Summary
Background
Observational studies show a strong association between delayed intestinal transit and the production of methane. Experimental data suggest a direct inhibitory activity of methane on the colonic and ileal smooth muscle and a possible role for methane as a gasotransmitter. Archaea are the only confirmed biological sources of methane in nature and Methanobrevibacter smithii is the predominant methanogen in the human intestine.
Aim
To review the biosynthesis and composition of archaeal cell membranes, archaeal methanogenesis and the mechanism of action of statins in this context.
Methods
Narrative review of the literature.
Results
Statins can inhibit archaeal cell membrane biosynthesis without affecting bacterial numbers as demonstrated in livestock and humans. This opens the possibility of a therapeutic intervention that targets a specific aetiological factor of constipation while protecting the intestinal microbiome. While it is generally believed that statins inhibit methane production via their effect on cell membrane biosynthesis, mediated by inhibition of the HMG‐CoA reductase, there is accumulating evidence for an alternative or additional mechanism of action where statins inhibit methanogenesis directly. It appears that this other mechanism may predominate when the lactone form of statins, particularly lovastatin lactone, is administered.
Conclusions
Clinical development appears promising. A phase 2 clinical trial is currently in progress that evaluates the effect of lovastatin lactone on methanogenesis and symptoms in patients with irritable bowel syndrome with constipation. The review concludes with an outlook for the future and subsequent work that needs to be done.
Introduction
Methane, which is now of great interest to climate researchers, plays an important if somewhat underappreciated role in gastrointestinal disorders. The link between this odourless gas and the intestine was first made in the early days of colonoscopy, when sparks from electrosurgical devices sometimes led to intracolonic explosions.1 Microbial metabolism of mannitol, a sugar alcohol that had been commonly used for colon lavage, led to increased concentrations of the flammable gases hydrogen and methane. Modern bowel preps do not carry that risk.
Methane can have inorganic (e.g. geothermal‐volcanic methane emissions) and organic sources. The only organisms known to produce methane are methanogenic archaea. Archaea are fascinating prokaryotes that were previously considered to be bacteria. However, these prokaryotes, while resembling bacteria on a superficial level, were found to have so many unique characteristics that they were given their own name, archaea, and were elevated to occupy a position as one of the three domains of life: archaea, bacteria, and eucarya.2, 3
Less spectacular than its explosive capacity but more important is recent evidence that links intestinal methane production to slowed intestinal transit.4 In this review, we will describe how large quantities of methane are produced in the colon and how statins may have therapeutic importance by inhibiting methane production through the selective suppression of the growth of methanogens.
Methane and constipation‐associated disorders
Methane and intestinal transit
Although 50–80% of methane in humans is passed as flatus, the presence of methane production can be accurately assessed by breath tests. A comprehensive review recently concluded that data from breath testing generally support the association between delayed intestinal transit and the production of methane.5 Methodological differences in study design precluded a formal meta‐analysis approach for studies that correlated diagnosis of constipation‐predominant IBS (IBS‐C) or chronic idiopathic constipation with methane production, nevertheless, the same report5 lists 14 studies supporting the association between methane production and constipation related disorders in adults. There are a few reports that primarily evaluated other hypotheses that did not show a link between methane on breath testing and constipation6, 7, 8 and one study that concludes that colonic methane production is not associated with clinical presentation in IBS patients.9
Two microbiome studies correlating methanogens and stool frequency have recently become available. One study evaluated the correlation between gut microbiota variation and stool consistency using the Bristol Stool Scale (BSS) classification. Enterotypes were distinctly distributed over the BSS scores: Within the RB enterotype, found in harder stool samples, the abundance of Methanobrevibacter and Akkermansia was positively correlated with colon transit time.10 The results of another microbiome study analysing 273 faecal samples showed that only patients with a constipation phenotype presented higher abundance of methanogenic archaea, confirming a link between low transit time and methane production capacity.11
Association does not prove causation, and it has long been known that intestinal dysmotility, as seen in scleroderma, leads to secondary small intestinal bacterial (and, presumably, archaeal) overgrowth. It could be argued that increased methane production is therefore not the cause but an effect of delayed intestinal transit. Fourteen healthy volunteers underwent four interventions: placebo, sulphate supplements, or sulphate supplements with either senna or loperamide. With faster intestinal transit a reduction in faecal methanogens and methane production was seen. The reverse effects were true with loperamide.12 However, there is experimental evidence that methane directly affects intestinal transit and contractility (methane‐first hypothesis).
Experimental evidence
While methane is predominantly produced in the colon, methanogens can also be demonstrated in the small intestine, especially when small bowel bacterial overgrowth exists.13 Pimentel and colleagues conducted a three‐part study consisting of two experiments – one in vivo (dogs) and one ex vivo (guinea pig ileum) – as well as a retrospective review of data in irritable bowel syndrome (IBS) patients who had previously undergone breath testing and antroduodenal motility studies.4 In dogs, small intestinal fistulae were created and transit of a radiotracer was measured during infusion of ambient air, followed by methane at a rate that was physiological (i.e. resulting in 50 ppm in exhaled air), a concentration that is typical for patients with irritable bowel syndrome with constipation (IBS‐C). The methane infusion resulted in decreased radiotracer recovery in each of five individual dogs compared to baseline. The average reduction was 59%, which was highly significant (P < 0.001).
In the second experiment, 3‐cm‐long terminal ileum preparations from sacrificed guinea pigs were oriented in an oral‐aboral fashion in a physiological buffer solution and stimulated before and after methane was bubbled through the tissue bath. Methane exposure at a concentration of 1000 ppm led to a significant augmentation of the contractile force in response to the applied brush stroke, consistent with hyperactivity.
Finally, after retrospective review, 11 methane‐producing and 12 sex‐matched hydrogen‐producing subjects were identified who met Rome I criteria for IBS and, in addition, had antroduodenal manometry data available. The fasting motility index, as a gross measure of forces (resistive and propulsive) in the small intestine, in methane‐producing subjects was noted to be significantly higher (1851 ± 861) compared with hydrogen‐producing subjects (1199 ± 301; P < 0.05).
The authors concluded that methane slows small bowel transit and augments contractile activity in the small bowel, and in addition, small intestinal contractile activity is increased in IBS patients who produce methane. They speculated that methane predisposes to constipation because it promotes segmental (nonpropagating contractions) and suggested that further work, especially involving the colon, was warranted.4
In similar experiments, a Korean group reproduced and expanded upon the results of the above studies by including colonic motility studies and comparing the effects of hydrogen gas with those of methane. Briefly, in an ex vivo experiment using guinea pig ileal, right and left colon segments in a peristaltic tissue bath, ileal contractile activity significantly decreased and the amplitude of peristaltic contractions increased in response to methane insufflation, while the opposite phenomenon was detected after hydrogen infusion. Colonic transit was shortened by hydrogen infusion, but this effect was blunted when methane was co‐administered, especially in the right colon.14
The preceding experimental and observational studies support a causative role for methane in constipation‐related disorders, especially IBS‐C. Ultimately the argument that slow transit could cause overgrowth of methanogenic bacteria is not necessarily a detractor from the methane‐first hypothesis. Perhaps a feedback loop exists where the terms cause and effect have less meaning and opportunities for therapeutic intervention exist in this circle: Slow transit promotes archaeal growth – archaea produce methane – methane produces better growth and survival conditions for archaea – more methane is produced further slowing down intestinal transit leading to symptomatic constipation – archaea thrive and multiply until a steady state is achieved.
