Abstract
Large dsRNA molecules can cause potent cytotoxic and immunostimulatory effects through the activation of pattern recognition receptors; however, synthetic versions of these molecules are mostly limited to simple sequences like poly-I:C and poly-A:U. Here we show that large RNA molecules generated by rolling circle transcription fold into periodic-shRNA (p-shRNA) structures and cause potent cytotoxicity and gene silencing when delivered to cancer cells. We determined structural requirements for the dumbbell templates used to synthesize p-shRNA, and showed that these molecules likely adopt a co-transcriptionally folded structure. The cytotoxicity of p-shRNA was robustly observed across four different cancer cell lines using two different delivery systems. Despite having a considerably different folded structure than conventional dsRNA, the cytotoxicity of p-shRNA was either equal to or substantially greater than that of poly-I:C depending on the delivery vehicle. Furthermore, p-shRNA caused greater NF-κB activation in SKOV3 cells compared to poly-I:C, indicating that it is a powerful activator of innate immunity. The tuneable sequence and combined gene silencing, immunostimulatory and cytotoxic capacity of p-shRNA make it an attractive platform for cancer immunotherapy.
INTRODUCTION
Synthetic RNAs with diverse biological activities can be designed by taking advantage of the structural and genetic information encoded in their sequences. siRNA provides a good example of the combined importance of structural and genetic information in governing RNA activity: the short double-helical structure of siRNA engages the cell's RNAi machinery, while its specific nucleotide sequence determines which mRNA is targeted for destruction (1). siRNA can be combined with other RNA motifs to produce more complex structures with fine-tuned activities. For example, siRNA units have been appended to RNA motifs that can self-assemble into well-defined nanostructures (2–4), while combining RNA aptamer sequences with siRNA can promote cellular targeting (5–7). Other modifications to siRNA have been used to link it together and increase its length, which can influence its physicochemical and biological properties by affecting intracellular processing, stability and immunogenicity (8–10).
Rolling circle transcription (RCT) provides a simple mechanism for amplifying circular DNA into complementary RNA up to ∼100× the length of the DNA template (11,12). This is analogous to a concatenation operation that pastes the sequence encoded in the circular DNA template into a linear, periodic RNA string. RCT from dumbbell templates–double stranded DNA sequences flanked on both sides by single stranded loops—can yield multiple siRNA/miRNA sequences linked together by single-stranded regions (13,14). These molecules could potentially fold into a periodic-shRNA (p-shRNA) structure that may resemble primary-miRNA; however, the folded structure of these molecules had yet to be elucidated.
The structural properties of p-shRNA could lead to biological activities in addition to gene silencing. Large dsRNA molecules are potent activators of the innate immune system through pattern recognition receptors (PRRs), and have shown promising anticancer activities arising from direct cytotoxicity to cancer cells and immunostimulatory effects (15,16). The typical dsRNA molecules used in these studies have simple sequences, like poly-I:C or poly-A:U, and thus do not encode genetically relevant information. On the other hand, modifications to siRNA, like the addition of a 5′-PPP or increasing its length through complementary overhangs, have yielded molecules capable of both immunostimulation and gene silencing (10,17,18). We hypothesized that the size and structure of p-shRNA would make it immunostimulatory, providing cytotoxicity against cancer cells independent from its capacity for RNAi.
Herein, we sought to better control and understand the synthesis and structural properties of p-shRNA molecules and test their cytotoxicity against a panel of cancer cell lines. We explored different modifications to the DNA template design and found parameters that influenced the productivity of the RCT reaction; we also studied the structure of p-shRNA through folding models, circular dichroism (CD) and enzymatic digests, which suggested that p-shRNA co-transcriptionally folds into a periodic hairpin structure. Compared to poly-I:C, p-shRNA showed equal or greater cytotoxicity against a panel of cancer cell lines when delivered with common transfection reagents (Lipofectamine 2000® and TransIT-X2®), induced greater NF-κB nuclear translocation in SKOV3 cells, and caused gene silencing. These findings suggest that p-shRNA is both a potent cytotoxic agent and innate immune activator capable of inducing RNAi.
MATERIALS AND METHODS
General
All reagents used for in vitro transcription were purchased from New England Biolabs (NEB). The transfection reagents used in this study were either Lipofectamine® 2000 (Life Technologies) or TransIT-X2® (Mirus). The poly-I:C used for all experiments was poly(I:C) HMW from Invivogen, which has an average size of 1.5–8 kb. Nucleic acid ladders were purchased from NEB and all gels were stained with GelRed (Biotium) for visualization and quantification. RNA quantification was carried out by UV-260 absorbance (Nanodrop) and Quant-iT Ribogreen assay (ThermoFisher). Antibodies used were NF-κB p65 (Abcam ab32536) and Alexa 488-labeled goat anti-rabbit 2° antibody (Thermo A-11008). Nuclear staining was carried out with NucBlue (Molecular Probes).
Circular DNA template design and synthesis
ssDNA oligos with a 5′-phosphate modification were purchased from Integrated DNA Technologies with polyacrylamide gel electrophoresis (PAGE) purification. For a complete list of template sequences, please refer to the Supplementary Information (Supplementary Table S1). All templates were designed to fold into dumbbell structures with the 5′ and 3′ ends within the ds-region. For circularization, the templates were first diluted to 3 μM in 1× quick ligation buffer (NEB) and heated to 95°C for 2 min followed by a gradual cooling to 25°C (∼1°C/min) to facilitate folding. T4 DNA ligase was next added to the DNA solution at a ratio of 400 U/ 0.4 nmol DNA, and the ligation reaction was left at 25°C for 2 h. Ligation was confirmed by denaturing PAGE (15% tris-borate-EDTA (TBE)-urea); the circularized templates were immediately used for RCT without purification.
Cell culture
Green fluorescent protein (GFP)-expressing HeLa and A549 cell lines were purchased form CellBioLabs and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Luciferase-expressing SKOV3 and UCI101 ovarian cancer cells were kind gifts from Drs. Lorenzo Ceppi, Wei Wei and Michael Birrer (Massachusetts General Hospital). These were maintained in RPMI with 10% FBS, 1% penicillin/streptomycin and 40 μg/ml blasticidin for selection. All cells were maintained at 37°C in a 5% CO2 humidified atmosphere.
Rolling circle transcription and isolation of p-shRNA
Optimized reaction conditions for RCT were found by varying the concentrations of nucleotide triphosphates (NTPs), circular DNA and T7 RNA polymerase according to a central composite design. Optimal conditions were determined by response surface modeling carried out in JMP Pro 11; these conditions were then used for all subsequent experiments (for additional details see the Supplemental Information). The optimized RCT reaction conditions for a typical 200 μl reaction were as follows: 120 μl of the DNA ligation reaction (3 μM DNA) was combined with 20 μl of 10× RNAPol reaction buffer, 12.5 μl of 100 mM mixed NTPs, 40 μl of T7 RNA polymerase (50 000 U/ml) and 7.5 μl of RNase-free water. The reaction mixture was incubated for 48 h at 37°C before quenching with 20 μl of 0.5 M ethylenediaminetetraacetic acid (EDTA) (this step also dissolves magnesium pyrophosphate/RNA microsponge particles, releasing all RNA into the solution (19)); 20 min later, the RNA was isolated by pressure-driven ultrafiltration (200 kDa MWCO), washed 3× with 1 ml of RNase-free water and resuspended in 200 μl of RNase-free water. p-shRNA concentration, quality and size distribution were determined by UV-absorbance (Nanodrop) and gel electrophoresis (1.5% agarose, 1× TBE). Denaturing gel electrophoresis was performed by heating the RNA samples in formamide-containing buffer (RNA loading buffer, NEB) for 10 min at 70°C then loading onto either a formaldehyde-agarose gel (1.5% agarose, 1× MOPS buffer, 7% formaldehyde) or a 15% PAGE-Urea gel (Bio-Rad).
