Summary
Plants display numerous strategies to cope with phosphate (Pi)‐deficiency. Despite multiple genetic studies, the molecular mechanisms of low‐Pi‐signalling remain unknown. To validate the interest of chemical genetics to investigate this pathway we discovered and analysed the effects of PHOSTIN (PSN), a drug mimicking Pi‐starvation in Arabidopsis.
We assessed the effects of PSN and structural analogues on the induction of Pi‐deficiency responses in mutants and wild‐type and followed their accumulation in plants organs by high pressure liquid chromotography (HPLC) or mass‐spectrophotometry.
We show that PSN is cleaved in the growth medium, releasing its active motif (PSN11), which accumulates in plants roots. Despite the overaccumulation of Pi in the roots of treated plants, PSN11 elicits both local and systemic Pi‐starvation effects. Nevertheless, albeit that the transcriptional activation of low‐Pi genes by PSN11 is lost in the phr1;phl1 double mutant, neither PHO1 nor PHO2 are required for PSN11 effects.
The range of local and systemic responses to Pi‐starvation elicited, and their dependence on the PHR1/PHL1 function suggests that PSN11 affects an important and early step of Pi‐starvation signalling. Its independence from PHO1 and PHO2 suggest the existence of unknown pathway(s), showing the usefulness of PSN and chemical genetics to bring new elements to this field.
Keywords: Arabidopsis thaliana, chemical genetics, Oryza sativa, phosphate homeostasis, phosphate starvation, PHOSTIN, phr1;phl1 mutant
Introduction
Phosphorus is a fundamental element for life. As for several other nutrients (nitrate, potassium, iron, etc.), spatial and temporal phosphate (Pi)‐deficiency triggers in plants local and long‐distance (systemic) molecular, biochemical and morphological responses, in order to adapt their physiology to the heterogeneous distribution and availability of Pi in soil.
Transcriptomic analysis in Arabidopsis has helped to decipher networks and clusters of target genes that are coordinately regulated by Pi‐starvation. For example, genes related to Pi‐homeostasis maintenance, involved in Pi absorption and acquisition, or in Pi retrieval are systemically upregulated. By contrast, genes locally induced are involved in stress responses (oxidative processes, defence responses and metal detoxification) (Thibaud et al., 2010).
There is accumulating evidence that these appropriate responses rely on sensing mechanisms and complex signalling cascades (see for review Chiou & Lin, 2011; Zhang et al., 2014). Local responses depend of the Pi concentration in the growth medium surrounding the tissues (Bates & Lynch, 1996; Svistoonoff et al., 2007; Thibaud et al., 2010), while systemic responses depend on the phosphorus status of the whole plant (Foehse & Jungk, 1983; Linkohr et al., 2002; Thibaud et al., 2010).
Because of their importance for plant acclimation to low‐Pi, numerous genetic studies have been done over the past 15 yr. Nevertheless, only few elements of these local and systemic signalling pathways have been unveiled (Yang & Finnegan, 2010; Abel, 2011).
Some transcription factors have been found controlling some of the Pi‐starvation responses. But only PHR1 and its paralogue PHL1 (PHR1‐like 1) regulate the expression of numerous genes involved in various responses induced by Pi‐starvation. Both are important for long‐distance Pi‐signalling regulating Pi transport and remobilization as well as the induction of anthocyanin biosynthesis and the changes in carbohydrate metabolism observed during Pi‐starvation (Rubio et al., 2001; Bustos et al., 2010). Interestingly, the loss of function of PHR1 also impairs some local responses to Pi‐starvation as the transcriptomic induction of the ribonuclease RNS1 (Rubio et al., 2001; Duan et al., 2008; Thibaud et al., 2010).
The regulation of PHO2, an ubiquitin‐conjugating E2 enzyme (UBC24) that tags for degradation of some proteins involved in Pi transport (Liu et al., 2012; Huang et al., 2013; Park et al., 2014), is by now the best described systemic signal linking leaves with roots that regulate Pi absorption (Fujii et al., 2005; Aung et al., 2006; Bari et al., 2006; Chiou et al., 2006). This regulation occurs through the action of the microRNA miR399 that targets and cleaves PHO2 mRNA in the roots. The expression of miR399 in the shoots depends on PHR1 and its transfer to the roots maintains an appropriate Pi‐homeostasis under Pi‐sufficient conditions. This shoot–roots movement makes of miR399 the first component of a systemic signal specifically controlling Pi‐homeostasis.
Finally, the signalling steps responsible for the elicitation of the local responses to Pi‐starvation are unknown. However, in Arabidopsis, the inhibition of the primary root meristem activity when encountering a Pi‐deficient medium (Sánchez‐Calderón et al., 2005) was found to control the elicitation of the local transcriptional responses to low‐Pi (Lai et al., 2007; Thibaud et al., 2010).
Our knowledge on mechanisms governing local and systemic responses to low‐Pi is rudimentary. The connection between the known actors of these pathways remains obscure and the sensing mechanism(s) controlling the local and long‐distance responses to Pi‐deficiency are unknown. Functional redundancy supported by multigenic families and lethal mutations are known to be limiting elements when it comes to elucidate signalling pathways and could explain the difficulties encountered by genetics approaches to elucidate the Pi‐sensing mechanism.
Chemical genetics has been proved to be a powerful approach for studying biological processes and identifying relevant elements in signalling pathways in plants (Blackwell & Zhao, 2003; McCourt & Desveaux, 2010; Tóth & van der Hoorn, 2010). For example, the small molecule Pyrabactin was instrumental to discover the abscisic acid (ABA) receptors PYR1 (Park et al., 2009). Like genetic mutations, the small molecules (drugs) used in chemical genetic approaches can perturb biologic processes and thus represent tools to dissect any biological function. Furthermore, one drug can alter the activity of several proteins of the same family, thereby offering an alternative strategy to overcome higher plants functional redundancy in multigenic families. In addition, the drugs dose‐effects relation, allow the characterization of genes that display lethal loss of function (Cutler & McCourt, 2005). Finding and characterizing mutants (Zhao et al., 2003, 2007; Rojas‐Pierce et al., 2007; Park et al., 2009; Rosado et al., 2011) or natural accessions (Zhao et al., 2007) with altered sensitivity to the drug allows the discovery of genes that escaped the classical phenotype‐based genetics screens. For these reasons, we have considered chemical genetics as a new approach to study the low‐Pi‐signalling.
In this paper, we demonstrate that chemical genetics is a valid method to investigate Pi‐homeostasis regulation. We found that a screen of a chemical library can lead to the identification of a drug eliciting Pi‐starvation responses. The PHOSTIN (PSN) is a small organic compound that induces local and systemic Pi‐starvation responses, including Pi‐uptake and consequently uncoupling Pi‐content and Pi‐homeostasis regulation. We show that the transcriptional effects of PSN depend of PHR1 and PHL1. Therefore, PSN seems to interfere with an important and early regulatory step of phosphate sensing or signalling. Interestingly, the effect of PSN on the Pi‐content of treated plant was shown to be independent on PHO2 or PHO1 suggesting the existence of unknown signalling pathways. We identify the chemical motif responsible for PSN effects by characterization of the effects of PSN analogues and the measure of PSN and analogues content in treated plants. Our results demonstrate the interest of both PSN and chemical genetic as tools to dissect the Pi‐starvation pathway.
Materials and Methods
Plant material and general growth conditions
All Arabidopsis thaliana L. (Heynh.) lines used are in the Columbia (Col‐0) or in the Coler105 backgrounds, a Columbia background with the null allele erecta‐105 (NASC reference N89504; Torii et al., 1996), except the pPHT1;4::β‐glucuronidase (GUS) line in WS background (Misson et al., 2004). Various Arabidopsis transgenic and mutant lines were used: pIPS1::GUS (Martín et al., 2000), pSPX1::GUS and pSPX3::GUS (Duan et al., 2008), pPLDζ2::GUS (Cruz‐Ramírez et al., 2006), pMGD3::GUS (Kobayashi et al., 2009), pSQD1::GUS (Hammond et al., 2003), pACP5::GUS (Del Pozo et al., 1999), pRNS1::GUS (Hillwig et al., 2008), pCHX17::GUS (Cellier et al., 2004), pSTP13::GUS (Schofield et al., 2009) the mutant lines pho1‐2 and pho2‐1 (Delhaize & Randall, 1995), PHO1‐B1 (Rouached et al., 2011) and phr1‐3;phl1‐2 (Bournier et al., 2013). Unless indicated, the Arabidopsis seedlings were grown as previously described (Svistoonoff et al., 2007).
The chemical library screen was performed in a modified liquid Murashige and Skoog medium (MS)/10 medium (+Pi medium) (0.15 mM MgSO4, 2.1 mM NH4NO3, 1.9 mM KNO3, 0.5 or 0.005 mM NaH2PO4, 0.34 mM CaCl2, 0.5 μM KI, 10 μM FeCl2, 10 μM H3BO3, 10 μM MnSO4, 3 μM ZnSO4, 0.1 μM CuSO4, 0.1 μM CoCl2, 1 μM Na2MoO4, vitamins: 5.9 μM thiamine, 4.9 μM pyridoxine, 8.1 μM nicotinic acid and 55 μM inositol in 3.4 mM MES (pH 5.8), 0.5% sucrose).
For the other in vitro analyses on Arabidopsis, seeds were grown on the previous medium without vitamins, supplemented with 0.8% agar.
For the low‐nitrate experiment, a nitrogen‐free MS/10 medium containing 75 μM of Pi was prepared, to which we added 50 μM KNO3 + 10 mM KCl or 10 mM KNO3 to make the low‐nitrate (−NO3 −) and high‐nitrate (+NO3 −) plates, respectively.