The origin and properties of methane production
The five major gases in the colon are: nitrogen (23–80%), hydrogen (0.06–47%), carbon dioxide (5.1–29%), oxygen (0.1–2.3%) and methane (0–26%).1
In addition, nitric oxide and hydrogen sulphide are found as well. These gases have great importance through their function as gasotransmitters and components of a complex microbial ecosystem.12, 15 All the methane in the intestine is produced by methanogenic bacteria of which Methanobrevibacter smithii, described in more detail below, is the most important contributor. Methane is overwhelmingly produced by strictly anaerobic archaea that produce methane from substrates such as hydrogen, carbon dioxide and certain other substrates. Emissions from agriculture represent around 40% of the methane emissions produced by human‐related activities, the single largest source is enteric fermentation in livestock.16
Archaea are a large and diverse class of prokaryotes that, together with bacteria and eukaryotes, make up the three domains of life.17 More recently evidence for direct methane production and emission by eukaryotes such as plants, animals and fungi has been presented. While this seems to have been confirmed for plants, mammalian endogenous (nonmicrobial) methane production remains controversial.18 These findings, if corroborated, could, however, support a gasotransmitter role for methane.19
Various archaeal species have been shown to inhabit distinct human body sites such as the intestine, the oral cavity, the vagina and, most recently, the skin.20 The majority of the so far detected archaea in humans, particularly from the gut and mouth, represent members of the family Methanobacteriaceae.21
Methane phenotype
Breath tests conducted in humans register either methane (30–50%) or hydrogen (50–70%) but only rarely both.22 This makes sense if one considers the stoichiometry of hydrogen removal by methanogens: The production of one mole of methane removes 4 moles of hydrogen (4H2 + CO2 → CH4 + 2H2O). The designation of subjects as ‘methane producers’ or ‘methane nonproducers’ based on breath testing only accounts for one way of escape but not the large amount that is passed directly as flatus (between 50% and 80% of the total). Indeed, one older report suggests that methane is not detected in breath until the methanogens reach a density of 108 methanogenic bacteria per gram of stool.23 However, as always, the situation could be more complex. A recent study from New Zealand using highly sensitive open‐circuit respiration chambers (that account for total animal methane emissions) identified a low‐ and high‐methane phenotype in sheep that was attributed to the composition of microbial communities with methanogen densities that were not significantly different between the high and low groups.24 Open‐circuit respiration chambers are not practical in humans and dividing subjects into methane producers and nonproducers by conventional breath test results is both convenient and clinically meaningful.
Methane as a gasotransmitter
Methane may be one of a series of gasotransmitters – endogenously generated, gaseous signalling molecules that do not require membrane receptors for their activity. Gasotransmitters are evolutionally conserved from bacteria and archaea to plant and mammalian cells. Nitric oxide (NO) was the first identified gasotransmitter; carbon monoxide and hydrogen sulphide have been added more recently, and ammonia, hydrogen and methane have achieved candidate status.25 The discovery of mammalian NO metabolism (‘endothelium‐derived relaxing factor’) has led to a whole class of drugs, the phosphodiesterase inhibitors.26 Drugs that release hydrogen sulphide are currently being developed for their possible anti‐neoplastic, chemoprotective and chemopreventive potential.27, 28
Some intriguing findings support the candidacy of methane as a gasotransmitter. Boros, et al. 29 demonstrated that the inhalation of 2.5% CH4 significantly ameliorated the extent of ischaemia‐reperfusion damage in dogs. A parallel series of experiments published in the same report showed that exogenous CH4 inhibited leucocyte infiltration in vitro, suggesting a connection between CH4 function and the immune system. Pimentel et al. showed that intestinal methane gas infusion slowed intestinal transit in dogs, and these experiments were reviewed in detail above.4 Boros et al.19 carefully examined to which extent methane fulfils the criteria proposed by Wang25 that characterise a gasotransmitters. The authors concluded that the data do not fully support the gasotransmitter concept, but suggested that methane liberation (from endogenous sources, not through the resident flora) may be linked to redox regulation connected to hypoxic events leading to, or associated with, mitochondrial dysfunction.
In order for methane to move from its status as candidate‐gasotransmitter to accepted gasotransmitter, direct endogenous production in mammalian organisms will need to be confirmed and the biochemistry and physiology clarified. As will be reviewed in the following, regardless of the eventual outcome of these investigations, sufficient evidence is accumulating to justify exploration of the methanogenesis pathway of intestinal archaea as a drug target in patients with certain constipation related disorders.
Methanogenesis in the human gut
Understanding the properties of methanogens in the gut and their physiology helps us to consider how their methane production might be curtailed by statins and other compounds.
Archaea and methanobacteria
Like bacteria, archaea are prokaryotes but many of their characteristic features are distinct from those of bacteria and eukaryota. Many archaea are best known for their ability to thrive in extreme environments. Others, such as the class called Methanobacteria (a name which was assigned before it was recognised that they are not ‘bacteria’), may live in less forbidding environments but are ‘extreme’ in their metabolic capabilities. They carry out what at first appears to be a contradiction in terms: anaerobic respiration. Aerobic respiration has oxygen as final electron acceptor. In contrast, anaerobic respiration relies on a final electron acceptor other than oxygen, in the case of M. smithii and other methanogenic species, carbon dioxide. Furthermore, M. smithii cannot use the conventional carbohydrate, lipid and protein electron donors for energy generation, instead it uses hydrogen gas. Biological methanogenesis is indeed an ancient ‘archaic’ process dating back 2.7 billion years when oxygen was still a trace element in the atmosphere and hydrogen was abundant.30 In summary, methanogens in the gut generate energy by ‘breathing’ the exhaust of other bacteria in the gut (hydrogen and carbon dioxide) and producing methane as waste: 4 H2 + CO2 → 2H2O + CH4.31
It should be noted that methanogenic archaea are not the only consumers of hydrogen (hydrogenotrophs) in the gut. Sulphate‐reducing bacteria can obtain energy by oxidising organic compounds or molecular hydrogen while reducing sulphate to hydrogen sulphide, previously mentioned as a gasotransmitter. Hydrogen sulphide is a colourless gas with the characteristic foul odour of rotten eggs. Hydrogen sulphide is toxic to colonocytes and can impair their metabolic function and has been implicated in the aetiology of ulcerative colitis.32 It is therefore conceivable that a decrease in methanogenic activity could favour sulphate‐reducing bacteria with a concomitant increase in sulphide production. Indeed, competition between hydrogenotrophs has long been of interest to microbial ecologists. However, according to a comprehensive review of this issue, it appears impossible to fully explain the outcome of the competition among hydrogenotrophs in the human colon by idealised theory or in vitro chemostat systems,33 and clinical studies will have to address this question.
M. smithii
The dominant methanogen in the human intestine is M. smithii. Methanosphaera stadtmanae, which transfers electrons from hydrogen to methanol instead of carbon dioxide, is found less frequently (Figure 1). M. smithii is well adapted to the human intestine. It produces surface glycans resembling those found in the gut mucosa, regulates the expression of adhesin‐like proteins, consumes fermentation products produced by saccharolytic bacteria, effectively competes for nitrogen compounds, and – crucial for being well adapted to the gut – can grow in the presence of bile salts.34
Figure 1.
