Abstract
Enhancing the nutritional value of food crops is a means for improving human nutrition and health. We report here the positional cloning of Gpc-B1, a wheat quantitative trait locus associated with increased grain protein, Zn, and Fe content. The ancestral wild wheat allele encodes a NAC transcription factor (NAM-B1) that accelerates senescence and increases nutrient remobilization from leaves to developing grains, whereas modern wheat varieties carry a non-functional NAM-B1 allele. Reduction in RNA levels of the multiple NAM homologs by RNA interference delayed senescence by over three weeks and reduced wheat grain protein, Zn, and Fe content by over 30%.
The World Health Organization estimates that over 2 billion people have deficiencies in key micronutrients such as Zn and Fe, and over 160 million children under the age of five lack adequate protein (1); leading to an economic burden for society (2). The two major types of wheat, tetraploid wheats (2n = 28), used for pasta, and hexaploid wheats (2n = 42), used primarily for bread, account for approximately 20% of all calories consumed worldwide. Annual wheat production is estimated at 620 million tons of grain (3), translating into approximately 62 million tons of protein. Increasing grain protein content (GPC) has been hindered by environmental effects, complex genetic systems governing this trait, and a negative correlation with yield (4). Less progress has been made in increasing Zn and Fe content; the focal point of the HarvestPlus global initiatives (5).
Wild emmer wheat (Triticum turgidum ssp. dicoccoides), abbreviated hereafter as DIC, is the ancestor of cultivated pasta wheat (T. turgidum ssp. durum) and a promising source of genetic variation in protein, Zn, and Fe content (6, 7). A quantitative trait locus (QTL) for GPC was mapped on chromosome arm 6BS in a population of recombinant inbred lines derived from the T. turgidum ssp. durum cultivar Langdon (LDN) and the chromosome substitution line LDN (DIC6B) (8). This locus was associated with GPC increases of approximately 14 g*kg−1 in both tetraploid and hexaploid lines (8–10). Olmos et al. (11) mapped this QTL as a simple Mendelian locus, Gpc-B1 (Fig. 1A), which was later located within a 0.3 cM interval (12). Molecular markers Xuhw89 and Xucw71 within this region flank a 245-kb physical contig including Gpc-B1 (13).
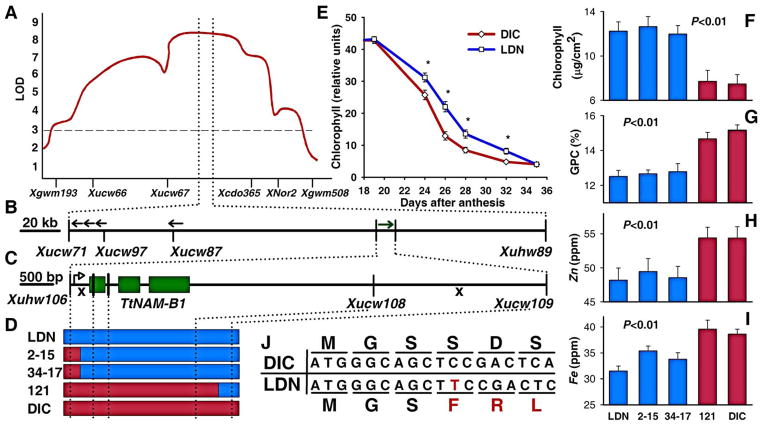
Fig. 1.
