Abstract
Stripe rust is a devastating fungal disease that afflicts wheat in many regions of the world. New races of Puccinia striiformis, the pathogen responsible for this disease, are virulent on most of the known race-specific resistance genes. We report here the map-based cloning of the gene Yr36 (WKS1), which confers non race-specific resistance to stripe rust at relatively high temperatures (25–35 °C). This gene includes a kinase and a putative START lipid-binding domain. Five independent mutations and transgenic complementation confirmed that both domains are necessary to confer resistance. Yr36 is present in wild wheat but absent in modern pasta and bread wheat varieties and therefore, can now be used to improve resistance to stripe rust in a broad set of varieties.
Bread wheat (Triticum aestivum L.) provides approximately 20% of the calories consumed by humankind. The increasing world demand for cereals requires improved strategies to reduce yield losses due to pathogens. Wheat stripe rust, caused by the fungus Puccinia striiformis f. sp. tritici (PST, table S1) affects millions of hectares of wheat and virulent races that appeared within the last decade are causing large yield losses (1–3). Historically, resistant varieties have provided an economical and “environmentally friendly” method to control stripe rust. Numerous race-specific resistance genes have been deployed by breeders, but each had limited durability presumably because of rapid pathogen evolution. In contrast, non race-specific (i.e. “slow-rusting”) genes generally have a broader spectrum of resistance, are more effective at adult plant stages, provide partial resistance, and usually confer more durable resistance than race-specific genes (1). Unfortunately, our understanding of non race-specific resistance is limited because none of these genes has yet been cloned.
We report here the positional cloning of the high-temperature stripe rust resistance gene Yr36. This gene was first discovered in wild emmer wheat (T. turgidum ssp. dicoccoides accession FA15-3) (DIC) (4). Analysis of Yr36 isogenic lines in different genetic backgrounds confirmed that this gene confers partial resistance to PST under field conditions, and is associated with significant yield increases when the pathogen is present. In controlled environments, plants with Yr36 are resistant at relatively high temperatures (25–35 °C) but susceptible at lower temperatures (e.g. 15 °C) (4). Yr36 resistance, originally discovered in adult plants, has some effectiveness in seedlings at high-temperatures (figs. S1). Other high-temperature non race-specific resistance genes have provided durable resistance to stripe rust and are used frequently in wheat breeding programs (5–8).
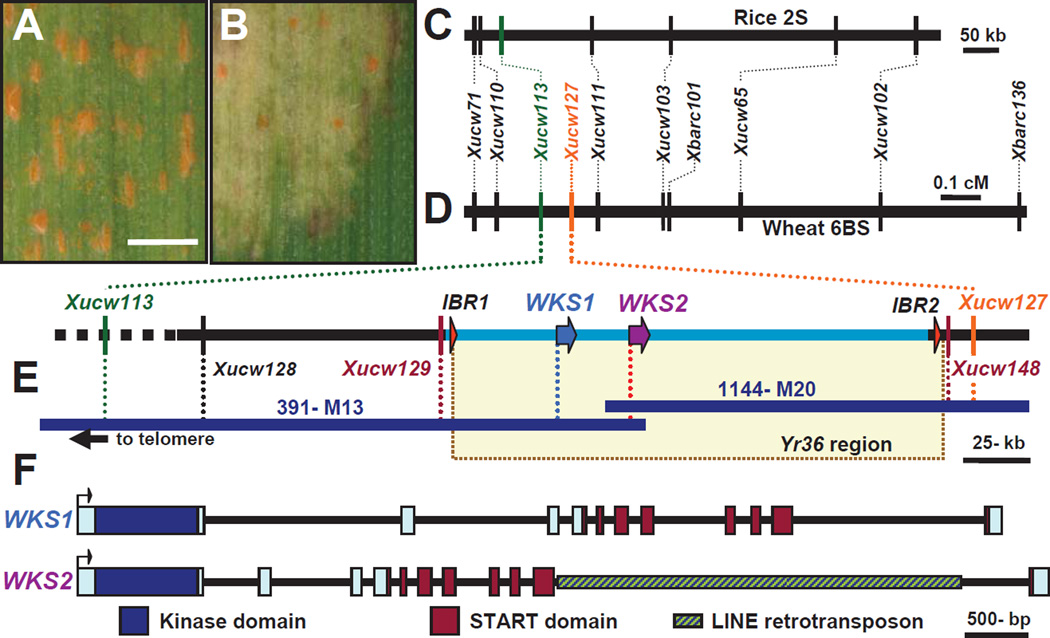
To clone Yr36, we crossed the susceptible durum wheat variety Langdon (LDN, Fig. 1A) with the resistant isogenic recombinant substitution line RSL65 (Fig. 1B), which carries Yr36 in a LDN genetic background. We screened a population of 4500 F2 plants using Yr36 flanking markers Xucw71 and Xbarc136 (4) and identified 121 lines with recombination events between these two markers. Based on genes from the rice colinear region (9), nine PCR markers were developed to construct a high-density map of Yr36 (Fig. 1C and D, table S2). Using replicated field trials and controlled environment inoculations (tables S3 and S4, figs. S2 and S3), Yr36 was mapped to a 0.14 cM interval delimited by markers Xucw113 and Xucw111 (Fig. 1D).
