Abstract
Urine has evolved as one of the most important biofluids in clinical proteomics due to its noninvasive sampling and its stability. Yet, it is used in clinical diagnostics of several disorders by detecting changes in its components including urinary protein/polypeptide profile. Despite the fact that majority of proteins detected in urine are primarily originated from the urogenital (UG) tract, determining its precise source within the UG tract remains elusive. In this article, we performed a comprehensive analysis of ureter proteome to assemble the first unbiased ureter dataset. Next, we compared these data to urine, urinary exosome, and kidney mass spectrometric datasets. Our result concluded that among 2217 nonredundant ureter proteins, 751 protein candidates (33.8%) were detected in urine as urinary protein/polypeptide or exosomal protein. On the other hand, comparing ureter protein hits (48) that are not shown in corresponding databases to urinary bladder and prostate human protein atlas databases pinpointed 21 proteins that might be unique to ureter tissue. In conclusion, this finding offers future perspectives for possible identification of ureter disease‐associated biomarkers such as ureter carcinoma. In addition, the ureter proteomic dataset published in this article will provide a valuable resource for researchers working in the field of urology and urine biomarker discovery. All MS data have been deposited in the ProteomeXchange with identifier PXD002620 (http://proteomecentral.proteomexchange.org/dataset/PXD002620).
Keywords: Biomarker, Cell biology, Dataset, OFFGel fractionation, Ureter, Urine
Abbreviations
- emPAI
Exponentially Modified Protein Abundance Index
- HPA
human protein atlas
- UG
urogenital
Screening of human urinary proteome for disease biomarkers is a subject of attractions in clinical proteomics 1, 2, 3, 4. Besides sampling simplicity and relative stability to other biofluids, urine proteome analysis is preferred over other complex biofluids (serum or plasma) because it is less complex especially when targeting protein/peptide biomarkers 5, 6. As a regular excretory process, urine is produced in the kidneys, to eliminate plasma wastes, and passed through a 25–30 cm long tube “ureters” to be stored in the urinary bladder 1. During this process, more than 90% of the “initial” urine is reabsorbed 1. The remaining “final” urine contains not only wasted molecules but also some protein molecules originated from urogenital (UG) tract and plasma as well. Theoretically, urinary proteins might be originated from renal tubular secretion of soluble proteins, sloughed cells of UG tract, glycosylphosphatidylinositol‐anchored protein detachment (such as uromodulin), membranous/cytosolic exosomal proteins, or glomerular‐filtrated plasma proteins 7, 8. It was reported that in a normal healthy individual, 70% of urinary proteome are originated from the UG tract, while remaining 30% are plasma proteins filtered by the glomerulus 1. Although several studies have been published describing the potential uses of urine proteome and peptidome in some UG diseases (acute transplant rejection 9, UG tract infection 10, chronic kidney disease 11, cancer 12), little is known regarding where these protein molecules originated from. More specifically within UG tract, if these proteins originated from the kidneys, ureter, or urinary bladder.
To disclose this fundamental question, we attempted to analyze the ureter proteome. Then, we proposed that some of these proteins might be released in urine during its passage within the ureter as entrained exosomes or as a process of urothelium turnover. In order to confirm the validity of our assumption, we compared ureter proteome with a comprehensive mass spectrometric urinary database, exosomal urinary database, and kidney database. Retained protein hits to ureter were further compared to urinary bladder and prostate databases from human protein atlas (HPA) [13].
