Abstract
Wheat was domesticated approximately 10,000 years ago and has since spread worldwide to become one of the major crops. Its adaptability to diverse environments and end-uses is surprising given the diversity bottlenecks expected from recent domestication and polyploid speciation events. Wheat compensates for these bottlenecks by capturing part of the genetic diversity of its progenitors and by generating new diversity at a relatively fast pace. Frequent gene deletions and disruptions generated by a fast replacement rate of repetitive sequences are buffered by the polyploid nature of wheat, resulting in subtle dosage effects on which selection can operate.
With 620 million tons produced annually worldwide, wheat provides approximately one fifth of the calories consumed by humans (1). Roughly 95% of the wheat crop is common wheat, used for making bread, cookies, and pastries, whereas the remaining 5% is durum wheat, used for making pasta and other semolina products. Einkorn wheat and other hulled wheats, namely emmer and spelt, are today relic crops of minor economic significance (2, 3).
While einkorn is a diploid species, durum and common wheat are polyploid species that originated by interspecific hybridization of two and three different diploid species, respectively (Fig. 1). The success of these domesticated polyploid species parallels the success of natural polyploid species, which represent more than 70% of the plant species (reviewed in (4)), and tend to have a more extended geographic distribution than their close diploid relatives (5). Consequently, recent advances in wheat genomics may shed light on the genetic causes of the broad adaptability of natural polyploid plant species.
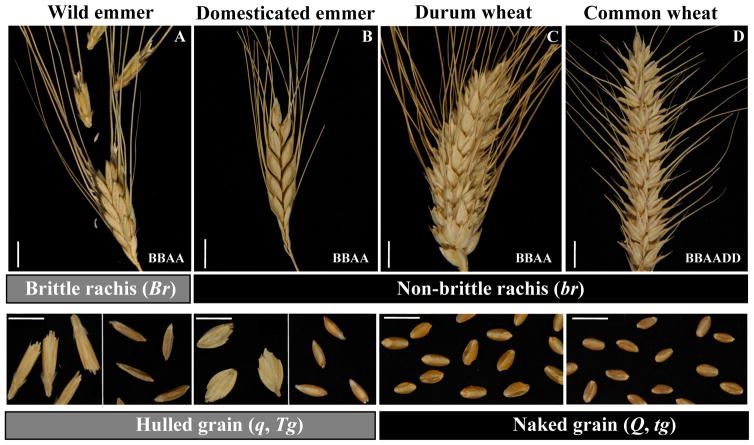
Fig. 1.
Wheat spikes showing A) brittle rachis, B–D) non-brittle rachis, A–B) hulled grain, C–D) naked grain. A) wild emmer wheat (T. turgidum ssp. dicoccoides), B) domesticated emmer (T. turgidum ssp. dicoccon), C) durum (T. turgidum ssp. durum), and D) common wheat (T. aestivum). White bars represent 1 cm. Letters at the lower right corner indicate the genome formula of each type of wheat. Gene symbols: Br = brittle rachis, Tg = tenacious glumes, and Q = square head. Photos by C. Uauy.