For the low‐sulphur experiment, a sulphur‐free MS/10 medium containing 75 μM of Pi was prepared to make the low‐sulphur (−SO4 2−) medium, to which we added 2 mM MgSO4 to make the high‐sulphur plates (+SO4 2−).
For the low‐potassium experiment, a potassium‐free MS/10 medium containing 75 μM of Pi was prepared to make the low‐potassium (−K+) medium, to which we added 1.9 mM KCl to make the high‐potassium plates (+K+).
Wild‐type seeds of Oriza sativa (L. cv Nipponbare) were cultivated on a KimuraB/2 medium (Tanoi et al., 2011).
For rice and Arabidopsis Pi‐deficient medium, an equivalent concentration of NaCl was used to replace the sodium provided by NaH2PO4.
For the split‐root experiments, the primary root tip of 5 d old after germination (5‐dag) seedlings was cut to induce the lateral roots growth. Four days later, plants were transferred in compartmented plates such that lateral roots lie on different media and grown 4 more days.
Drugs were dissolved in dimethyl sulfoxide (DMSO) and added to the growth medium before pouring the plates, and mocked controls were done with the same DMSO final concentration (+Pi DMSO).
Chemical library screen and search for analogues
The Library of AcTive Compound on Arabidopsis (LATCA) (Zhao et al., 2007) was provided by Sean Cutler (UC Riverside) as 2.5 mM stock solutions dissolved in DMSO (Schreiber et al., 2008). Three seeds per well of Arabidopsis pPHT1;4::GUS lines were sown in 96‐wells plates containing 100 μl of MS/10 liquid medium. Five days after germination, drugs were added at 25 μM. After 2 d, the growth medium was replaced by the GUS staining solution and incubated overnight at 37°C. Plants were screened under a dissecting microscope. A secondary screen, performed in the same condition, on the first hits compounds (18 drugs) confirmed the activity of eight.
Structural analogues of the PSN were searched online in the Chembridge (http://chembridge.com) and Maybridge (http://www.maybridge.com) collections. Table 1 indicates their chemical formula, molecular weight, name, supplier and product code.
Table 1.
Phostin (PSN) and PSN analogues Maybridge and Chembridge references
| Name | Supplier | Product code | Molecular formula | MW | Product name |
|---|---|---|---|---|---|
| PSN | Maybridge | CD02621 | C19H14ClN3O5 | 400 | O5‐{[3‐(2‐chlorophenyl)‐5‐methylisoxazol‐4‐yl]carbonyl}‐1,3‐benzodioxole‐5‐carbohydroximamide |
| PSN2 | Maybridge | SPB0273 | C16H13ClN4O3S | 377 | O4‐{[3‐(2‐chlorophenyl)‐5‐methylisoxazol‐4‐yl]carbonyl}‐2‐methyl‐1,3‐thiazole‐4‐carbohydroximamide |
| PSN3 | Maybridge | CD03092 | C16H16ClN3O3 | 334 | 2‐[({[3‐(2‐chlorophenyl)‐5‐methyl‐4‐isoxazolyl]carbonyl}oxy)imino]piperidine |
| PSN4 | Maybridge | CD02622 | C15H10Cl2N2O4 | 353 | O5‐(3,4‐dichlorobenzoyl)‐1,3‐benzodioxole‐5‐carbohydroximamide |
| PSN5 | Maybridge | CD02598 | C8H8N2O3 | 180 | N′‐hydroxy‐1,3‐benzodioxole‐5‐carboximidamide |
| PSN6 | Maybridge | SEW05933 | C13H12ClN3O3 | 294 | 4‐chloro‐N′‐{[(3,5‐dimethylisoxazol‐4‐yl)carbonyl]oxy}benzenecarboximidamide |
| PSN7 | Maybridge | SPB02369 | C18H13Cl2N3O3 | 390 | O1‐{[5‐(4‐chlorophenyl)‐3‐methylisoxazol‐4‐yl]carbonyl}‐4‐chlorobenzene‐1‐carbohydroximamide |
| PSN8 | Chembridge | # 7991375 | C18H14ClN3O3 | 356 | N′‐({[3‐(2‐chlorophenyl)‐5‐methyl‐4‐isoxazolyl]carbonyl}oxy)benzenecarboximidamide |
| PSN9 | Chembridge | # 5919370 | C18H13ClN2O4 | 357 | N‐1,3‐benzodioxol‐5‐yl‐3‐(2‐chlorophenyl)‐5‐methyl‐4‐isoxazolecarboxamide |
| PSN10 | Chembridge | # 7984462 | C19H14ClNO5 | 372 | 1,3‐benzodioxol‐5‐ylmethyl 3‐(2‐chlorophenyl)‐5‐methyl‐4‐isoxazolecarboxylate |
| PSN11 | Maybridge | SB01885 | C11H8ClNO3 | 238 | 3‐(2‐chlorophenyl)‐5‐methylisoxazole‐4‐carboxylic acid |
Microscopy and root length measurement
GUS activity was assayed as described (Sarrobert et al., 2000). Pictures were taken on a transmitted light microscope (DMRXA; Leica Microsystems, Wetzlar, Germany) (×10 or ×20 magnification) or on dissecting microscope (MZ16A, Leica Microsystems) (×6 for leaves, ×25 for roots) (Leica Microsystems). Ten plantlets per condition were imaged and the experiments were done in triplicate.
The primary root length was measured using the software ImageJ (US National Institutes of Health, Bethesda, MD, USA). The average lengths were calculated from three independent experiments on 15 plantlets.
Gene expression analysis
Total RNA was extracted from 8 to 12 roots of 14 dag plantlets with the RNeasy Mini Kit (Qiagen, http://www.qiagen.com). RNAs were treated with DNase (TURBO DNase Ambion, http://www.ambion.com) and the cDNA were produced using the Superscript III reverse transcriptase (Superscript VILO cDNA synthesis kit, http://www.invitrogen.com). Quantitative real‐time reverse transcription (qRT)‐PCR were performed on a 480 LightCycler thermocycler (Roche) using the manufacturer's instructions with Light cycler 480sybr green I master (Roche). We used ROC3 (AT2G16600) in Arabidopsis and eEF‐1α (AK061464) in rice (Jain 2006) as reference genes for normalization. Primers sequences are described in Table 2.
Table 2.
List of the forward and reverse primers (5′ to 3′)
| Gene | AGI | F | R |
|---|---|---|---|
| Arabidopsis | |||
| ACP5 | AT3G17790 | TTTGACATAAGAGTTGCGAGATG | GTGAGCTTCAGAGATTTATAGAGCC |
| AMT1.1 | AT4G13510 | ACACTGTGGCCAGTTAGGCG | CCGTGGGGATGTCTTTGAGA |
| HAK5 | AT4G13420 | TCTCGTAGCGCTCCTCAAAT | GCTGTGTGGTTGGAAGTTCA |
| IPS1 | AT3G09922 | CGAAGCTTGCCAAAGGATAG | TGAAGACTGCAGAAGGCTGA |
| LPR1 | AT1G23010 | GCACCATCAAAACTTCGCAGAGATCG | CCGGGCTATGTCTACCATTGTCAC |
| NRT2.1 | AT1G08090 | AGTCGCTTGCACGTTACCTG | ACCCTCTGACTTGGCGTTCTC |
| NRT2.4 | AT5G60770 | CCGTCTTCTCCATGTCTTTC | CTGACCATTGAACATTGTGC |
| PHO1‐H1 | AT1G68740 | GCCACCACAAGACAGAGCCAACCAAGGC | AGCAATGCAGTGTGCAAGGAGGTGGT |
| PHR1 | AT4G28610 | GCTCTTTCACTACCGCCAAG | GTTCAGCAGCAACCTTCTCC |
| PLDζ2 | AT3G05630 | TTTTGAAGCCGTTTCTTGCT | TGCATTGCTGGAGACAAAAG |
| ROC3 | AT2G16600 | TCGGTGAAAGCTTGATCCTT | ATCGTGATGGAGCTTTACGC |
| SDI1 | AT5G48850 | GTCAGAGCCAAACATGCTCA | ACGAGGACGGAAAGATTTGA |
| SULTR1;1 | AT4G08620 | GCGAGAGGAGCAAGAAAATG | TCACCACTGGTCCTGGATTT |
| Rice | |||
| eEF‐1α | Os03g08020.1 | TTTCACTCTTGGTGTGAAGCAGAT | GACTTCCTTCACGATTTCATCGTAA |
| OsPT2 | Os03g05640.1 | GACGAGACCGCCCAAGAAG | TTTTCAGTCACTCACGTCGAGAC |
Pi‐content and Pi influx
The Arabidopsis Pi‐content and Pi‐uptake experiments were assayed as previously (Misson et al., 2004).
Detection of 33Pi accumulation was performed with imaging plates as described (Kanno et al., 2012). The data were analysed using the Image Gauge software (Fujifilm, Tokyo, Japan). Preliminary experiments have shown that Pi‐uptake was linear at least from 30 min to 2 h (data not shown).
Starch and anthocyanin assay
Anthocyanin were extracted from 10 plants and quantified as described (Ticconi et al., 2001). Starch was stained with Lugol (Sigma). These experiments were done three times independently on five biological replicates.
Phosphatase activity
The acid phosphatases were extracted from 5 mg (FW) of roots of 12 dag plants grown the last 7 d on the medium to test. We measured the phosphatase activity of the extract with a p‐nitrophenylphosphate colorimetric method (Kolari & Sarjala, 1995). The extract activity for each condition was calculated from three technical measures of three biological replicates and the experiment was conducted three times.