Circular dichroism
CD was measured with an Aviv Model 202 CD spectrometer. RNA samples were dissolved at 60 ng/ml in tris-EDTA (TE) buffer and spectra were collected from 200–300 nm with a 1 nm bandwidth and 5 s averaging. After collecting an initial spectrum at 25°C, the CD signal at 210 nm was monitored while heating to 95°C (CD signal measured at 2°C intervals with 0.25 min equilibration time). A final spectrum was then collected at 95°C.
Enzymatic digests and serum stability
RNase I digests were performed with recombinant RNase If (New England Biolabs). p-shRNA samples were dissolved at a final concentration of 60 ng/μl in the recommended buffer (NEBuffer 3) and sonicated for 5 min prior to the addition of 1 μl of enzyme solution (50 000 Units/ml). The reaction was incubated at 37°C for 10 min, then inactivated through the addition of 3 μl of 0.5 M EDTA and heating for 10 min at 70°C. For analysis, 10 μl aliquots from each reaction were run on a non-denaturing 15% TBE-PAGE gel for 1.5 h at 80 V, stained with GelRed® (Biotium) and imaged under UV illumination.
RNase T1 digests were performed using recombinant RNase T1 (New England Biolabs) following a similar procedure except that 50 mM Tris buffer containing 2 mM EDTA was used with 7 Units of enzyme and the digests were carried out for 5 h at 37°C. For analysis, 10 μl aliquots were immediately loaded onto a non-denaturing PAGE gel (15% TBE), or for denaturing PAGE analysis, samples were mixed with formamide-containing denaturing buffer and heated to 70°C for 10 min prior to loading onto a denaturing PAGE gel (15% TBE-urea). Band quantification was performed using Image J and the ratios of large to small fragments are reported as the average of three experiments ± s.e.m.
Serum degradation studies were carried out using 50% human serum (Corning Cellgro). p-shRNA or siRNA samples were first diluted to 25 ng/μl in phosphate buffered saline (PBS) then mixed with an equal volume of human serum and incubated at 37°C. Aliquots were removed at the indicated time points, combined with RNaseOut (to inactivate serum nucleases) and stored at −80°C until analysis. Samples were run on a non-denaturing PAGE gel (15% TBE) at 80 V for 1.5 h, stained with GelRed and imaged under UV illumination. Band quantification was performed using Image J and half-lives were calculated by fitting an exponential decay function in Prism. An additional gel showing a serum only control is provided in the Supplemental Information (Supplementary Figure S6C).
Structural prediction
Folding predictions were performed using 2–6 repeat sequences of p-shRNA, assuming that transcription was initiated at one of the single-stranded loops. The minimum free energy (MFE) and co-transcriptionally folded structures were determined using the RNAfold (20) and CoFold (21) web servers, respectively. Both predictions were carried out using the energy/folding parameters indicated in the text. 3D models based on MFE or co-transcriptional folding were generated using RNAComposer (22) and measurements/rendering were performed using PyMol.
p-shRNA complexation with transfection reagents
Complexation with Lipofectamine® 2000: RNA was dissolved in Opti-MEM at a concentration of 6 μg/ml and combined with Lipofectamine® 2000 reagent dissolved in an equal volume of Opti-MEM (1.7 μl of Lipofectamine/μg RNA). After mixing, the sample was allowed to sit for 15 min at room temperature prior to use, giving a final RNA concentration of 3 μg/ml.
Complexation with Mirus TransIT-X2®: TransIT-X2® reagent was added to a 5.5 μg/ml solution of RNA in Opti-MEM at a ratio of 3.7 μl of reagent/μg of RNA. The solution was mixed thoroughly and allowed to sit for 25 min before diluting with Opti-MEM to a final RNA concentration of 3 μg/ml. Lipofectamine® and TransIT-X2® only samples were both prepared following the above procedures but excluding RNA.
Cell viability and knockdown assays
Cell viability: experiments were carried out 24 h after seeding cells in a 96-well plate with 100 μl of complete DMEM (10% FBS), at which point the cells were ∼50% confluent. Complexes formed in serum-free Opti-MEM were added to the cells at volumes ranging from 1 μl to 100 μl (each condition was performed in triplicate); following the addition of RNA, well volumes were all adjusted to 200 μl final volume with Opti-MEM and left to incubate for 48 h (final [FBS] = 5%). Cell viability was then assayed with CellTiter-Glo reagent (Promega) according to the manufacturer's instructions and IC50 values were calculated by fitting a 3-parameter log-logistic model to the dose-response data via the DRC package in R (23).
Caspase activity: caspase activation was measured using the Caspase-Glo 3/7, Caspase-Glo 8 and Caspase-Glo 9 assay systems (Promega). Cells were incubated with p-shRNA or poly-I:C/Lipofectamine complexes for 14 h prior to measuring caspase activity according to the manufacturer's protocol.
GFP knockdown: cells were prepared in a 96-well plate as described above. RNA complexes formed in Opti-MEM were added to the cells to give a final RNA concentration of 10 or 30 nM (with respect to dsRNA units; this corresponds to ∼180–540 ng/ml RNA); after 24 h, the media was replaced with fresh DMEM and the cells were left for an additional 48 h before analyzing reporter gene expression (i.e. 72 h after adding RNA). GFP was analyzed by flow cytometry using a FACSCalibur flow cytometer equipped with a high-throughput sampler (λEx = 488 nm; λEm = 530/30 nm). To prepare the cells for flow cytometry, they were first washed with PBS, disassociated with trypsin and resuspended in DMEM containing 10% FBS. The resulting data was analyzed using FloJo and presented as mean fluorescence averaged over three independent samples, normalized to the average fluorescence of untreated cells.
Luciferase knockdown: cells were prepared in a 96-well plate as described above. RNA complexes formed in Opti-MEM were added to cells to give a final concentration of 15 nM and the medium was replaced with fresh RPMI medium 24 h after transfection. After an additional 48 h, the cells were lysed with 75 μl 1× Glo Lysis Buffer (Promega). For each well, a 25 μl aliquot of the cell lysate was transferred to another 96-well plate and assayed for total protein content with the Pierce BCA Protein Assay Kit. Luciferase expression in the remaining cell lysate was measured with the Steady-Glo luciferase assay system (Promega) using a Tecan Infinite M200 Pro 96-well plate reader. Luminescence readings were first normalized to total protein and presented as the mean for three independent samples normalized to the corresponding value for untreated cells.
NF-κB nuclear translocation assay
p-shRNA or poly-I:C/Lipofectamine complexes were added at varying concentrations to SKOV3 cells in a 24 well plate (∼50% confluent) 24 h after seeding. After 1 h, the media was removed and cells were fixed with 3.7% formaldehyde in PBS (10 min), permeabilized with 0.1% Triton-X in PBS (5 min), incubated with 1° NF-κB antibody (350× dilution) for 1 h at RT, then stained with 2° antibody (500× dilution) for 1hr in the dark at RT. During the final 10 min of staining, DAPI (NucBlue) was added to each well (one drop/well) prior to washing the cells with PBS and adding mounting medium. Cells were imaged on an Olympus IX51 epifluorescence microscope using appropriate filter sets for DAPI and AF488. Image analysis was carried out using Cell Profiler (24).
Statistical analysis
All pairwise statistical comparisons were made using a two-sided Welch's test. A P-value cutoff of 0.05 was used for determining statistical significance. Where appropriate, P-values were corrected for multiple comparisons using Holm's method. Results are presented as mean or geometric mean ± s.d., s.e.m., or 95% C.I., as specified in the figure legends.