Fig. 1. Map-based cloning of Yr36.
(A and B) Phenotype of susceptible parent Langdon with PST sporulation (A) and partially resistant parent RSL65 (B). Bar = 1mm. (C and D) Genetic maps of colinear regions of rice chromosome 2 (C) and wheat chromosome 6B (D). (E) Physical map of the Yr36 region. Genes are represented by colored arrows and the deleted region in Langdon by a light blue line. (F) Structure of the WKS genes. Exons are represented by rectangles and the kinase and START domains are shown in different colors.
Screening the RSL65 BAC library (10) with the distal marker Xucw113 yielded six BACs (fig. S4). BAC ends were used to re-screen the library and extend the contig by chromosome walking. BAC-end marker Xucw127 (table S2, fig. S4) was mapped proximal to Yr36, thereby completing the physical map (Fig. 1E). BAC clones 391M13 and 1144M20 were sequenced and a contiguous 314-kb sequence including the flanking markers was annotated and deposited in GenBank (EU835198, fig. S5). New markers were developed from the sequence (table S2) and Yr36 resistance (eight PST races, table S5) was mapped between Xucw129 and Xucw148 (0.02 cM).
This region has two pairs of duplicated genes (fig. S5). The first pair includes two short putative genes (IBR1 and IBR2) with an ‘in between RING finger’ domain (IBR, pfam01485). The two other duplicated genes, which we designated WHEAT KINASE-START 1 and 2 (WKS1 and WKS2, Fig. 1F), encode 86% identical proteins that have a predicted kinase domain followed by a predicted steroidogenic acute regulatory protein-related lipid transfer domain (START, pfam01852). WKS1, WKS2 and IBR1 are deleted in the susceptible parent (Fig. 1E). The WKS genes were prioritized for functional characterization because their domains have been associated with plant responses to pathogens in other species (11–13).
Primers specific for WKS1 and WKS2 putative kinase and START domains (table S6) were used to screen a population of 1,536 ethyl methanesulphonate (EMS) mutagenized M2 lines from the common wheat breeding line UC1041+Yr36 (14). Of the 117 mutants found in the TILLING screen (15), we selected for functional characterization 6 mutants with changes in conserved amino acids in WKS1 (figs. S6 and S7A) and 3 with premature stop codons in WKS2 (table S7).