PHOSTIN extraction and detection
PSN were extracted from shoots or roots of 7–10 dag plants transferred at 5 dag for 1–72 h on +Pi medium containing 25 μM of PSN2 (+Pi PSN2), or 48 h on the described compartmented conditions. Experiments were done twice for the kinetics of accumulation and once for the compartmented plants. PSN‐like compound were extracted by hydroalcoholic extraction done as described for metabolites (Moing et al., 2004) with two minor modifications: extractions were done on fresh material (50 mg−1 sample, c. 30–40 plantlets); samples were extracted successively with 300 μl of ethanol 100%, ethanol 50% (v: v) and pure water at 80°C for 15 min. Supernatants were combined and centrifuged at 25 000 g for 10 min. The resulting supernatant was lyophilized for 24 h. Extracts were resuspended in 200 μl of pure water and centrifuged at 21 000 g before separation by HPLC on a C18 Acclaim PA2 column (Dionex) (2.1 × 150 mm, 3 μm).
For detection of PSN and its derivatives in the medium, chemicals were added at 25 μM in +Pi liquid medium in sterile condition. Media were incubated in the culture chamber (16 h 24°C : 8 h 21°C, light : dark) and reactions were stopped at indicated times (30 min, 3, 16, 24 or 48 h) by freezing in liquid nitrogen. Samples (10 μl) were directly injected in the HPLC after thawing.
For ultraviolet (UV) light detection, the medium component and the drugs were separated by HPLC as described (Lecoq et al., 2001). We identified the absorption peaks of the drugs with their UV signature (data not shown) by using an UV spectrophotometer (Ultimate 3000 VWD; Dionex, Sunnyvale, CA, USA) and an UV diode array detector (Ultimate 3000 RS; Dionex) between 220 and 340 nm.
For mass spectrometry detection, metabolites and PSN of the plant extracts were separated on an Ultimate 3000 HPLC system (Dionex). Solvents A and B were respectively H2O with 0.1% formic acid and acetonitrile. Flow rate was 0.25 ml min−1. Separation gradient was 0–5 min 97% A; 5–30 min from 97% A to 5% A; 30–35 min 5% A; 35–36 min from 5% A to 97% A; 36–51 min 97% A. Mass spectrometry detection was performed using a micrOTOF‐Q (Bruker Daltonics, Bremen, Germany). The mass spectrometer was equipped with an electrospray ionization probe operated in positive mode. Spray voltage was 4.5 kV. Mass range was 70–1500 Th and acquisition rate was 1 spectrum 2 s−1. MS spectra were calibrated using a 10 μM lithium formiate solution. Comparing retention time, accurate mass and fragments with the commercial drug standard identified PSN2 and PSN11. Elemental formulas were calculated using the generate molecular formula module of the Data Analysis software (Bruker Daltonics). Extracted ion chromatograms at m/z = 377.04 ± 0.3 Th and 238.02 ± 0.3 Th were used for quantification of PSN2 and PSN11, respectively.
UV and mass detection quantification were done by injection of standards (2.5–100 μM) for PSN and its analogues and peaks areas integration. Linear correlations were observed between peaks areas and drugs concentration (data not shown). Calculations of the measure and the technical error were done by injecting three times 100 μl of solution containing 500 nM, 1, 2.5, 5, 10, 25 and 50 μM of PSN and its analogues (data not shown).
Results
PHOSTIN induces Pi‐starvation responses
Pi‐starvation induces the expression of hundreds of genes involved in various adaptive responses to low‐Pi (Hammond et al., 2003; Misson et al., 2005; Morcuende et al., 2007; Thibaud et al., 2010). To find drugs altering the Pi‐starvation signalling we screened the LATCA chemical library (Zhao et al., 2007) for chemicals inducing several of the Arabidopsis Pi‐starvation responses tested. We chose the LATCA library because this medium‐size library of structurally diverse compounds contains only biologically active molecules and therefore potentially increasing the chance to find interesting drugs by screening < 4000 different molecules.
PHT1;4 encodes an Arabidopsis high‐affinity Pi‐transporter (Nussaume et al., 2011) specifically and strongly induced in response to low‐Pi in the root epidermis and tip (Misson et al., 2004). Therefore, we used a PHT1;4::GUS Arabidopsis gene‐trap line (Misson et al., 2004; Hirsch et al., 2011) as a visual reporter tool to identify candidate elicitors of Pi‐starvation responses within the LATCA chemicals. The responding drugs were then tested on several transcriptional, physiologic and morphologic Arabidopsis responses to Pi‐starvation.
Less than 0.25% of the LATCA chemicals (eight over 3580) induced the expression of the PHT1;4::GUS marker (data not shown). Only one of them, that we named PHOSTIN (PHOsphate STarvation responses INductor), was found to stimulate the expression of all the low‐Pi markers tested (see later) and therefore selected.
As shown in Fig. 1(a), using GUS transcriptional reporter lines, the presence of PSN in a +Pi growth medium stimulates the expression of numerous genes that are known to respond to low‐Pi: PHT1;4, IPS1, SPX1, SPX3, SQD1, RNS1, ACP5, PLDζ2, CHX17 and STP13 (Essigmann et al., 1998; Martín et al., 2000; Gonzalez et al., 2005; Nakamura et al., 2005; Cruz‐Ramírez et al., 2006; Franco‐Zorrilla et al., 2007; Duan et al., 2008; Bayle et al., 2011). This observation was validated by the measure of the expression of the genes ACP5 and PLDζ2 by qRT‐PCR (Fig. 1b).
Figure 1.
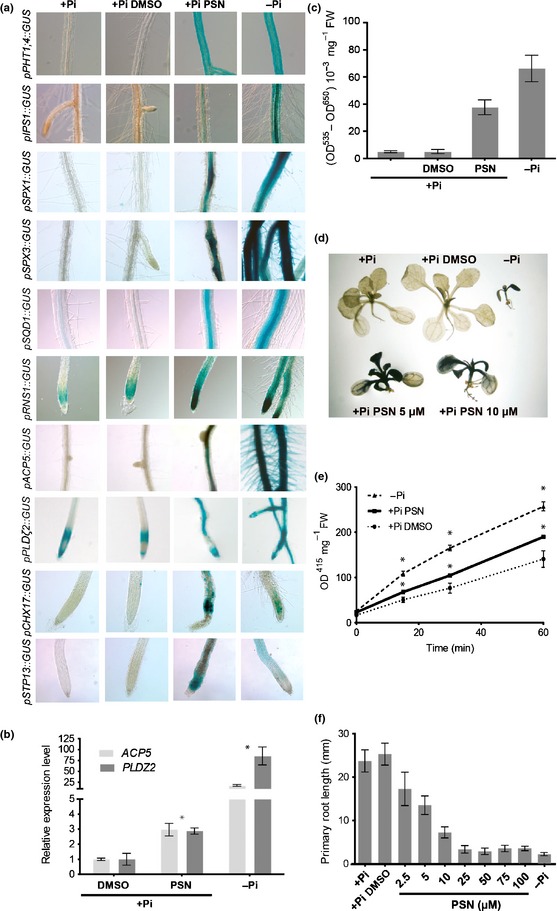
Phostin (PSN) induces phosphate starvation responses. (a) Effect of PSN on the expression of various phosphate (Pi)‐starvation reporter genes. (b) Relative expression of ACP5 and PLDζ2. Five days after germination wild‐type Arabidopsis seedlings were transferred on the indicated media for 4 d before total RNA extraction for quantitative real‐time reverse transcription‐PCR analysis. (c) Effect of PSN on anthocyanin accumulation. (d) Effect of PSN on starch accumulation. (e) Effect of PSN on phosphatase activity. (f) Primary root length of Arabidopsis seedlings grown in +Pi media. Seedlings were grown (a) 8 d, (c–e) 12 d or (f) 7 d in the indicated conditions. Growth conditions: +Pi; +Pi dimethyl sulfoxide (DMSO): (a, b) 0.25% DMSO or (d–f) 0.1% DMSO; +Pi PSN: (a, b) 25 μM, (d, e) 10 μM or (f) 2.5–100 μM and −Pi. For all quantitative experiments, averages ± SE were calculated from three independent experiments, except (b) (one experiment with two biological replicates). (b, e) *, Significantly different from +Pi DMSO treatment value (bilateral t‐test for sample of equal variance, P < 0.05).
PSN also stimulates typical physiologic responses to Pi‐starvation as the accumulation of starch and anthocyanin in leaves (Plaxton & Carswell, 1999; Wu et al., 2003; Misson et al., 2005) (Fig. 1c, d) or the root expression and secretion of phosphatases such as ACP5 (Duff et al., 1994; Köck et al., 1998; Trull & Deikman, 1998; Del Pozo et al., 1999; Haran et al., 2000). Indeed, in agreement with the higher expression of ACP5 (Fig. 1b), the roots of PSN‐treated seedlings contain a phosphatase activity 1.35 times stronger than the mocked control (Fig. 1e). In addition, PSN has a similar inhibiting effect on the primary root growth as Pi‐starvation (Péret et al., 2011, 2014) (Fig. 1f).
These results show that PSN stimulates the expression of a large set of low‐Pi genes and induces typical physiological and morphological Pi‐starvation responses suggesting that PSN either affect the Pi‐starvation signal or homeostasis. Moreover, in rice, PSN treatment doubled the expression level of OsPT2 (Ai et al., 2009), an homologue of the Arabidopsis PHT1;4, suggesting that the PSN target is conserved in different plant species (Supporting Information Fig. S1).
PHOSTIN mimics the local and long‐distance Pi‐starvation regulations
As shown earlier, PSN induces the expression of genes known to be regulated either locally (RNS1, STP13 and CHX17) or at long‐distance (PHT1;4, IPS1, ACP5, PLDζ2, SQD1) by Pi‐starvation (Thibaud et al., 2010). To test whether this induction by PSN follows the genuine local or systemic pattern of these genes, we performed a split‐root experiment in which we divided the root system of plants in two parts, each one fed with a different media.