RESULTS
p-shRNA synthesis
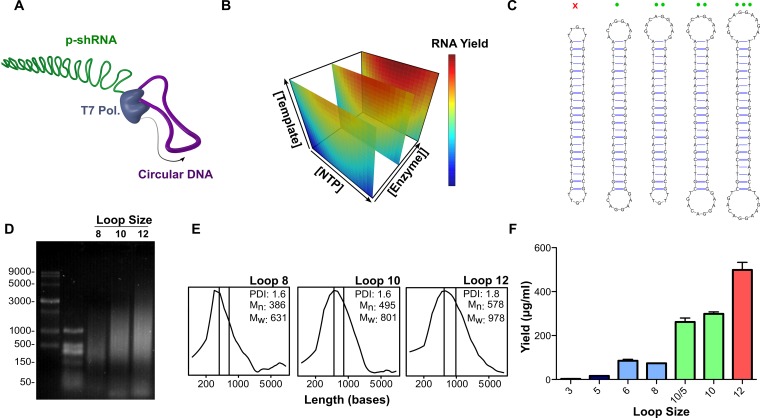
A schematic of the RCT reaction used in this study is illustrated in Figure 1A. We adopted a dumbbell template design (13) predicted to introduce alternating double-stranded and single-stranded regions into the transcribed RNA. We first optimized the RCT reaction from a DNA template with two equivalent 10-base loops (template 6: loop sequence corresponds to the miR-23a loop) and a 21 bp ds stem by varying the concentrations of T7 RNA polymerase, DNA and NTPs according to a central composite design screen. The response surface model generated from the screen identified that [Enzyme], [Template] and [NTP]*[NTP] significantly influenced (P < 0.05) the reaction yield (Figure 1B; for further details see the Supplemental Information, Tables S2-S3 and Figure S1). The second-order response to [NTP] was confirmed in a follow-up experiment, where [NTP] was varied from 2–10 mM at a constant concentration of template and enzyme, which demonstrated inhibition of the RCT reaction at [NTP] greater than ∼7 mM (Supplementary Figure S2).
Figure 1.
Optimization of rolling circle transcription (RCT) reaction. (A) Schematic representation of RCT from a dumbbell-shaped circular DNA template. (B) Response surface model of RNA yield from RCT as a function of template, NTP and T7 RNA polymerase concentrations. RNA yield increased linearly as a function of template and polymerase concentrations, but showed a second order response to NTP concentration, with a maximum yield at ≈7 mM total NTPs. (C) RCT was carried out from a series of DNA templates with variable loop sizes. The red x indicates that no RCT occurred from the template with two 5-base loops; green circles indicate RCT yield increased as a function of loop size for loops >5 bases. (D) Denaturing agarose gel electrophoresis (1.5%, MOPS, 7% formaldehyde) of RCT reaction products obtained from the templates with 8-base, 10-base, and 12-base loops (templates 4, 6 and 11). (E) Size distributions with corresponding number avg. molecular weight (Mn), weight avg. molecular weight (Mw) and PDI, calculated from the gel in (D). (F) Quant-iT Ribogreen quantification of the RNA yield from the different dumbbell templates. Yields are reported as the mean with s.d. for two independent reactions.
To assess the scope of useable dumbbell templates for RCT, we varied the lengths and sequences of both the single-stranded and double-stranded regions of DNA dumbbells and analyzed the RNA products by gel electrophoresis and Ribogreen assays after 24 h in vitro transcription reactions (Figure 1C–F; for a list of all sequences used in this study see the Supplemental Information, Table S1). We initially investigated a series of templates containing a constant 21 bp double-stranded stem corresponding to an siGFP sequence, and loops with variable lengths ranging from 3 to 12 bases (templates with 3- and 6-base loops had one additional base pair). These loop sequences were derived by adding or subtracting bases to the 10-base miR-23a loop sequence while avoiding the introduction of secondary structure. Dumbbell templates that contained two identical loops shorter than 6 bases did not yield any concatenated RNA product (Figure 1F and Supplementary Figure S3). We observed trends of increasing RNA size and yield with increasing loop size, with the average size for the RNA corresponding to 6–10 repeats based on denaturing agarose (formaldehyde/MOPS) gel analysis (Figure 1D–F and Supplementary Figure S3). A similar trend was previously reported for transcription from unstructured circular templates, where an increase in transcript size/yield was observed in going from a 13- to 18-base circular template with T7 RNA polymerase (25).
To test whether the effect of loop size on RCT productivity was primarily due to transcription initiation or elongation, we carried out RCT from an asymmetric DNA dumbbell containing 5- and 10-base loops (GFP-5/10). RCT from GFP-5/10 yielded only slightly less RNA than the symmetric template with two 10-base loops (262 versus 299 μg/ml), with a similar RNA size distribution (Figure 1D–F); a single loop of 5 bases is thus tolerated during the elongation phase of transcription, suggesting that loop size mainly influences transcription initiation.
Transcription initiation by T7 RNA polymerase is known to heavily favor incorporating GTP as the first nucleotide (26). We therefore tested whether the presence of Cs in the template loops had an influence on transcription initiation. We compared five DNA dumbbell templates with 10 base loops containing either no Cs, 1 C in one of the loops, 1 C in both loops (i.e. the original loop sequence), 5 Cs in both loops and 10 Cs in both loops (i.e. all Cs). Agarose gel electrophoresis of the products from these templates showed no difference in size distribution when comparing 0–1 C in the loop sequence; however, a notable decrease in transcript size was observed upon increasing to 5 Cs in the loops, and the 10-C template gave only a short transcript at roughly 100 bases (Supplementary Figure S4A). We also compared the yields from these different templates, and found minimal differences for loops with 0–5 Cs, but a significant drop-off in yield (>50%) for the 10-C loop (Supplementary Figure S4B).
We also investigated the influence of the double-stranded stem length on the yield of RCT. It was previously reported that a stem length of 29 bp required the introduction of mismatches for efficient transcription, whereas stems of ≤25 bp gave efficient transcription without mismatches (13). We confirmed these findings and observed similar p-shRNA products from templates with 25 and 27 bp stems (with 10-base loops; templates 12–16) compared to the analogous 21 bp template, with the introduction of three mismatches appearing to improve the reaction yield. In contrast, we observed a sharp drop-off in yield for a template containing a 29 bp stem with three mismatches (Supplementary Figure S5).
p-shRNA structure and stability
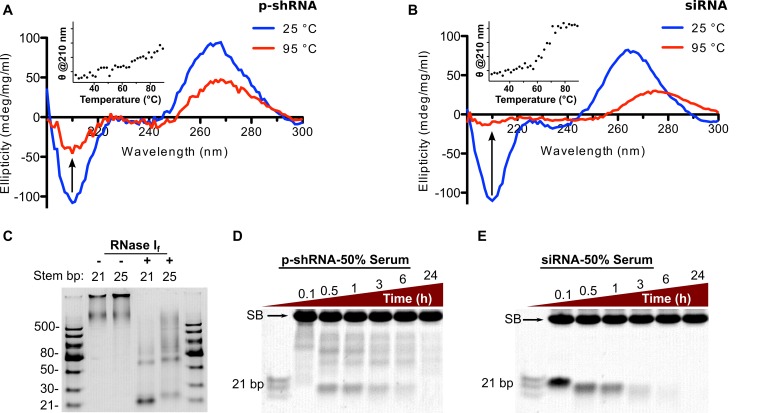
We characterized the secondary structure of the RNA molecules produced by RCT using CD and enzymatic digests, focusing on RNA produced from template 6 (GFP-10a). Figure 2A and B shows CD spectra for p-shRNA and a 21 b siRNA for comparison. The spectra for the two samples have similar shapes and normalized peak intensities, which are consistent with the A-form double helical structure expected for dsRNA. However, differences between the p-shRNA and siRNA spectra were observed upon heating the samples to 95°C. As expected, the siRNA sample underwent a sharp melting transition at ∼65°C (monitored at 210 nm) (27); in contrast, p-shRNA showed a linear decrease of CD signal with increased temperature, and appeared to retain some of its secondary structure up to 95°C. Digesting p-shRNA with RNase I, which is selective for ssRNA, yielded products consistent with the predicted secondary structure of alternating single-stranded and double-stranded regions. p-shRNAs predicted to have 21 and 25 bp double-strand regions (p-shRNA-21 and p-shRNA-25; synthesized from templates 6 and 12) were treated with RNase I: following 15 min, we observed prominent bands at 21 and 25 bp, respectively (Figure 2C). Treatment of the 21 bp p-shRNA with 50% human serum resulted in a similar banding pattern compared with RNase I treatment. Figure 2D shows the appearance of a prominent 21 bp band–along with other larger fragments–within 30 min; the 21 bp band subsequently decreased in intensity over the course of the experiment. Degradation half-lives were calculated for siRNA and the 21 bp fragment derived from p-shRNA by image densitometry, which gave values of 2.0 and 5.0 h, respectively (Supplementary Figure S6). Although the 21 bp p-shRNA fragment is expected to degrade at a similar rate as 21 bp siRNA, p-shRNA must first be broken down from larger fragments: these appear to be degraded more slowly than siRNA, and thereby prolong the effective serum half-life of p-shRNA compared to siRNA.