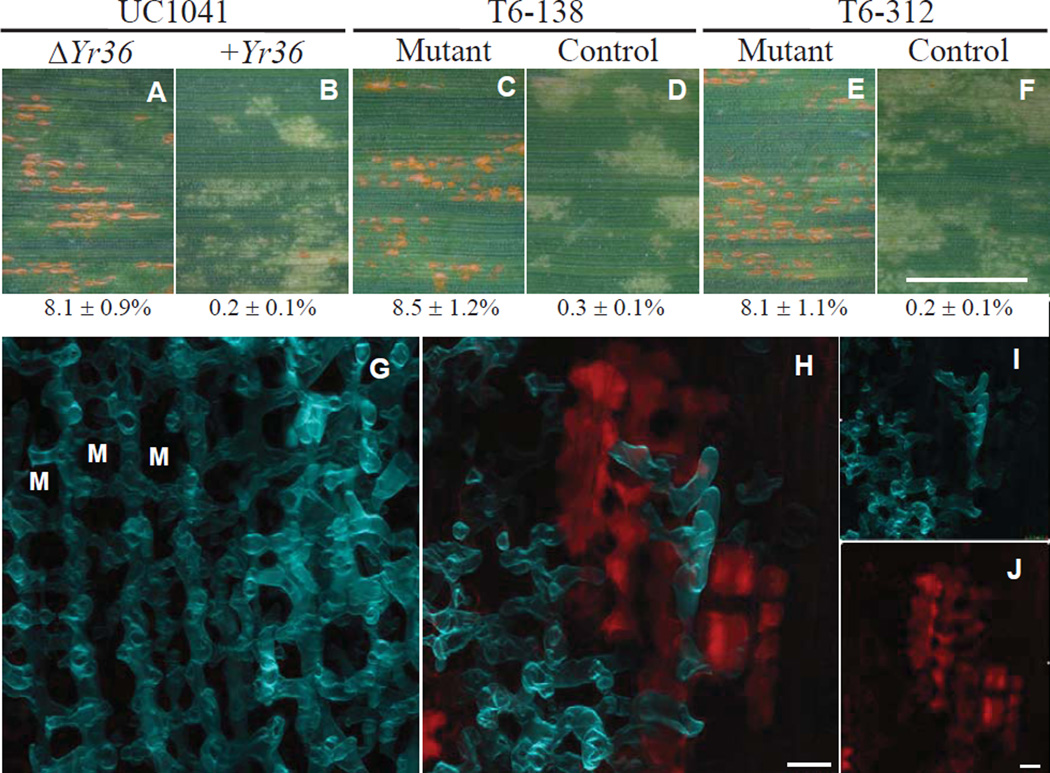
Of the 6 WKS1 mutants, 5 showed susceptible reactions similar to the susceptible UC1041 control line (Fig. 2A to F, and figs. S8 and S9). In contrast, none of the WKS2 truncation mutants was susceptible (fig. S8), suggesting that WKS1 is Yr36. Both the kinase (fig. S8) and START domains (fig. S9) were necessary for the resistance response. Laser point scanning confocal microscopy showed that the T6-312 mutant had an unrestricted network of fungal growth, whereas the control line with a functional WKS1 gene had a resistance response inside the leaf with reduced fungal growth delimited by autofluorescing plant cells (Fig. 2G to J).
Fig. 2. Functional validation of Yr36 by mutational analysis.
(A to F) Leaf surfaces 11 days after PST inoculation. Bar = 5 mm. Numbers below leaves are average percent leaf area with pustules ± SEM (N=8, fig. S2). An ANOVA of the log-transformed data showed significant differences (P<0.01) between mutant and control lines. (A) UC1041 without Yr36. (B) UC1041+Yr36 isogenic line used for mutagenesis. (C and E) Lines T6-138 and T6-312 with homozygous mutations in the WKS1 kinase domain. (D and F) Sister lines without the mutations. These and additional mutant lines are described in table S7, and figs. S6 to S9. (G and H) A dual-channel, confocal microscopic z-series inside a wheat leaf 13 days after PST inoculation. Bar = 20 µm. The uvitex-stained fungus (false-color blue) and autofluorescing wheat leaf cells (false color red) are visible. (G) The susceptible T6-312 mutant has an extensive mycelial network in which each (invisible) plant mesophyll cell (selected cells shown as M) is encircled by a hypha. (H) The T6-312 control line has a poorly developed fungal network surrounded by autofluorescent mesophyll cells that presumably were involved in the resistance response. (I and J) Separate channels of panel (H).