We chose the pRNS1::GUS and pCHX17::GUS lines to test the effect of PSN on local induction and the pACP5::GUS and pPLDζ2::GUS lines for PSN effect on the long‐distance induction (Fig. 2). In +Pi/−Pi split conditions, RNS1 and CHX17 are induced in roots lying on the −Pi medium but not in roots on the +Pi medium, as expected for local markers, whereas the long‐distance markers ACP5 and PLDζ2 are induced in roots lying in both −Pi and +Pi medium. Interestingly in +Pi/+Pi +PSN split plants, PSN induces the same pattern of expression as does −Pi for these local and systemic genes. Remarkably, in split plants, the level of expression of the systemic genes is higher in roots directly in contact with PSN than in roots without. This pattern of expression is identical to the one observed in the +Pi/−Pi split control plants.
Figure 2.
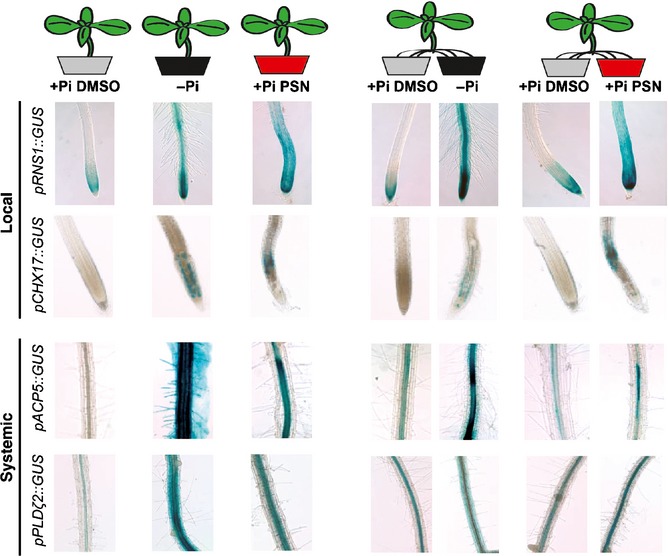
Phostin (PSN) treatment mimics local and systemic phosphate starvation regulation. Expression of genes locally regulated (RNS1 and CHX17) or systemically regulated (ACP5 and PLDζ2) by phosphate (Pi)‐starvation thanks to the promoter::β‐glucuronidase (GUS) fusion Arabidopsis reporter lines: pRNS1::GUS,pCHX17::GUS,pACP5::GUS and pPLDζ2::GUS. For control plants (left panel), lateral roots were grown on two identical media: +Pi dimethyl sulfoxide (DMSO)/+Pi DMSO (0.25% DMSO), −Pi/−Pi and +Pi PSN/+Pi PSN (25 μM PSN). For the other plants (right panel), lateral roots were grown on two different medium: +Pi DMSO/+Pi PSN or +Pi DMSO/−Pi.
This striking result shows that PSN mimics both local and systemic effects of Pi‐starvation and therefore suggests that PSN affects an early step of the low‐Pi signal or Pi‐homeostasis.
PHOSTIN uncouples Pi content and Pi‐starvation responses
The stimulation of Pi‐starvation responses in PSN‐treated plants could be the consequence of a reduced Pi‐uptake and content. To test the possibility that PSN represses Pi‐uptake, we measured the absorption of 33Pi in shoots and roots (Fig. 3a). Interestingly, PSN‐treated plants absorb 33Pi twice more rapidly than mocked plants (DMSO). These results corroborate the inductive effect of PSN on the expression of PHT1;4 (Fig. 1a), a crucial transporter for Pi absorption (Misson et al., 2004; Shin et al., 2004; Ayadi et al., 2015).
Figure 3.
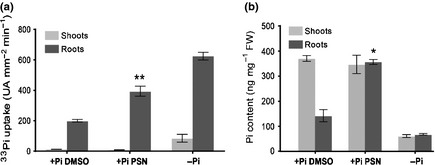
Phostin (PSN) increases phosphate uptake and accumulation. (a) Measure of 33phosphate (Pi) absorption and accumulation in PSN‐treated and untreated Arabidopsis Columbia (Col) seedlings in 15 min. **, 33Pi total absorption and 33Pi specific accumulation in roots are respectively significantly different from the 33Pi total absorption and root accumulation in untreated plants (+Pi dimethyl sulfoxide; DMSO) (bilateral t‐test for sample with unequal variance): P < 0.05. (b) Pi concentrations in shoots and roots. *, Significantly different from the Pi‐content in root of −Pi and +Pi DMSO plants (bilateral t‐test for sample with identical variance, P < 0.05). (a, b) Five days after germination wild‐type‐seedlings (Col) were transferred in the indicated growth conditions for seven more days before transfer on 33Pi medium: +Pi DMSO (0.1% DMSO); +Pi PSN (10 μM PSN) and −Pi. Averages ± SE for 33Pi uptake and Pi‐content were calculated from triplicates of three independent experiments.
Surprisingly, and by contrast to Pi‐starved plants, this increased absorption is limited to the root system. The speed of 33Pi accumulation in the shoots is identical in the PSN‐treated plant and mocked plant (Fig. 3a). This asymmetric effect of PSN on the speed of Pi absorption in shoots and roots is correlated to the Pi‐content of these tissues (Fig. 3b). Thus, although roots of PSN‐treated plants absorb more Pi than the untreated control, the amount of Pi transferred and stored in shoots is identical to the control plants. It seems therefore that PSN stimulates Pi‐uptake in the roots but does not increase Pi‐transfer to, or accumulation in the shoot.
These results show that PSN does not reduce the Pi‐content in plants but heightens it by increasing their Pi‐uptake. This effect is presumably a consequence of the induction of the expression of the genes involved in Pi absorption as the high‐affinity Pi‐transporters in PSN‐treated plants. Therefore, PSN artificially uncouples the signalling of Pi‐starvation from the actual Pi‐content in cells, possibly by targeting a central component of the Pi‐signalling.
PHOSTIN effects are independent of PHO1 and PHO2
PHO1 and PHO2 are two proteins controlling the balance of Pi between roots and shoots. The pho1 loss‐of‐function mutant over‐accumulates Pi in roots when grown on a Pi‐rich medium (Poirier et al., 1991; Rouached et al., 2011). By contrast, the pho2 loss‐of‐function mutant transfers more Pi from the roots to the leaves, resulting in a decrease of the root Pi‐content and the overaccumulation of Pi in the shoot. To alter the shoot to root Pi‐balance PSN might depend on PHO1 and or on PHO2.
To test this idea, we measured the Pi‐contents in two pho1 impaired lines (the pho1 mutant and the PHO1 under‐expresser PHO1‐B1), and in the pho2 mutant (Delhaize & Randall, 1995; Rouached et al., 2011) treated with PSN. Unexpectedly, like in wild‐type (WT) plants, PSN increases the Pi‐content in both the shoot and roots of the PHO1 deficient lines (Fig. 4). PSN also substantially reverts the reduced Pi‐content of pho2 roots (Fig. 4).
Figure 4.
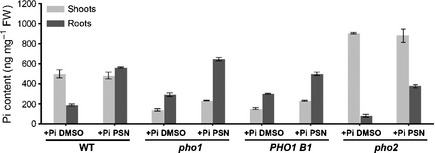
Effects of Phostin (PSN) on Pi accumulation in the pho1 and pho2 deficient lines. Five days after germination Arabidopsis seedlings were transferred on the indicated growth media. After 7 d, the free inorganic phosphate (Pi) concentration in roots and leaves was measured. Averages ± SE were calculated from triplicates of three independent experiments.
These results show that PSN does not need PHO1 and PHO2 activities to modify the shoot to root Pi‐balance.
The biological activity of the PHOSTIN relies on the hydrolytic release of its active motif PSN11
A mean toward understanding PSN activity is to determine the active part of the molecule. For this purpose, we tested the effect of 10 structural analogues of PSN (PSN2 to PSN11, Fig. 5a) on WT plants.
Figure 5.
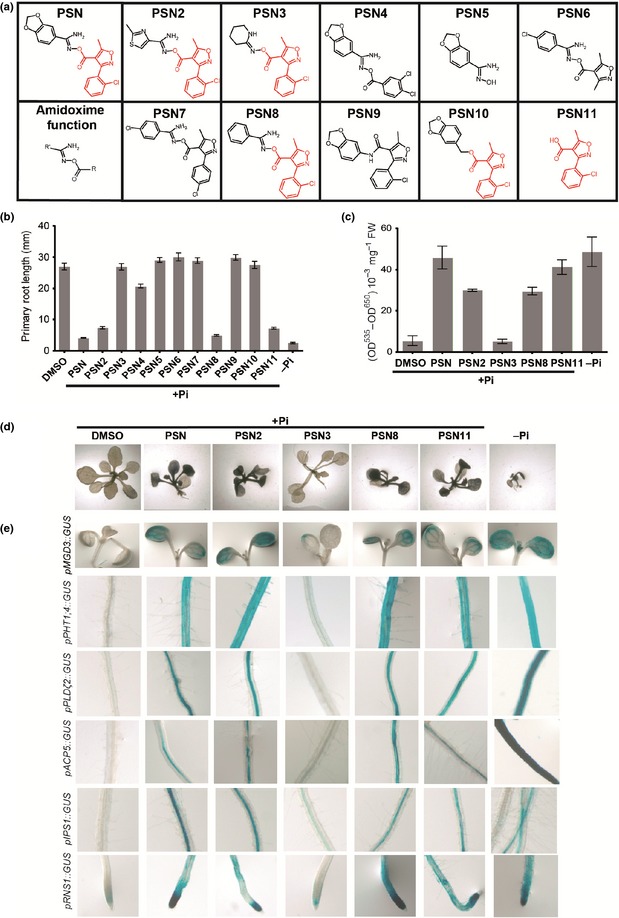
Structure–activity relationship of Phostin (PSN) structural analogues. (a) Chemical structure of PSN structural analogues. In red is drawn the motif corresponding to PSN11. (b) Primary root length of 8 d after germination (dag) Arabidopsis wild‐type seedlings treated with PSN and PSN analogues. (c) Anthocyanins accumulation in leaves of 12 dag seedlings. (d) Starch staining in 12 dag seedlings grown in presence of the indicated PSN analogues. (e) Expression of the indicated promoter‐β‐glucuronidase (GUS) fusion in seedlings. Plants were grown directly on the indicated medium: +phosphate (Pi) dimethyl sulfoxide (DMSO): (a, d) 0.1% or (b, c) 0.25%; +Pi supplemented with (a, d) 10 μM or (b, c) 25 μM of PSN or PSN analogues and −Pi. For (b, c) averages ± SE were calculated from 15 plants for each condition of three independent experiments.