Wheat domestication
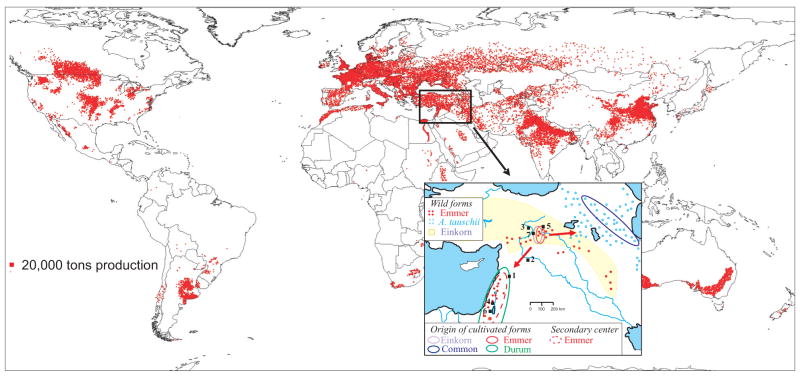
The transition from hunting and gathering to agrarian lifestyles in western Asia was a threshold in the evolution of human societies. Domestication of three cereals – einkorn, emmer, and barley – marked the beginning of that process (6). Genetic relationships between wild and domesticated einkorn and emmer suggest that the region west of Diyarbakir in southeastern Turkey is the most likely site of their domestication (Fig. 2) (7–9). From this area, the expansion of agriculture lead to the dissemination of domesticated einkorn (T. monococcum, genomes AmAm) and domesticated emmer (T. turgidum ssp. dicoccon, genomes BBAA) across Asia, Europe, and Africa. Southwestern expansion of domesticated emmer cultivation resulted in sympatry with the southern subpopulation of wild emmer (T. turgidum ssp. dicoccoides, genomes BBAA). Gene exchanges between the northern domesticated emmer with southern wild emmer populations or with emmer domesticated in the southern region resulted in the formation of a center of emmer diversity in Southern Levant (Fig. 2) (9). The consequence was a subdivision of domesticated emmer into northern and southern subpopulations with an increase in gene diversity in the latter (9). Northeast expansion of domesticated emmer cultivation resulted in sympatry with Aegilops tauschii (genomes DD) and the emergence of hexaploid common wheat (T. aestivum, genomes BBAADD) (10) within the corridor stretching from Armenia to the southwestern coastal area of the Caspian Sea (11) (Fig. 2).
Fig. 2.
The origin and current distribution of wheat. The wheat production map was provided by Dave Hodson, CIMMYT (20). The solid line ovals in the inset indicate the geographic regions of origin of the cultivated forms, while the dotted red line indicates a southern center of emmer diversity. The approximate distributions of wild emmer and Ae. tauschii are indicated by dots and that of wild einkorn by yellow shading (3). Numbers indicate archeological sites where remains of domesticated cereals dating back more than 9000 years BP were found: 1) Tell Aswad, 2) Abu Hureyra, 3) Cafer Höyük, 4) Jericho, 5) Cayönü, 6) Nahal Hemar, 7) Nevali Cori (from (2)).
The genetic changes responsible for the suite of traits that differentiate domesticated plants from their wild ancestors are referred to as the “domestication syndrome” (12). In wheat, as in other cereals, a primary component of this syndrome was the loss of spike shattering, preventing the grains to be scattered by the wind and facilitating harvesting (Fig. 1). Abscission scars of einkorn remains from archeological sites in northern Syria and southeastern Turkey revealed a gradual increase of non-shattering einkorn spikes from 9250 to 6500 years BP, a discovery interpreted as evidence of a prolonged domestication period of cereals (13). The chromosome locations of the genes controlling shattering in einkorn are unknown, but in emmer wheat it is determined by the Br (brittle rachis) loci on chromosomes 3A and 3B (14) (Fig. 1).
Another important trait for wheat domestication was the loss of tough glumes, converting hulled wheat into free-threshing wheat (Fig. 1). The primary genetic determinants of the free-threshing habit are recessive mutations at the Tg (tenacious glume) loci (15), accompanied by modifying effects of the dominant mutation at the Q locus and mutations at several other loci (15). The recent cloning of Q, which also controls the square spike phenotype in common wheat, showed that it encodes an AP2-like transcription factor. The mutation that gave rise to the Q allele is the same in tetraploid and hexaploid free-threshing wheats suggesting that it occurred only once (16).
Seeds of free-threshing wheat began to appear in archeological sites about 10,000 years before present (BP) (17). The tetraploid forms of these Neolithic free-threshing wheat may be the ancestor of the modern large-seeded, free-threshing durum (Fig. 1), which is genetically most closely related to the Mediterranean and Ethiopian subpopulations of domesticated emmer (Fig. 2) (9). The first archeological records of durum appeared in Egypt during the Greco-Roman times (reviewed in (2)).