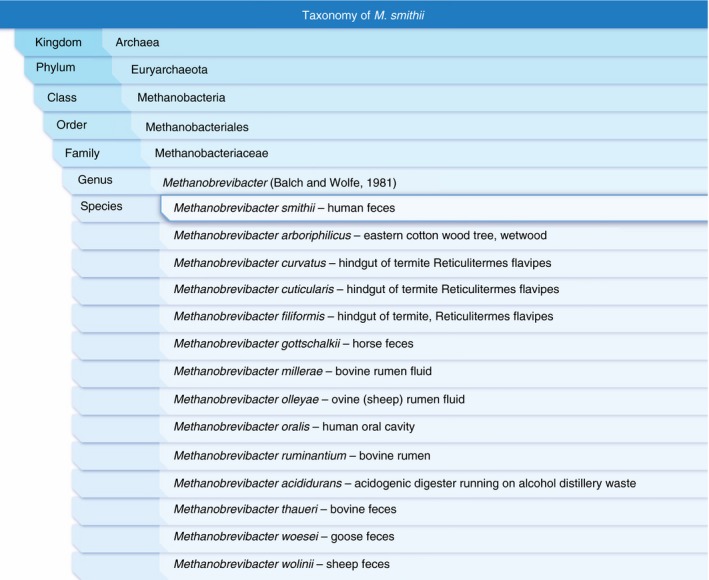
Taxonomy of Methanobrevibacter smithii. Only two methanogenic species have so far been isolated from the human colon: M. smithii is the predominant methanogen in the human gut. Methanosphaera stadtmanae, family Methanobacteriaceae, is less abundant. Note the Methanobrevibacter species in bovine and sheep rumen and mammalian faeces. The official taxonomy does not necessarily correlate with current molecular phylogenetics. Data from the Integrated Taxonomic Information System (ITIS)52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64 with isolate information from the Global Catalogue of Microorganisms.65
The cytoplasm of M. smithii is enclosed by a cell membrane and a cell wall35 that is decorated with the glycans (mentioned above), an arrangement that is familiar to us from the Gram‐positive bacteria. Indeed, M. smithii stains positive with the Gram stain.34 However, compared to bacteria, there are significant differences, both in cell wall and in cell membrane composition. Cell wall peculiarities have relevance to susceptibility to common antibiotics. The cell membrane of most bacteria is synthesised by the MEP (methylerythritol 4‐phosphate) pathway.36 In contrast, the isoprenoid biosynthesis for the main cell membrane components in archaea (archaeol) relies on the same enzyme that catalyses the biosynthesis of the isoprenoid cholesterol in humans, the HMG‐CoA reductase (mevalonate pathway).37 This obviously raises the possibility that statins – which are HMG‐CoA reductase inhibitors – could be used to interfere with the biosynthesis of the archaeal cell membrane and thus to inhibit archaeal growth, as described by Miller and Wolin in 2001.38
Archaeal cell wall
The cell wall of archaea is composed of pseudomurein (pseudopeptidoglycan). There are significant differences between pseudomurein and murein such as the occurrence of talosaminuronic acid instead of muramic acid, the presence of β (1→3) linkage instead of β (1→4) linkage of the glycan components, the partial replacement of glucosamine by galactosamine, the lack of D‐amino acids, and the accumulation of ε‐ and γ‐peptide bonds.35 Together these differences are responsible for resistance to cell wall antibiotics such as β‐lactams and to lysozyme and common proteases.35 In addition, archaea are resistant to a number of other cell wall‐altering antibiotics with the notable exception of squalene. They are susceptible to a variable degree to anti‐microbials that interfere with DNA, including the broad‐spectrum imidazoles (e.g. metronidazole).39
Archaeal cell membrane
Perhaps, more important for our discussion is the fact that the cell membrane of archaea contains unique polar lipids. Recall that the limiting membrane of cells and various organelles (in eukaryotes) is composed of a fluid phospholipid bilayer with intercalating proteins (and cholesterol in animals). In organisms other than archaea, this phospholipid bilayer is composed of glycerol‐3‐phosphate linked as an ester to fatty acids. In contrast, archaeal lipids are not composed of fatty acid esters; instead, they are saturated, branched, repeating isoprenoid subunits that attach to glycerol via an ether linkage (Figure 2). Isoprenoids are a ubiquitous and ancient class of biomolecules found in all living organisms that exhibit a remarkable diversity of structures and functions. Isoprenoids are derived from a basic five‐carbon precursor unit, isopentenyl diphosphate (IPP), and its isomer dimethylallyl diphosphate (DMAPP); these isomeric phosphates are the activated forms of isoprene.40 M. smithii has a cell membrane whose phospholipid fraction contains isoprenoids typical for archaea, archaeol and caldarcheol, the latter is arranged in a monolayer rather than a bilayer in the cell membrane.41, 42
Figure 2.
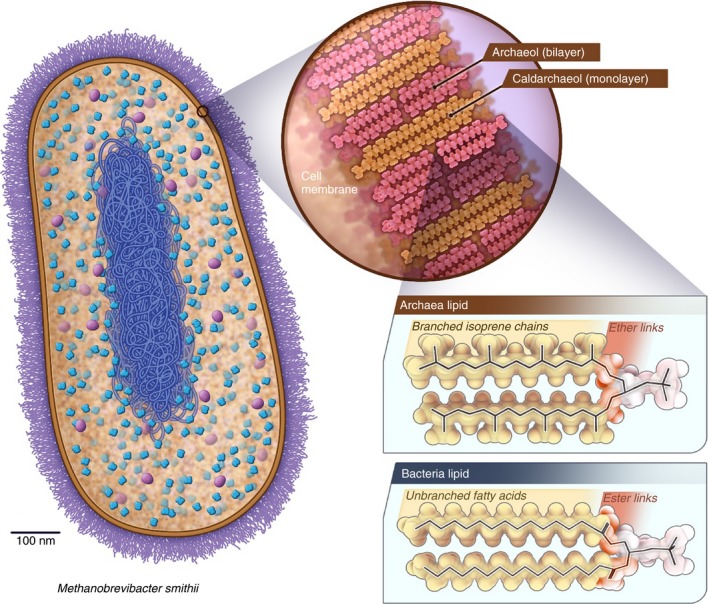
Methanobrevibacter smithii cell wall and cell membrane determine susceptibility to antibiotics and statins. The cell wall (violet) is composed of pseudomurein (and not murein as in bacteria) which makes archaea resistant to lysozyme and many antibiotics that interfere with cell wall synthesis. The cell membrane (ochre) consists of a lipid bilayer or monolayer the backbone of which composed of isoprene units that are linked to glycerol by ether bonds. In contrast, the lipid bilayer of bacteria consists of a fatty acid backbone that is linked to glycerol by an ester bond. The presence of statin‐sensitive isoprene units in the cell membrane of archaea allows statins to selectively interfere with the growth of archaea while leaving the cell membrane of bacteria unaffected. While bacteria do not use isoprene units in their cell membrane they are still required elsewhere. These bacterial isoprene units are, however, synthesised by a pathway (MEP) that is not inhibited by statins. See also Figure 3.
Archaea and humans use the MVA and bacteria the MEP pathway for isoprenoid biosynthesis
For many years, it was believed that IPP was synthesised by all organisms from acetyl‐CoA through the well‐known mevalonate (MVA) or HMG‐CoA reductase pathway. However, it is now well established that an alternative, MVA‐independent pathway is used in the majority bacteria, algae and plants, for the biosynthesis of IPP and DMAPP, the MEP pathway mentioned earlier.40 This is relevant for two reasons: (i) Statins can interfere with the cell membrane synthesis of archaea and will leave the overwhelming majority of the gut microbiome untouched. (ii) Interference with cell wall function interferes with the function of the cell membrane‐bound enzymes that are responsible for methanogenesis (Figure 3).
Figure 3.
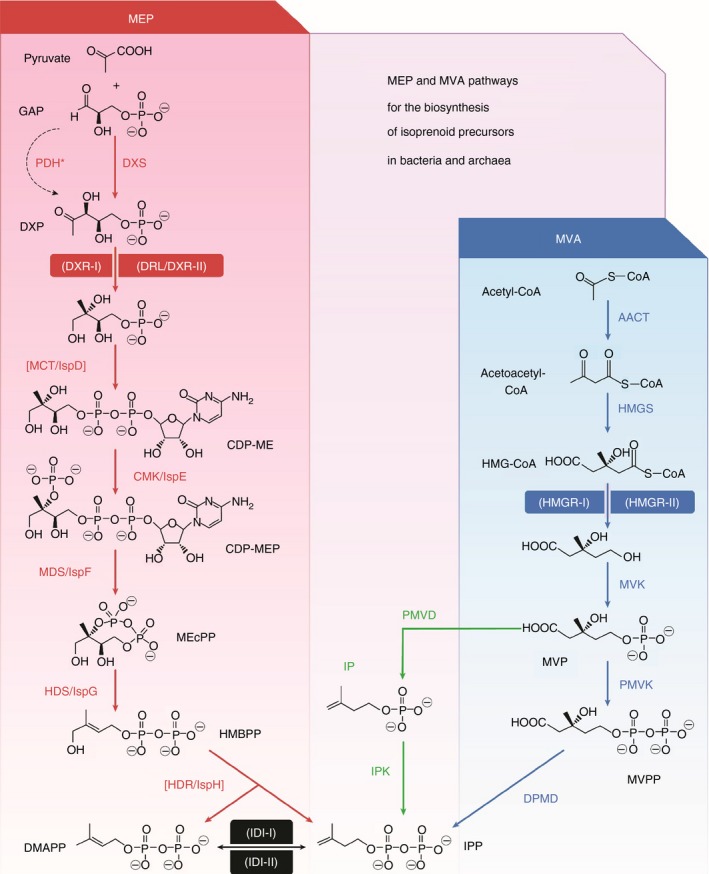
Most bacteria are not affected by statins. Most bacteria produce isoprenoids by the MEP pathway (red) that is not affected by HMGR‐inhibitors (statins), most other organisms including humans and archaea use the MVA pathway (blue). MEP: 2‐C‐methyl‐D‐erythritol 4‐phosphate. MVA: mevalonic acid. Humans and archaea utilise the HMGR‐I isoform (inhibited by statins) for isoprenoid biosynthesis. GAP, D‐glyceraldehyde 3‐phosphate; DXP, 1‐deoxy‐D‐xylulose 5‐phosphate; CDP‐ME, 4‐diphosphocytidyl‐2‐C‐methyl‐d‐erythritol; MEcPP, 2‐C‐methyl‐D‐erythritol 2,4‐cyclodiphosphate; HMBPP, 4‐hydroxy‐3‐methylbut‐2‐enyl diphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate. HMG‐CoA, 3‐hydroxy‐3‐methylglutaryl‐CoA; MVP, 5‐phosphomevalonate; MVPP, 5‐diphosphomevalonate. The alternative steps described in archaea are shown in green (IP, isopentenyl phosphate). Enzymes catalysing other reactions besides those in the MEP pathway are between brackets. Steps catalysed by different types of enzymes (shown within parentheses) are highlighted. Enzyme acronyms are given in the paper by Perez‐Gil and Rodriguez‐Concepcion 36 from which this illustration has been adapted.