Figure 2.
Characterizing the secondary structure and stability of p-shRNA. (A and B) CD spectra of p-shRNA (21 bp stem/10 base loops) and siRNA (21 bp with same sequence) measured at 25 and 95°C. The insets plot the CD signal at 210 nm as a function of temperature. (C) p-shRNAs with 21 or 25 bp stems (from templates 6 and 12, respectively) were treated with RNase I for 15 min then run on a native 15% TBE-PAGE gel. The observed banding pattern confirms the predicted alternating single-/double-stranded structure of p-shRNA. (D and E) p-shRNA (21 bp stem/10 base loops; from template 6) and siRNA (21 bp) were treated with 50% human serum for 0.1–24 h and run on a native 15% TBE-PAGE gel (SB indicates background serum band). Ladders = dsRNA ladder (NEB) and siRNA marker (NEB).
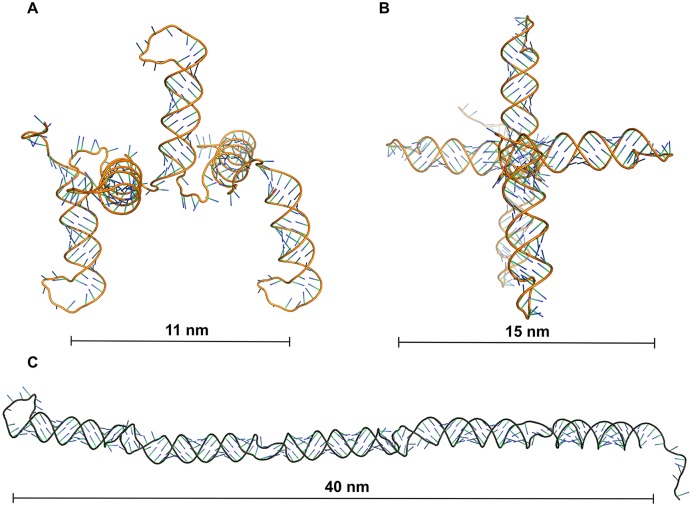
p-shRNA molecules can potentially fold into several different structures that are consistent with the above results. The predicted MFE structure for p-shRNAs involves the entire molecule folded back on itself, as depicted in Figure 3C; however, the realization of this MFE structure is unlikely since p-shRNA folding is expected to occur co-transcriptionally. We used CoFold–a free energy minimization algorithm that takes folding kinetics into account–to model p-shRNA folding (21). Folding predictions were performed for 2–6 repeats of a p-shRNA sequence with symmetric 10-base loops while varying the folding parameter τ (lower values of τ place a greater emphasis on kinetic control of folding) and using the Turner (1999) energy parameters. The simple periodic hairpin structure shown in Figure 3A was predicted across a wide range of τ values including the default value of 640, which was previously optimized for predicting the folded structure of large RNAs (21). We note that this structure is predicted to be only ∼1 kcal/mol/repeat higher in energy than the MFE prediction using the same energy parameters. The 3D structures for the periodic hairpin and MFE structures are unique in their dimensions, with five repeats of the co-transcriptionally folded structure measuring 11 × 15 nm compared to 40 × 3 nm for the MFE structure. Notably, the MFE structure appears similar to a conventional dsRNA, whereas the co-transcriptionally folded structure is considerably different. Increasing τ to roughly double or triple the default value gave structures with double- and triple-hairpins through further back-folding of the RNA molecule. This suggests that the simple periodic hairpin arrangement likely predominates, but a mixture of further back-folded structures could be present depending on the rates of transcription and folding.
Figure 3.
Comparison of 3D models of p-shRNA fragments based on co-transcriptionally folded (kinetic) or minimum free energy (MFE) secondary structures. (A and B) Side and front views of a 3D model based on the secondary structure of p-shRNA (5 repeats) predicted to form through co-transcriptional folding. (C) Side view of a 3D model based on the predicted MFE structure of p-shRNA (5 repeats). Co-transcriptional and MFE secondary structures were predicted using CoFold and RNAFold, respectively. 3D models were generated using RNAComposer and rendered with PyMol.
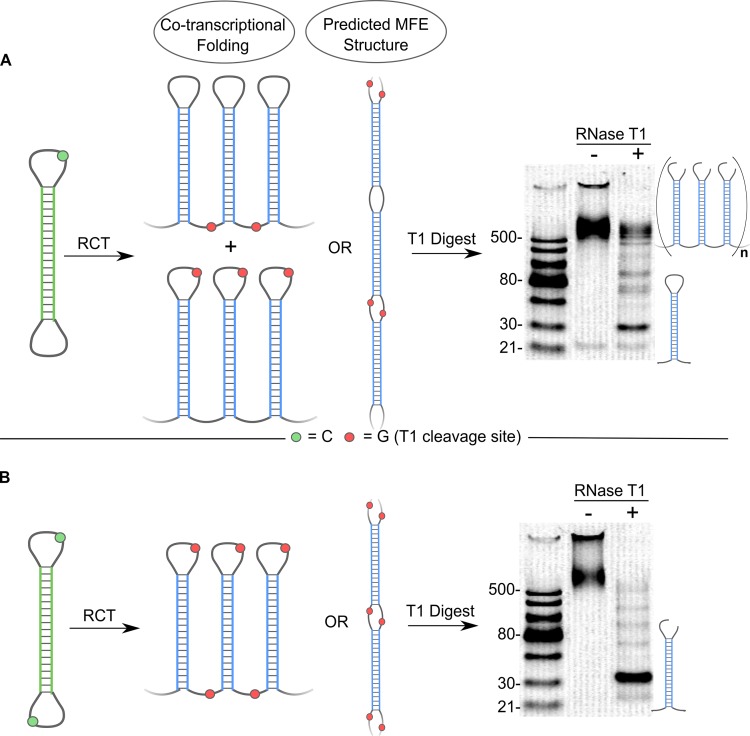
RNase T1–which selectively cleaves ssRNA 3′ to guanines–was next used to distinguish between the different predicted folded structures of p-shRNA. We used a DNA template with two different 10-base loops–one containing a single cytosine and the other lacking cytosines (template 7)–to give p-shRNA with one loop containing a single guanine and the other loop without any guanines. Since this template had equally sized loops with nearly identical sequences, we predicted that transcription would initiate randomly from either loop leading to the two co-transcriptionally folded structures shown in Figure 4A. Treatment of these RNA products with RNase T1 would then either cleave at the top of the hairpin loop or at the bottom of the stem, theoretically yielding a 1:1 mixture of intact p-shRNA and a band near 30 bp under non-denaturing conditions. In contrast, periodic double- or triple-hairpin structures would lead to a larger fraction of ∼60 bp fragments and smaller amounts of single hairpin fragments and ‘trimmed’ periodic RNA; RNase T1 treatment of the predicted MFE structure should lead almost entirely to ∼60 bp fragments. The results of the experiment are shown in Figure 4 and agree well with the predictions for the simple co-transcriptionally folded structure. Following treatment with RNase T1, we observed a prominent band at 30 bp (corresponding to a single hairpin), two faint bands at ∼60 and 90 bp (corresponding to two/three hairpins), and a group of bands >500 bp that migrated alongside the untreated sample. Integration of these bands (>500 versus 30/60/90 bp bands) gave close to the predicted 1:1 ratio (measured = 0.9 ± 0.2). An otherwise identical RNA with a G in both loops (from template 6) was almost completely broken down into 30 bp fragments under the same conditions; some higher order bands were again apparent, suggesting that these are the result of incomplete digestion (Figure 4B). Denaturing PAGE gels of the above products were virtually identical, both showing one major band corresponding to the length of a single hairpin repeat (Supplementary Figure S7); this result confirms that the apparently intact p-shRNA in Figure 4A was indeed nicked. Lastly, we attempted to convert the p-shRNA from Figure 4A to a lower energy structure by heating to 95°C and slowly cooling back to room temperature. Treating this refolded p-shRNA with RNase T1 gave a similar banding pattern as before, however, the relative intensity of the intact p-shRNA band compared to the shorter bands was around three times less than observed for the original p-shRNA sample (ratio = 0.3 ± 0.1, Supplementary Figure S8). While this result is inconsistent with the predicted MFE structure, it suggests that some structural rearrangement of p-shRNA occurred upon heating (see Supplemental Information for further discussion).