Map-based cloning of Gpc-B1. (A) QTL for grain protein on wheat chromosome arm 6BS (11). (B) Sequenced B-genome physical contig. The position and orientation of five genes is indicated by arrows. (C) Fine mapping of Gpc-B1. The x’s indicate the positions of critical recombination events flanking Gpc-B1. Vertical lines represent polymorphism mapped in the critical lines. A single gene with three exons (green rectangles) was annotated within the 7.4 kb region flanked by the closest recombination events. The open arrowhead indicates the transcription initiation site. (D) Graphical genotypes of critical recombinant substitution lines used for fine-mapping of Gpc-B1. Blue bars represent LDN markers; red bars represent DIC markers. (E) Flag leaf chlorophyll content of recombinant substitution lines segregating for Gpc-B1 (14). Asterisks indicate significant differences (P<0.01). Phenotypes of critical recombinant substitution lines: (F) chlorophyll at 20 days after anthesis (DAA), (G) grain protein, (H) Zn, and (I) Fe concentrations. Blue and red bars indicate the presence of the LDN and DIC alleles at TtNAM-B1, respectively. (J) First 18 nucleotides of DIC and LDN TtNAM-B1 alleles and their corresponding amino acid translation. The LDN allele carries a 1-bp insertion (red T) that disrupts the reading frame (indicated by red amino acid residues). Error bars represent standard error of the means (E–I).
Tetraploid and hexaploid wheat lines carrying this 245-kb DIC segment show delayed senescence and increased GPC and grain micronutrients (14–15). The complete sequencing of this region (DQ871219) revealed five genes (Fig. 1B) (16). A high-resolution genetic map, based on approximately 9,000 gametes and new molecular markers (table S1), was used to determine the linkage between these genes and the Gpc-B1 locus. Three recombinant substitution lines with recombination events between markers Xuhw106 and Xucw109 delimited a 7.4-kb region (Fig. 1, C–D) (16). The recombinant lines carrying this DIC segment senesced on average 4 to 5 days earlier (P<0.01, Fig. 1, E–F) and exhibited a 10% to 15% increase in GPC (Fig. 1G), Zn (Fig. 1H), and Fe (Fig. 1I) concentrations in the grain (P<0.01). Complete linkage of the 7.4-kb region with the different phenotypes suggests that Gpc-B1 is a single gene with multiple pleiotropic effects.
The annotation of this 7.4-kb region (Fig. 1C) identified a single gene encoding a NAC domain protein, characteristic of the plant specific family of NAC transcription factors (17). NAC genes play important roles in developmental processes, auxin signaling, defense and abiotic stress responses, and leaf senescence (18, 19). Phylogenetic analyses revealed that the closest plant proteins were the rice gene ONAC010 (NP_911241) and a clade of three Arabidopsis proteins including No Apical Meristem (NAM) (figs. S1–2). On the basis of these similarities, the gene was designated NAM-B1 (DQ869673). To indicate the species source we have added a two letter prefix (e.g. Ta and Tt for T. aestivum and T. turgidum genes, respectively).
Comparison of the parental TtNAM-B1 sequences revealed a 1-bp substitution within the first intron and a thymine residue insertion at position 11, generating a frame shift mutation in the LDN allele (DQ869674, Fig. 1J). This frame shift resulted in a predicted protein having no similarity to any GenBank sequence and lacking the NAC domain.
The wild type TtNAM-B1 allele was found in all 42 wild emmer accessions examined (T. turgidum ssp. dicoccoides) (table S2), and in 17 of the 19 domesticated emmer accessions (T. turgidum ssp. dicoccum). However, 57 cultivated durum lines (T. turgidum ssp. durum) (20) (table S3) lack the functional allele suggesting that the 1-bp frame shift insertion was fixed during the domestication of durum wheat. The wild type TaNAM-B1 allele was also absent from a collection of 34 varieties of hexaploid wheat (T. aestivum ssp. aestivum), representing different market classes and geographic locations. Twenty-nine of these showed no PCR amplification products of the TaNAM-B1 gene suggesting that it is deleted, while the remaining five lines have the same 1-bp insertion observed in the durum lines (table S4).
