Abstract
The PC (proprotein convertase) furin cleaves a large variety of proproteins and hence plays a major role in many pathologies. Therefore furin inhibition might be a good strategy for therapeutic intervention, and several furin inhibitors have been generated, although none are entirely furin-specific. To reduce potential side effects caused by cross-reactivity with other proteases, dromedary heavy-chain-derived nanobodies against catalytically active furin were developed as specific furin inhibitors. The nanobodies bound only to furin but not to other PCs. Upon overexpression in cell lines, they inhibited the cleavage of two different furin substrates, TGFβ (transforming growth factor β) and GPC3 (glypican 3). Purified nanobodies could inhibit the cleavage of diphtheria toxin into its enzymatically active A fragment, but did not inhibit cleavage of a small synthetic peptide-based substrate, suggesting a mode-of-action based on steric hindrance. The dissociation constant of purified nanobody 14 is in the nanomolar range. The nanobodies were non-competitive inhibitors with an inhibitory constant in the micromolar range as demonstrated by Dixon plot. Furthermore, anti-furin nanobodies could protect HEK (human embryonic kidney)-293T cells from diphtheria-toxin-induced cytotoxicity as efficiently as the PC inhibitor nona-d-arginine. In conclusion, these antibody-based single-domain nanobodies represent the first generation of highly specific non-competitive furin inhibitors.
Keywords: furin, diphtheria toxin, nanobody, proprotein convertase
INTRODUCTION
Furin belongs to a family of seven closely related subtilisin-like serine endoproteases, known as PCs (proprotein convertases), PC1/3, PC2, PC4, PC5/6, PACE4 and PC7 [1]. The physiological role of PCs is to cleave and hence activate a large variety of proproteins, such as growth factors, receptors, enzymes and cell-adhesion molecules. Furin and its family members have similar substrate specificities and cleave C-terminal of basic amino acid motifs. For this reason, it is impossible to demonstrate physiological enzyme–substrate pairs and/or redundancy on the basis of overexpression studies only. Therefore knockout mouse models have been generated to investigate the roles of various PCs [2]. Embryos lacking furin die between days 10.5 and 11.5, most probably due to impaired processing of factors from the TGFβ (transforming growth factor β) superfamily [3]. In contrast with the indispensable role of furin during embryogenesis, its function in adult life appears to be partially redundant, as shown by conditional furin inactivation in mice in specific organs [4–8]. Processing of several substrates, for example the precursors of albumin, αV integrin, lipoprotein receptor-related protein, vitronectin and α1-microglobulin/bikunin, was found to be reduced but not blocked in these conditional knockouts. On the other hand, processing of TGFβ and PCSK9 (PC subtilisin/kexin type 9) seems to be entirely dependent on furin.
Furin has also been implicated in several pathological processes. First, furin expression is increased in many cancers, such as breast, ovarian, and head and neck cancers, and furin expression and tumour aggressiveness are highly correlated [9–11]. In addition, several bacterial toxins require furin-mediated activation, such as Pseudomonas exotoxin A [12], diphtheria toxin [13], Shiga toxin [14], anthrax toxin [15] and the lytic toxin aerolysin [16]. Furthermore, a broad range of pathogenic viruses require furin cleavage of their envelope glycoproteins to be able to fuse with the host cell membranes, such as HIV-1 [17], influenza A virus [18], RSV (respiratory syncitial virus) [19], paramyxovirus [20], CMV (cytomegalovirus) [21] and Ebola [22]. In conclusion, the broad range of substrates gives furin a central role in not only many physiological processes but also in several pathologies.
The absence of a severe phenotype in the tissue-specific furin-knockout models raises the possibility of using furin as a therapeutic target. Several in vivo studies have provided proof-of-concept that furin inhibition might give therapeutic benefit. Mice injected with tumour cell lines with reduced furin activity showed reduced tumour invasion, metastasis, proliferation and angiogenesis [23]. Furthermore, the development and progression of PLAG1 (pleiomorphic adenoma gene 1)-induced pleomorphic adenomas of the salivary glands was either absent or significantly delayed by the genetic ablation of furin [5]. Finally, furin inhibitors show a protective effect in vivo against Pseudomonas exotoxin and anthrax infection [24–26]. Taken together, this suggests that furin might be a possible therapeutic target in a diverse range of pathologies.
Several effective furin inhibitors have been developed to date, although none are entirely furin-specific. There are peptide-based furin inhibitors such as polyarginines, peptidyl-chloroalkanes and peptidyl-aminobenzylamides, as well as engineered serpins which are mutants of α1-proteinase inhibitor, α2-macroglobulin and α1-antitrypsin [17,21,27–30]. All of these inhibitors are pseudosubstrates containing an Arg-X-X-Arg motif, or variants thereof. Given the highly conserved substrate-binding region of the catalytic domains of PCs [31], it is not surprising that these competitive inhibitors have limited specificity. Small-molecule inhibitors such as 2,5-dideoxystreptamine-derived molecules and dicoumarol derivatives are also potent competitive inhibitors of PCs, but with limited specificity as well [32,33].
To obtain highly specific inhibitors, antibodies, and especially the dromedary-derived single-domain antigen-binding fragments, also known as nanobodies, have been demonstrated to have great potential as enzyme inhibitors [34,35]. Nanobodies comprise the recombinant variable fragment of the heavy chain of camelid heavy-chain antibodies that lack light chains. They are perfectly soluble and stable polypeptides harbouring the full antigen-binding capacity of the original heavy-chain antibody. Owing to the extended CDRs (complementarity-determining regions), and the convex shape of the antigen-binding site (the paratope) these recombinant antibodies are frequently found to have enzyme-inhibiting activity [35–37].