The physiology of methane production by Archaea
The importance of HMG‐CoA reductase
HMG‐CoA reductase (HMGR, 3‐Hydroxy‐3‐methylglutaryl coenzyme A reductase) can be found in two different forms: eukaryotes and archaea use HMGR class I (membrane‐bound) and a few bacteria use HMGR class II (cytosolic).36 As was mentioned, the majority of bacteria do not employ HMGR but the enzymes of the MEP pathway for isoprenoid biosynthesis. HMGR catalyses the first step in the biosynthesis of cholesterol in humans and also the first step in the biosynthesis of the archaeal isoprene polymers (polyprenols) archaeol and caldarchaeol, which are the main constituents of the M. smithii cell membrane.
Anaerobic respiration by methanogenesis
Anaerobic respiration by methane biosynthesis is more complex than many other common respiratory processes because it requires the biosynthesis of six unusual coenzymes, a multistep pathway to methane, and a number of unique membrane‐bound enzyme complexes for coupling to the protein motive‐force17 (Figure 4). The inhibitory effects of HMGR‐inhibitors are generally thought to be related to the disruption of the cell membrane by interfering with isoprenoid biosynthesis, thereby disturbing the membrane‐bound respiratory chain of methanogenesis; however, alternative possibilities exist, and these could be relevant in explaining some of the experimental results observed with different types of statins.43 A brief review of the key steps in methanogenesis is therefore necessary.
Figure 4.
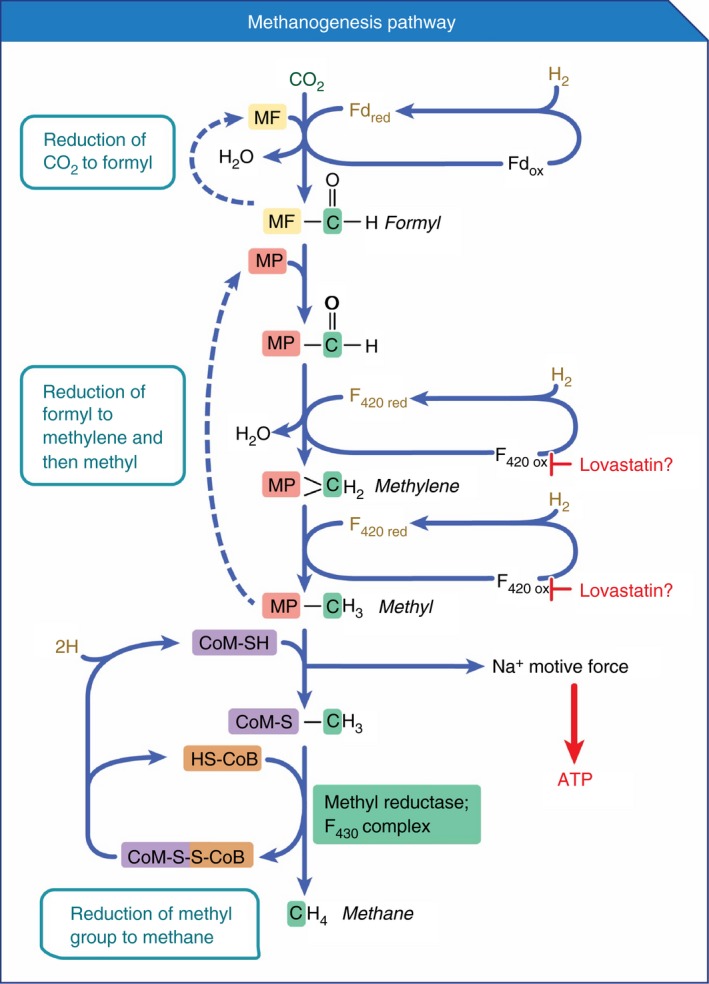
Methanogenesis pathway and potential role of lovastatin. Coenzymes F430 and F420 play crucial roles in the methanogenesis pathway (see also Figure 5). In silico molecular docking of the methanogenic enzyme F420‐dependent NADP oxidoreductase (fno) showed that both lovastatin and mevastatin had higher affinities for the F420 binding site on fno than did F420 itself. As such, lovastatin may act as inhibitor of fno. Fno is not shown in the diagram because it catalyses an alternative pathway of methanogenesis that utilises alcohol and methanol as substrates. However, if inhibition of fno by lovastatin can be experimentally confirmed, it is likely that other enzymes, such as those in the hydrogen‐CO 2‐methanogenesis pathway depicted here that require the coenzyme F420 would also be inhibited by lovastatin (as suggested in the diagram). The carbon atom reduced is highlighted in green. MF, methanofuran; MP, methanopterin; CoM, coenzyme M; Fd, ferredoxin; CoB, coenzyme B. This figure was adapted from reference.66
The final common pathway of methanogenesis
Early pathways in methanogenesis funnel into the same final step catalysed by the key enzyme methyl‐coenzyme M reductase (mcr): methyl‐coenzyme M (CH3‐S‐CoM) reacts with a second thiol coenzyme called coenzyme B (CoB‐SH) to form methane and the heterodisuphide of coenzyme M and coenzyme B (CoM‐S‐S‐CoB) (Figure 4). In its active site, mcr contains a unique active group, a nickel porphinoid, as prosthetic group called coenzyme F430 17 (Figure 5). The characteristic auto‐fluorescence of methanogens, is, however, largely caused by coenzyme F (for Fluorescence) F420 [with an absorption maximum at 420 nm emitting at 480 nm (blue)], which participates in earlier steps of methanogenesis44 (Figure 5).
Figure 5.
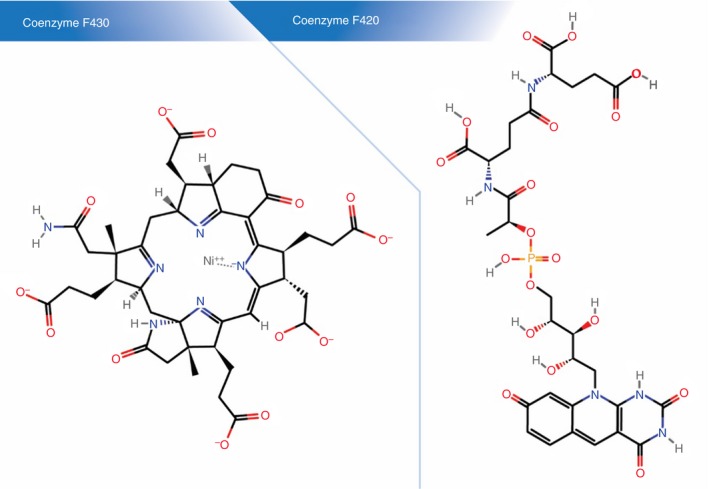
Coenzymes in the methanogenesis pathway. The final step in the methanogenesis pathway is catalysed by the key enzyme methyl‐coenzyme M reductase (Mcr). In its active site, Mcr contains a unique active group, a nickel porphinoid (left panel), called coenzyme F430. Coenzyme F420 (right panel) participates in two earlier steps in the methanogenesis pathway (see Figure 4) and is also responsible for the characteristic fluorescence of methanogens. In silico protein–ligand docking experiments suggest that lovastatin may have higher affinity for the F420 binding site than F420 itself.