In addition to the mutant TtNAM-B1 copy, the durum wheat genome includes an orthologous copy (TtNAM-A1) on chromosome arm 6AS and a paralogous one (TtNAM-B2) 91% identical at the DNA level to TtNAM-B1 on chromosome arm 2BS (21) (fig. S3, table S5). These two copies have no apparent mutations. Comparisons at the protein level of the five domains characteristic of NAC transcription factors (17) revealed 98% to 100% protein identity (fig. S2) between barley, wheat, rice, and maize homologs.
Quantitative PCR (16) showed transcripts from the three TtNAM genes at low levels in flag leaves prior to anthesis, after which their levels increased significantly towards grain maturity (Fig. 2A). Transcripts were also detected in green spikes and peduncles. The similar transcription profiles and near identical sequences of TtNAM-A1, B1 and B2 suggest that the 4–5 day delay in senescence and the 10% to 15% decrease in grain protein, Zn, and Fe content observed in LDN are likely the result of a reduction in the amount of functional protein rather than the complete loss-of-function of a unique gene.
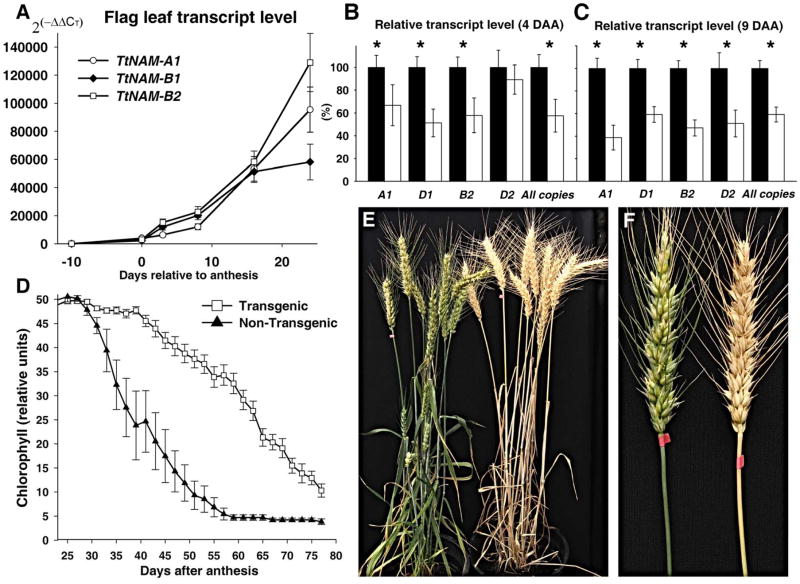
Fig. 2.
(A) Expression profile of the different TtNAM genes relative to ACTIN in tetraploid wheat recombinant substitution line 300 carrying a functional TtNAM-B1 gene. Units are values linearized with the 2(−Δ ΔCT) method, where CT is the threshold cycle. (B) Relative transcript level of endogenous TaNAM genes in T2 plants (L19-54) segregating for transgenic (n = 12, white) and non-transgenic (n = 11, black) TaNAM RNAi constructs at 4 and (C) 9 days after anthesis. Asterisks indicate significant differences (P<0.05). (D) Flag leaf chlorophyll content profile of transgenic (n = 22 T1 plants) and non-transgenic controls (n = 10 T1 plants). (E) Representative transgenic (left) and non-transgenic (right) plants 50 DAA. (F) Main spike and peduncles of representative transgenic and non-transgenic plants 50 DAA. Error bars represent standard error of the means.
To test this hypothesis, we reduced the transcript levels of all NAM copies using RNA interference (RNAi). An RNAi construct (16) was transformed into the hexaploid wheat variety Bobwhite, selected for its higher transformation efficiency relative to tetraploid wheat. The RNAi construct targeted the 3′ end of the four TaNAM genes found in hexaploid wheat (TaNAM-A1, D1, B2 and D2), outside the NAC domain, to avoid interference with other NAC transcription factors (fig. S4, table S6) (22).