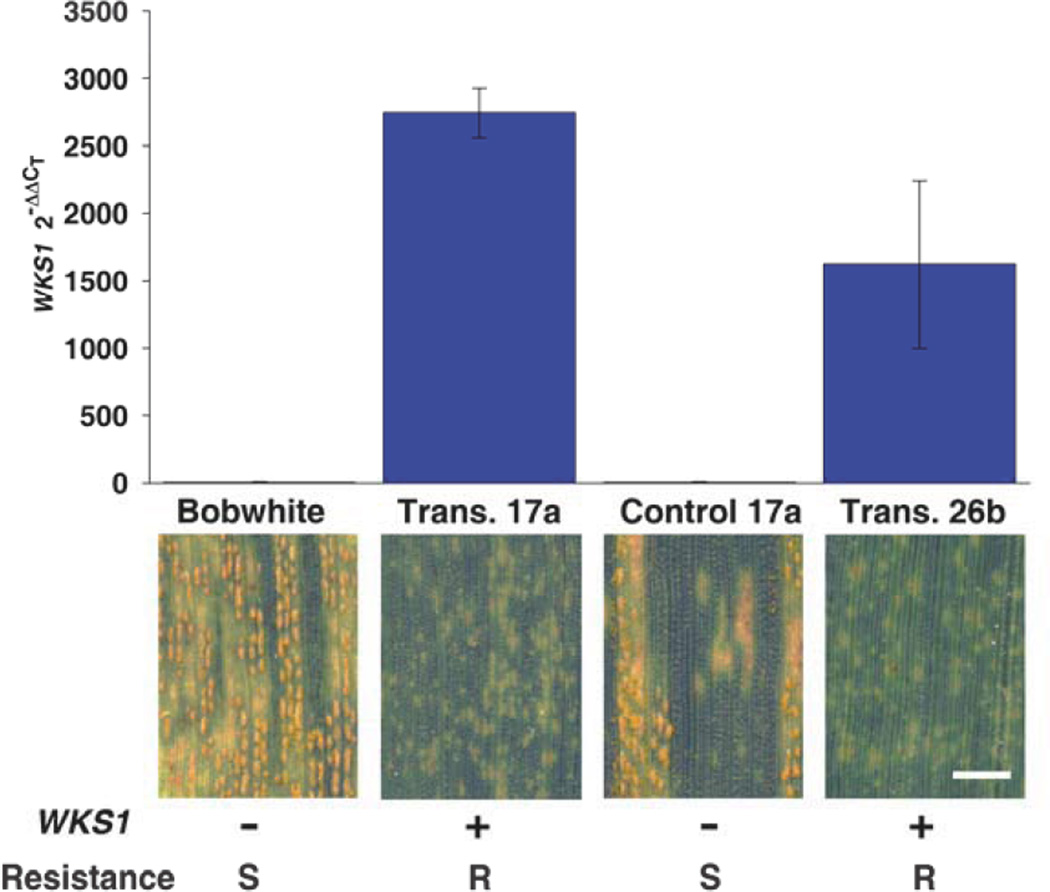
To confirm the identity between WKS1 and Yr36, we transformed the susceptible wheat variety ‘Bobwhite’ with a 12.2-kb genomic fragment that includes the complete WKS1 coding and flanking regions (14). Only two of the nine independent T1 transgenic lines had complete WKS1 transcripts and they were both resistant to stripe rust (Fig. 3 and fig. S10) demonstrating that WKS1 is Yr36.
Fig. 3. WKS1 transcript levels and resistance phenotype in transgenic wheat plants.
Average WKS1 transcript levels (± SEM) in independent transgenic events 17a (5 plants) and 26b (7 plants) were determined by quantitative RT-PCR. Negative controls are the un-transformed variety Bobwhite and the average of three T1 sister lines of 17a without the transgene. Leaf phenotypes: S = susceptible, R = resistant. Bar = 2 mm. Southern blots and transcription profiles of individual T1 plants are shown in fig. S10.
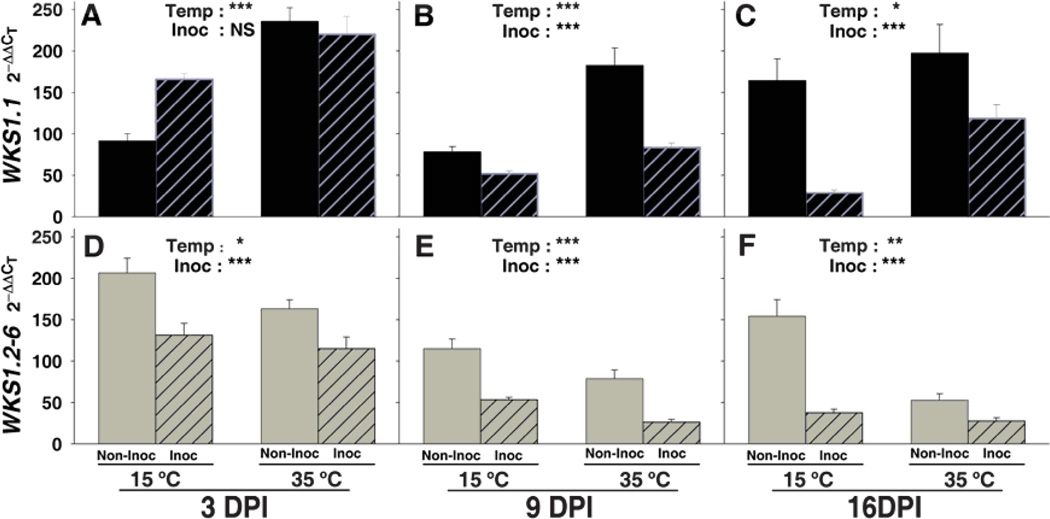
The cloning and sequencing of 56 full-length WKS1 cDNAs revealed six alternative transcript variants (WKS1.1-6, fig. S11). WKS1.1 encodes a complete WKS1 protein, whereas the other five (WKS1.2-6) lack exon 11 and encode proteins with truncated START domains. Some of the missing amino acids are well conserved across the plant kingdom (fig. S7b). Quantitative PCR showed that even the lowest transcript levels of WKS1.1 and WKS1.2-6 are only 3-fold lower than those of ACTIN, indicating relatively high transcript levels. Overall, high temperature up-regulates WKS1.1 (Fig. 4A to C and fig. S12) and down-regulates WKS1.2-6 (Fig. 4D to F and fig. S12) (P<0.0001, table S8).