Coenzyme F420 and NADP‐dependent oxidoreductase
Coenzyme F420, a hydride carrier, is found in archaea and some bacteria and has, in addition to methanogenesis, crucial roles in antibiotic biosynthesis, DNA repair, and activation of anti‐tubercular compounds 45 (Figure 5). While methanogenesis in M. smithii can proceed with carbon dioxide and hydrogen alone, there is evidence that M. smithii can utilise the bacterial fermentation products alcohol and methanol for methanogenesis using an inducible NADP‐dependent oxidoreductase with a coenzyme F420 prosthetic group (F420‐dependent NADP oxidoreductase, fno).46 Fno is mentioned here in anticipation of a review of molecular docking experiments with fno that suggest an additional or alternative mechanism of action for statins.
How statins inhibit methanogenesis
Statins are fungal secondary metabolites which inhibit hydroxy‐methylglutaryl coenzyme A (HMG‐CoA) reductase as the first committed enzyme of cholesterol biosynthesis in humans (Figure 3, MVA pathway). Statins lower blood cholesterol by inhibiting cholesterol biosynthesis in the liver; the reduced hepatic cholesterol concentration leads to a compensatory increase in expression of LDL receptors in liver cell membranes, which enhances the clearance of the circulating LDL cholesterol particles in the blood.47 Here we are, however, interested in the methane lowering activity of statins, and to better understand the mechanism of action, some statin pharmacology will need to be reviewed.
Lipid and water solubility of statins
In general, statins have hydrophilic and hydrophobic regions, and are classed as amphiphilic drugs. Amphiphilic drugs do not require specific transport mechanisms to cross membranes. As they are soluble in aqueous biological fluids and lipid membranes, they can simply diffuse through the body,48 at least in theory. Based on water‐octanol partition coefficients, a differentiation into fat soluble or hydrophobic (atorvastatin, simvastatin, fluvastatin, lovastatin, cerivastatin) and water soluble or hydrophilic statins (rosuvastatin, pravastatin) is made. Lipophilic statins undergo hepatic and enteric metabolism via cytochrome P450 (the CYP450 family of enzymes), whereas the water soluble statins are excreted largely unchanged.48 Fat soluble statins are known to cross the blood brain barrier, whereas water soluble statins are often thought not to cross the barrier. However, the situation is more complicated, and this may have relevance for the crossing other lipid membranes.
Lactone and hydroxyacid forms of statins
Another important aspect of statin chemistry is the existence of closed‐ring lactone and hydroxyacid forms (Figure 6). All statins can be converted from one to the other, but simvastatin and lovastatin are the commercially available statins that come in the lactone form; they are prodrugs and to be active (i.e. inhibit HMGR) they require opening of the lactone ring into the hydroxyacid form.47, 49 The lactones of cerivastatin, simvastatin, atorvastatin, lovastatin, fluvastatin, and pravastatin, ordered according to decreasing hydrophobicity, are three to four times more lipophilic as compared to their acid forms.50 In other words, the lactones of lipophilic statins are best equipped to cross lipid membranes. At one stage or another, the lactone ring would need to be broken up to inhibit HMGR; this will be discussed shortly.
Figure 6.
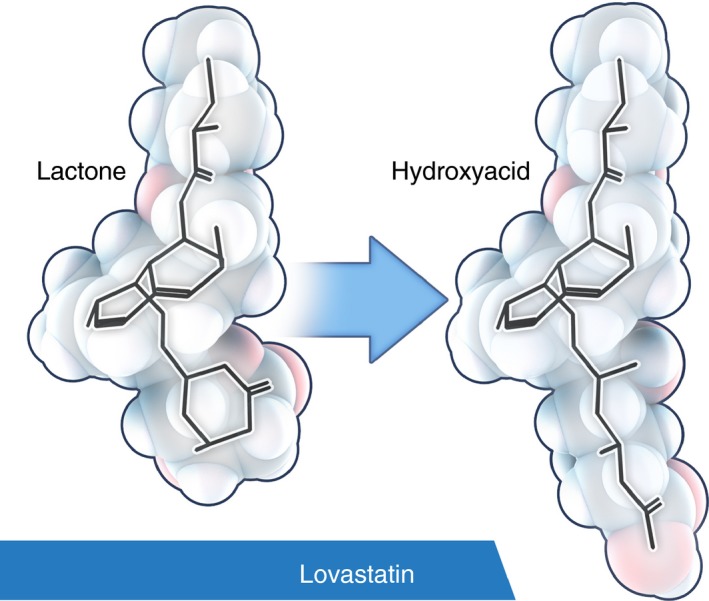
Lovastatin lactone may have a different target in archaea than the hydroxyacid. Simvastatin and lovastatin are the commercially available statins that come in the lactone form. Their cholesterol‐lowering effect and the impairment of archaeal membrane synthesis through inhibition of HMGR requires activation, i.e. the lactone ring needs to be opened to result in the hydroxyacid form. As can be seen, the stereochemistry of lovastatin lactone and hydroxyacid is significantly different. Recent evidence suggests that methanogenesis is preferentially inhibited by the lactone form of lovastatin. This and other evidence would suggest that lovastatin may have a different or an additional target other than HMGR. A possible target for the lactone form are enzymes in the methanogenesis pathway that have F420 as coenzyme. See Figure 5. The 3‐D was model generated with CORINA (http://www.molecular-networks.com/).
Hydrophilic statins
Pravastatin, whether lactone or acid, has the lowest oil‐water distribution coefficients when compared with the other statins and would not normally be expected to cross the blood–brain barrier, however, it does, at least in mice, perhaps aided by the same organic anion transporter polypeptide (OATP) that allows it to enter liver cells.49 Atorvastatin acid, lovastatin acid, and simvastatin acid, all more hydrophilic than their respective lactone forms, were also found to be transported into cells by OATP2. Other transporters such as the monocarboxylic acid transporters (MCT) may play a role as well.49
Considerations for the uptake of statins by M. smithii
Let us now return to M. smithii. The ideal statin to get absorbed by M. smithii would be a lactone prodrug such as simvastatin and lovastatin. For example, the degree of hydrophobicity of imidazole derivatives correlates with improved activity against human methanogenic archaea,51 and the situation should be analogous for statins. However, if a lactone statin is given orally and expected to inhibit the archaeal HMGR, the lactone ring needs to be broken up once inside the cell – for example, by a carboxylesterase or paroxonase, as is the case in human plasma and the liver.52 Indeed, M. smithii has a predicted acetylesterase (NCBI Ref Seq WP_019264088.1) which could catalyse this reaction. Pravastatin, the most hydrophilic of all statins, would be expected to be the least likely to cross the archaeal cell wall. In view of this, it is surprising that pravastatin remarkably inhibited the growth of M. thermautotrophicus and methanogenesis.53 Whether this can be ascribed to active transport or not, is not known.
In summary, the activity of statins in relation to methanogens can apparently not be predicted based on simple pharmacological principles alone and requires empiric evaluation. What is, then, the experimental evidence?