In the present study, a dromedary was immunized with active furin to raise a specific immune response in the heavy-chain antibody class and with the objective to obtain specific furin-inhibiting nanobodies. Furin-binding nanobodies were isolated from a nanobody library generated from DNA isolated from dromedary lymphocytes. The identified nanobodies were tested for the ability to inhibit furin in vitro and in cell lines using a variety of substrates. Furthermore, the protection against the toxic effect of diphtheria toxin was tested in cellulo. Finally, specificity and inhibitor kinetics were determined.
EXPERIMENTAL
Generation of anti-furin nanobodies
Nanobodies were generated as described previously [34]. Briefly, a female dromedary (Camelus dromedarius) kept at the Central Veterinary Research Laboratory (Dubai, United Arab Emirates) was injected subcutaneously on days 0, 7, 14, 21, 28 and 35 with soluble human furin. The injected furin was more than 95% pure as determined by SDS/PAGE and had an activity of 88 units/µl [where 1 unit represents 1 pmol of AMC (7-amino-4-methylcoumarin) generated/min]. For each injection, 1 ml of antigen sample (90 µg) was emulsified using 1 ml of Gerbu adjuvant LQ #3000 (GERBU Biotechnik) and the emulsion was injected subcutaneously. On day 39, approximately 50 ml of blood was collected on EDTA anticoagulating medium and used for the preparation of plasma and peripheral blood lymphocytes using Leucosep tubes (Greiner Bio-One) as instructed by suppliers. The animal experimentation followed the guidelines published by the regional United Arab Emirates government.
To analyse the immune response, IgG subclasses were obtained by successive affinity chromatography on HiTrap Protein A and HiTrap Protein G columns. A VHH library was constructed in the phagemid vector pHEN4 [16] and screened for the presence of furin-specific nanobodies [34]. Four consecutive rounds of panning were performed in microtitre plates coated with furin (10 µg/well).
Expression and purification of the nanobody fragments
To purify the nanobodies, the nanobody sequences were cloned into the pHEN6 vector [34], which is equivalent to the pHEN4 vector, except that the HA (haemagglutinin) tag and geneIII have been replaced by a His6 detection and purification tag. The purification of recombinant nanobodies was performed as described previously [34].
For overexpression studies in mammalian cell lines, the nanobody sequences were cloned into the pcDNA3 vector (Invitrogen). A sequence encoding an HA epitope tag was added before the stop codon and the signal peptide of PC5/6 was introduced in front of the nanobody sequence. In brief, the signal peptide of PC5/6was amplified from the pGEMPC6A vector [38], using the following primers 5′-ACCTGTAAGCTTACCATGGACTGGGGGAAC-3′and 5′-ACCTGTGGATCCCCAGTGGTTGGTGTAGACG-3′, introducing a HindIII and BamHI site respectively. The nanobody sequences were amplified with the following primers 5′-CCTGTGGATCCCAGGTGCAGCTGCAGGAG-3′and 5′-GCCAGTGAATTCTATTAGTGATGG-3′, introducing a BamHI and EcoRI site respectively. Both PCR products were digested with the appropriate restriction enzymes and ligated into the pcDNA3 host vector.
Co-immunoprecipitation of furin and other PCs with nanobodies
Furin-deficient RPE.40 cells were transfected in six-well plates with mouse and human furin, and FLAG-tagged PC1, PC2, PC4, PACE4, PC6A and PC7 all in the pcDNA3 vector [39] with or without the different nanobodies, using FuGENE® 6 (Roche) according to the manufacturer’s protocol. The cells were lysed in TNT buffer [50 mM Tris/HCl, 150 mM NaCl, 0.5% Triton X-100 (pH 7.0), containing Complete™ protease inhibitor cocktail (Roche). The lysate was precleared with Protein G–Sepharose beads and incubated overnight at 4°C with Protein G–Sepharose beads coated with 1 µg of mouse monoclonal anti-HA antibody (16B12, Covance). The beads were boiled in sample buffer and analysed by Western blotting. The mouse monoclonal antibodies MON-148 and MON-152 were used to detect mouse and human furin (Enzo Life Sciences). Rabbit polyclonal antibodies against the other PCs (Enzo Life Sciences) were used at a dilution of 1:1000.
Western blot analysis to investigate inhibition of furin-mediated cleavage of different furin substrates
HEK (human embryonic kidney)-293T cells or, in the case of renin, RPE.40 cells were transfected in 12-well plates with TGFβ [40], HA-tagged GPC3 (glypican 3) [41] or the mutant construct RVRTKR of renin-2 (kindly provided by Dr Nakayama, Institute of Biological Sciences, University of Tsukuba, Tsukuba, Japan) [42], together with empty vector, vectors encoding α1-PDX (α-1 antitrypsin Portland variant) [43] or the different nanobodies, using FuGENE® 6. For analysis of GPC3, the cells were lysed after 24 h. In the case of TGFβ, the cells were incubated in serum-free medium and the medium was collected 24 h later. Immunoprecipitation was performed to precipitate secreted pro- and mature TGFβ. Subsequently, Western blotting was performed to visualize pro- and mature TGFβ in the medium, using the anti-LAP (latency-associated protein) polyclonal antibody (0.5 µg for immunoprecipitation, 1:2000 for Western blot analysis, R&D Systems). The bands were quantified using ImageJ software (NIH) and the ratio of mature/precursor protein was calculated.
In case of renin, the cells were incubated in serum-free medium and the medium was collected 24 h later. After methanol precipitation, Western blot analysis was performed using the mouse monoclonal anti-renin antibody (1:200 dilution, Santa Cruz Biotechnology).
Cleavage of the fluorigenic furin substrate by purified furin in vitro
Purified soluble furin (20 ng) was pre-incubated at room temperature (20°C) with either different concentrations of D9R (nona-d-arginine; Pepscan) or purified nanobody in Hepes buffer [100 mM Hepes, 0.5% Triton X-100 and 0.5 mM CaCl2 (pH 7.0)] for 30 min in a volume of 80 µl. After pre-incubation, the pyr-Arg-Thr-Lys-Arg-AMC substrate (Bachem) was added to a final concentration of 100 µMand incubated for 1 h at 37°C. The fluorescence (390 nm excitation, 460 nm emission) was measured using a Fluostar Galaxy fluorimeter.