Fig. 4. Effect of temperature and PST inoculation on transcript levels of WKS1 transcript variants WKS1.1 and WKS1.2-6 in RSL65.
Quantitative RT-PCR transcripts of WKS1.1 are indicated in black and those of WKS1.2-6 in gray. PST-inoculated plants are indicated by stripes and non-inoculated controls by solid colors. No significant interactions between temperature and inoculation were detected in the individual 2-way ANOVAs except for panel A (significant differences between inoculation classes only at low temperature). *: P<0.05, **: P<0.01, ***: P<0.001, and NS: not significant. Each data point is an average based on six replicates (± SEM). Overall ANOVAs are presented in table S8 and comparisons between WKS1.1 and WKS1.2-6 transcript levels are shown in fig. S12.
PST inoculation consistently down-regulated WKS1.2-6 across temperature and time, but the effect on WKS1.1 transcript levels varied with sampling times (Fig. 4A to C). Comparisons between WKS1.1 and WKS1.2-6 transcript levels in PST-inoculated plants (fig. S12, A to C) showed no significant differences at low temperature (susceptible response, P>0.55), and significantly higher values of WKS1.1 relative to WKS1.2-6 at high temperature (resistant response, P<0.01) for all three days.
The relative increase in transcript levels of the variant with the complete START domain (WKS1.1) at high temperature parallels the observed high-temperature resistance conferred by Yr36. START domain proteins in humans are known to play important roles in lipid trafficking, metabolism and sensing, and their binding with sterols and ceramides result in protein conformational changes (reviewed in 16). If the putative WKS1 START domain has the ability to bind lipids from, or redirected by PST at high-temperature and change its conformation, this may cause the kinase domain to initiate a signaling cascade leading to the observed programmed cell death (Fig. 2 and fig. S8). The WKS1 Ser/Thr kinase domain (pfam00069) was confirmed to have kinase activity (fig. S13).
The combination of the kinase and START domains in WKS1 apparently is the result of a novel domain shuffling because these two domains are not found together in other organisms (supporting online text). The most similar protein in Arabidopsis to the putative WKS1 START domain is EDR2, a protein that negatively regulates plant defense to the powdery mildew pathogen Golovinomyces cichoracearum (12, 13) (supporting online text). EDR2 has a PH (pfam00169) and a DUF1336 (pfam07059) domain, which are absent in WKS1. The WKS1 kinase has high similarity to several Arabidopsis WAK-like kinases (fig. S6), but WKS1 lacks the additional domains characteristic of WAK-like kinases (17). The WKS1 kinase belongs to the non-RD kinases, which are frequently involved in the early steps of the innate immune response (11).
The appearance of this novel gene architecture preceded the origin of the Triticeae since WKS1 and WKS2 were detected in several species from this tribe (table S9, fig. S14). However, the presence of these two genes was rare among Triticeae species and varied across accessions within those species where they were detected. This suggests that WKS1 and WKS2 were lost repeatedly in several grass lineages, including the diploid donors of the A and D genomes of polyploid wheat (table S9). Amongst 131 wild and cultivated tetraploid wheat accessions, WKS1 was detected only in wild wheat (24% of accessions) suggesting that WKS1 was not incorporated into the initial domesticated forms. In hexaploid wheat WKS1 was present only in five accessions where the DIC segment was incorporated recently (table S10).