Figure 4.
Distinguishing between kinetic and thermodynamic folded structures of p-shRNA. (A) Schematic and native PAGE gel (15% TBE) for T1 digest of p-shRNA derived from template 7 (one C in one loop). The gel shows prominent bands corresponding to knicked, intact p-shRNA (>500 bp) and a single hairpin unit (∼30 bp). (B) Schematic and native PAGE gel (15% TBE) for T1 digest of p-shRNA derived from template 6 (one C in both loops). The gel shows a single prominent band at ∼30 bp following the T1 digest, corresponding to a knicked single hairpin unit of p-shRNA. Ladders = dsRNA ladder (NEB).
Cytotoxicity, silencing activity and immunostimulation
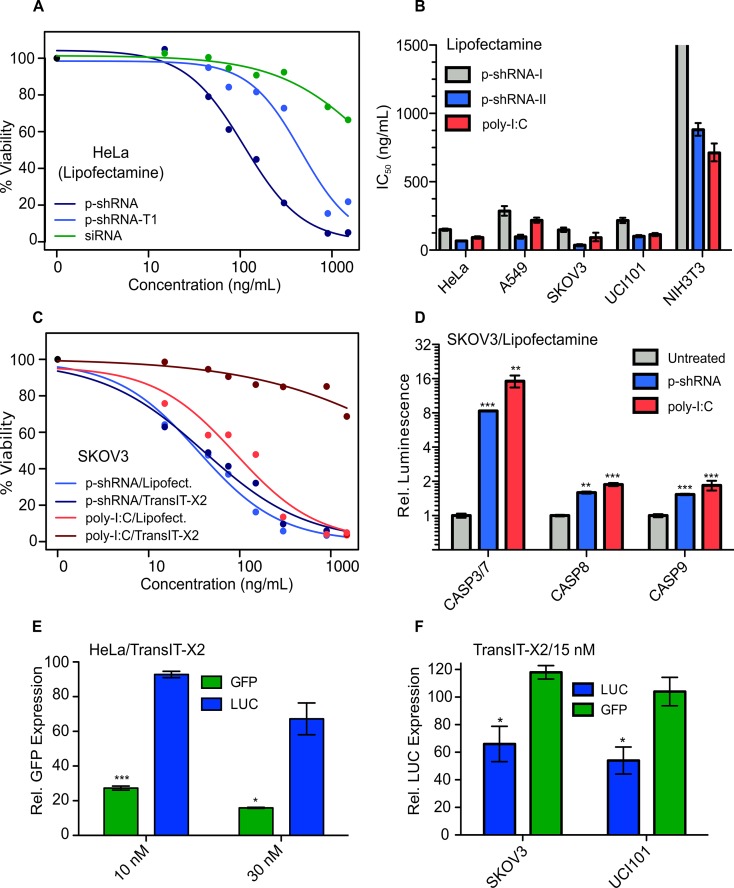
The toxicity of large dsRNA in cancer cells has been attributed to TLR3 (28,29) and the cytosolic PRRs RIG-I/MDA5, particularly when dsRNA is delivered with transfection reagents (18,30–32). We first tested the cytotoxicity of p-shRNA in HeLa cells when delivered with Lipofectamine as a function of p-shRNA length (Figure 5A). Intact p-shRNA (21 bp) delivered with Lipofectamine caused a concentration-dependent decrease in cell viability with an IC50 value of 110 ng/ml. When p-shRNA was first broken down into small fragments (by treating with RNase T1) it was ∼4-fold less potent, with an IC50 value of 461 ng/ml. For comparison, a 21 bp siRNA with the same dsRNA sequence as the p-shRNA was much less toxic than the enzymatically digested p-shRNA when delivered with Lipofectamine (IC50 > 1500 ng/ml). Lipofectamine on its own was also found to be non-toxic to HeLa cells when administered at the same concentrations without RNA (data not shown).
Figure 5.
Cytotoxicity and gene knockdown studies with p-shRNA in different cell lines. (A) Cell viability dose-response curves for p-shRNA, RNase T1-digested p-shRNA and siRNA delivered with Lipofectamine to HeLa cells. Viability was measured by the CellTiterGlo assay and normalized to untreated cells. p-shRNA for this experiment was derived from template 6. (B) IC50 values ± 95% C.I. (based on three parameter log-logistic model) calculated from viability curves for cell lines treated with p-shRNA (I = p-shRNA-21, II = p-shRNA-25; derived from templates 17 and 18) or HMW poly-I:C complexed with Lipofectamine. (C) Cell viability dose-response curves for p-shRNA-25 and poly-I:C delivered with either Lipofectamine or TransIT-X2 to SKOV3 cells. (D) Caspase activation measured using CaspaseGlo luminescence assays 14 h after treating SKOV3 cells with p-shRNA-25- or poly-I:C-Lipofectamine complexes at 300 ng/ml. Values correspond to the means of three biological replicates (±s.e.m.). (E) Mean GFP expression determined by flow cytometry was measured in HeLa cells 72 h following transfection with 10 nM p-shRNA (p-shGFP-21 or p-shLUC-21) complexed with TransIT-X2. Values were derived from mean fluorescence and correspond to the mean of three biological replicates (±s.e.m.) relative to untreated cells. (F) Luciferase expression was measured as bioluminescence normalized to total protein in SKOV3 and UCI101 cells following treatment with 15 nM p-shRNA complexed with TransIT-X2 (p-shLUC and p-shGFP used in this experiment were synthesized from templates 19 and 20). Values are presented as means for three biological replicates (±s.e.m.) relative to untreated cells. For all graphs: P < 0.05 (*), P < 0.01 (**), P < 0.001 (***).