Experimental evidence for use of statins to inhibit methanogenesis
Interest in statins as potential inhibitors of methanogenesis originated with work in ruminants and recognition that the rate‐limiting step in the synthesis of the isoprene lipid membranes of archaea is catalysed by HMGR.46, 54 In the earliest reported studies, mevastatin and lovastatin were found to inhibit the growth of Methanobrevibacter species isolated from bovine rumen. A mevastatin concentration of 5.6 μmol/L inhibited Methanobrevibacter growth in vitro by 80–100% but did not inhibit the growth of rumen bacteria responsible for the fermentation of polysaccharides and starch in these animals (i.e. the bacteria providing methanogenic substrates), as would be expected. In subsequent studies, a lovastatin concentration of 4 μmol/L caused a 50% inhibition of Methanobrevibacter growth in vitro and 100% inhibition of growth and CH4 production was observed at a lovastatin concentration of 10 μmol/L.38
The effects of lovastatin were reiterated in studies using a rumen simulation technique (Rusitec). Lovastatin (150 mg/L; 371 μmol/L) supplementation to the Rusitec fermentation medium reduced overall CH4 production by approximately 42% without altering bacterial counts or nutrient fermentation (including concentrations of short‐chain fatty acids) in the medium. Garlic oil (300 mg/L) was more effective than lovastatin as an inhibitor of CH4 production in this study (91% reduction); however, garlic oil also inhibited bacterial growth, which likely reduced the availability of methanogenesis substrates.55 Diallyl disulphide, the main ingredient of garlic oil, is known to inhibit HMGR.56 However, garlic at the doses employed has unwanted effects on the gut microbiome, and this may be one of the reasons that this approach is apparently not being actively pursued. A study comparing diallyl disulphide and lovastatin as feed additives in sheep found that neither additive caused an absolute change in CH4 emissions; however, each treatment modestly reduced CH4 produced per g of dietary fibre consumed by these animals.57
The potential expense of utilising purified statins led to a number of studies evaluating natural sources of statins as agents to mitigate ruminant CH4 production. Lovastatin is a secondary metabolite produced during fungal growth and can be produced by cultures of Penicillium species, Aspergillus terreus, Monascus species (e.g. red yeast rice Monascus purpureus), Hypomyces, Doratomyces, Phoma, Eupenicillium, Gymnoascus and Trichoderma and is found at concentrations up to 2.8% of the dry weight of oyster mushrooms (Pleurotus ostreatus).58
Fermentation of rice straw (an agricultural waste product) with A. terreus produced lovastatin with a yield of 260.8 mg/kg dry matter. The lovastatin produced was found predominantly in the β‐hydroxyacid form (rather than the lactone), the hydroxyacid form being the active HMGR‐inhibiting species.59 A methanolic extract of the fermented rice straw (FRSE) containing lovastatin (97 mg/g dry mass) significantly reduced total gas and CH4 production by a mixed culture of ruminant organisms in vitro but did not alter H2 production. In addition, the FRSE reduced the total population of methanogens in the ruminant microbial culture, specifically lowering Methanobacteriales and aerobic fungi. FRSE also increased the expression of the HMGR gene (hmg), but had no effect on expression of methyl‐coenzyme M reductase subunit A (mcrA), the key enzyme for the common final pathway in methanogenesis.59 These findings suggest that the mechanism of action is related to an impaired biosynthesis of the cell membrane isoprenoids as we have discussed before.
A subsequent study explored the effects of lovastatin‐containing FRSE on methanogenesis in more detail by comparing the direct effects of FRSE and commercial lovastatin on M. smithii. Commercial lovastatin (administered as the lactone) and FRSE (in which approximately 75% of lovastatin was the β‐hydroxyacid) both significantly inhibited CH4 production and M. smithii growth in vitro in a dose‐dependent manner. At equivalent concentrations of lovastatin, the effect of FRSE on CH4 production was greater than observed for commercial lovastatin, suggesting the β‐hydroxyacid form of lovastatin is more active. M. smithii morphology was significantly altered by both commercial lovastatin and FRSE, resulting in abnormal membrane formation during mitosis and asymmetric (off‐centred) cell divisions.60 It is well established that the β‐hydroxyacid form of lovastatin is the active HMGR‐binding form of the molecule and, as described above, HMGR catalyses the rate‐limiting step in the synthesis of membrane lipids in archaea. Increased anti‐methanogenic activity of the FRSE (containing the β‐hydroxyacid) relative to commercial lovastatin (lactone) and alteration to M. smithii membranes are both consistent with an inhibition of cell membrane synthesis via inhibition of HMGR.
However, a comparison of the effects of FRSE and commercial lovastatin on gene expression suggests that lovastatin may directly modulate methanogenic processes. Consistent with the known action of statins, both FRSE and commercial lovastatin were found to increase HMGR gene expression. FRSE also modulated expression of several genes associated with methanogenesis, increasing expression of mtr, mta and mcr while decreasing expression of hmd and fno. Commercial lovastatin increased mtr and mta expression and decreased fno expression but has no effect on hmd.60
Other potential mechanisms of action for statins in the inhibition of methanogenesis
If gene expression of methanogenesis enzymes is affected differently by FRSE (contains lovastatin hydroxyacid) and lovastatin lactone, as suggested above, the question becomes: Is this due to the lactone and hydroxyacid species or perhaps an unknown ingredient in FRSE?
Let us assume for now that the differences are indeed entirely attributable to the lactone vs. hydroxyacid forms. In this case one would think that the lactone is not simply converted into the hydroxyacid form to exert its effect but instead has a different target or more than one target besides the HMG‐CoA reductase. Indeed, an intriguing addition to the mechanistic discussion of statin anti‐methanogenic activity was provided in a hypothesis paper comparing the structures of lovastatin and mevastatin to that of coenzyme F420. Computational modelling (in silico molecular docking) of the methanogenic enzyme F420‐dependent NADP oxidoreductase determined that both lovastatin and mevastatin had higher affinities for the F420 binding site on fno than did F420 itself. As such, lovastatin and mevastatin may act as inhibitors of fno 43 and could inhibit other enzymes that require F420 for their activity such as those that are crucial to the main methanogenesis pathway (Figure 4).
Recent experimental results seem to support a direct anti‐methanogenic effect of lovastatin lactone in humans that is not shared by other statins. Marsh et al. assessed nine statins for methane inhibition at a concentration of 5 mg/g in homogenised human stool; lovastatin lactone and hydroxyacid, pravastatin lactone and hydroxyacid, simvastatin lactone, mevastatin lactone, rosuvastatin hydroxyacid, atorvastatin lactone and hydroxyacid. Lovastatin lactone was identified as the only effective methane inhibitor, significantly inhibiting methane levels by – 65% of the control stool. Lovastatin lactone at 5 mg/g produced the maximum inhibiting effect, providing an average methane level of 3% of the control over time. In a final validation comparison of three lovastatin species (5 mg/g), both lactone and lactone‐diol types proved to be effective. In all assessments, statins of the hydroxyacid form were least able to inhibit methane production in fresh stool samples.61
Clinical development programmes
To summarise, there is an association between irritable bowel syndrome with constipation and high breath methane levels, and there is experimental evidence suggesting that methane may slow down intestinal transit. Statins, specifically lovastatin, have been shown to lower methanogenesis in human stool samples. In addition, recent research examining the effects of lovastatin in a rat model of diet‐induced constipation and M. smithii proliferation showed that gavage with lovastatin significantly reduced the ratio of M. smithii to total bacteria in the ileum. Most importantly, there was an increase in stool wet‐weight (a proxy for soft stools in rats) noted in rats receiving lovastatin lactone gavage.62 Naturally, a clinical development programme that evaluates the ability of statins to suppress methane production and associated symptoms in humans makes sense and we have consequently embarked on a proof‐of‐concept clinical trial.
To our knowledge, this is the only such programme in humans. This Phase 2, randomised, multi‐centre, multi‐dose study has enrolled sixty subjects with irritable bowel syndrome with constipation who were between the ages of 18 and 65, with a trial duration of up to 43 days.63 The study has one placebo and two active comparator arms: Low Dose 21 mg and High Dose 42 mg with an assignment of 1:1:1. The main outcome measure is change from baseline in the area under the curve (AUC) of breath CH4 production at day 7. The active drug, SYN‐010, is a proprietary lovastatin dual pulse dosage form. Our company, Synthetic Biologics hopes to release preliminary results towards the end of 2015.
Conclusion and outlook for the future
Many issues will need to be addressed on both basic science and clinical science levels. Most of the knowledge gaps are apparent from this review, and we will highlight some which we think are particularly important. Further research needs to be conducted to elucidate methane's role as potential gasotransmitter. Like research conducted around nitrous oxide, the payoffs may go beyond a better understanding of animal physiology. Next we need to understand the conditions which determine the ‘methane phenotype’ in both farm animals and humans. M. smithii is obviously tightly integrated into a network of microbial neighbours, and its own fate depends to a large extent on the composition of the local microbiome or symbiome. Next, we need to better understand the mechanism of action of statins as it pertains to archaeal growth and methanogenesis. Research in this direction could not only benefit individual humans directly by reducing their own intestinal methane production but also H. sapiens as a species, if this research leads to methods by which the production of the greenhouse gas methane can be curbed in an economical fashion. Lastly, clinical studies that evaluate statins for the treatment of IBS‐C are currently in the proof‐of‐concept stage, and further clinical trials need to be conducted.