Our results show that only three analogues (PSN2, PSN8 and PSN11) have a PSN‐like activity (i.e. induce all PSN responses as strongly as PSN) (Table 3). They repress the primary root growth (Figs 5b, S2a) and induce the accumulation of anthocyanin (Fig. 5c) and starch in leaves (Fig. 5d) of plants grown in +Pi. They also stimulate the expression of −Pi inducible genes by contrast to PSN3 taken as a negative control (Table 3; Figs 5e, S2b). Strikingly, these three actives (PSN‐like) analogues and PSN share a common structural motif corresponding to PSN11, the smallest analogue tested (Fig. 5a). Indeed, PSN11 is sufficient to display a PSN activity (Table 3). However, PSN3 and PSN10 also display the PSN11 motif (Fig. 5a), but they are not or poorly active in our biological assays (Fig. 5b,e; Table 3).
Table 3.
Summary of the effects of the Phostin (PSN) analogues on the induction of phosphate (Pi)‐starvation markers in Arabidopsis seedlings
| Drug | Primary root | Starch | Anthocyanin | PHT1;4 | IPS1 | MGD3 Roots | MGD3 Shoots | PLDζ2 | ACP5 | RNS1 |
|---|---|---|---|---|---|---|---|---|---|---|
| DMSO | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PSN | XXX | XXX | XX | XXX | X | XXX | X | XX | XX | XX |
| PSN2 | XX | X | XX | XXX | XXX | XXX | X | XX | XX | XX |
| PSN3 | 0 | 0 | 0 | XX | X | X | X | 0 | 0 | 0 |
| PSN4 | 0 | 0 | 0 | X | X | 0 | 0 | 0 | 0 | 0 |
| PSN5 | 0 | 0 | 0 | X | X | X | XX | 0 | X | 0 |
| PSN6 | 0 | 0 | 0 | X | 0 | XX | XX | 0 | 0 | 0 |
| PSN7 | 0 | 0 | 0 | X | X | X | 0 | 0 | 0 | 0 |
| PSN8 | XXXX | XXX | XXX | XXX | XXX | XX | 0 | XXX | XXX | XX |
| PSN9 | 0 | 0 | 0 | X | XX | X | 0 | 0 | X | 0 |
| PSN10 | 0 | 0 | 0 | 0 | XX | X | X | 0 | 0 | 0 |
| PSN11 | XX | X | XXXX | XXX | XX | XX | 0 | XXX | XX | XX |
Effect of the PSN analogues (PSN2–11) treatments at 10 μM in +Pi medium were tested on the primary root growth inhibition (Primary root), starch accumulation in leaves (Starch), anthocyanin accumulation in leaves (Anthocyanin), induction of PHT1;4, PHF1, IPS1, MGD3, PLDζ2, ACP5 and RNS1 expression in roots and induction of MGD3 expression in shoots or roots. Gene expression was monitored with promoter::β‐glucuronidase (GUS) reporter lines. 0, no effect (+Pi 0.1% dimethyl sulfoxide (DMSO) treatment phenotype); X, low; XX, medium; XXX, high; XXXX, highest effect found for these drugs.
By comparing the structures of the active analogues with the inactive ones, we found that the PSN11 motif is covalently linked to an aromatic group by an amidoxime function in the active molecules (benzene in PSN and PSN8; thiazole in PSN2) (Fig. 5a). Amidoxime functions linking two aromatic cycles are prone to hydrolysis in acidic conditions (Clayden et al., 2001; see the scheme in Fig. S3a). Thus, by allowing the release of the PSN11, the hydrolysis of the amidoxime function provides a possible explanation for the biological activity of PSN, PSN2 and PSN8, and the absence of PSN‐like activity of PSN3 and PSN10 on seedlings in our growth conditions (pH 5.8).
To test this hypothesis we followed by HPLC‐UV spectrophotometry the evolution of the concentrations of PSN, PSN2, PSN3 or PSN11 when supplemented to our plant growth liquid media (Fig. S3b). In the medium supplemented with PSN11, the concentration of PSN11 remained stable. By contrast, in the media supplemented with PSN or PSN2, we observed a biphasic decrease of the concentration of these drugs (fast during the first 30 min and then slow), mirrored by a concomitant accumulation of PSN11. These kinetics show that PSN and PSN2 are unstable in our growth medium, and release PSN11. Note that PSN3 is also unstable but does not release PSN11.
We then tested the role of the pH of the growth medium on the hydrolytic release of PSN11 from PSN2. As shown in Fig. S3(c), the disappearance of PSN2 and the accumulation of PSN11 are faster at pH 5.0 than at pH 6.0, therefore confirming the acidic hydrolysis hypothesis. Finally, we tested the biological activity of PSN, PSN2, 8 and 11 at pH 5.0 and 6.0 (Fig. S3d). As measured on the repression of the primary root growth, the activity of PSN, PSN2 and PSN8 is higher at pH 5.0 than at pH 6.0, whereas the activity of PSN11 is identical at both pH.
Collectively these results support the view that the biological activity of the PSN‐like compounds relies on the hydrolytic release of the active chemical PSN11. The lack of an aromatic cycle linked to the amidoxime function in PSN3 (apart those of PSN11) and the lack of an amidoxime function in PSN10, prevent the release of PSN11 and explain their biological inactivity.
The induction of systemic and local Pi‐starvation responses by PHOSTIN depends of the accumulation of PSN11 in the roots of treated plants
While PSN induces the systemic expression of long‐distance responses to low‐Pi, it does not promote at long distance the expression of the local response. This suggests that PSN11, the PSN bioactive derivative, does not move from the PSN‐treated roots to the untreated roots of split plants.
To know if and in which organ PSN11 and the PSN‐like drugs enters and accumulates, we used HPLC coupled with mass spectrometry. Among the PSN‐like drugs, PSN2 was the most stable in nonhydrolytic condition, for this reason plants were treated with PSN2 for different times and PSN2 and PSN11 were quantified in their shoot and roots separately.
The concentration of PSN2 in roots and shoots of treated plants reaches its maximum 1 h after transfer but decreased to 25% of its maximum only 24 h after transfer and is undetectable 48 h after transfer (Fig. 6a). The kinetics of the decrease of the PSN2 in the plant resembles to the kinetics of its degradation in the medium (Fig. S3b, c).
Figure 6.
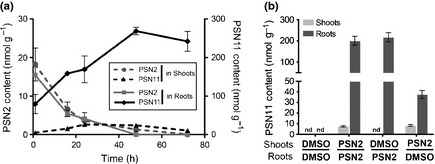
Phostin (PSN)11 enters and accumulates essentially in roots. (a) PSN2 and PSN11 content in plants. At 7 d after germination Arabidopsis wild‐type (WT)‐seedlings (Col) were transferred onto +phosphate (Pi) medium supplemented with 25 μM of PSN2. (b) PSN2 and PSN11 content in compartmented plants. At 7 dag WT‐seedlings (Col) were transferred onto compartmented plates such as the roots lies on another medium than the shoots. We used +Pi medium supplemented with 0.25% of dimethyl sulfoxide (DMSO) or 25 μM of PSN2 (PSN2). nd, not detected. For (a, b) PSN2 and PSN11 content were assayed in roots and shoots separately after (a) 1, 16, 24, 48 and 72 h of transfer or only after (b) 48 h of transfer by HPLC‐MS. Error bars, averages ± SD.
By contrast, PSN11 progressively accumulates in the plant (Fig. 6a). Its concentration reaches a plateau c. 48 h after transfer, in both shoots and roots. The PSN11 concentration is 10 times higher in roots than in shoots already 24 h after transfer (Fig. 6a). All through the experiment, the concentration of PSN11 in roots is 5 to 10 times higher than the highest concentration of PSN2 in shoots or roots. The differences in the concentrations of PSN2 and PSN11 and their kinetic of accumulation in the plant reinforce the hypothesis that the active compound is PSN11 (or a PSN11 derivative).
To test whether PSN11 moves between root and shoot, we followed its accumulation in both organs of plants grown such that the roots and the shoot lie on different media. When only the roots of the plants are in contact with PSN2, we observe the accumulation of the PSN11 in roots but not in shoots (Fig. 6b). In addition, the PSN11 concentration in the roots of these plants is identical to the one of plants grown both roots and shoot in contact with the PSN2 medium. By contrast, when only the shoot lies on the PSN2 medium, accumulations of PSN11 is almost undetectable in the shoot and six times lower in roots compared with plants for which both shoots and roots were in contact with PSN2 (Fig. 6b). This indicates that PSN11 enters into the plants preferably by the root system, moves poorly from the shoot to roots and not in a detectable quantity from roots to the shoot or that PSN11 is quickly metabolized in the shoots.