We next compared the dose-response behavior for the lipofection of p-shRNA and HMW poly-I:C in a panel of cancer cell lines (HeLa, A549, SKOV3 and UCI101) and NIH-3T3 fibroblasts. We tested two different p-shRNAs with 21 or 25 bp ds-regions (from templates 17 and 18; referred to here as p-shRNA-21 and p-shRNA-25). Overall, poly-I:C and p-shRNA-25 were found to be similarly cytotoxic (Figure 5B), and p-shRNA25 was ∼2-fold more potent than p-shRNA-21. Out of the different cell lines tested, the lowest p-shRNA and poly-I:C toxicities were observed in 3T3 fibroblast cells. The IC50 values for p-shRNA-21, p-shRNA-25 and poly-I:C in 3T3 cells were >1500, 880 and 711 ng/ml, respectively; in contrast the corresponding IC50 values averaged across the cancer cell lines tested were 200, 75 and 129 ng/ml. It is important to note that in addition to having different ds-lengths, p-shRNA-21 and p-shRNA-25 also had unrelated sequences. To test whether ds-length was an important factor in determining p-shRNA cytotoxicity, we tested another pair of 21 and 25 bp p-shRNA sequences in SKOV3 cells (p-shGFP-21 and p-shGFP-25; from templates 6 and 12), which had identical sequences except for the four extra base pairs in the 25 bp p-shRNA. Of these two p-shRNAs, p-shGFP-21 was slightly more toxic to SKOV3 cells (IC50 = 103 versus 168 ng/ml, see Supplementary Figure S9), suggesting that ds-length alone is not a strong predictor of cytotoxicity; however, different double-strand sequences do appear to have different potencies.
The cytotoxicity of lipofected poly-I:C was previously shown to proceed through a caspase-dependent apoptotic pathway in cancer cells (18). We compared caspase activation in SKOV3 cells 14 h following treatment with p-shRNA-25 or poly-I:C delivered with Lipofectamine (Figure 5D). Both poly-I:C and p-shRNA-25 caused significant increases in caspase 3/7, caspase 8 and caspase 9 relative to untreated cells, suggesting that p-shRNA activates apoptotic pathways in a similar manner to poly-I:C following lipofection.
To test the generality of p-shRNA toxicity in cancer cells we treated the same four cancer cell lines with p-shRNA-25 complexed to a polymeric transfection reagent (TransIT-X2). Figure 5C shows the dose-response curves for p-shRNA-25 and poly-I:C delivered with TransIT-X2 or Lipofectamine in SKOV3 ovarian cancer cells. Notably, p-shRNA showed nearly identical toxicity when delivered with TransIT-X2, whereas poly-I:C was more than an order of magnitude less toxic when transfected with TransIT-X2 (>1500 ng/ml) compared to Lipofectamine. The average IC50 value for p-shRNA-25 delivered with TransIT-X2 in the four cancer cell lines was 239 ng/ml; poly-I:C delivered with TransIT-X2 was moderately toxic to A549 cells (IC50 = 947 ng/ml) and relatively nontoxic to all other cell lines (IC50 > 1500 ng/ml).
Reporter gene knockdown using p-shRNA was initially measured in GFP-expressing HeLa cells (Figure 5E). Using TransIT-X2, we observed 73 and 84% GFP knockdown relative to untreated cells for 10 and 30 nM p-shGFP-21 doses at 72 h (mean fluorescence measured by flow cytometry). We observed minimal non-specific knockdown with the negative control p-shRNA (p-shLUC-21) at 10 nM; however, a 33% decrease in GFP expression was seen at the higher 30 nM dose. p-shRNA delivered with Lipofectamine also caused knockdown in HeLa cells, however, the effect size was smaller than with TransIT-X2 (48 and 56% knockdown at 10 and 30 nM doses; see Supplementary Figure S10A). p-shRNA with a 25 bp stem (p-shGFP-25) was also effective at causing knockdown in HeLa cells, with no significant differences in activity observed relative to p-shGFP-21 (Supplementary Figure S10B and C). We also tested knockdown with TransIT-X2 in luciferase-expressing SKOV3 and UCI101 ovarian cancer cells using 25 bp p-shRNA derived from template 19. In both cell lines, we observed 50–60% knockdown relative to untreated cells at a 15 nM dose (measured as luminescence signal normalized to total protein); this knockdown was significant (P < 0.05) compared to cells treated with negative control p-shRNA (containing an siGFP sequence transcribed from template 20).
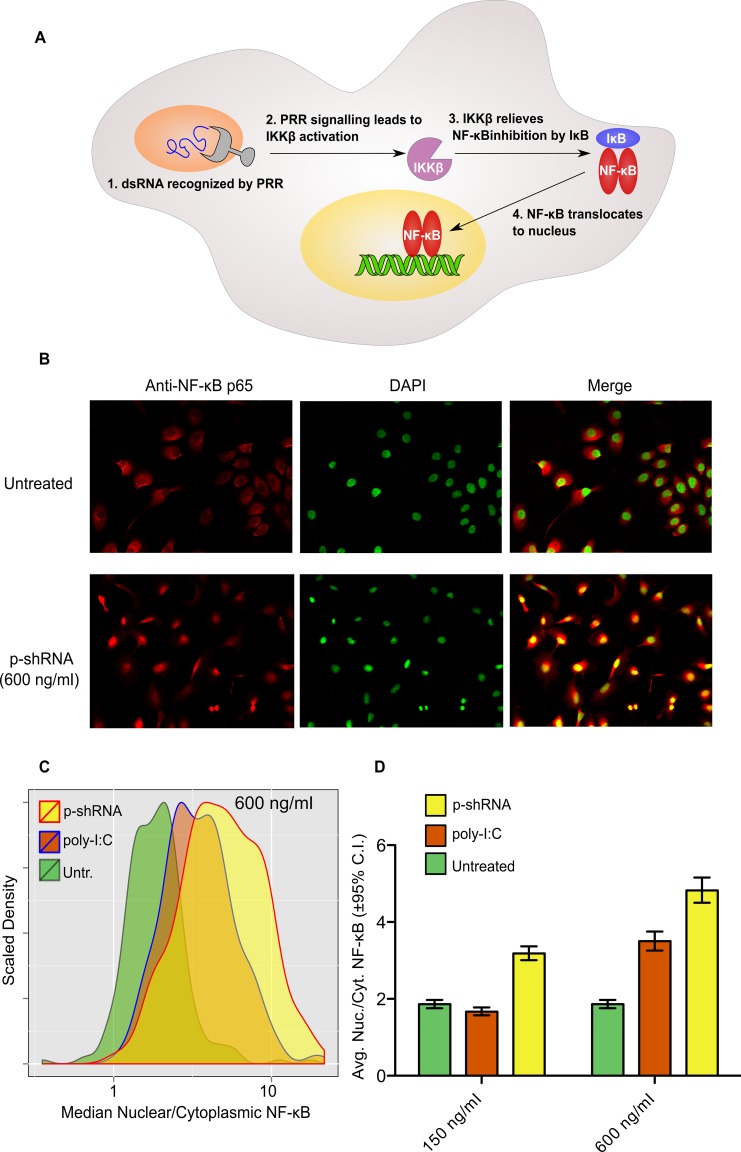
The activation and nuclear translocation of NF-κB is a canonical marker of innate immune stimulation (Figure 6A). To test if p-shRNA transfection leads to NF-κB activation, we stained SKOV3 cells with NF-κB antibody before and after transfection with p-shRNA-25/Lipofectamine and compared with poly-I:C as a positive control. Following stimulation with p-shRNA-25/Lipofectamine, an increase in nuclear NF-κB signal was apparent after 1 h, as evidenced by co-localization of the NF-κB stain with the DAPI nuclear stain (Figure 6B; yellow color indicates overlap). We next quantified the extent of NF-κB nuclear translocation for untreated, p-shRNA treated and poly-I:C treated cells by taking the ratio of median fluorescence inside the nucleus to median fluorescence in the cytoplasm. Based on this metric we observed a concentration-dependent shift in the cell population to higher nuclear localization following p-shRNA transfection, with a 2.6-fold increase relative to untreated cells for cells treated at 600 ng/ml (Figure 6C and D). The nuclear localization of NF-κB following p-shRNA treatment was greater than for poly-I:C (1.9-fold increase at 600 ng/ml), indicating that p-shRNA is a potent activator of innate immune pathways.