We identified two independent transgenic plants (L19-54 and L23-119) with an expected stay-green phenotype. Quantitative PCR analysis of transgenic L19-54 plants showed a significant reduction in the endogenous RNA levels of the different TaNAM copies (22) at four and nine days after anthesis (Fig. 2, B–C, P<0.05) compared to control lines. Transgenic plants reached 50% chlorophyll degradation in flag leaves 24 days later than their non-transgenic sibs (Fig. 2D, P<0.001), and their main spike peduncles turned yellow more than 30 days later than the controls (Fig. 2, E–F).
The presence of the RNAi transgene also had significant effects on grain protein, Zn and Fe concentrations. Transgenic plants showed a reduction of over 30% in GPC (P<0.001), 36% in Zn (P<0.01), and 38% in Fe (P<0.01) concentration compared to the non-transgenic controls (Table 1). No significant differences were observed in grain size (P=0.41), suggesting that the extra days of grain filling conferred by the reduced TaNAM transcript level did not translate into larger grains in our greenhouse experiments (23). Similar results were obtained for the second transgenic event, L23-119 (fig. S5, table S7).
Table 1.
Characterization of grain and senescence related traits of transgenic Bobwhite T1 plants (event L19-54) segregating for the presence (transgenic, n = 22 plants) or absence (non-transgenic, n = 10 plants) of the TaNAM RNAi construct.
| GPC (%) | Zn (ppm) | Fe (ppm) | TKW* (g) | Dry Peduncle (DAA)* | Dry Spike (DAA) | |
|---|---|---|---|---|---|---|
| Transgenic | 13.27 | 52.45 | 37.40 | 30.23 | 72.5 | 53.0 |
| Non-transgenic | 19.08 | 82.50 | 60.83 | 31.27 | 38.4 | 37.2 |
| Difference | −5.81 | −30.09 | −23.42 | −1.04 | + 34.1 | + 15.8 |
| P value | <0.001 | <0.01 | <0.01 | 0.41 | <0.001 | <0.001 |
TKW = Thousand kernel weight, DAA = Days after anthesis.
These results suggest that the reduced grain protein, Zn, and Fe concentrations were the result of reduced translocation from leaves, rather than a dilution effect caused by larger grains. This hypothesis was confirmed by analyzing the residual nitrogen (N), Zn, and Fe content in the flag leaves. We analyzed both transgenic events together (due to greater variability in flag leaves compared to the grains) and confirmed higher levels of N (P=0.01), Zn (P<0.01), and Fe (P<0.01) in the flag leaves of transgenic plants compared to the non-transgenic sister lines (table S8). This supports a more efficient N, Zn, and Fe remobilization in plants with higher levels of functional TaNAM transcripts.
These results confirm that a reduction in RNA levels of the TaNAM genes is associated with a delay in whole plant senescence, a decrease in grain protein, Zn and Fe concentrations, and an increase in residual N, Zn and Fe in the flag leaf. These multiple pleiotropic effects suggest a central role for the NAM genes as transcriptional regulators of multiple processes during leaf senescence, including nutrient remobilization to the developing grain.
The differences observed between the transgenic and non-transgenic plants for these traits were larger than those observed between the LDN and DIC alleles. The RNA interference on all functional TaNAM homologs may result in a larger reduction of functional transcripts than the single non-functional TtNAM-B1 allele in tetraploid recombinant lines carrying the LDN allele.
The cloning of Gpc-B1 provides a direct link between the regulation of senescence and nutrient remobilization and an entry point to characterize the genes regulating these two processes. This may contribute to their more efficient manipulation in crops and translate into food with enhanced nutritional value.
Supplementary Material
Acknowledgments
We thank Drs. G. Hart and L. Joppa for the original mapping materials, L. Li, L. Valarikova, and L. Beloborodov for expert technical assistance, R. Thilmony for critical reading of the manuscript and the National Small Grain Collection, M. Sanguineti, J. Dvorak, and E. Nevo for germplasm used in this study. This research was supported by the National Research Initiative of USDA’s Cooperative State Research, Education and Extension Service, CAP Grant No. 2006-55606-16629, by BARD, the United States – Israel Binational Agricultural Research and Development Fund, Grant No. US-3573-04C and by a Vaadia-BARD Fellowship. GenBank accession numbers are DQ871219 and DQ869672 to DQ869679.