Cleavage of diphtheria toxin by purified furin in vitro
Purified soluble furin (0.3 ng) was pre-incubated with either different concentrations of D9R or purified nanobody in Hepes buffer at room temperature for 30 min. After the pre-incubation, 3 µg of uncleaved diphtheria toxin (Merck) was added and incubated at 37°C for 1 h. The samples were analysed by SDS/PAGE and visualized by Coomassie Blue staining.
Measurement of cytotoxicity of diphtheria and anthrax toxin
The protective effect of the nanobodies on diphtheria toxin cytotoxicity was determined either by overexpression of the nanobodies or by pre-incubation with purified nanobodies. For the overexpression studies, HEK-293T cells were transfected in 24-well plates with empty vector, the different nanobodies or α1-PDX using FuGENE® 6. After 24 h, 3 µg of diphtheria toxin were added and incubation continued for 3 h. Cell viability was then measured using MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide; Sigma] according to the manufacturer’s protocol. This is a colorimetric assay that measures the reduction of yellow MTT by mitochondrial succinate dehydrogenase. HEK-293T cells were pre-incubated in 24-well plates with 10 µM purified nanobodies or 10 µM D9R. After a 2 h pre-incubation at 37°C, 3 µg of diphtheria toxin was added and incubation continued for 1.5 h. The cell viability was then measured using the MTT assay described above.
To determine the protective effect of the purified nanobodies on anthrax toxin cytotoxicity, RAW cells were pre-incubated with 20 µM purified nanobodies or 10 µM D9R in 48-well plates (Costar) for 30 min at 37 °C. Subsequently, 200 ng/ml PA (protective antigen; List Biological Labs) and 400 ng/ml LF (lethal factor; List Biological Labs) were added and the cells were incubated for 1.5 h, with one medium change after 45 min. The cell viability was determined using the MTT assay described above.
Nanobody uptake experiment
HEK-293T cells were transfected with FLAG-tagged furin in a pcDNA3 vector [44]. The next day the cells were placed in serum-free medium with 0.3 M sucrose for 30 min at 37°C and washed with ice-cold PBS. Subsequently the cells were incubated either with conditioned serum-free medium from HEK-293T cells transfected with HA-tagged Nb6 or with serum-free medium containing 1 µg/ml anti-FLAGM2 antibody (Sigma) for 30 min at 4°C, followed by 15 min at 37°C. After incubation with antibody, the cells were washed in ice-cold PBS, fixed in 4% formaldehyde, permeabilized with PBS containing 0.2% Triton X-100 for 15 min and incubated with 1 µg/ml mouse anti-HA or mouse anti-FLAG M2 antibodies dissolved in PBS+10% FBS (fetal bovine serum) for 1 h at room temperature respectively. Finally, the cells were incubated with Alexa Fluor® 555-conjugated goat anti-mouse antibody for 30 min at room temperature. Slides were analysed with a Zeiss Axioimager microscope.
Determination of Kd, kon and koff
The equilibrium dissociation constant (Kd) and the association (kon) and dissociation rates (koff) were determined using surface plasmon resonance detection on a BIACore T200 (GE Healthcare). Furin was immobilized on to a CM5 S series sensor chip (GE Healthcare) using standard amine coupling. After activation of the carboxy moieties on the matrix on the chip surface with a 7-min injection of a 1:1 mixture of 0.4 M EDC [N-ethyl-N′-(3-dimethylaminopropyl)carbodi-imide] and 0.1 M NHS (N-hydroxysuccinimide), the furin (10 µg/ml in 10 mM acetate, pH 4.5) was immobilized to a predefined level of RU (response units). Approximately 320 and 560 RU of furin was bound on to Fc (flow channels) 4 and 2 respectively. The remaining activated carboxy groups were blocked by injecting 1 M ethanolamine (pH 8.5) for 7 min. Flow rate was kept constant at 10 µl/min. Fc 1 and 3 served as reference channels (activated with EDC/NHS and immediately afterwards blocked by ethanolamine). Throughout the analysis 10 mM Hepes, 150 mM NaCl, 1 mM CaCl2 and 0.01% Triton X-100 (pH 7.35) was used as running buffer. Because regeneration of the immobilized furin (removing all bound nanobody without losing native furin) was impossible, single cycle kinetics were used to determine the Kd, kon and koff values. Then 50, 150, 300, 450 and 600 nM of Nb14 diluted in running buffer was injected (75 µl/min during 120 s) consecutively over Fc 2 using Fc 1 as a reference. This analysis was performed twice. Using the same conditions, 5, 20, 60, 180 and 500 nM were assayed over Fc 4 using Fc 3 as a reference. Kinetic parameters were determined using Biacore T200 evaluation software (GE Healthcare).
Ki determination
Furin (20 ng) was pre-incubated with various concentrations of four different nanobodies or D9R (2, 6, 10, 14, 20 and 28 µM) in buffer solution [0.5% Triton X-100, 0.5 mM CaCl2 and 100 mM Hepes solution (pH 7.0)] for 30 min at room temperature. Subsequently, different concentrations of diphtheria toxin (4.2, 8.4, 12.8 and 17 mM) were added as substrate for 1 h at 37°C. The samples were analysed by SDS/PAGE and visualized by Krypton protein staining, following the manufacturer’s protocol (Thermo Scientific). The bands were quantified using ImageJ software (NIH) and a Dixon plot was used to determine the Ki.