Other traits of the wheat domestication syndrome shared by all domesticated wheats are increased seed size (Fig. 1A–B), reduced number of tillers, more erect growth, and reduced seed dormancy. One gene affecting seed size is GPC-B1, an early regulator of senescence with pleiotropic effects on grain nutrient content (18). In some genotypes and environments the accelerated grain maturity conferred by the functional GPC-B1 allele is associated with smaller seeds (19). Therefore, indirect selection for large seeds may explain the fixation of the nonfunctional GPC-B1 allele in both durum and T. aestivum (18). Except for Q and GPC-B1, no other genes relevant to the wheat domestication syndrome have been isolated so far, and a systematic effort to do so is long overdue. Not only is this knowledge critical for understanding the genetic and molecular mechanisms of domestication, it is also possible that genetic variation at these same loci plays an important role in the success of wheat as a modern crop.
Success of wheat as a crop
Domesticated wheat exemplifies the positive correlation between ploidy and success as a crop. In almost all areas where domesticated einkorn and domesticated emmer were cultivated together, it was domesticated emmer that became the primary cereal (2). Emmer remained the most important crop in the Fertile Crescent until the early Bronze Age, when it was replaced by free-threshing wheat (2). Even though a free-threshing form of einkorn has been identified, it is not widely cultivated because of the association between soft glumes and reduced ear length in this diploid species (17).
The story repeated itself with hexaploid T. aestivum expanding further than durum. Today, hexaploid T. aestivum accounts for most of the global wheat crop and is grown from Norway and Russia at 65° N to Argentina at 45° S (Fig. 2) (20). However, in tropical and subtropical regions wheat is restricted to higher elevations. Although the dominance of tetraploid wheat over diploid wheat potentially could be attributed to the greater robustness of tetraploid wheat, this does not explain the dominance of T. aestivum over durum. Durum often has larger seeds than hexaploid wheat (Fig. 1C–D) and similar yield potential as hexaploid wheat under optimum growth conditions (Table S1).
The vast majority of polyploid plants, including wheat, originated by hybridization between different species (allopolyploidy). Allopolyploidy results in the convergence in a single organism of genomes previously adapted to different environments, thus creating the potential for the adaptation of the new allopolyploid species to a wider range of environmental conditions. This has clearly been the case for hexaploid wheat, which combines the D genome from Ae. tauschii with the AB genomes from tetraploid wheat. Compared to tetraploid wheat, hexaploid T. aestivum has broader adaptability to different photoperiod and vernalization requirements, improved tolerance to salt, low pH, aluminum, and frost, better resistance to several pests and diseases, and extended potential to make different food products (Table S2).
This does not mean, however, that gene expression in an allopolyploid is the summation of gene expression in its diploid ancestors. Non-additive gene expression has been reported in numerous artificial allopolyploids (reviewed in (4, 21)). Rapid and stochastic processes of differential gene expression (22) provide an additional source of genetic variation which could be important for the successful adaptation of new allopolyploids.
There are detrimental aspects to polyploidy as well. Polyploid speciation is accompanied by a “polyploidy bottleneck” (5), in which the small number of plants contributing to the formation of a new polyploid species constrains its initial gene diversity. As only a few Ae. tauschii genotypes participated in the origin of T. aestivum (23, 24), its D-genome diversity is expected to be limited.
Recent advances in the understanding of the dynamics of gene diversity during domestication and the subsequent evolution of polyploid wheat are reviewed in the following sections to reconcile these opposing effects of polyploidy and shed light on the mechanisms by which T. aestivum come to be one of humankind’s most important crops (Fig. 2).
The capture of pre-existing diversity
Domestication is accompanied by “domestication bottlenecks” resulting in reduced gene diversity (reviewed by (25)). A study utilizing 131 RFLP loci showed that gene diversity values in cultivated emmer were 58% of those observed in wild emmer across its entire geographic distribution (9). For comparison, gene diversity values in domesticated maize and pearl millet are 57% (26) and 67% (27), respectively, of those present in their wild progenitors. It is surprising that self-pollinating emmer has an approximately equivalent proportion of the genetic diversity of its wild ancestor as cross-pollinating maize and pearl millet. Several lines of evidence indicate that gene flow between wild and domesticated emmer occurred in all places where the two were sympatric (9). Additionally, if the emmer domestication process took as long as that of einkorn domestication (13), even a slow rate of gene flow would probably be sufficient for domesticated emmer to capture a significant proportion of the genetic diversity of its wild relative.