Normal healthy ureter sample (4–5 cm in length) was dissected from diseased kidney and obtained from individuals with informed consent and under the approval of Committee of Ethics for Life and Genes of the Graduate School of Medical and Dental Sciences, Niigata University. Protein extracts were obtained by placing dissected ureter tissues in protein OFFGel prefractionation buffer supplied by the manufacturer (containing urea, thiourea, DTT, glycerol, and buffer with ampholytes pH [3–10]) 14, 15, 16. Complete ultraproteases (Roche, Mannheim, Germany) were added to the buffer. Precellys 24 tissue homogenizer was used for protein extraction at 4°C (Precellys, Bertin Technologies). Two milligram of recovered protein extract was subjected to OFFGel fractionation using 3100 OFFGel fractionator (Agilent Technologies, Japan) as previously described 14. Twelve fraction platforms were used in the current study. Following successful prefractionation, acetone precipitation, and protein quantification, 80 μg from each OFFGel fractions (n = 12) were subjected to reduction and alkylation, and digested with trypsin as described elsewhere 14, 17. Digested peptide solution was acidified using 90% formic acid to a final pH 3 and enriched using stage tip 18, 19. Efficiency of fractionation and digestion was confirmed as shown in Supporting Information 1. Chromatography of purified peptides was performed using Thermo Q‐Exactive and separation was applied using a binary gradient for 120 min with ACN as mobile phase. The precursor full MS scan ranged from 40–1200 m/z. Dynamic exclusion setting used were as follows: repeat count, 1; repeat duration, 30 s; exclusion list size, 450; and exclusion duration 60 s. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD002620 and 10.6019/PXD002620. In addition, all raw data (Thermo.RAW) are also available in the Peptide Atlas repository at ftp://PASS00641:ZI6249ae@ftp.peptideatlas.org/ 20.
Protein and peptide identification were searched by MASCOT in addition to ProluCID search engine implemented in the integrated proteomics pipeline; IP2 (http://integratedproteomics.com/, version 1.01) 14. Tandem mass spectra were generated using RawExtract (version 1.9.9) and the MS/MS spectra were searched against updated UniProtKB/TrEMBL (Homo sapiens, 935 651 entries, released in February, 2015). The spectral search space included all fully and half‐tryptic peptide candidates within a 5‐ppm window surrounding the peptide candidate precursor mass. Carbamidomethylation (+57.02146) of cysteine was considered a static modification and oxidation at MHW (+15.995) as variable modification. Peptide candidates were filtered to 0.1% FDR and proteins candidates to 1% FDR using DTASelect 21 with a 10‐ppm window.
Our data analysis reported 2217 nonredundant protein representing ureter proteome (after omitting interfractional redundancy; see Fig. 1). Full list of protein description is available in Supporting Information 2. Interestingly, when we compared the ureter dataset against urine 22, urinary exosome 23, 24, and kidney databases 25, 26, 27, 28, 29, result showed that 24.3% of the ureter annotated proteins (540 candidates) were detected in earlier urine database reports (see Fig. 2A; zone E–H). Moreover, 21% of ureter proteins (467 proteins) were excreted as urinary exosomes (zone B, C, F, and G; details in Supporting Information 2). Validating these proteins in urine might allow more precise and supportive diagnostic determination of ureter disorders. Taken together, we conclude that over one‐third (33.8%; 751 proteins) of ureter proteome could be detected in urine as protein/polypeptide or in a vesicular exosomal form. Forty eight proteins were retained for ureter proteome (Fig. 2A, zone A); These hits were further compared to urinary bladder and prostate database from HPA. We found that 21 protein candidates were either in a pending state, not found in HPA, or unique to ureter (Fig. 2B, Supporting Information 3). Further investigations are required for these proteins. As illustrated in Fig. 3, the Exponentially Modified Protein Abundance Index 30, 31 (emPAI) values reflected the overwhelming abundance of these proteins in the urine. Consequently, these proteins may provide clues for monitoring the pathophysiology of the ureter. In conclusion, differentiating the precise origin of urinary proteins is still at its earliest stages and the full picture of urine biomarkers will be completed when we know not only the signature proteins of the UG tract tissues, but also plasma proteins as well. The ureter database generated in the current study will be initiative for further studies aimed at discriminating proteome of the UG tract and urine biomarkers precisely.
Figure 1.