RESULTS
Generation of nanobodies against catalytically active human furin
The aim of the present study was to generate and characterize new inhibitors of furin which do not inhibit the activity of other PCs. For this objective, nanobodies against catalytically active human furin were generated. A dromedary was injected subcutaneously at regular intervals with active soluble furin. On day 39 the immune response to furin was analysed and blood was collected for the isolation of lymphocytes to generate a VHH library. Total serum and three purified IgG subclasses (IgG1, IgG2 and IgG3) were tested by ELISA to assess the immune response to furin. There was immune response in all IgG subclasses although, overall, the immune response was very low (results not shown). A VHH library was generated using RNA obtained from lymphocytes and screened for the presence of furin-specific nanobodies. Four consecutive rounds of phage panning were performed on solid-phase coated furin. The enrichment for antigen-specific phages was assessed after each round of panning by polyclonal phage ELISA. There was a clear enrichment after the 3rd and 4th round of panning. In total, 190 individual colonies (95 from the 3rd round and 95 from the 4th round) were randomly selected and analysed by ELISA for the presence of antigen-specific VHHs in periplasmic extracts. In total 67 colonies (26 from the 3rd round and 41 from the 4th round) scored positively in this assay. The VHHs from 31 clones were sequenced, revealing many identical nanobodies. Then 13 unique nanobody gene sequences were cloned into the pcDNA3 vector for expression in mammalian cells. Sequences encoding a signal peptide were added for translocations of the nanobody into the secretory pathway and an HA tag for nanobody detection. The binding of the 13 nanobodies to human furin was first assessed by co-immunoprecipitation under non-denaturing conditions. In total, 11 out of the 13 nanobodies bound to furin at variable levels (Figure 1). None of the nanobodies detected furin after SDS/PAGE and blotting on to a nitrocellulose membrane (results not shown), indicating that only native furin is recognized.
Figure 1. Co-immunoprecipitation of human furin with the different nanobodies.
RPE.40 cells were transfected with human furin with or without expression vectors encoding the different nanobodies tagged with HA and immunoprecipitation was performed using an anti-HA antibody. Subsequently, Western blot analysis was performed using the anti-furin antibody MON152. Lanes 1, beads without anti-HA antibody; lanes 2, beads with anti-HA antibody; lanes 3, total cell lysate. Untransfected cells were included as a negative control in the last lane (−)
Four nanobodies inhibit furin-mediated cleavage of two different furin substrates
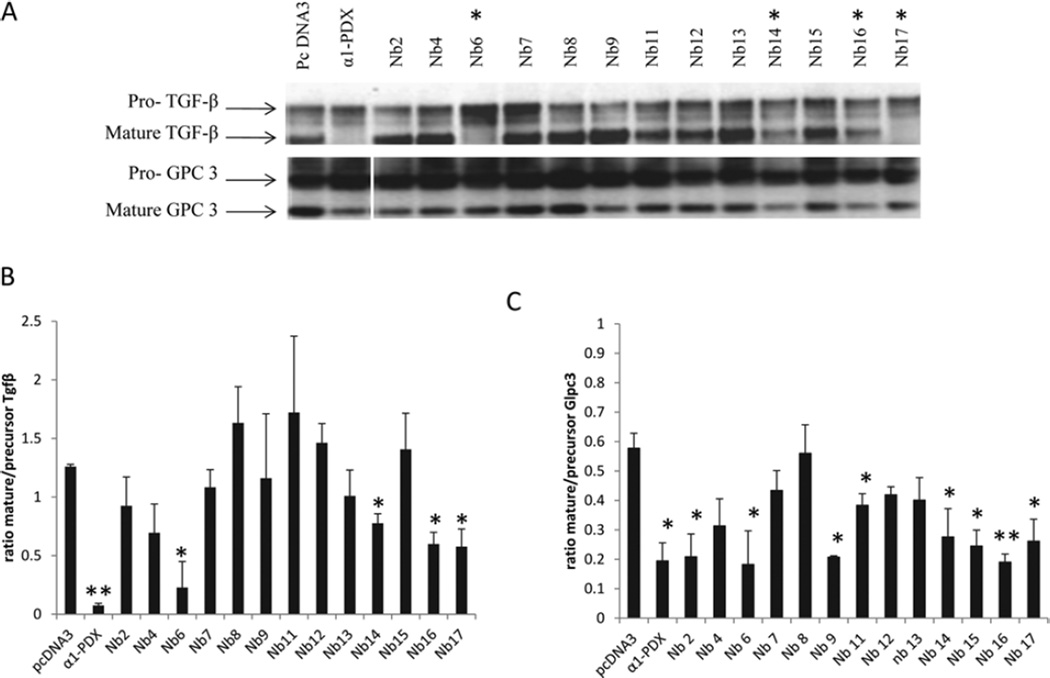
Binding of the nanobodies to native furin does not imply inhibition of the proteolytic activity of furin. To test this, we investigated the cleavage of two different furin substrates, GPC3 and TGFβ in a co-expression system. The different nanobodies and the well-characterized PC inhibitor α1-PDX were co-transfected with each of the furin substrates into HEK-293T cells. Western blot analysis revealed decreased processing of pro-TGFβ in the medium of the cells transfected with Nb6, Nb14, Nb16 and Nb17, as determined by the ratio of mature/precursor protein (Figure 2). Similarly, cleavage of GPC3was decreased by expression of Nb2, Nb6, Nb9, Nb11, Nb14, Nb15, Nb16 and Nb17 (Figure 2). We conclude that although most nanobodies bound to furin, only a subset were able to inhibit furin activity. Interestingly, the inhibitory capacity of the nanobodies was different for different furin substrates. Only four nanobodies, Nb6, Nb14, Nb16 and Nb17, affected the cleavage of the two different furin substrates and were chosen for further investigation in the present study.
Figure 2. Four different nanobodies inhibit the cleavage of the furin substrates TGFβ and GPC3.
HEK-293T cells were transfected with an expression vector encoding TGFβ or GPC3 together with empty vector, α1-PDX or an expression vector encoding the different nanobodies. (A) The inhibition of the furin-mediated cleavage of those substrates was analysed by Western blotting. Nb6, Nb14, Nb16 and Nb17 inhibited the cleavage of the two furin substrates (indicated with *). (B) The ratio of mature/precursor TGFβ was calculated from three different experiments using ImageJ software. Results are represented as means ± S.E.M. (n =3) *P ≤0.05, **P ≤0.005. (C) The ratio of mature/precursor GPC3 was calculated from three different experiments using ImageJ software. Results are represented as means ± S.E.M. (n =3) *P ≤0.05, **P ≤0.005.