Additional diversity bottlenecks occurred during the transition from hulled to free-threshing wheat (Fig. 1) and during the polyploid speciation of T. aestivum. A study based on 27 RFLP loci showed that diversity values in T. aestivum D genome are less than 15% of those present in populations of Ae. tauschii from Transcaucasia, reflecting the severity of the initial polyploidy bottleneck (11). However, in the A and B genomes of T. aestivum, the average diversity at the nucleotide level was found to be 30% of that present in wild emmer (28). This result suggests that difference in ploidy has presented only a weak barrier to gene flow from tetraploid wheat, including wild emmer, to hexaploid wheat (29), a result also supported by the discovery of hybrid swarms between wild emmer and common wheat (30). In summary, hexaploid wheat has captured a larger portion of the natural gene diversity present in its tetraploid ancestor than in its diploid ancestor.
The proportion of diversity captured by T. aestivum from both ancestors is likely to increase in the future as modern wheat breeders, realizing the importance of expanding diversity for successful crop improvement, are starting to use synthetic wheats in their breeding programs (31). Synthetic wheats are produced by hybridizing different tetraploid wheats and Ae. tauschii genotypes and then doubling the genomes by colchicine.
New sources of diversity
J. Doebley (32) analyzed mutations in plant genes contributing to domestication. He pointed out that none of the mutations were null alleles and suggested that domestication was achieved mostly through “tinkering” rather than “disassembling” or “crippling” wild relatives. Although this is a valid conclusion for the diploid and ancient polyploid species (maize) included in his study, null mutations that are lethal or have strong effects in diploid species may have only subtle dosage effects in young polyploid species like wheat, hence appearing as “tinkering” mutations with a potential to generate adaptive variation.
A null mutation of the GPC-B1 gene in the B genome of polyploid wheat illustrates this point. In tetraploid wheat, the GPC-B1 mutation caused a few days difference in maturity, while in diploid rice, RNAi of the rice GPC gene brings about almost complete seed sterility (supporting online text). Mutations in one of the three functional copies of a gene in hexaploid wheat are expected to have more subtle effects than in tetraploid wheat. This fact is illustrated by the higher tolerance of hexaploid wheat to induced mutations than tetraploid wheat (33). The fact that most of the 21 T. aestivum chromosomes can be removed to produce nullisomic plants exhibiting only minor phenotypic effects leaves no doubt of the buffering effect of polyploidy on gene deletions. This buffering effect is eroded in ancient polyploid species (supporting online text).
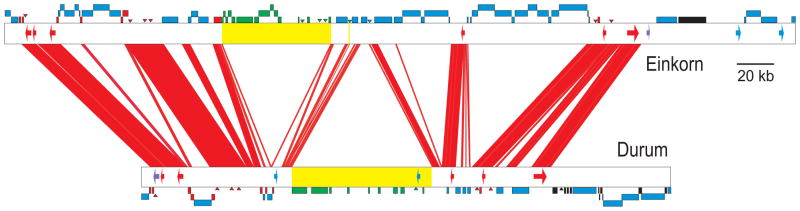
The abundance of repetitive elements in the wheat genomes (approximately 83% repetitive) (34)) greatly facilitates the generation of null mutations either by insertion of repetitive elements into genes (35) or by gene deletions (36, 37). As in maize, genes in wheat are embedded within long stretches of nested retroelements and other mobile sequences (Fig. 3). Studies of microsynteny among orthologous chromosomal regions across the tribe Triticeae showed that the intergenic space is subject to an exceedingly high rate of turnover (38). For example, 69% of the intergenic space within orthologous VRN2 regions from T. monococcum and the A genome of tetraploid wheat (Fig. 3) has been replaced over the course of the last 1.1 MYA (supporting online text).