To test whether PSN11 accumulation in roots or putative metabolization by shoots have a prevalent role in PSN effects we monitored the induction of three PSN‐inducible responses (MGD3 expression, increase of Pi and anthocyanin contents) when PSN is specifically delivered to the shoot or the roots. When only the root system is in contact with PSN the plant displays similar responses than when the whole plant is treated with PSN: roots accumulates as much as Pi (Fig. 7a), the anthocyanin content is increased in the shoot (Fig. 7b), and MGD3 expression is induced in the whole plant (Fig. 7c). By contrast, when only the shoot lies on a PSN medium, the Pi‐content in roots is only slightly increased (Fig. 7a) and the anthocyanin content (Fig. 7b) and MGD3 expression (Fig. 7c) are similar to the untreated controls.
Figure 7.
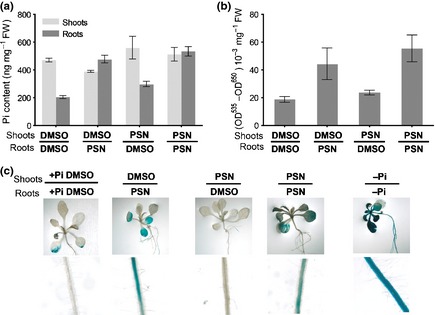
Phostin (PSN) effects depend on the root system. (a) Phosphate content in shoots and roots of compartmented Arabidopsis plants. (b) Anthocyanin content in shoots of compartmented plants. (c) pMGD3::β‐glucuronidase (GUS) expression in shoots and roots of compartmented plants. Five days after germination (a, b) wild‐type‐seedlings (Col) or (c) pMGD3::GUS were transferred onto compartmented plates such as the shoot and roots lie on different media: +phosphate (Pi), +Pi dimethyl sulfoxide (DMSO) (0.1% DMSO), +Pi PSN (10 μM PSN) and −Pi. After (c) 4 d or (a, b) 7 d plants were (c) GUS stained or harvested for (a) phosphate and (b) anthocyanin extraction and assay. For (a, b), averages ± SE for phosphate and anthocyanin were calculated from tree independent experiments with tree biological replicates per condition.
In conclusion plants slightly respond to PSN when it is delivered to the shoot, whereas they strongly respond to PSN when it is supplied to the roots. This suggests that the root system is the major path of entry of PSN11 into the plant and that its direct contact with the drug is necessary for the PSN‐like effects.
Transcriptional induction of low‐Pi markers by PSN11 depend of PHR1 and PHL1
PSN‐likes drugs elicit effects on transcriptional, physiologic and morphologic, local or systemic low‐Pi Arabidopsis responses suggesting that PSN11 affect an early and important step of the low‐Pi‐signalling pathway. Only the paralogous transcription factors PHR1 and PHL1 are known to be involved in the regulation of numerous local and systemic responses to Pi‐starvation in Arabidopsis. To test if the PSN‐like drugs targets the PHR1 and PHL1 pathway we compared the effect of PSN11 on the expression of typical low‐Pi marker genes in roots of WT and double loss‐of‐function mutant phr1;phl1 plants. As shown in Fig. 8, the low‐Pi induced genes ACP5, IPS1, PHO1‐H1 and PLDζ2 are induced by PSN11 in WT plants grown in high‐Pi but not in the phr1;phl1 plants. These results clearly show that the transcriptional effect of PSN11 relies on the intact master transcriptional regulators PHR1 and PHL1 and therefore that PSN11 probably elicit the low‐Pi‐signalling pathway upstream PHR1 and PHL1 functions. Note that LPR1 is not induced by PSN11, showing that PSN, like Pi‐starvation, do not inhibit root growth by increasing transcriptional expression of LPR1.
Figure 8.
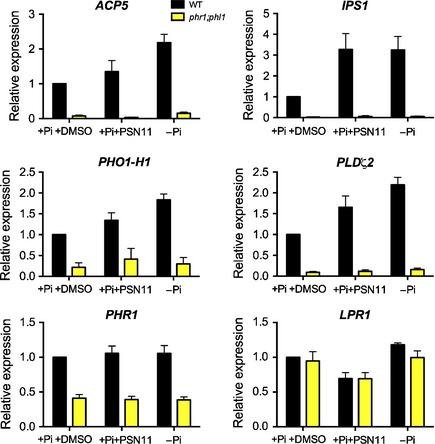
Relative expression level of genes related to the low‐Pi response. Five days after germination Arabidopsis wild‐type (WT) and phr1‐3;phl1‐2 double mutant seedlings were transferred on the indicated media (−phosphate (Pi) = 0 μM Pi; +Pi = 75 μM Pi; + Phostin (PSN)11 = 25 μM PSN11; + dimethyl sulfoxide (DMSO) = 0.1% DMSO) for 4 d before root RNA extraction for quantitative real‐time reverse transcription‐PCR analysis. Error bars, averages ± SE from three independent experiments.
Discussion
Chemical genetic has allowed us to find a drug affecting many aspects of the Pi‐starvation signalling
As a strategy complementary to the classical genetics in the elucidation of the molecular component of the Pi‐starvation signalling, we have tried chemical genetics. Indeed, this new, large scale pharmacological approach has been successful in the elucidation of important steps of plant signalling pathways such as the ABA perception (Park et al., 2009) or brassinosteroid signalling (De Rybel et al., 2009). Nevertheless, at the beginning of this work it was uncertain whether chemical genetics could be successfully applied to such an elusive signalling pathway underlying plant Pi‐homeostasis.
We show here the identification of PSN, the first small synthetic molecule mimicking Pi‐starvation in plants. PSN has the remarkable ability to mimic Pi‐starvation at several levels.
The PSN and the PSN‐like drugs interfere with morphologic (primary root growth inhibition), physiologic (increase of phosphatase activity and anthocyanin and starch accumulation in leaves) and transcriptional plant responses to Pi‐starvation and mimic the effect of Pi‐starvation on their regulation (Figs 1, 5, 8, S2). All the genes, which are systemically regulated by Pi‐deficiency, were found induced systemically by the PSN, while the local markers genes responded only locally (Fig. 2).
The activation of gene expression is observed for several typical molecular markers of Pi‐starvation and it is robust. As a matter of fact, these markers belong to different classes of responses (local and long‐distance) and different biochemical pathways (Pi‐transporters, Pi‐signalling, lipids synthesis, phosphatases). Importantly, the expressions of NRT2;4 and AMT1;1, induced in the root during nitrogen deficiency, and of SDI1 and SULTR1;1, induced in the root during sulphur deficiency (Howarth et al., 2009; Hubberten et al., 2012), are not affected by PSN11 (Fig. S4a,b). By contrast, the expressions of NRT2;1, another maker of nitrogen deficiency, and of HAK5, a marker of potassium deficiency (Ahn et al., 2004), known to be upregulated during Pi‐starvation (Lejay et al., 1999; Tian et al., 2009; Zheng et al., 2013; Misson et al., 2005; Shin et al., 2005; Lei et al., 2011) are induced by the presence of PSN11 in the medium (Fig. S4a,c). Therefore we conclude that PSN11 is not a general inducer of transcriptional responses to starvation.
The active motif of PHOSTIN and PSN‐like molecules is PSN11
By characterizing the PSN and several of its structural analogues, we have shown that their activities depend on the release of the active motif, PSN11. This release is low pH‐dependent and occurs in our growth medium (pH 5.8), most probably by the hydrolysis of the amidoxime function linking PSN11 to the remaining part of the compounds (Figs 6, S3). Indeed, we also observed that the proclivity of PSN and PSN‐like molecules to release PSN11 at acidic pH determines their biological activity. Correlated with this, PSN11 accumulates in plants by contrast to the other PSN‐like compound tested.
The effect of PHOSTIN on the local and systemic responses depends on PSN11 accumulation in root
The accumulation of PSN11 in roots appears responsible of most of the effects displayed by the PSN‐like analogues. The very low level of PSN11 detected in roots when PSN2 is only provided to the shoot suggests that the roots play a major role in PSN11 entry inside the plant and consequently in its activity. The presence of PSN11 in the roots of plants receiving PSN2 from their leaves, associated with their absence of PSN responses suggests the existence of a content threshold of PSN11 in roots necessary to activate these responses (Figs 6, 7).
Since PSN11 is detected in roots when supplied to the shoot (Fig. 6), PSN11 traffic from the shoots to the roots has to be considered. However, the opposite experiment indicates that PSN11 does not or poorly move from the root system to the shoot. We formally cannot exclude that PSN11 moves to the shoot where it is rapidly degraded or metabolized. Nevertheless, the PSN stimulation of local markers only in the treated root in split‐root experiments suggests that the active chemical does not move inside the plant, from root to root or remains below its active level. Consequently, the long‐distance induction of responses by the PSN‐like drugs is most probably not due to the accumulation of PSN11 in the untreated organs. This raises the question of the moving component: a chemical derivative of PSN11 or an endogenous Pi‐starvation signal that move from roots to the shoot? Despite several attempts, the detection of a molecule derived from PSN11 in extracts of PSN‐treated plants has failed (data not shown). Further work is necessary to identify the moving signal.
PHOSTIN target a component of the Pi signalling pathway controlled by PHR1 and PHL1
PSN increases the Pi influx and consequently the Pi‐content in roots (Fig. 3). It is puzzling that PSN‐treated plants contain higher levels of Pi and express the low‐Pi markers. The expression of these markers is thus uncoupled from the internal Pi‐content, suggesting that PSN disrupts the activity of a mechanism allowing the plant to adjust its low‐Pi molecular and physiological responses to its Pi‐content. The chemical structure of PSN11 does not inform us about its molecular target, and we are not aware of natural compounds sharing the PSN11‐structure. Interestingly, in an opposite way than PSN treatment, the under‐expression of PHO1 (in the PHO1‐B1 lines), encoding an SPX protein involved in the root to shoot Pi translocation, uncouples the low‐Pi responses from internal Pi‐content. Despite a reduced Pi‐content of the shoot as the pho1 loss‐of‐function mutant, PHO1‐B1 plants display a transcriptomic profile resembling to the one of WT plants grown on +Pi (Rouached et al., 2011). However, since the pho1 mutant and PHO1‐B1 lines still respond to PSN, the function of PHO1 is not essential for the PSN‐like activity (Fig. 4).