Figure 6.
Activation of NF-κB in SKOV3 cells by p-shRNA-25. (A) NF-κB is activated by dsRNA through pattern recognition receptor (PRR)-mediated signaling, resulting in nuclear translocation and transcriptional activation of myriad genes. (B) Fluorescence imaging of SKOV3 cells stained with anti-NF-κB (red) and DAPI (green). Poly-I:C and p-shRNA were complexed with Lipofectamine and incubated with cells at the indicated concentrations for 1 h prior to fixation and staining. The yellow color in the merged images indicates nuclear localization of NF-κB. (C and D) Nuclear localization of NF-κB was quantified using Cell Profiler by taking the ratio of median NF-κB fluorescence in the nucleus divided by median NF-κB fluorescence in the cytoplasm for untreated cells or cells treated with 150 or 600 ng/ml RNA/Lipofectamine complexes (for histograms of samples treated with 150 ng/ml see Supplementary Figure S11). Values in (D) correspond to the geometric means of the histograms ±95% confidence intervals.
DISCUSSION
In this study, we used rolling circle transcription (RCT) to concatenate shRNA units into large periodic RNA molecules that we refer to as p-shRNA. This was achieved using T7 RNA polymerase, which can transcribe p-shRNA from simple dumbbell templates that lack a promoter sequence. Our findings suggest that the loop size in these templates has a profound effect on p-shRNA yield and a moderate effect on transcript length, with larger loops causing both to increase. The importance of loop size appears to be mostly related to transcription initiation, where the larger loop may mimic a ‘transcription complex bubble’ similar to what has been described in other promoterless systems (11,33,34). Large loops (i.e. 11–22 bases) are also required by RNA Pol III for promoterless transcription from circular DNA templates; however, in the case of Pol III, RCT does not take place and short well-defined transcripts are obtained instead (35,36). For RCT with T7 polymerase, we found that although 6 base loops were sufficient for obtaining p-shRNA, the presence of at least one loop ≥10 bases greatly improved the RNA yield. With this in mind, the optimized RCT conditions described in this paper can yield quantities of RNA sufficient for in vivo work and other material-intensive experiments. Although T7 RNA polymerase preferentially initiates transcription with GTP, we found that cytosines were not required in the loops for RCT from dumbbell templates. One possibility is that the loops are important for T7 polymerase binding, but that initiation occurs at a cytosine in the adjacent double-stranded region. On the other hand, the inclusion of 5–10 cytosines in the dumbbell loops led to shorter transcripts. Previous work demonstrated that long stretches of Cs can halt transcription by T7 RNA polymerase, which was proposed to be due to the unusual stability of dC/rG base pairs (37).
The folded structure of p-shRNA is predicted to be very different depending on whether it assumes the minimum free-energy structure or a co-transcriptionally folded (kinetic) structure. Our results suggest that it adopts the latter–i.e. a repeating hairpin structure reminiscent of the typical models presented for primary miRNA. An interesting feature of this type of structure is the asymmetry of the single-stranded regions: one acts as the hairpin loop and the other links the hairpins together. When transcription is initiated from a dumbbell template, the initiating loop should end up in the latter position; since transcription initiation is determined by loop size, this should enable specific sequences to be placed in either the loop or linker regions as desired. The spatial positioning of different sequences could then be used to further optimize the properties of p-shRNA.
Cell viability assays in four different cancer cell lines revealed potent cytotoxicity for p-shRNA delivered with cationic lipid (Lipofectamine) and, to a slightly lesser extent, cationic polymer (TransIT-X2) transfection reagents. The large size of p-shRNA appeared to contribute to its toxicity along with the sequence of its double-stranded region. The influence of other factors, like the presence of the 5′ PPP moiety and the sequence of single-stranded regions, were not investigated in this study, but could also potentially contribute to p-shRNA toxicity. We also demonstrated that p-shRNA can cause gene silencing in multiple cancer cell lines. In the future, combining the knockdown of therapeutically relevant genes with the potent toxicity of p-shRNA could lead to synergistic activity.
We found that p-shRNA's toxicity was very similar to that of poly-I:C–a canonical innate immune activator with established anti-cancer activity–when both were delivered with Lipofectamine. In contrast, we were surprised to find that p-shRNA was orders of magnitude more toxic than poly-I:C following transfection with the polymeric TransIT-X2 reagent. Although the reason for this difference remains to be determined, it perhaps indicates differences in how and where p-shRNA and poly-I:C are recognized by PRRs. Poly-I:C is best known as a TLR3 agonist but can also activate RIG-I/MDA5 when delivered to the cytoplasm (38). In this context, poly-I:C induces an innate immune response characterized by IRF-3 and NF-κB activation, with the downstream production of type-I interferon. Interestingly, the apoptotic effects of poly-I:C in melanoma cells were shown to be mediated by the mitochondrial pathway, requiring caspase-9 activation and independent of type-I interferons (18). We also observed a significant induction of caspase-9 by p-shRNA in this study, indicating that a similar mechanism is at play. p-shRNA's observed cytotoxicity and activation of NF-κB suggest that it is a potent activator of PRRs, however, further work is needed to determine which PRRs are involved and to what extent.
Targeted gene silencing and immunostimulation can both be exploited for cancer therapy. We have demonstrated that p-shRNA can cause gene silencing, cytotoxicity and innate immune activation in various cancer cell lines. p-shRNA's double stranded regions, single stranded regions and 5′-PPP moiety could all influence PRR activation and cytotoxicity: these structures and their exact sequences are readily varied in p-shRNA, which could make it a flexible immunotherapy platform with gene silencing capabilities. The large size and flexible structure of p-shRNA is expected to enhance self-assembly behavior compared to siRNA, which could enable new avenues for in vivo delivery. Future work will aim to further elucidate the mechanism of action for p-shRNA and optimize its delivery for cancer therapy.