Authorship
Guarantor: K. Gottlieb
Author contributions: K.G. initiated the work on the manuscript, performed the literature searches, wrote the first and subsequent drafts, commissioned and revised illustrations. V. W. wrote white papers related to the subject matter which were in part used for this publication. In addition, he reviewed and revised all drafts. J. S. reviewed and revised all drafts. M. P. advised on the overall content of the manuscript, reviewed and revised all drafts.
All authors approved the final version of the manuscript.
Acknowledgements
The authors thank Chris Edwards, Ph.D., for critical review of the manuscript and helpful suggestions regarding organisation and readability.
Declaration of personal interests: K.G. and J. S. are employees of Synthetic Biologics, Inc. V. W. is a paid consultant to Synthetic Biologics. M. P. is a compensated advisor to Synthetic Biologics, Inc.
Declaration of funding interests: None.
This uncommissioned review article was subject to full peer‐review.
References
- 1. Ladas SD. Colonic gas explosion during therapeutic colonoscopy with electrocautery. World J Gastroenterol 2007; 13: 5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci 1990; 87: 4576–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caetano‐Anollés G, Kim KM. The origin and evolution of the Archaeal domain. Archaea 2014; 2014: doi:10.1155/2014/915828 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pimentel M, Lin HC, Enayati P, et al Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. Am J Physiol ‐ Gastrointest Liver Physiol 2006; 290: G1089–95. [DOI] [PubMed] [Google Scholar]
- 5. Triantafyllou K, Chang C, Pimentel M. Methanogens, methane and gastrointestinal motility. J Neurogastroenterol Motil 2014; 20: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vega AB, Perelló A, Martos L, et al Breath methane in functional constipation: response to treatment with Ispaghula husk. Neurogastroenterol Motil 2015; 27: 945–53. [DOI] [PubMed] [Google Scholar]
- 7. Kang D‐W, DiBaise JK, Ilhan ZE, et al Gut microbial and short‐chain fatty acid profiles in adults with chronic constipation before and after treatment with lubiprostone. Anaerobe 2015; 33: 33–41. [DOI] [PubMed] [Google Scholar]
- 8. Wilder‐Smith CH, Materna A, Wermelinger C, Schuler J. Fructose and lactose intolerance and malabsorption testing: the relationship with symptoms in functional gastrointestinal disorders. Aliment Pharmacol Ther 2013; 37: 1074–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Di Stefano M, Mengoli C, Bergonzi M, et al Breath methane excretion is not an accurate marker of colonic methane production in irritable bowel syndrome. Am J Gastroenterol 2015; 110: 891–8. [DOI] [PubMed] [Google Scholar]
- 10. Vandeputte D, Falony G, Vieira‐Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 2015; doi:10.1136/gutjnl‐2015‐309618 [epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pozuelo M, Panda S, Santiago A, et al Reduction of butyrate‐ and methane‐producing microorganisms in patients with Irritable Bowel Syndrome. Sci Rep 2015; 5: 12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lewis S, Cochrane S. Alteration of sulfate and hydrogen metabolism in the human colon by changing intestinal transit rate. Am J Gastroenterol 2007; 102: 624–33. [DOI] [PubMed] [Google Scholar]
- 13. Majewski M, McCallum RW. Results of small intestinal bacterial overgrowth testing in irritable bowel syndrome patients: clinical profiles and effects of antibiotic trial. Adv Med Sci 2007; 52: 139–42. [PubMed] [Google Scholar]
- 14. Jahng J, Jung IS, Choi EJ, Conklin JL, Park H. The effects of methane and hydrogen gases produced by enteric bacteria on ileal motility and colonic transit time. Neurogastroenterol Motil 2012; 24: 185–e92. [DOI] [PubMed] [Google Scholar]
- 15. Vermeiren J, Van de Wiele T, Van Nieuwenhuyse G, Boeckx P, Verstraete W, Boon N. Sulfide‐ and nitrite‐dependent nitric oxide production in the intestinal tract: NO release from nitrite by sulfide. Microb Biotechnol 2012; 5: 379–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leahy SC, Kelly WJ, Altermann E, et al The genome sequence of the rumen methanogen methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS ONE 2010; 5: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hedderich R, Whitman WB. Physiology and biochemistry of the methane‐producing archaea In: Dworkin M, Falkow S, Rosenberg E, Schleifer K‐H, Stackebrandt E, eds. The Prokaryotes. New York: Springer, 2006; 1050–79. [Google Scholar]
- 18. Liu J, Chen H, Zhu Q, et al A novel pathway of direct methane production and emission by eukaryotes including plants, animals and fungi: an overview. Atmos Environ 2015; 115: 26–35. [Google Scholar]
- 19. Boros M, Tuboly E, Mészáros A, Amann A. The role of methane in mammalian physiology—is it a gasotransmitter? J Breath Res 2015; 9: 014001. [DOI] [PubMed] [Google Scholar]
- 20. Probst AJ, Auerbach AK, Moissl‐Eichinger C. Archaea on human skin. PLoS ONE 2013; 8: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bang C, Schmitz RA. Archaea associated with human surfaces: not to be underestimated. FEMS Microbiol Rev 2015; doi: 10.1093/femsre/fuv010 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 22. de Lacy Costello BPJ, Ledochowski M, Ratcliffe NM. The importance of methane breath testing: a review. J Breath Res 2013; 7: 024001. [DOI] [PubMed] [Google Scholar]
- 23. Weaver GA, Krause JA, Miller TL, Wolin MJ. Incidence of methanogenic bacteria in a sigmoidoscopy population: an association of methanogenic bacteria and diverticulosis. Gut 1986; 27: 698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kittelmann S, Pinares‐Patiño CS, Seedorf H, et al Two different bacterial community types are linked with the low‐methane emission trait in sheep. PLoS ONE 2014; 9: e103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang R. Gasotransmitters: growing pains and joys. Trends Biochem Sci 2014; 39: 227–32. [DOI] [PubMed] [Google Scholar]
- 26. Boswell‐Smith V, Spina D, Page CP. Phosphodiesterase inhibitors. Br J Pharmacol 2006; 147(Suppl. 1): S252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gemici B, Elsheikh W, Feitosa KB, Costa SKP, Muscara MN, Wallace JL. H2S‐releasing drugs: anti‐inflammatory, cytoprotective and chemopreventative potential. Nitric Oxide 2015; 46: 25–31. [DOI] [PubMed] [Google Scholar]
- 28. Wallace JL, Wang R. Hydrogen sulfide‐based therapeutics: exploiting a unique but ubiquitous gasotransmitter. Nat Rev Drug Discov 2015; 14: 329–45. [DOI] [PubMed] [Google Scholar]
- 29. Boros M, Ghyczy M, Érces D, et al The anti‐inflammatory effects of methane*. Crit Care Med 2012; 40: 1269–78. [DOI] [PubMed] [Google Scholar]
- 30. Gribaldo S, Brochier‐Armanet C. The origin and evolution of Archaea: a state of the art. Philos Trans R Soc Lond B Biol Sci 2006; 361: 1007–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Whitman WB, Bowen TL, Boone DR. The methanogenic bacteria In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, eds. The Prokaryotes. Berlin Heidelberg: Springer, 2014; 123–63. [Google Scholar]
- 32. Lewis S, Brazier J, Beard D, Nazem N, Proctor D. Effects of metronidazole and oligofructose on faecal concentrations of sulphate‐reducing bacteria and their activity in human volunteers. Scand J Gastroenterol 2005; 40: 1296–303. [DOI] [PubMed] [Google Scholar]