What could be the signalling pathway targeted by PSN11? The induction of local and systemic responses to Pi‐starvation by the PSN‐like drugs (Figs 1, 2, 5, 7) suggests that PSN11 affects an early and important step of the Pi‐starvation signalling or homeostasis pathways. Among the known systemic regulators of the −Pi responses only PHR1 and PHL1 regulates systemic as well as some local −Pi markers (Rubio et al., 2001; Thibaud et al., 2010). The insensitivity of the double loss‐of‐function mutant phr1;phl1 to PSN11 shows that these two master regulatory genes are necessary for PSN transcriptional activation of low‐Pi genes (Fig. 8). This suggests that PSN11 targets an effector located upstream of PHR1 and PHL1 in the Pi‐starvation pathway. This is in contrast with the unaltered effect of PSN in the pho2 mutant (Fig. 4). It seems therefore that the phenotype induced by PSN only needs a subset of the known low‐Pi‐signalling proteins and that its effect on Pi accumulation, at least, is independent on the PHO2 systemic pathway (this suggests the existence of additional pathways controlling Pi‐homeostasis during a Pi‐stress signal).
In addition, PSN stimulates the transcription of PHT1;4 (in Arabidopsis) and OsPT2 (in rice) encoding high‐affinity Pi‐transporters in two plant species belonging to different clades. Although preliminary, this result suggests a phylogenetic conservation of the molecular target of PSN on Pi‐homeostasis, further supporting the idea that the PSN target is crucial for Pi‐homeostasis in plants.
In conclusion, our discovery of PSN‐like drugs show that chemical genetics can be used to dissect the Pi‐starvation responses and signalling. These drugs seem to target an early step of the Pi‐starvation signal. Therefore, their use will help us to identify new components of the Pi‐starvation signalling pathway. This is complementary to another drug (Phosphatin) identified in our laboratory that alleviates low‐Pi response (Arnaud et al., 2014).
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Phostin (PSN) effect on the rice OsPT2 expression.
Fig. S2 Effects of Phostin (PSN) structural analogues on the induction of phosphate starvation markers.
Fig. S3 Acidic hydrolysis release of Phostin (PSN)11 from the active PSN analogues.
Fig. S4 Relative expression level of genes related to the low‐nitrate, low‐sulphur and low‐potassium responses.
Acknowledgements
We thank S. Cutler for providing the LATCA library; Y. Poirier (PHO1‐B1); R. Paz‐Ares (pPHF1::GUS, pACP5::GUS, pIPS1::GUS), F. Gaymard (phr1‐3;phl1‐2), G. C. MacIntosh (pRNS1:GUS), P. Wu (pSPX1::GUS & pSPX3::GUS), L. Herrera‐Estrella (pPLDζ2::GUS), H. Ohta (pMGD3::GUS), P. J. White (pSQD1::GUS), J. Rothstein (pSTP13::GUS), F. Cellier (pCHX17::GUS); the Groupe de Recherches Appliquées en Phytotechnologie for taking care of the plants; E. and C. Girard‐Smith for their advice regarding the chemical properties of the PSN‐like compounds. H. Javot, E. Marin, B. Péret, M‐C. Thibaud and anonymous referees for their comments on a first version of the manuscript. Support was provided by the HélioBiotec platform, funded by the European Regional Development Fund, the Région Provence‐Alpes‐Côte‐d'Azur, the French Ministry of Research, and the Commissariat à l'Energie Atomique et aux Energies Alternatives. Financial support: CEA‐IRTELIS PhD grant, ANR‐09‐BLAN‐0118, ANR‐08 KBBE‐041451.
References
- Abel S. 2011. Phosphate sensing in root development. Current Opinion in Plant Biology 14: 303–309. [DOI] [PubMed] [Google Scholar]
- Ahn SJ, Shin R, Schachtman DP. 2004. Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiology 134: 1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai P, Sun S, Zhao J, Fan X, Xin W, Guo Q, Yu L, Shen Q, Wu P, Miller AJ. 2009. Two rice phosphate transporters, OsPht1; 2 and OsPht1; 6, have different functions and kinetic properties in uptake and translocation. Plant Journal 57: 798–809. [DOI] [PubMed] [Google Scholar]
- Arnaud C, Clément M, Thibaud MC, Javot H, Chiarenza S, Delannoy E, Revol J, Soreau P, Balzergue S, Block MA et al 2014. Identification of phosphatin, a drug alleviating phosphate starvation responses in Arabidopsis. Plant Physiology 166: 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su C, Chiou TJ. 2006. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiology 141: 1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayadi A, David P, Arrighi JF, Chiarenza S, Thibaud MC, Nussaume L, Marin E. 2015. Reducing the genetic redundancy of Arabidopsis PHT1 transporters to study phosphate uptake and signaling. Plant Physiology 172: 1511–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. 2006. PHO2, microRNA399, and PHR1 define a phosphate‐signaling pathway in plants. Plant Physiology 141: 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TR, Lynch JP. 1996. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant, Cell & Environment 19: 529–538. [Google Scholar]
- Bayle V, Arrighi JF, Creff A, Nespoulous C, Vialaret J, Rossignol M, Gonzalez E, Paz‐Ares J, Nussaume L. 2011. Arabidopsis thaliana high‐affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell HE, Zhao Y. 2003. Chemical genetic approaches to plant biology. Plant Physiology 133: 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournier M, Tissot N, Mari S, Boucherez J, Lacombe E, Briat JF, Gaymard F. 2013. Arabidopsis ferritin 1 (AtFer1) gene regulation by the phosphate starvation response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. The Journal of Biological Chemistry 288: 22670–22680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez‐Pérez J, Solano R, Leyva A, Paz‐Ares J. 2010. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genetics 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellier F, Conéjéro G, Ricaud L, Luu DT, Lepetit M, Gosti F, Casse F. 2004. Characterization of AtCHX17, a member of the cation/H+ exchangers, CHX family, from Arabidopsis thaliana suggests a role in K+ homeostasis. Plant Journal 39: 834–846. [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su C. 2006. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. 2011. Signaling network in sensing phosphate availability in plants. Annual Review of Plant Biology 62: 185–206. [DOI] [PubMed] [Google Scholar]
- Clayden J, Greeves N, Warren S, Wothers P. 2001. Organic chemistry. New York, NY, USA: Oxford University Press. [Google Scholar]
- Cruz‐Ramírez A, Oropeza‐Aburto A, Razo‐Hernández F, Ramírez‐Chávez E, Herrera‐Estrella L. 2006. Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proceedings of the National Academy of Sciences, USA 103: 6765–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S, McCourt P. 2005. Dude, where's my phenotype? Dealing with redundancy in signaling networks. Plant Physiology 138: 558–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Vert G, Rozhon W, Mayerhofer J, Peelman F, Coutuer S, Denayer T, Jansen L, Nguyen L. 2009. Chemical inhibition of a subset of Arabidopsis thaliana GSK3‐like kinases activates brassinosteroid signaling. Chemistry & Biology 16: 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pozo JC, Allona I, Rubio V, Leyva A, De La Peña A, Aragoncillo C, Paz Ares J. 1999. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant Journal 19: 579–589. [DOI] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ. 1995. Characterization of a phosphate‐accumulator mutant of Arabidopsis thaliana . Plant Physiology 107: 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. 2008. Characterization of a sub family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant Journal 54: 965–975. [DOI] [PubMed] [Google Scholar]
- Duff SM, Sarath G, Plaxton WC. 1994. The role of acid phosphatases in plant phosphorus metabolism. Physiologia Plantarum 90: 791–800. [Google Scholar]
- Essigmann B, Güler S, Narang RA, Linke D, Benning C. 1998. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 95: 1950–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foehse D, Jungk A. 1983. Influence of phosphate and nitrate supply on root hair formation of rape, spinach and tomato plants. Plant and Soil 74: 359–368. [Google Scholar]
- Franco‐Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio‐Somoza I, Leyva A, Weigel D, García JA, Paz‐Ares J. 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. 2005. A miRNA involved in phosphate‐starvation response in Arabidopsis. Current Biology 15: 2038–2043. [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Solano R, Rubio V, Leyva A, Paz‐Ares J. 2005. Phosphate Transporter Traffic Facilitator1 is a plant‐specific SEC12‐related protein that enables the endoplasmic reticulum exit of a high‐affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ. 2003. Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiology 132: 578–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran S, Logendra S, Seskar M, Bratanova M, Raskin I. 2000. Characterization of Arabidopsis acid phosphatase promoter and regulation of acid phosphatase expression. Plant Physiology 124: 615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig MS, LeBrasseur ND, Green PJ, MacIntosh GC. 2008. Impact of transcriptional, ABA‐dependent, and ABA‐independent pathways on wounding regulation of RNS1 expression. Molecular Genetics and Genomics 280: 249–261. [DOI] [PubMed] [Google Scholar]
- Hirsch J, Misson J, Crisp PA, David P, Bayle V, Estavillo GM, Javot H, Chiarenza S, Mallory AC, Maizel A et al 2011. A novel fry1 allele reveals the existence of a mutant phenotype unrelated to 5′–>3′ exoribonuclease (XRN) activities in Arabidopsis thaliana roots. PLoS ONE 6: e16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth JR, Parmar S, Barraclough PB, Hawkesford MJ. 2009. A sulphur deficiency‐induced gene, sdi1, involved in the utilization of stored sulphate pools under sulphur‐limiting conditions has potential as a diagnostic indicator of sulphur nutritional status. Plant Biotechnology Journal 7: 200–209. [DOI] [PubMed] [Google Scholar]