Fig. 3.
DNA insertions and deletions in orthologous VRN2 regions from the Am genome of T. monococcum (AY485644) and the A genome of durum wheat variety Langdon (new sequence EF540321), which diverged 1.1 ± 0.1 MYA. The red lines connect orthologous regions (>96% identical). Arrows represent genes: red = orthologous, blue = ortholog absent, violet = pseudogene. Rectangles represent repetitive elements in their actual nested structure: red = orthologous, blue = insertions after divergence, green = deletion in the opposite genome (yellow region), black = not determined. Only 31% of the orthologous intergenic regions have not been replaced. See supporting online text for details.
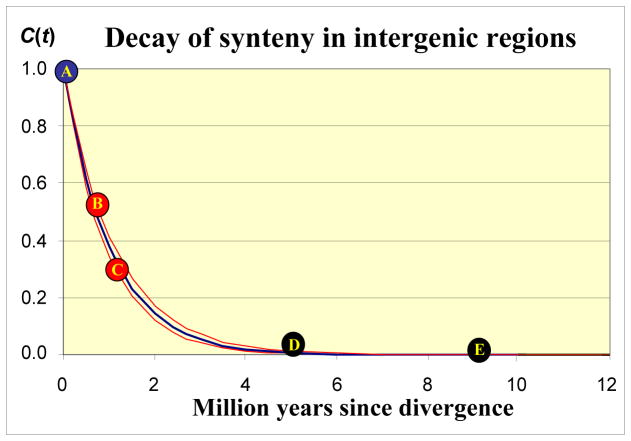
These data, along with a comparison of orthologous regions in T. urartu and the A genome of tetraploid wheat (29) yield an average replacement of 62% ± 3% of the intergenic regions during the first million years of divergence (Fig. 4, and supporting online text). The model in Fig. 4 predicts correctly the very low levels of conservation observed among orthologous intergenic regions in the A, B and D genomes of wheat (29, 39) and the complete divergence observed in comparisons of orthologous regions between wheat and barley (40, 41) (Fig. 4). To put the magnitude of this rate into perspective, indel polymorphisms from both chimpanzee and human genomes (6 to 7 MYA divergence time) equal less than 4% of the intergenic regions from these genomes (42, 43).
Fig. 4.
Decay of the proportion of conserved sequences (C(t)) in orthologous intergenic regions with divergence time. The upper and lower red curves were calculated using two independent decay rate constants (K1 and K2), and the blue curve using an average rate constant. A) The blue circle represents identical sequences at the initial time of divergence. B) The comparison between T. urartu and durum A genome PSR920 regions was used to estimate K1 (upper red curve) (29). C) The comparison between einkorn and durum A genome VRN2 regions was used to estimate K2 (lower red curve). D) Comparison of orthologous intergenic regions between wheat B genome (AY368673) and D genome (AF497474) GLU1 regions (58). E) Comparison of orthologous intergenic regions between wheat (AF459639) and barley (AY013246) VRN1 regions (40, 41). See supporting online text for details.
Studies documenting the impact of this remarkably high rate of DNA replacement on wheat genes are starting to accumulate. Insertions of repetitive elements within regulatory regions of the wheat VRN1 and VRN3 vernalization genes as well as four large independent deletions within the VRN1 first intron have been associated with the elimination of the vernalization requirement (44–47). A deletion upstream of the PPD-D1 photoperiod gene is associated with the widely distributed photoperiod insensitive allele (48). Such diversity in genes regulating flowering time is particularly relevant due to its large impact on wheat adaptability to different environments. Deletions have also provided increased diversity in wheat products. Puroindoline A and B gene deletions, which have become fixed in the A and B genomes, are responsible for the hard grain texture of pasta wheat. A polymorphism for a Puroindoline A deletion (or for a point mutation in Puroindoline B) in the hexaploid wheat D genome dramatically affects grain hardness, dividing wheat into those used for bread (hard texture) or for cookies and pastries (soft texture) (49).