The furin-inhibiting nanobodies do not bind to other PCs
To address whether Nb6, Nb14, Nb16 and Nb17 were specific for human furin, cross-reactivity with the six other closely related PC family members and with mouse furin was investigated. Nanobodies were overexpressed together with each PC in furin-deficient RPE.40 cells. After co-immunoprecipitation using an antibody against the HA tag within the nanobodies, Western blot analysis for the different PCs was performed. Only human and mouse furin co-immunoprecipitated with the nanobodies (Figure 3A). This indicates that the nanobodies are specific for furin and do not bind to related family members.
Figure 3. Nanobodies bind to mouse and human furin, but not to the other PC family members.
(A) RPE.40 cells were transfected with furin cDNA or cDNAs encoding closely related PCs with or without cDNAs encoding the different nanobodies and immunoprecipitation was performed using an anti-HA antibody. Only mouse and human furin, but not family members, co-immunoprecipitated with Nb6, Nb14, Nb16 and Nb17, indicating their specificity for furin. A representative image for Nb14 is shown. Lane 1, beads without anti-HA antibody; lane 2, beads with anti-HA antibody; lane 3, total cell lysate. (B) RPE.40 cells were transfected with a construct encoding the mutant RVRTKR of renin-2 and furin cDNA or cDNAs encoding closely related PCs together with the empty pcDNA3 vector (lane 1), α1-PDX (lane 2) or cDNA encoding Nb6 (lane 3). Subsequently, Western blot analysis was performed using the anti-renin antibody. h, human; m, mouse.
The furin-inhibiting nanobodies inhibit the furin-mediated processing of renin, but not the cleavage by other PCs
The nanobodies only bind to furin but not to other PCs. This suggests that they are specific furin inhibitors. To prove that the nanobodies can only inhibit furin activity and not that of other PCs, RPE.40 cells were transfected with the PC substrate renin with or without the nanobodies or α1-PDX using FuGENE as a transfection reagent. Pro-renin was efficiently processed by most PCs except PC4 (Figure 3B). This PC-mediated processing could be inhibited by α1-PDX, except for PC7 and PACE4 (Figure 3B), consistent with the reported limited specificity of α1-PDX in co-transfection experiments [45]. In contrast, the nanobodies could inhibit only the furin-mediated processing of renin and not that of other PCs. These experiments substantiate that the furin-inhibiting nanobodies are specific furin inhibitors (Figure 3B).
Purified nanobodies cannot inhibit the cleavage of the small pyr-Arg-Thr-Lys-Arg-AMC substrate, but do inhibit the cleavage of diphtheria toxin and anthrax toxin
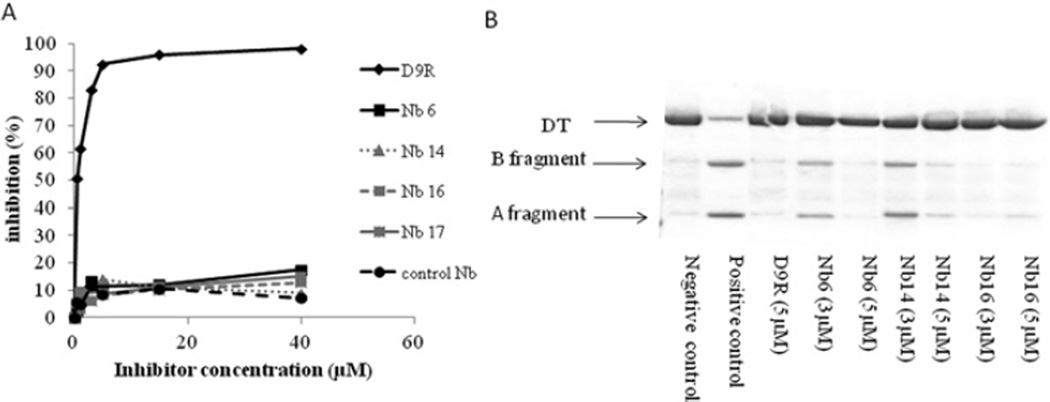
Subsequently, we analysed the furin-inhibiting capacity of purified nanobodies in vitro. Nanobodies were produced using a prokaryotic expression system and purified via their C-terminal His6 tag. Subsequently, the capacity of the nanobodies to inhibit furin-mediated cleavage of the small pyr-Arg-Thr-Lys-Arg-AMC fluorigenic substrate was evaluated. No inhibition of the furin-mediated cleavage of this substrate could be observed for any of the nanobodies, whereas the well-characterized PC inhibitor D9R did inhibit the furin-mediated cleavage of this substrate (Figure 4A). A small reduction in activity was observed upon addition of the nanobodies, but this was interpreted as a nonspecific effect as a similar reduction was observed upon addition of a control nanobody that was not directed against furin. These results showing that nanobodies cannot inhibit the processing of small peptides by furin suggest that the nanobodies do not bind to the catalytic site of furin, but inhibit furin activity by steric hindrance. In other words, small substrates can still enter the catalytic cleft of furin, but larger proteins cannot.
Figure 4. Purified nanobodies cannot inhibit the cleavage of the small pyr-Arg-Thr-Lys-Arg-AMC substrate, but do inhibit the cleavage of diphtheria toxin.
(A) Purified Nb6, Nb14, Nb16 and Nb17 and a control nanobody cannot inhibit furin-mediated cleavage of pyr-Arg-Thr-Lys-Arg-AMC, as shown by a fluorimetric assay. In contrast, the well-characterized furin inhibitor D9R effectively inhibits the cleavage of this substrate. (B) D9R and purified Nb6, Nb14 and Nb16 inhibit the furin-mediated cleavage of diphtheria toxin (DT), as shown by Coomassie Brilliant Blue staining.