- Huang TK, Han CL, Lin SI, Chen YJ, Tsai YC, Chen YR, Chen JW, Lin WY, Chen PM, Liu TY et al 2013. Identification of downstream components of ubiquitin‐conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25: 4044–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubberten HM, Drozd A, Tran BV, Hesse H, Hoefgen R. 2012. Local and systemic regulation of sulfur homeostasis in roots of Arabidopsis thaliana . Plant Journal 72: 625–635. [DOI] [PubMed] [Google Scholar]
- Kanno S, Yamawaki M, Ishibashi H, Kobayashi NI, Hirose A, Tanoi K, Nussaume L, Nakanishi TM. 2012. Development of real‐time radioisotope imaging systems for plant nutrient uptake studies. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 367: 1501–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Awai K, Nakamura M, Nagatani A, Masuda T, Ohta H. 2009. Type B monogalactosyldiacylglycerol synthases are involved in phosphate starvation induced lipid remodeling, and are crucial for low phosphate adaptation. Plant Journal 57: 322–331. [DOI] [PubMed] [Google Scholar]
- Köck M, Theierl K, Stenzel I, Glund K. 1998. Extracellular administration of phosphate‐sequestering metabolites induces ribonucleases in cultured tomato cells. Planta 204: 404–407. [Google Scholar]
- Kolari KK, Sarjala T. 1995. Acid phosphatase activity and phosphorus nutrition in Scots pine needles. Tree Physiology 15: 747–752. [DOI] [PubMed] [Google Scholar]
- Lai F, Thacker J, Li Y, Doerner P. 2007. Cell division activity determines the magnitude of phosphate starvation responses in Arabidopsis. Plant Journal 50: 545–556. [DOI] [PubMed] [Google Scholar]
- Lei M, Liu Y, Zhang B, Zhao Y, Wang X, Zhou Y, Raghothama KG, Liu D. 2011. Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiology 156: 1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoq K, Belloc I, Desgranges C, Daignan Fornier B. 2001. Role of adenosine kinase in Saccharomyces cerevisiae: identification of the ADO1 gene and study of the mutant phenotypes. Yeast 18: 335–342. [DOI] [PubMed] [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive F, Filleur S, Daniel‐Vedele F, Gojon A. 1999. Molecular and functional regulation of two NO3‐ uptake systems by N‐ and C‐status of Arabidopsis plants. Plant Journal 18: 509–519. [DOI] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HM. 2002. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant Journal 29: 751–760. [DOI] [PubMed] [Google Scholar]
- Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ. 2012. PHO2‐dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín AC, Del Pozo JC, Iglesias J, Rubio V, Solano R, De La Peña A, Leyva A, Paz Ares J. 2000. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant Journal 24: 559–567. [DOI] [PubMed] [Google Scholar]
- McCourt P, Desveaux D. 2010. Plant chemical genetics. New Phytologist 185: 15–26. [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N et al 2005. A genome‐wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proceedings of the National Academy of Sciences, USA 102: 11934–11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Thibaud MC, Bechtold N, Raghothama K, Nussaume L. 2004. Transcriptional regulation and functional properties of Arabidopsis Pht1; 4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Molecular Biology 55: 727–741. [DOI] [PubMed] [Google Scholar]
- Moing A, Maucourt M, Renaud C, Gaudillere M, Brouquisse R, Lebouteiller B, Gousset‐Dupont A, Vidal J, Granot D, Denoyes‐rothan B. 2004. Quantitative metabolic profiling by 1‐dimensional 1H‐NMR analyses: application to plant genetics and functional genomics. Functional Plant Biology 31: 889–902. [DOI] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M. 2007. Genome wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant, Cell & Environment 30: 85–112. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Awai K, Masuda T, Yoshioka Y, Takamiya K, Ohta H. 2005. A novel phosphatidylcholine‐hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. Journal of Biological Chemistry 280: 7469–7476. [DOI] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC. 2011. Phosphate import in plants: focus on the PHT1 transporters. Frontiers in Plant Science 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua NH. 2014. Nitrogen Limitation Adaptation recruits Phosphate2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T. 2011. Root developmental adaptation to phosphate starvation: better safe than sorry. Trends in Plant Science 16: 442–450. [DOI] [PubMed] [Google Scholar]
- Péret B, Desnos T, Jost R, Kanno S, Berkowitz O, Nussaume L. 2014. Root architecture responses: in search of phosphate. Plant Physiology 166: 1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Carswell MC. 1999. Metabolic aspects of the phosphate starvation response in plants In: Lerner HR, ed. Plant responses to environmental stresses: from phytohormones to genome reorganization. New York, NY, USA: Dekker, 349–372. [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J. 1991. Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiology 97: 1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas‐Pierce M, Titapiwatanakun B, Sohn EJ, Fang F, Larive CK, Blakeslee J, Cheng Y, Cuttler S, Peer WA, Murphy AS. 2007. Arabidopsis P‐glycoprotein19 participates in the inhibition of gravitropism by gravacin. Chemistry & Biology 14: 1366–1376. [DOI] [PubMed] [Google Scholar]
- Rosado A, Hicks GR, Norambuena L, Rogachev I, Meir S, Pourcel L, Zouhar J, Brown MQ, Boirsdore MP, Puckrin RS. 2011. Sortin1‐hypersensitive mutants link vacuolar‐trafficking defects and flavonoid metabolism in Arabidopsis vegetative tissues. Chemistry & Biology 18: 187–197. [DOI] [PubMed] [Google Scholar]
- Rouached H, Stefanovic A, Secco D, Bulak Arpat A, Gout E, Bligny R, Poirier Y. 2011. Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. Plant Journal 65: 557–570. [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz‐Ares J. 2001. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes & Development 15: 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Calderón L, López‐Bucio J, Chacón‐López A, Cruz‐Ramírez A, Nieto‐Jacobo F, Dubrovsky JG, Herrera‐Estrella L. 2005. Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana . Plant and Cell Physiology 46: 174–184. [DOI] [PubMed] [Google Scholar]
- Sarrobert C, Thibaud M‐C, Contard‐David P, Gineste S, Bechtold N, Robaglia C, Nussaume L. 2000. Identification of an Arabidopsis thaliana mutant accumulating threonine resulting from mutation in a new dihydrodipicolinate synthase gene. Plant Journal 24: 357–368. [DOI] [PubMed] [Google Scholar]
- Schofield RA, Bi YM, Kant S, Rothstein SJ. 2009. Over‐expression of STP13, a hexose transporter, improves plant growth and nitrogen use in Arabidopsis thaliana seedlings. Plant, Cell & Environment 32: 271–285. [DOI] [PubMed] [Google Scholar]
- Schreiber K, Ckurshumova W, Peek J, Desveaux D. 2008. A high throughput chemical screen for resistance to Pseudomonas syringae in Arabidopsis. Plant Journal 54: 522–531. [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ. 2004. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low‐ and high‐phosphate environments. Plant Journal 39: 629–642. [DOI] [PubMed] [Google Scholar]
- Shin R, Berg RH, Schachtman DP. 2005. Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant & Cell Physiology 46: 1350–1357. [DOI] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot‐Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T. 2007. Root tip contact with low‐phosphate media reprograms plant root architecture. Nature Genetics 39: 792–796. [DOI] [PubMed] [Google Scholar]
- Tanoi K, Saito T, Iwata N, Kobayashi NI, Nakanishi TM. 2011. The analysis of magnesium transport system from external solution to xylem in rice root. Soil Science and Plant Nutrition 57: 265–271. [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz Ares J, Poirier Y, Nussaume L. 2010. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant Journal 64: 775–789. [DOI] [PubMed] [Google Scholar]
- Tian QY, Sun P, Zhang WH. 2009. Ethylene is involved in nitrate‐dependent root growth and branching in Arabidopsis thaliana . New Phytologist 184: 918–931. [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Abel S. 2001. Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiology 127: 963–972. [PMC free article] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. 1996. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine‐rich repeats. Plant Cell 8: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth R, van der Hoorn RA. 2010. Emerging principles in plant chemical genetics. Trends in Plant Science 15: 81–88. [DOI] [PubMed] [Google Scholar]
- Trull MC, Deikman J. 1998. An Arabidopsis mutant missing one acid phosphatase isoform. Planta 206: 544–550. [DOI] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW. 2003. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiology 132: 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Finnegan PM. 2010. Regulation of phosphate starvation responses in higher plants. Annals of Botany 105: 513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liao H, Lucas WJ. 2014. Molecular mechanisms underlying phosphate sensing, signaling and adaptation in plants. Journal of Integrative Plant Biology 56: 192–220. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chow TF, Puckrin RS, Alfred SE, Korir AK, Larive CK, Cutler SR. 2007. Chemical genetic interrogation of natural variation uncovers a molecule that is glycoactivated. Nature Chemical Biology 3: 716–721. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J. 2003. SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301: 1107–1110. [DOI] [PubMed] [Google Scholar]
- Zheng D, Han X, An Y, Guo H, Xia X, Yin W. 2013. The nitrate transporter NRT2.1 functions in the ethylene response to nitrate deficiency in Arabidopsis. Plant, Cell & Environment 36: 1328–1337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Phostin (PSN) effect on the rice OsPT2 expression.
Fig. S2 Effects of Phostin (PSN) structural analogues on the induction of phosphate starvation markers.
Fig. S3 Acidic hydrolysis release of Phostin (PSN)11 from the active PSN analogues.
Fig. S4 Relative expression level of genes related to the low‐nitrate, low‐sulphur and low‐potassium responses.