Supplementary Material
Acknowledgments
The authors thank the Koch Institute Swanson Biotechnology Center for technical support and Drs L. Ceppi, W. Wei and M. Birrer (MGH) for generously providing luciferase-expressing UCI101 and SKOV3 cell lines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Defense Teal Innovator Award (to P.T.H); Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship (to K.E.S.); National Science Foundation Graduate Research Fellowship (to C.W.); National Institutes of Health [NIBIB 1F32EB017614- 02 to E.C.D.]; Koch Institute Support Grant from the National Cancer Institute [P30-CA14051, in part]. Funding for open access charge: Department of Defense Teal Innovator Award.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wilson R.C., Doudna J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shu Y., Cinier M., Fox S.R., Ben-Johnathan N., Guo P. Assembly of therapeutic pRNA-siRNA nanoparticles using bipartite approach. Mol. Ther. 2011;19:1304–1311. doi: 10.1038/mt.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shu Y., Haque F., Shu D., Li W., Zhu Z., Kotb M., Lyubchenko Y., Guo P. Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA. 2013;19:767–777. doi: 10.1261/rna.037002.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grabow W.W., Zakrevsky P., Afonin K.A., Chworos A., Shapiro B.A., Jaeger L. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett. 2011;11:878–887. doi: 10.1021/nl104271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNamara J.O., Andrechek E.R., Wang Y., Viles K.D., Rempel R.E., Gilboa E., Sullenger B.A., Giangrande P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 6.Zhou J., Li H., Li S., Zaia J., Rossi J.J. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol. Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler L.A., Trifonova R., Vrbanac V., Basar E., McKernan S., Xu Z., Seung E., Deruaz M., Dudek T., Einarsson J.I., et al. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J. Clin. Invest. 2011;121:2401–2412. doi: 10.1172/JCI45876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolcato-Bellemin A.-L., Bonnet M.-E., Creusat G., Erbacher P., Behr J.-P. Sticky overhangs enhance siRNA-mediated gene silencing. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16050–16055. doi: 10.1073/pnas.0707831104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mok H., Lee S.H., Park J.W., Park T.G. Multimeric small interfering ribonucleic acid for highly efficient sequence-specific gene silencing. Nat. Mater. 2010;9:272–278. doi: 10.1038/nmat2626. [DOI] [PubMed] [Google Scholar]
- 10.Sajeesh S., Lee T.Y., Hong S.W., Dua P., Choe J.Y., Kang A., Yun W.S., Song C., Park S.H., Kim S., et al. Long dsRNA-mediated RNA interference and immunostimulation: a targeted delivery approach using polyethyleneimine based nano-carriers. Mol. Pharm. 2014;11:872–884. doi: 10.1021/mp400541z. [DOI] [PubMed] [Google Scholar]
- 11.Daubendiek S.L., Ryan K., Kool E.T. Rolling-circle RNA synthesis: circular oligonucleotides as efficient substrates for T7 RNA polymerase. J. Am. Chem. Soc. 1995;117:7818–7819. doi: 10.1021/ja00134a032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daubendiek S.L., Kool E.T. Generation of catalytic RNAs by rolling transcription of synthetic DNA nanocircles. Nat. Biotechnol. 1997;15:273–277. doi: 10.1038/nbt0397-273. [DOI] [PubMed] [Google Scholar]
- 13.Seyhan A.A., Vlassov A.V., Johnston B.H. RNA interference from multimeric shRNAs generated by rolling circle transcription. Oligonucleotides. 2006;363:353–363. doi: 10.1089/oli.2006.16.353. [DOI] [PubMed] [Google Scholar]
- 14.Seidl C.I., Ryan K. Circular single-stranded synthetic DNA delivery vectors for microRNA. PLoS One. 2011;6:e16925. doi: 10.1371/journal.pone.0016925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubbell H.R., Liu R.-S., Maxwell B.L. Independent sensitivity of human tumor cell lines to interferon and double-stranded RNA. Cancer Res. 1984;44:3252–3257. [PubMed] [Google Scholar]
- 16.Ewel C.H., Urba W.J., Kopp W.C., Smith J.W. II, Steis R.G., Rossio J.L., Longo D.L., Jones M.J., Alvord W.G., Pinsky C.M., et al. Polyinosinic-polycytidylic acid complexed with poly-l-lysine and carboxymethylcellulose in combination with interleukin 2 in patients with cancer: clinical and immunological effects. Cancer Res. 1992;52:3005–3010. [PubMed] [Google Scholar]
- 17.Poeck H., Besch R., Maihoefer C., Renn M., Tormo D., Morskaya S.S., Kirschnek S., Gaffal E., Landsberg J., Hellmuth J., et al. 5′-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat. Med. 2008;14:1256–1263. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- 18.Besch R., Poeck H., Hohenauer T., Senft D., Häcker G., Berking C., Hornung V., Endres S., Ruzicka T., Rothenfusser S., et al. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J. Clin. Invest. 2009;119:2399–2411. doi: 10.1172/JCI37155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shopsowitz K.E., Roh Y.H., Deng Z.J., Morton S.W., Hammond P.T. RNAi-microsponges form through self-assembly of the organic and inorganic products of transcription. Small. 2014;10:1623–1633. doi: 10.1002/smll.201302676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:70–74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Proctor J.R., Meyer I.M. CoFold: An RNA secondary structure prediction method that takes co-transcriptional folding into account. Nucleic Acids Res. 2013;41:e102. doi: 10.1093/nar/gkt174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popenda M., Szachniuk M., Antczak M., Purzycka K.J., Lukasiak P., Bartol N., Blazewicz J., Adamiak R.W. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40:e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritz C., Streibig J.C. Bioassay analysis using R. J. Stat. Softw. 2005;12:1–22. [Google Scholar]
- 24.Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frieden M., Pedroso E., Kool E.T. Tightening the belt on polymerases: evaluating the physical constraints on enzyme substrate size. Angew. Chem. Int. Ed. 1999;38:3654–3657. [PubMed] [Google Scholar]
- 26.Chamberlin M., Ring J. Characterization of T7-specific ribonucleic acid polymerase. J. Biol. Chem. 1973;248:2235–2244. [PubMed] [Google Scholar]
- 27.Lesnik E.A., Freier S.M. Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry. 1995;34:10807–10815. doi: 10.1021/bi00034a013. [DOI] [PubMed] [Google Scholar]
- 28.Salaun B., Coste I., Rissoan M.-C., Lebecque S.J., Renno T. TLR3 can directly trigger apoptosis in human cancer cells. J. Immunol. 2006;176:4894–4901. doi: 10.4049/jimmunol.176.8.4894. [DOI] [PubMed] [Google Scholar]
- 29.Jiang Q., Wei H., Tian Z. Poly I:C enhances cycloheximide-induced apoptosis of tumor cells through TLR3 pathway. BMC Cancer. 2008;8:12. doi: 10.1186/1471-2407-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kübler K., tho Pesch C., Gehrke N., Riemann S., Dassler J., Coch C., Landsberg J., Wimmenauer V., Pölcher M., Rudlowski C., et al. Immunogenic cell death of human ovarian cancer cells induced by cytosolic poly(I:C) leads to myeloid cell maturation and activates NK cells. Eur. J. Immunol. 2011;41:3028–3039. doi: 10.1002/eji.201141555. [DOI] [PubMed] [Google Scholar]
- 31.Liu J., Guo Y.-M., Hirokawa M., Iwamoto K., Ubukawa K., Michishita Y., Fujishima N., Tagawa H., Takahashi N., Xiao W., et al. A synthetic double-stranded RNA, poly I:C, induces a rapid apoptosis of human CD34+ cells. Exp. Hematol. 2012;40:330–341. doi: 10.1016/j.exphem.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Duewell P., Steger A., Lohr H., Bourhis H., Hoelz H., Kirchleitner S. V, Stieg M.R., Grassmann S., Kobold S., Siveke J.T., et al. RIG-I-like helicases induce immunogenic cell death of pancreatic cancer cells and sensitize tumors toward killing by CD8(+) T cells. Cell Death Differ. 2014;21:1825–1837. doi: 10.1038/cdd.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daube S.S., von Hippel P.H. Functional transcription elongation complexes from synthetic RNA-DNA bubble duplexes. Science. 1992;258:1320–1324. doi: 10.1126/science.1280856. [DOI] [PubMed] [Google Scholar]
- 34.Møllegaard N.E., Buchardt O., Egholm M., Nielsen P.E. Peptide nucleic acid.DNA strand displacement loops as artificial transcription promoters. Proc. Natl. Acad. Sci. U.S.A. 1994;91:3892–3895. doi: 10.1073/pnas.91.9.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seidl C.I., Lama L., Ryan K. Circularized synthetic oligodeoxynucleotides serve as promoterless RNA polymerase III templates for small RNA generation in human cells. Nucleic Acids Res. 2013;41:2552–2564. doi: 10.1093/nar/gks1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lama L., Seidl C.I., Ryan K. New insights into the promoterless transcription of DNA coligo templates by RNA polymerase III. Transcription. 2014;5:1–14. doi: 10.4161/trns.27913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belotserkovskii B.P., Liu R., Tornaletti S., Krasilnikova M.M., Mirkin S.M., Hanawalt P.C. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.