The example in Fig. 3 shows two genes affected by deletions within a small genomic region, providing an additional example of the high frequency of gene deletions. Such deletions are fixed in polyploid wheat with an initial rate of 1.8 × 10−2 locus−1 MY−1, ten times faster than in wheat’s diploid ancestors (50). However, most deletions are still polymorphic and represent, together with point mutations, an important component of genetic diversity in polyploid wheat (51).
Evidence is accumulating that the creation of artificial allopolyploids can be immediately followed by reactivation of mobile elements (52, 53). In one Arabidopsis allotetraploid, these changes were associated with genomic rearrangements, chromosomal abnormalities, DNA deletions (1% of the genome), and pollen sterility (52). A higher level of DNA deletions (12–14%) was found in two wheat artificial allotetraploids involving different diploid species than the ones that originated tetraploid wheat (54). An association of these deletions with chromosomal abnormalities would limit the chances of these diploid combinations to generate new successful allopolyploid species. Examination of polymorphisms for gene deletions in the D genome of T. aestivum showed that only 0.17% of the D genome has been deleted during the last 8000 years and that deletions are present at low frequencies, suggesting a gradual accumulation of gene deletions rather than a burst of deletions immediately following the hexaploid what polyploidization event (51).
Repetitive DNA can also facilitate gene duplication. A study tracing the evolution of a dispersed multigene family in wheat showed that duplication of a gene into the intergenic space accelerated its subsequent duplication rate 20-fold (55). Additionally, a promoter supplied by a neighboring mobile sequence facilitated the expression of one of the duplicated gene copies as well as the generation of a new gene (55). This study suggests that wheat intergenic DNA facilitates both gene duplication and novel expression of duplicated genes. Studies in rice and maize provide extreme examples of mobile repetitive elements duplicating gene fragments and, occasionally, complete genes across the genome (reviewed by (56)). The importance of gene duplication in wheat is exemplified by the recently isolated wheat VRN2 and GPC1 genes, both of which likely originated as dispersed duplications after the wheat-rice divergence (18, 57).
Although more research is needed to refine our understanding of the specific mechanisms by which repetitive sequences affect gene content in wheat, evidence already available indicates that the dynamic nature of wheat repetitive sequences readily generate new genetic variation which may facilitate the success of polyploid wheat as a crop.
Concluding remarks
Polyploid wheat has been able to compensate for diversity bottlenecks caused by domestication and polyploidy by capturing a relatively large proportion of the variability of its tetraploid wild progenitor. In addition, new variation is rapidly generated in the dynamic wheat genomes through gene deletions and insertions of repetitive elements into coding and regulatory gene regions. These mutations can then be expressed as quantitative gene dosage differences due to the polyploid nature of wheat. Synergy between the high mutation rates and the buffering effects of polyploidy makes it possible for polyploid wheat to capitalize on the diversity generated by its dynamic genomes.
Supplementary Material
Acknowledgments
We thank Drs. L. Yan, W. Ramakrishna, P. SanMiguel and J. Bennetzen for their help to sequence the VRN2 region. We also thank M. Feldman, A. Levy, P. Morell, P. McGuire, M. Nesbitt, C. Uauy, E. Akhunov, and I. Lowe for their valuable suggestions. This research was supported by NRI USDA-CSREES Grants No. 2007-35301-17737 and 2006-55606-16629 and by NSF Grant No. DBI-0321757.