To confirm this hypothesis, we investigated whether purified nanobodies could inhibit furin-cleavage of diphtheria toxin, which is a much larger furin substrate (63 kDa). In vitro incubation of the diphtheria toxin with furin results in the cleavage of the toxin into its A and B fragments. This furin-mediated cleavage of the diphtheria toxin can be dose-dependently inhibited by the three purified nanobodies tested, as well as by D9R (Figure 4B). Steric hindrance might therefore explain the cleavage inhibition of big substrates and the lack of similar inhibition of small substrates, since the latter can still enter the catalytic site of the furin enzyme.
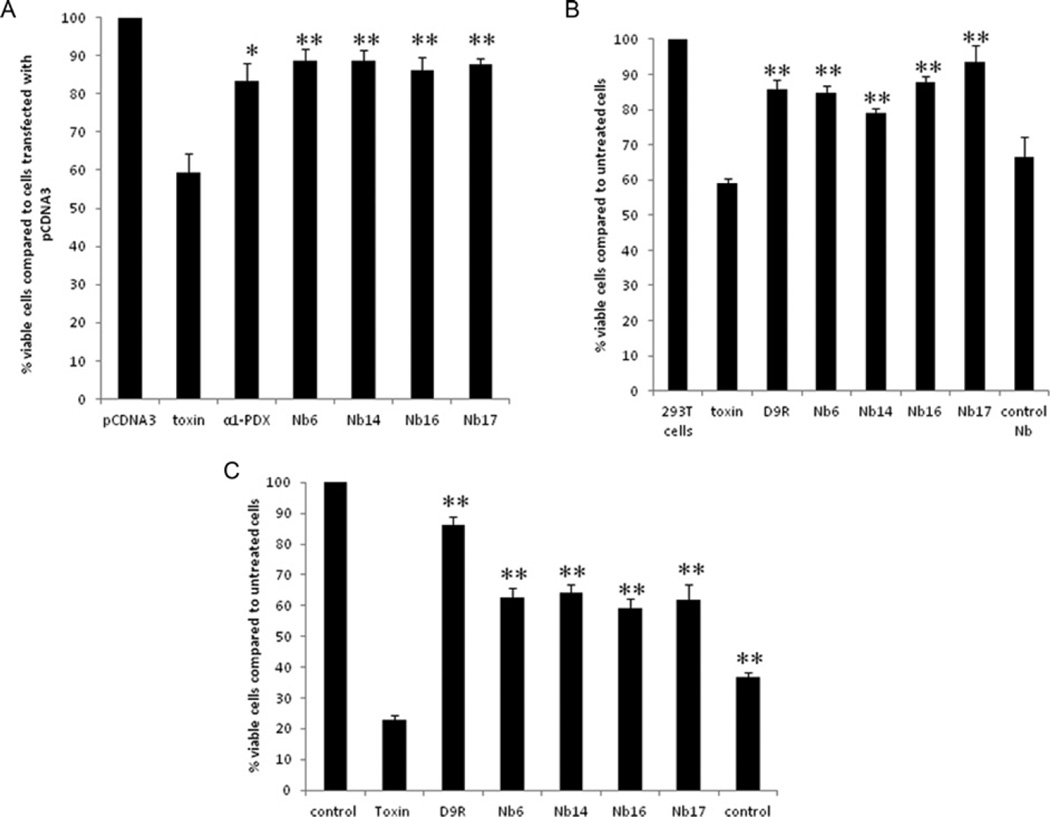
Proteolytic cleavage of diphtheria toxin is required to release the enzymatically active A fragment which is required for its cytotoxic action [13]. Furthermore, furin plays an essential role in the cleavage of diphtheria toxin, since furin-deficient LoVo cells are resistant to intact diphtheria toxin [14]. Since purified nanobodies can inhibit the cleavage of diphtheria toxin in vitro, we next studied whether they can also decrease the cellular toxicity of diphtheria toxin in HEK-293T cells. To achieve this goal, HEK-293T cells were transfected with expression vectors encoding different nanobodies 24 h before they were exposed to diphtheria toxin (Figure 5A). Alternatively, the cells were incubated with the purified nanobodies 2 h before adding the toxin. Cell viability was measured using an MTT assay. As positive controls, α1-PDX (Figure 5A) or D9R (Figure 5B) were used. In both assays, nanobodies protected HEK-293T cells from diphtheria toxin-induced cell toxicity as efficiently as α1-PDX and D9R (Figure 5). Cells transfected with empty plasmid, or cells exposed to nanobodies which do not bind to furin, could not protect HEK-293T cells from diphtheria toxin-induced cytotoxicity (Figure 5B, last bar).
Figure 5. The nanobodies inhibit diphtheria-toxin-mediated cytotoxicity as efficiently as α1-PDX and D9R.
(A) HEK-293T cells were transfected with empty vector, an expression vector encoding α1-PDX or the different nanobodies 24 h before exposure to diphtheria toxin. Then, 3 h after adding diphtheria toxin, cell viability was assessed by the MTT assay. The nanobodies protected the cells from cytotoxicity as efficiently as the well-characterized furin inhibitor α1-PDX. The viability of cells treated with the diphtheria toxin together with α1-PDX or the different nanobodies is significantly higher when compared with cells only treated with the diphtheria toxin. (B) At 2 h prior to the exposure to diphtheria toxin, HEK-293T cells were incubated with 10 µM of purified nanobodies or with D9R. Then 1.5 h after adding the diphtheria toxin, cell viability was assessed by the MTT assay. The nanobodies targeting furin significantly protected the cells from cytotoxicity; protection was as efficient as the well-characterized furin inhibitor D9R, whereas control nanobodies which do not target furin did not inhibit cell toxicity. (C) At 30 min prior to the exposure to anthrax toxin (200 ng/ml PA and 400 ng/ml LF), RAW cells were incubated with 20 µM purified nanobodies or with 10 µM D9R. Then, 1.5 h after adding the toxins cell viablility was assayed using the MTT assay. Nanobodies targeting furin significantly protected the cells from cytotoxicity although less efficient than D9R, whereas control nanobodies which do not target furin only showed a mild effect on the cell toxicity. Results are presented as means ± S.E.M. (n =3) *P ≤0.05, **P ≤0.005.