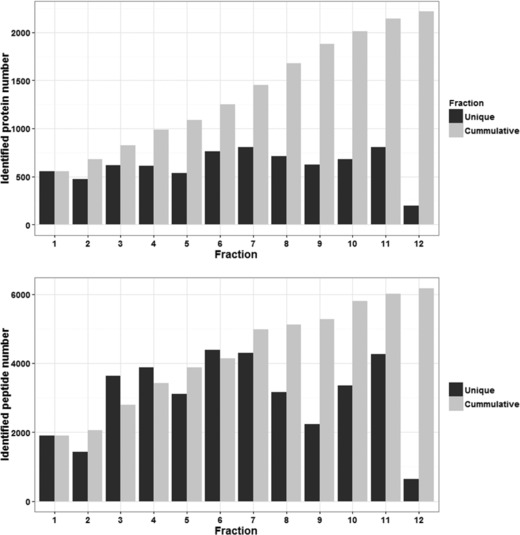
Identified proteins and peptides of the human ureter proteome. Upper figure, identified nonredundant proteins (within fraction) in 12 OFFGel fractions. Lower figure, identified nonredundant peptides (within fraction) in 12 OFFGel fractions. Black bars represent unique protein or peptide candidate within fraction. Gray bars represent newly added proteins/peptides from the subsequent fraction.
Figure 2.
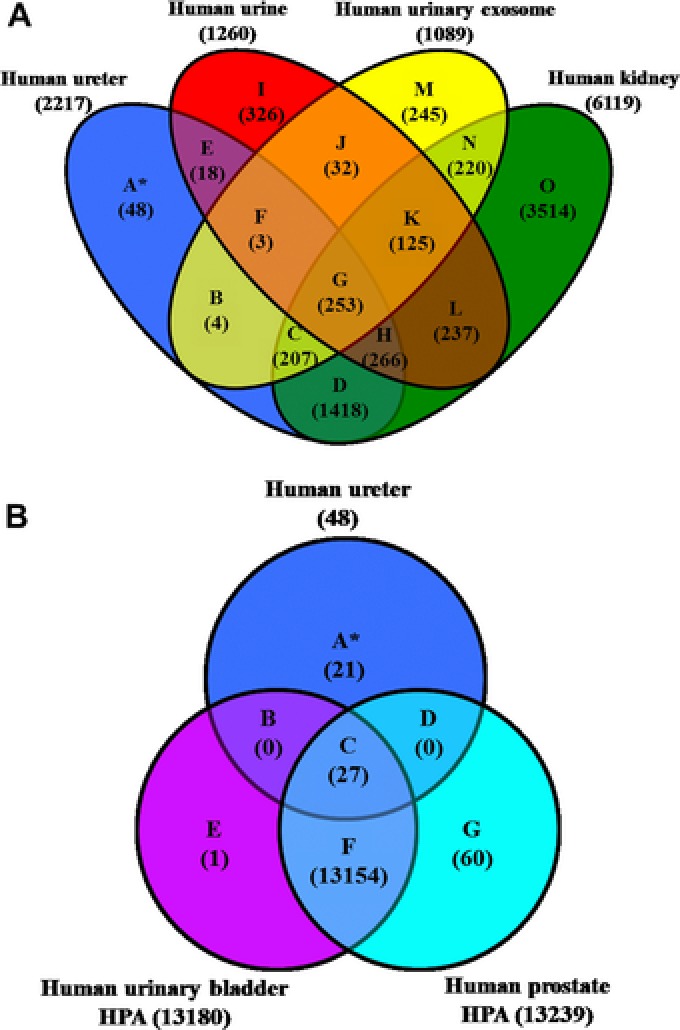
Venn diagrams of unique and shared ureter proteome with other databases. (A) Venn diagram of human ureter proteome overlapped with urinary, urinary exosomal, and kidney mass spectrometric databases. (B) Nonshared proteins in panel A (zone A) was further compared to human urinary bladder and prostate databases retrieved from HPA (based on immunohistochemistry) where 27 proteins were shared and 21 [see Supporting Information 3].
Figure 3.
