. Phosphorylation of TPP1 regulates cell cycle-dependent telomerase recruitment. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5457–5462. doi: 10.1073/pnas.1217733110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sexton A.N., Regalado S.G., Lai C.S., Cost G.J., O'Neil C.M., Urnov F.D., Gregory P.D., Jaenisch R., Collins K., Hockemeyer D. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev. 2014;28:1885–1899. doi: 10.1101/gad.246819.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brochu C., Cabrita M.A., Melanson B.D., Hamill J.D., Lau R., Pratt M.A., McKay B.C. NF-kappaB-dependent role for cold-inducible RNA binding protein in regulating interleukin 1beta. PloS One. 2013;8:e57426. doi: 10.1371/journal.pone.0057426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cong Y.S., Wright W.E., Shay J.W. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jady B.E., Richard P., Bertrand E., Kiss T. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol. Biol. Cell. 2006;17:944–954. doi: 10.1091/mbc.E05-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tomlinson R.L., Ziegler T.D., Supakorndej T., Terns R.M., Terns M.P. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tomlinson R.L., Li J., Culp B.R., Terns R.M., Terns M.P. A Cajal body-independent pathway for telomerase trafficking in mice. Exp. Cell Res. 2010;316:2797–2809. doi: 10.1016/j.yexcr.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stern J.L., Zyner K.G., Pickett H.A., Cohen S.B., Bryan T.M. Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol. Cell. Biol. 2012;32:2384–2395. doi: 10.1128/MCB.00379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kappei D., Butter F., Benda C., Scheibe M., Draskovic I., Stevense M., Novo C.L., Basquin C., Araki M., Araki K., et al. HOT1 is a mammalian direct telomere repeat-binding protein contributing to telomerase recruitment. EMBO J. 2013;32:1681–1701. doi: 10.1038/emboj.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cioce M., Lamond A.I. Cajal bodies-a long history of discovery. Annu. Rev. Cell Dev. Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- 92.Ye Z.W., Chen X.C., Ping H., Yang X.P., Yang Y., Hou L., Lu G.C. Expression of telomerase gene hTERT in testes of infertile male and its significance. Zhonghua Nan ke Xue = Natl. J. Androl. 2003;9:16–19. [PubMed] [Google Scholar]
- 93.Siderakis M., Tarsounas M. Telomere regulation and function during meiosis. Chromosome Res. 2007;15:667–679. doi: 10.1007/s10577-007-1149-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weise J.M., Gunes C. Differential regulation of human and mouse telomerase reverse transcriptase (TERT) promoter activity during testis development. Mol. Reprod. Dev. 2009;76:309–317. doi: 10.1002/mrd.20954. [DOI] [PubMed] [Google Scholar]
- 95.Sun D., Lopez-Guajardo C.C., Quada J., Hurley L.H., Von Hoff D.D. Regulation of catalytic activity and processivity of human telomerase. Biochemistry. 1999;38:4037–4044. doi: 10.1021/bi982249n. [DOI] [PubMed] [Google Scholar]
- 96.Al-Fageeh M.B., Smales C.M. Cold-inducible RNA binding protein (CIRP) expression is modulated by alternative mRNAs. RNA. 2009;15:1164–1176. doi: 10.1261/rna.1179109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saito K., Fukuda N., Matsumoto T., Iribe Y., Tsunemi A., Kazama T., Yoshida-Noro C., Hayashi N. Moderate low temperature preserves the stemness of neural stem cells and suppresses apoptosis of the cells via activation of the cold-inducible RNA binding protein. Brain Res. 2010;1358:20–29. doi: 10.1016/j.brainres.2010.08.048. [DOI] [PubMed] [Google Scholar]
- 98.Tong G., Endersfelder S., Rosenthal L.M., Wollersheim S., Sauer I.M., Buhrer C., Berger F., Schmitt K.R.L. Effects of moderate and deep hypothermia on RNA-binding proteins RBM3 and CIRP expressions in murine hippocampal brain slices. Brain Res. 2013;1504:74–84. doi: 10.1016/j.brainres.2013.01.041. [DOI] [PubMed] [Google Scholar]
- 99.Fujita J. Cold shock response in mammalian cells. J. Mol. Microbiol. Biotechnol. 1999;1:243–255. [PubMed] [Google Scholar]
- 100.Achi M.V., Ravindranath N., Dym M. Telomere length in male germ cells is inversely correlated with telomerase activity. Biol. Reprod. 2000;63:591–598. doi: 10.1095/biolreprod63.2.591. [DOI] [PubMed] [Google Scholar]
- 101.Ozturk S. Telomerase Activity and Telomere Length in Male Germ Cells. Biol. Reprod. 2015;92:53. doi: 10.1095/biolreprod.114.124008. [DOI] [PubMed] [Google Scholar]
- 102.Meeker A.K., Coffey D.S. Telomerase: a promising marker of biological immortality of germ, stem, and cancer cells. A review. Biochem. Biokhim. 1997;62:1323–1331. [PubMed] [Google Scholar]
- 103.Shay J.W., Bacchetti S. A survey of telomerase activity in human cancer. Eur. J. Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 104.Lleonart M.E. A new generation of proto-oncogenes: cold-inducible RNA binding proteins. Biochimica et Biophysica Acta. 2010;1805:43–52. doi: 10.1016/j.bbcan.2009.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.