Introgression of WKS1 in transgenic Bobwhite wheat and in susceptible varieties by backcrossing (4) improved their resistance to stripe rust. This indicates that either WKS1 is sufficient to improve resistance, or that WKS1 can trigger intermediate genes that initiate the hypersensitive response, and that these genes are still present in the tested varieties. This, together with the absence of WKS1 in almost all modern commercial varieties of pasta and bread wheat (table S10), suggests that the introgression of Yr36 could have a broad impact in improving resistance to this pathogen. Yr36 resistance has remained effective against the numerous stripe rust races present in California (2004 – 2008 field tests) and to all races tested so far in controlled environments (table S5). Moreover, Yr36 has improved resistance in a variety carrying the non race-specific Yr18 resistance gene (4), which suggests that pyramiding appropriate combinations of non race-specific resistance genes may provide adequate resistance against this pathogen. The discovery of different proteins and resistance mechanisms for non-race specific resistance genes Yr36 and Yr18/Lr34 (18) suggests that this type of resistance may involve a heterogeneous group of genes and mechanisms.
Supplementary Material
Acknowledgments
This project was funded by the USDA-CSREES grants 2005-00975 and 2006-55606-16629, and the US and Israel BARD Fund grant US-4024-07. We thank: C. Li, F. Paraiso, L. Penman, K.L. Richardson, S. Bassein, M.R. Paddy, Q. Lam and M. Jindal for their technical assistance, and R. Thilmony and M. Whalen for critical reading of the manuscript. Gene sequences have been deposited in GenBank with accession numbers: EU835198-EU835200, FJ154103-FJ154118 and FJ155069-FJ155070.
Footnotes
References and Notes
- 1.Singh RP, William HM, Huerta-Espino J, Rosewarne G. Proc. 4th Int. Crop Science Congress; 26 Sep – 1 Oct (2004); Brisbane, Australia. [Google Scholar]
- 2.Chen XM. Aust. J. Agric. Res. 2007;58:648. [Google Scholar]
- 3.Wan AM, Chen XM, He ZH. Aust. J. Agric. Res. 2007;58:605. [Google Scholar]
- 4.Uauy C, et al. Theor. Appl. Genet. 2005;112:97. doi: 10.1007/s00122-005-0109-x. [DOI] [PubMed] [Google Scholar]
- 5.Qayoum A, Line RF. Phytopathology. 1985;75:1121. [Google Scholar]
- 6.Shang HS. Scientia Agricultura Sinica. 1998;31:46. [Google Scholar]
- 7.Line RF. Annu. Rev. Phytopathol. 2002;40:75. doi: 10.1146/annurev.phyto.40.020102.111645. [DOI] [PubMed] [Google Scholar]
- 8.Ma Q, Shang HS. J. Plant Pathol. 2004;86:19. [Google Scholar]
- 9.Distelfeld A, et al. Funct. Integr. Genomics. 2004;4:59. doi: 10.1007/s10142-003-0097-3. [DOI] [PubMed] [Google Scholar]
- 10.Cenci A, et al. Theor. Appl. Genet. 2003;107:931. doi: 10.1007/s00122-003-1331-z. [DOI] [PubMed] [Google Scholar]
- 11.Dardick C, Ronald P. PLOS Pathog. 2006;2:14. doi: 10.1371/journal.ppat.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang DZ, Ade J, Frye CA, Innes RW. Plant J. 2005;44:245. doi: 10.1111/j.1365-313X.2005.02523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vorwerk S, et al. BMC Plant Biology. 2007;7:1. doi: 10.1186/1471-2229-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Material and methods are presented in detail in the Supplemental Online Materials (SOM).
- 15.McCallum CM, Comai L, Greene EA, Henikoff S. Nat. Biotechnol. 2000;18:455. doi: 10.1038/74542. [DOI] [PubMed] [Google Scholar]
- 16.Alpy F, Tomasetto C. J. Cell Sci. 2005;118:2791. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- 17.Verica JA, He ZH. Plant Physiol. 2002;129:455. doi: 10.1104/pp.011028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krattinger SG, et al. Science. In press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.