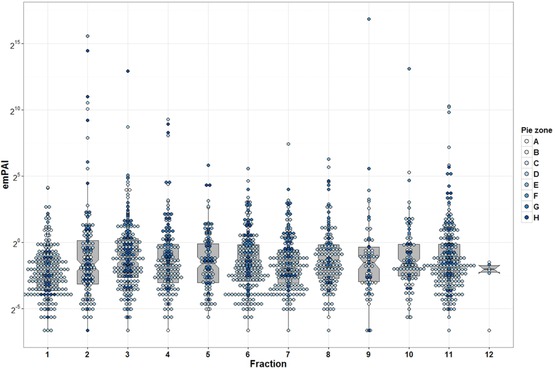
Whisker and box plot overlaid with dot plot showing median and quartile values of the emPAI for ureter proteome database. Dots color represents pie zone location based on Fig. 2A sorting. The y‐axis shows emPAI value in log2 view.
The authors have declared no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Supporting Information
Acknowledgments
The MS proteomics data in this paper have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [20]: dataset identifier PXD002620.
This work was financially supported by Ministry of Education, Culture, Sports, Science, and Technology of Japan to S.M. under the standard JSPS grant for foreign researcher (P 14105) and by the Center of Innovation Program from Japan Science and Technology Agency, JST to T.Y. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Colour Online: See the article online to view Fig. 2 in colour.
References
- 1. Decramer, S. , Gonzalez de Peredo, A. , Breuil, B. , Mischak, H. et al., Urine in clinical proteomics. Mol. Cell. Proteomics 2008, 7, 1850–1862. [DOI] [PubMed] [Google Scholar]
- 2. Zhang, X. , Jin, M. , Wu, H. , Nadasdy, T. et al., Biomarkers of lupus nephritis determined by serial urine proteomics. Kidney Int. 2008, 74, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu, J. , Chen, Y. D. , Gu, W. , Urinary proteomics as a novel tool for biomarker discovery in kidney diseases. J. Zhejiang Univ. Sci. B 2010, 11, 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. von zur Muhlen, C. , Schiffer, E. , Sackmann, C. , Zurbig, P. et al., Urine proteome analysis reflects atherosclerotic disease in an ApoE‒/‒ mouse model and allows the discovery of new candidate biomarkers in mouse and human atherosclerosis. Mol. Cell. Proteomics 2012, 11, M111.013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roelofsen, H. , Alvarez‐Llamas, G. , Schepers, M. , Landman, K. , Vonk, R. J. , Proteomics profiling of urine with surface enhanced laser desorption/ionization time of flight mass spectrometry. Proteome Sci. 2007, 5, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moon, P. G. , You, S. , Lee, J. E. , Hwang, D. , Baek, M. C. , Urinary exosomes and proteomics. Mass Spectrom. Rev. 2011, 30, 1185–1202. [DOI] [PubMed] [Google Scholar]
- 7. Iorember, F. M. , Vehaskari, V. M. , Uromodulin: old friend with new roles in health and disease. Pediatr. Nephrol. 2014, 29, 1151–1158. [DOI] [PubMed] [Google Scholar]
- 8. Hoorn, E. J. , Pisitkun, T. , Zietse, R. , Gross, P. et al., Prospects for urinary proteomics: exosomes as a source of urinary biomarkers. Nephrology 2005, 10, 283–290. [DOI] [PubMed] [Google Scholar]
- 9. Schaub, S. , Rush, D. , Wilkins, J. , Gibson, I. W. et al., Proteomic‐based detection of urine proteins associated with acute renal allograft rejection. J. Am. Soc. Nephrol. 2004, 15, 219–227. [DOI] [PubMed] [Google Scholar]
- 10. Wittke, S. , Haubitz, M. , Walden, M. , Rohde, F. et al., Detection of acute tubulointerstitial rejection by proteomic analysis of urinary samples in renal transplant recipients. Am. J. Transplant. 2005, 5, 2479–2488. [DOI] [PubMed] [Google Scholar]
- 11. Meguid El Nahas, A ., Bello, A. K. , Chronic kidney disease: the global challenge. Lancet 2005, 365, 331–340. [DOI] [PubMed] [Google Scholar]
- 12. Hernandez, J. , Thompson, I. M. , Prostate‐specific antigen: a review of the validation of the most commonly used cancer biomarker. Cancer 2004, 101, 894–904. [DOI] [PubMed] [Google Scholar]
- 13. Uhlen, M. , Oksvold, P. , Fagerberg, L. , Lundberg, E. et al., Towards a knowledge‐based human protein atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [DOI] [PubMed] [Google Scholar]
- 14. Magdeldin, S. , Yamamoto, K. , Yoshida, Y. , Xu, B. et al., Deep proteome mapping of mouse kidney based on OFFGel prefractionation reveals remarkable protein post‐ translational modifications. J. Proteome Res. 2014, 13, 1636–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Magdeldin, S. , Elguoshy, A. , Yoshida, Y. , Hirao, Y. et al., Complementary protein and peptide OFFGEL fractionation for high‐throughput proteomic analysis. Anal. Chem. 2015, 87, 8481–8488. [DOI] [PubMed] [Google Scholar]
- 16. Magdeldin, S. , Moser, A. , Affinity Chromatography: Principles and Applications, InTech, Rijeka 2012, pp. 1–16.