Footnotes
References and Notes
- 1.World Health Organization. Meeting of interested parties: Nutrition. 2001 http://www.who.int/mipfiles/2299/MIP_01_APR_SDE_3.en.pdf.
- 2.Welch RM, Graham RD. J Exp Bot. 2004;55:353. doi: 10.1093/jxb/erh064. [DOI] [PubMed] [Google Scholar]
- 3.UN Food and Agriculture Organization. Food Outlook. 2005;4 http://www.fao.org/documents. [Google Scholar]
- 4.Simmonds N. J Sci Food Agric. 1995;67:309. [Google Scholar]
- 5.http://www.harvestplus.org/index.html
- 6.Avivi L. High protein content in wild tetraploid Triticum dicoccoides Korn. In: Ramanujam S, editor. Indian Soc Genet, Plant Breed; 5th Int. Wheat Genet. Symp; New Delhi. 1978. [Google Scholar]
- 7.Cakmak I, et al. Soil Sci Plant Nut. 2004;50:1047. [Google Scholar]
- 8.Joppa LR, Du C, Hart GE, Hareland GA. Crop Sci. 1997;37:1586. [Google Scholar]
- 9.Mesfin A, Frohberg R, Anderson J. Crop Sci. 1999;39:508. [Google Scholar]
- 10.Chee PW, Elias EM, Anderson J, Kianian SF. Crop Sci. 2001;41:295. [Google Scholar]
- 11.Olmos S, et al. Theor Appl Genet. 2003;107:1243. doi: 10.1007/s00122-003-1377-y. [DOI] [PubMed] [Google Scholar]
- 12.Distelfeld A, et al. Funct Integr Genomics. 2004;4:59. doi: 10.1007/s10142-003-0097-3. [DOI] [PubMed] [Google Scholar]
- 13.Distelfeld A, Uauy C, Fahima T, Dubcovsky J. New Phytol. 2006;169:753. doi: 10.1111/j.1469-8137.2005.01627.x. [DOI] [PubMed] [Google Scholar]
- 14.Uauy C, Brevis J, Dubcovsky J. J Exp Bot. 2006;57:2785. doi: 10.1093/jxb/erl047. [DOI] [PubMed] [Google Scholar]
- 15.Distelfeld A, et al. Physiol Plant. in press. [Google Scholar]
- 16.Materials and methods are available as supporting material on Science Online.
- 17.Ooka H, et al. DNA Res. 2003;10:239. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 18.Olsen AN, Ernst HA, Leggio LL, Skriver K. Trends Plant Sci. 2005;10:79. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 19.Guo Y, Gan S. Plant J. 2006;46:601. doi: 10.1111/j.1365-313X.2006.02723.x. [DOI] [PubMed] [Google Scholar]
- 20.Maccaferri M, Sanguineti MC, Donini P, Tuberosa R. Theor Appl Genet. 2003;107:783. doi: 10.1007/s00122-003-1319-8. [DOI] [PubMed] [Google Scholar]
- 21.The paralogous TtNAM-A2 copy was not detected in the LDN tetraploid BAC library nor by PCR with 2A genome specific primers derived from T. urartu, where the TuNAM-A2 gene is present.
- 22.The Bobwhite TaNAM-B1 gene is deleted as determined by PCR with four sets of independent NAM-B1 specific primers (table S5). Therefore, no expression data is included for TaNAM-B1 in the transgenic plants.
- 23.Field experiments including Gpc-B1 isogenic lines showed a more variable effect of the DIC chromosome region (including TtNAM-B1) on grain size (14).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.