Footnotes
References and Notes
- 1.FAO. Statistical Yearbook 2005–2006. United Nations: 2006. WEB Edition. [Google Scholar]
- 2.Nesbitt M, Samuel D. In: Padulosi S, Hammer K, Heller J, editors. From staple crop to extinction? The archaeology and history of hulled wheats; Proc. 1st Int. Workshop on hulled wheats; Castelvecchio, Pacoli, Italy. Rome, Italy: Int. Plant Genet. Res. Inst; 1996. [Google Scholar]
- 3.Zohary D, Hopf M. Domestication of plants in the Old World. 3. Oxford University Press; Oxford, Oxford: 2000. [Google Scholar]
- 4.Wendel JF. Plant Mol Biol. 2000;42:225. [PubMed] [Google Scholar]
- 5.Stebbins GL. Variation and Evolution in Plants. Columbia University Press; New York: 1950. [Google Scholar]
- 6.Harlan JR, Zohary D. Science. 1966;153:1074. doi: 10.1126/science.153.3740.1074. [DOI] [PubMed] [Google Scholar]
- 7.Heun M, et al. Science. 1997;278:1312. [Google Scholar]
- 8.Ozkan H, et al. Theor Appl Genet. 2005;110:1052. doi: 10.1007/s00122-005-1925-8. [DOI] [PubMed] [Google Scholar]
- 9.Luo MC, et al. Theor Appl Genet. 2007;114:947. doi: 10.1007/s00122-006-0474-0. [DOI] [PubMed] [Google Scholar]
- 10.Kihara H. Agric and Hort (Tokyo) 1944;19:13. [Google Scholar]
- 11.Dvorak J, Luo MC, Yang ZL, Zhang HB. Theor Appl Genet. 1998;97:657. [Google Scholar]
- 12.Hammer K. Kulturpflanze. 1984;32:11. [Google Scholar]
- 13.Tanno K, Willcox G. Science. 2006;311:1886. doi: 10.1126/science.1124635. [DOI] [PubMed] [Google Scholar]
- 14.Nalam VJ, Vales MI, Watson CJW, Kianian SF, Riera-Lizarazu O. Theor Appl Genet. 2006;112:373. doi: 10.1007/s00122-005-0140-y. [DOI] [PubMed] [Google Scholar]
- 15.Jantasuriyarat C, Vales MI, Watson CJW, Riera-Lizarazu O. Theor Appl Genet. 2004;108:261. doi: 10.1007/s00122-003-1432-8. [DOI] [PubMed] [Google Scholar]
- 16.Simons KJ, et al. Genetics. 2006;172:547. doi: 10.1534/genetics.105.044727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salamini F, Ozkan H, Brandolini A, Schafer-Pregl R, Martin W. Nat Rev Genet. 2002;3:429. doi: 10.1038/nrg817. [DOI] [PubMed] [Google Scholar]
- 18.Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. Science. 2006;314:1298. doi: 10.1126/science.1133649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uauy C, Brevis JC, Dubcovsky J. J Exp Bot. 2006;57:2785. doi: 10.1093/jxb/erl047. [DOI] [PubMed] [Google Scholar]
- 20.Lantican MA, Dubin HJ, Morris ML. Impacts of international wheat breeding research in the Developing World, 1988–2002. CIMMYT. 2005 [Google Scholar]
- 21.Chen ZJ, Ni ZF. Bioessays. 2006;28:240. doi: 10.1002/bies.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang JL, et al. Genetics. 2004;167:1961. doi: 10.1534/genetics.104.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dvorak J, Luo MC, Yang ZL. In: The origins of agriculture and crop domestication. Damania AB, Valkoun J, Willcox G, Qualset CO, editors. ICARDA; Aleppo, Syria: 1998. p. 235. [Google Scholar]
- 24.Talbert LE, Smith LY, Blake MK. Genome. 1998;41:402. [Google Scholar]
- 25.Buckler ES, Thornsberry JM, Kresovich S. Genet Res. 2001;77:213. doi: 10.1017/s0016672301005158. [DOI] [PubMed] [Google Scholar]
- 26.Wright SI, et al. Science. 2005;308:1310. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- 27.Gaut BS, Clegg MT. Genetics. 1993;135:1091. doi: 10.1093/genetics/135.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haudry A, et al. Mol Biol Evol. 2007 doi: 10.1093/molbev/msm077. [DOI] [PubMed] [Google Scholar]
- 29.Dvorak J, Akhunov ED, Akhunov AR, Deal KR, Luo MC. Mol Biol Evol. 2006;23:1386. doi: 10.1093/molbev/msl004. [DOI] [PubMed] [Google Scholar]
- 30.Zohary D, Brick Z. Wheat Inf Service. 1961;13:6. [Google Scholar]