- 17. Magdeldin, S. , Moresco, J. J. , Yamamoto, T. , Off‐line multidimensional liquid chromatography and auto sampling result in sample loss in LC/LC‐MS/MS. J. Proteome Res. 2014, 13, 3826–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rappsilber, J. , Ishihama, Y. , Mann, M. , Stop and go extraction tips for matrix‐assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003, 75, 663–670. [DOI] [PubMed] [Google Scholar]
- 19. Rappsilber, J. , Mann, M. , Ishihama, Y. , Protocol for micro‐purification, enrichment, pre‐fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [DOI] [PubMed] [Google Scholar]
- 20. Vizcaino, J. A. , Cote, R. G. , Csordas, A. , Dianes, J. A. et al., The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013, 41, D1063–D1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tabb, D. L. , McDonald, W. H. , Yates, J. R., 3rd , DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002, 1, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marimuthu, A. , O'Meally, R. N. , Chaerkady, R. , Subbannayya, Y. et al., A comprehensive map of the human urinary proteome. J. Proteome Res. 2011, 10, 2734–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pisitkun, T. , Shen, R. F. , Knepper, M. A. , Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gonzales, P. A. , Pisitkun, T. , Hoffert, J. D. , Tchapyjnikov, D. et al., Large‐scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 2009, 20, 363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilhelm, M. , Schlegl, J. , Hahne, H. , Moghaddas Gholami, A. et al., Mass‐spectrometry‐based draft of the human proteome. Nature 2014, 509, 582–587. [DOI] [PubMed] [Google Scholar]
- 26. Pinto, S. M. , Manda, S. S. , Kim, M. S. , Taylor, K. et al., Functional annotation of proteome encoded by human chromosome 22. J. Proteome Res. 2014, 13, 2749–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miyamoto, M. , Yoshida, Y. , Taguchi, I. , Nagasaka, Y. et al., In‐depth proteomic profiling of the normal human kidney glomerulus using two‐dimensional protein prefractionation in combination with liquid chromatography‐tandem mass spectrometry. J. Proteome Res. 2007, 6, 3680–3690. [DOI] [PubMed] [Google Scholar]
- 28. Cui, Z. , Yoshida, Y. , Xu, B. , Zhang, Y. et al., Profiling and annotation of human kidney glomerulus proteome. Proteome Sci. 2013, 11, 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang, Y. , Yoshida, Y. , Xu, B. , Magdeldin, S. et al., Comparison of human glomerulus proteomic profiles obtained from low quantities of samples by different mass spectrometry with the comprehensive database. Proteome Sci. 2011, 9, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ishihama, Y. , Oda, Y. , Tabata, T. , Sato, T. et al., Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 2005, 4, 1265–1272. [DOI] [PubMed] [Google Scholar]
- 31. Magdeldin, S. , Yoshida, Y. , Li, H. , Maeda, Y. et al., Murine colon proteome and characterization of the protein pathways. BioData Min. 2012, 5, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Supporting Information