- 33. Nakamura N, Lin HC, McSweeney CS, Mackie RI, Gaskins HR. Mechanisms of microbial hydrogen disposal in the human colon and implications for health and disease. Annu Rev Food Sci Technol 2010; 1: 363–95. [DOI] [PubMed] [Google Scholar]
- 34. Oren A. The family methanobacteriaceae In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, eds. The Prokaryotes. Berlin Heidelberg: Springer, 2014; 165–93. [Google Scholar]
- 35. Claus H, König H. Cell envelopes of methanogens In: König H, Claus H, Varma A, eds. Prokaryotic Cell Wall Compounds. Berlin Heidelberg: Springer, 2010; 231–51. [Google Scholar]
- 36. Perez‐Gil J, Rodriguez‐Concepcion M. Metabolic plasticity for isoprenoid biosynthesis in bacteria. Biochem J 2013; 452: 19–25. [DOI] [PubMed] [Google Scholar]
- 37. Jain S, Caforio A, Driessen AJM. Biosynthesis of archaeal membrane ether lipids. Microb Physiol Metab 2014; 5: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miller TL, Wolin MJ. Inhibition of growth of methane‐producing bacteria of the ruminant forestomach by hydroxymethylglutaryl∼SCoA reductase inhibitors. J Dairy Sci 2001; 84: 1445–8. [DOI] [PubMed] [Google Scholar]
- 39. Khelaifia S, Drancourt M. Susceptibility of archaea to antimicrobial agents: applications to clinical microbiology. Clin Microbiol Infect 2012; 18: 841–8. [DOI] [PubMed] [Google Scholar]
- 40. Rodríguez‐Concepción M, Boronat A. Isoprenoid biosynthesis in prokaryotic organisms In: Bach TJ, Rohmer M, eds. Isoprenoid Synthesis in Plants and Microorganisms. New York: Springer, 2012; 1–16. [Google Scholar]
- 41. Sprott GD, Brisson J‐R, Dicaire CJ, et al A structural comparison of the total polar lipids from the human archaea Methanobrevibacter smithii and Methanosphaera stadtmanae and its relevance to the adjuvant activities of their liposomes1. Biochim Biophys Acta BBA – Mol Cell Biol Lipids 1999; 1440: 275–88. [DOI] [PubMed] [Google Scholar]
- 42. Bauersachs T, Weidenbach K, Schmitz RA, Schwark L. Distribution of glycerol ether lipids in halophilic, methanogenic and hyperthermophilic archaea. Org Geochem 2015; 83–84: 101–8. [Google Scholar]
- 43. Sharma A, Chaudhary PP, Sirohi SK, Saxena J. Structure modeling and inhibitor prediction ofNADP oxidoreductase enzyme from Methanobrevibacter smithii . Bioinformation 2011; 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bashiri G, Rehan AM, Greenwood DR, Dickson JMJ, Baker EN. Metabolic engineering of cofactor F420 production in Mycobacterium smegmatis . PLoS ONE 2010; 5: e15803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Forouhar F, Abashidze M, Xu H, et al Molecular insights into the biosynthesis of the F420 coenzyme. J Biol Chem 2008; 283: 11832–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Samuel BS, Hansen EE, Manchester JK, et al Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci USA 2007; 104: 10643–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sirtori CR. The pharmacology of statins. Pharmacol Res 2014; 88: 3–11. [DOI] [PubMed] [Google Scholar]
- 48. Fong CW. Statins in therapy: understanding their hydrophilicity, lipophilicity, binding to 3‐hydroxy‐3‐methylglutaryl‐CoA reductase, ability to cross the blood brain barrier and metabolic stability based on electrostatic molecular orbital studies. Eur J Med Chem 2014; 85: 661–74. [DOI] [PubMed] [Google Scholar]
- 49. Wood WG, Mΰller WE, Eckert GP. Statins and neuroprotection: basic pharmacology needed. Mol Neurobiol 2014; 50: 214–20. [DOI] [PubMed] [Google Scholar]
- 50. Ishigami M, Honda T, Takasaki W, et al A comparison of the effects of 3‐hydroxy‐3‐methylglutaryl‐coenzyme a (HMG‐CoA) reductase inhibitors on the CYP3A4‐dependent oxidation of mexazolam in vitro. Drug Metab Dispos 2001; 29: 282–8. [PubMed] [Google Scholar]
- 51. Khelaifia S, Brunel JM, Raoult D, Drancourt M. Hydrophobicity of imidazole derivatives correlates with improved activity against human methanogenic archaea. Int J Antimicrob Agents 2013; 41: 544–7. [DOI] [PubMed] [Google Scholar]
- 52. Tang B‐K, Kalow W. Variable activation of lovastatin by hydrolytic enzymes in human plasma and liver. Eur J Clin Pharmacol 1995; 47: 449–51. [DOI] [PubMed] [Google Scholar]
- 53. Nováková Z, Blasko J, Hapala I, Smigá P. Effects of 3‐hydroxy‐3‐methylglutaryl‐coenzyme a reductase inhibitor pravastatin on membrane lipids and membrane associated functions of Methanothermobacter thermautotrophicus. Folia Microbiol (Praha) 2010; 55: 359–62. [DOI] [PubMed] [Google Scholar]
- 54. Dridi B, Raoult D, Drancourt M. Archaea as emerging organisms in complex human microbiomes. Anaerobe 2011; 17: 56–63. [DOI] [PubMed] [Google Scholar]
- 55. Soliva CR, Amelchanka SL, Duval SM, Kreuzer M. Ruminal methane inhibition potential of various pure compounds in comparison with garlic oil as determined with a rumen simulation technique (Rusitec). Br J Nutr 2011; 106: 114–22. [DOI] [PubMed] [Google Scholar]
- 56. Calsamiglia S, Busquet M, Cardozo PW, Castillejos L, Ferret A. Invited review: essential oils as modifiers of rumen microbial fermentation. J Dairy Sci 2007; 90: 2580–95. [DOI] [PubMed] [Google Scholar]
- 57. Klevenhusen F, Duval S, Zeitz JO, Kreuzer M, Soliva CR. Diallyl disulphide and lovastatin: effects on energy and protein utilisation in, as well as methane emission from, sheep. Arch Anim Nutr 2011; 65: 255–66. [DOI] [PubMed] [Google Scholar]
- 58. Faseleh Jahromi M, Liang JB, Ho YW, Mohamad R, Goh YM, Shokryazdan P. Lovastatin production by Aspergillus terreus using agro‐biomass as substrate in solid state fermentation. J Biomed Biotechnol 2012; 2012: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Faseleh Jahromi M, Liang JB, Mohamad R, Goh YM, Shokryazdan P, Ho YW. Lovastatin‐enriched rice straw enhances biomass quality and suppresses ruminal methanogenesis. BioMed Res Int 2013; 2013: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Faseleh Jahromi M, Liang JB, Ho YW, et al Lovastatin in Aspergillus terreus: fermented rice straw extracts interferes with methane production and gene expression in Methanobrevibacter smithii . BioMed Res Int. 2013; 2013: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marsh E, Morales W, Marsh Z, et al Lovastatin lactone inhibits methane production in human stool homogenates. Poster Presentation presented at: American College of Gastroenterology Annual Meeting; 16 October 2015; Honolulu, HI.
- 62. Morales W, Marsh E, Yu A, et al Mo2051 lovastatin improves stool form in Methanobrevibacter smithii colonized rats with constipation. Gastroenterology 2015; 148: S–779. [Google Scholar]
- 63. Synthetic Biologics . A Randomized, Double‐Blind, Parallel‐Group, Placebo‐Controlled, Study of the Effect of SYN‐010 on Subjects With IBS‐C [Internet]. Clinicaltrials.gov, 2015. June. Report No.: SB‐2‐010‐001. Available at: https://clinicaltrials.gov/ct2/show/NCT02495623. Accessed 3 November 2015.
- 64. ITIS Standard Report Page: Methanobrevibacter [Internet]. Integrated Taxonomic Information System. Available at: http://www.itis.gov/servlet/SingleRpt/SingleRpt (accessed 30 June 2015).
- 65. Global Catalogue of Microorganisms [Internet]. Available at: http://gcm.wfcc.info/ (accessed 30 June 2015).
- 66. Madigan MT, Clark DP, Stahl D, Martinko JM. Brock Biology of Microorganisms. 13th ed. San Francisco: Benjamin Cummings, 2010: 1155 pp. [Google Scholar]