Similarly, the purified furin-inhibiting nanobodies could also inhibit anthrax toxin-induced cytotoxicity of RAW cells, although less efficiently than D9R (Figure 5C).
Purified nanobodies can enter the cells via endocytosis
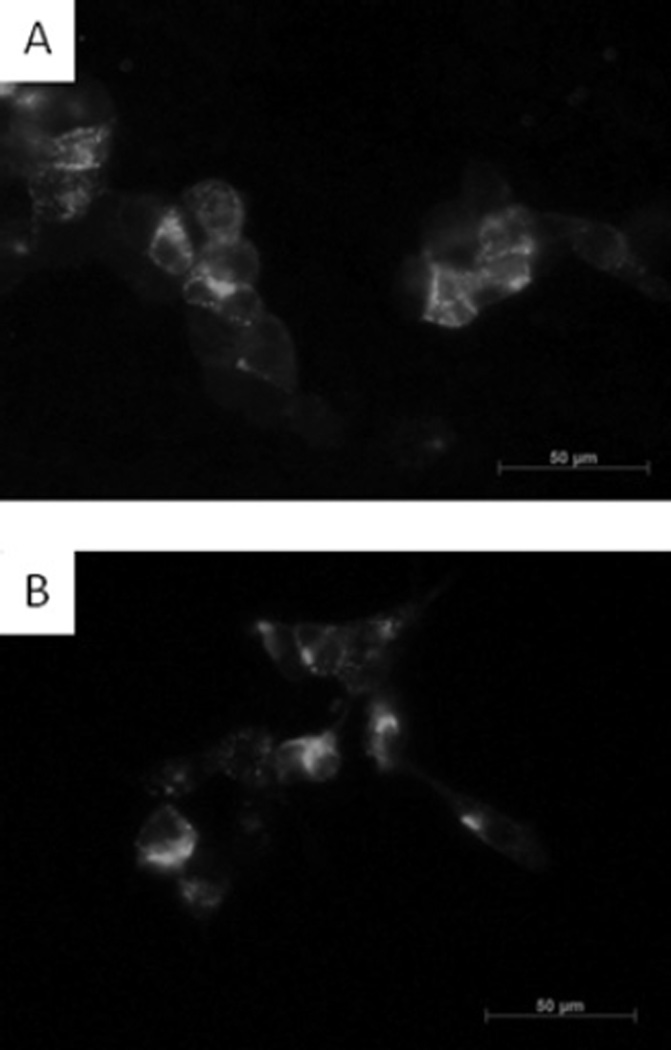
The purified nanobodies effectively reduce the cleavage of extracellular furin substrates such as diphtheria toxin and anthrax toxin. We subsequently analysed whether they are also taken up by the cells by endocytosis and hence are able to inhibit intracellular furin substrates. Previously, it has been shown that furin cycles between the plasma membrane and the TGN (trans-Golgi network). Indeed antibody uptake experiments have shown that anti-FLAG antibodies can bind to surface furin and recycle to the TGN [44]. Figure 6 shows that the nanobodies can also bind to surface furin and be recycled to similar compartments as anti-FLAG antibodies which bind to FLAG-tagged furin overexpressed in HEK-293T cells.
Figure 6. Antibody uptake experiments.
Antibody uptake experiments (30 min at 4°C, 15 min at 37°C) using the anti-HA antibody to detect the HA-tagged nanobodies (A) and anti-FLAG antibody M2 (B) were performed on HEK-293T cells transfected with FLAG-tagged furin.
The nanobodies are non-competitive furin inhibitors
The Ki values of the purified nanobodies were determined and compared with that of D9R. A Dixon plot was generated to calculate the different Ki values. The different nanobodies inhibited the cleavage of intact diphtheria toxin with Ki values of 28, 25, 23 and 22 µM for Nb6, Nb14, Nb16 and Nb17 respectively (Figures 7A–7D). The Dixon plot also revealed that these nanobodies are non-competitive inhibitors, since the lines drawn for each concentration of substrate meet in a single point on the x-axis. Nevertheless, the nanobodies represent less potent furin inhibitors as compared with D9R, which has a Ki value of 0.14 µM (Figure 6E). In contrast, D9R is a competitive inhibitor, in agreement with previous work [46].
Figure 7. The nanobodies are non-competitive inhibitors with Ki values in the micromolar range.
(A–D) Dixon plot for inhibition of furin-mediated cleavage of diphtheria toxin by Nb6 (A), Nb14 (B), Nb16 (C) and Nb17 (D). The reciprocal velocity is plotted against the inhibitor concentration. The trendlines drawn for each substrate concentration intersect in a single point on the x-axis, indicating non-competitive inhibition. (E) Dixon plot for inhibition of furin-mediated cleavage of diphtheria toxin by D9R. The reciprocal velocity is plotted against the inhibitor concentration. The trendlines drawn for each concentration of substrate intersect in a single point above the x-axis, indicating competitive inhibition. (F) Single cycle kinetic analysis for Nb14-furin interaction. Nb14 (50, 150, 300, 450 and 600 nM) was injected consecutively over the immobilized furin. Binding of Nb14 (in RU) is shown in grey as a function of time. The start and end of the injections are indicated with closed and open arrows respectively. The fitted model is indicated with a broken black line. One out of three experiments is shown as a representative Figure.