- 31.Warburton ML, et al. Euphytica. 2006;149:289. [Google Scholar]
- 32.Doebley J. Science. 2006;312:1318. doi: 10.1126/science.1128836. [DOI] [PubMed] [Google Scholar]
- 33.Slade A, Fuerstenberg S, Loeffler D, Steine M, Facciotti D. Nat Biotechnol. 2005;23:75. doi: 10.1038/nbt1043. [DOI] [PubMed] [Google Scholar]
- 34.Flavell RB, Bennett MD, Smith JB, Smith DB. Biochem Genet. 1974;12:257. doi: 10.1007/BF00485947. [DOI] [PubMed] [Google Scholar]
- 35.Harberd NP, Flavell RB, Thompson RD. Mol Gen Genet. 1987;209:326. doi: 10.1007/BF00329661. [DOI] [PubMed] [Google Scholar]
- 36.Chantret N, et al. Plant Cell. 2005;17:1033. doi: 10.1105/tpc.104.029181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kidwell MG, Lisch D. Proc Natl Acad Sci USA. 1997;94:7704. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wicker T, et al. The Plant Cell. 2003;15:1186. doi: 10.1105/tpc.011023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu YQ, et al. Genetics. 2006;174:1493. doi: 10.1534/genetics.106.060756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.SanMiguel P, Ramakrishna W, Bennetzen JL, Busso CS, Dubcovsky J. Funct Integr Genomics. 2002;2:70. doi: 10.1007/s10142-002-0056-4. [DOI] [PubMed] [Google Scholar]
- 41.Ramakrishna W, et al. Genetics. 2002;162:1389. doi: 10.1093/genetics/162.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mikkelsen TS, et al. Nature. 2005;437:69. [Google Scholar]
- 43.It would be interesting to compare mammalian genomes from species with more similar generation times to those of annual cereals, to determine the effect of generation time on these differences.
- 44.Yan L, et al. Theor Appl Genet. 2004;109:1677. doi: 10.1007/s00122-004-1796-4. [DOI] [PubMed] [Google Scholar]
- 45.Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J. Plant Physiol. 2005;138:2364. doi: 10.1104/pp.105.064287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan L, et al. Proc Natl Acad Sci USA. 2006;103:19581. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu D, et al. Mol Gen Genomics. 2005;273:54. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- 48.Faure S, Turner A, Beales J, Higgins J, Laurie DA. Photoperiodic control of flowering time in barley and wheat. Plant & Animal Genome XV; San Diego, CA. January 2007; 2007. [Google Scholar]
- 49.Giroux MJ, Morris CF. Proc Natl Acad Sci USA. 1998;95:6262. doi: 10.1073/pnas.95.11.6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dvorak J, Akhunov ED. Genetics. 2005;171:323. doi: 10.1534/genetics.105.041632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dvorak J, Yang ZL, You FM, Luo MC. Genetics. 2004;168:1665. doi: 10.1534/genetics.103.024927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madlung A, et al. Plant J. 2005;41:221. doi: 10.1111/j.1365-313X.2004.02297.x. [DOI] [PubMed] [Google Scholar]
- 53.Kashkush K, Feldman M, Levy AA. Nat Genet. 2003;33:102. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]
- 54.Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA. Plant Cell. 2001;13:1749. doi: 10.1105/TPC.010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akhunov ED, Akhunova AR, Dvorak J. Mol Biol Evol. 2007;24:539. doi: 10.1093/molbev/msl183. [DOI] [PubMed] [Google Scholar]
- 56.Morgante M. Curr Opin Biotech. 2006;17:168. doi: 10.1016/j.copbio.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 57.Yan L, et al. Science. 2004;303:1640. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kong XY, Gu YQ, You FM, Dubcovsky J, Anderson OD. Plant Mol Biol. 2004;54:55. doi: 10.1023/B:PLAN.0000028768.21587.dc. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.