The interaction kinetics were determined for Nb14. Nb14 binds to furin with an overall Kd of 20.0 ± 2.9 nM. A two-state reaction model (A+B↔AB↔AB*) was most appropriate to evaluate the kinetics (Figure 7F). The association (kon1) and dissociation rates (koff1) for the first equation are (9.06 ± 0.93)×105 M−1 · s−1 and (3.31 ± 0.24)×10−2 s−1. The second ‘forward’ (kon2) and ‘reverse’ rate constant (koff2) are (3.02 ± 0.62)×10−3 s−1 and (3.55 ± 0.59)×10−3 s−1.
DISCUSSION
In the present study we describe the first antibody-based furin-inhibiting nanobodies. Consistent with the specificity typical for antibodies, no cross-reactivity with other PCs was observed. Furthermore, the furin-inhibiting nanobodies can only inhibit the cleavage of prorenin by furin, but not by other PCs. As such they are the first specific furin inhibitors.
The results of the present study suggest that the nanobodies do not bind directly to the catalytic pocket of furin, but instead inhibit the proteolytic cleavage of furin substrates by steric hindrance. Indeed, small peptide substrates such as the pyr-Arg-Thr-Lys-Arg-AMC substrate can still enter the catalytic cleft of furin to be cleaved. This is supported by the interaction kinetics. Although showing rather high affinity, Nb14 has a Kd of 20 nM, the Ki of Nb14 is 800-fold higher (25 µM). Such a difference can be explained by binding of the nanobody at some distance of the catalytic pocket. In addition, the interaction kinetics suggest a two-state reaction model for Nb14 (A+B↔AB↔AB*). This model describes a 1:1 binding of Nb14 to furin followed by a conformational change in the complex. Of course, only biophysical approaches (e.g. spectroscopy or NMR) can provide conclusive evidence on the nature and the extent of this alteration in the interaction complex. A modest conformational change induced by the nanobody could also influence efficient binding of larger substrates. Furthermore, the cleavage of different furin substrates was differentially inhibited depending on the particular nanobody used. This suggests that entrance to the catalytic cleft depends on the shape of the substrate and the binding sites of the different nanobodies and on the intrinsic kinetic binding rate constants as well. Four out of 13 nanobodies affected processing of all of the protein substrates investigated. Certain nanobodies only inhibited the cleavage of some substrates (GPC3 or diphtheria toxin), but had no effects on other substrates (TGFβ). This substrate-specificity of the inhibitor might constitute an advantage for some therapeutic applications. Indeed, these particular nanobodies might be used to interfere with certain pathways without interruption of other signal transduction cascades. This would be useful not only in scientific studies which examine pathway-specific effects, but also might lead to fewer side effects when used for therapy. This possibility needs to be explored further.
In the future, these nanobodies might be developed for therapeutic applications as furin is involved in many diseases, such as infectious diseases and cancer. Previous data demonstrate that genetic ablation of furin in the salivary glands in a mouse model for pleomorphic adenomas prevents or significantly delays tumour formation without obvious side effects in salivary glands [5]. Although this work [5] supports the general feasibility of targeting furin in cancer, crucial hurdles remain to be overcome. The greatest concern regards potential side effects of such a therapy. Indeed, furin and several other PCs play an essential role during embryogenesis, and genetic ablation of furin during embryogenesis leads to embryonic lethality [3]. The role of furin during adult life is less clear and largely remains to be elucidated. Using conditional knockout mice, it has been demonstrated that genetic ablation of furin in the liver [6,47] and in the salivary glands [5] does not lead to any obvious histological abnormalities in those organs. In contrast, genetic ablation of furin in T-cells permits normal T-cell development, but impairs the function of regulatory and effector T-cells, which produce less active TGFβ1 [8]. As a result, immune responses are activated, which can be beneficial in the short term, but may eventually result in autoimmune responses. Conditional inactivation of furin in the pancreas leads to impaired acidification of dense core granules, without resulting in a pathological condition [7]. Nevertheless it should be noted that in those mouse models, furin was inactivated during embryogenesis in those particular organs, and it is still not clear whether similar problems would occur if furin were inactivated only during adulthood. In any case, methods to locally inactivate furin activity might be feasible with limited side effects. Recently, a Phase I clinical trial with a short hairpin RNAi (RNA interference) vector targeting furin has been performed in patients with advanced cancer [48]. No serious adverse events were observed, whereas an immune response correlating with prolonged survival was achieved. The use of a vector expressing a furin-inhibiting nanobody, as described in the present study, would have the advantage that not only furin in the transfected cells would be inhibited, but the secreted nanobody can inhibit furin in adjacent cells. Several furin inhibitors have been developed, but so far none are entirely furin-specific. To reduce potential side effects, the generation of specific furin inhibitors is of great interest. The newly generated nanobodies described in the present study do not bind to any other family members and therefore cannot inhibit the proteolytic activity of these PCs. The cross-reactivity with mouse furin is not surprising as the conservation of amino acids in the catalytic domain is >99%. Such a high conservation of furin between species might also explain the low immune response observed in camelids. Previous attempts to raise antibodies in a camel and a llama were unsuccessful and the observed immune response in the dromedary was low.
Although purified nanobodies cannot penetrate the cell membrane, they effectively reduce the cleavage of extracellular furin substrates such as diphtheria toxin. Nevertheless, the furin-inhibiting nanobodies do enter into cells via antibody uptake following binding to surface furin, and are recycled to the TGN, similar to epitope tag-directed antibodies [44]. In this way the nanobodies might also affect the cleavage of intracellular furin substrates. To allow more efficient interference with the cleavage of intracellular substrates, nanobodies could be modified to allow penetration across the cell membrane. Manipulation of nanobodies is facilitated by the small size and single exon nature of the encoding gene fragment (360–380 bp). Furthermore, nanobodies have unique structural and functional properties and have many advantages compared with conventional antibodies. First of all, they are much smaller (12–15 kDa) than common antibodies (150 kDa) and highly soluble [35,36,49]. Moreover, they are not immunogenic, because their sequence shares a high degree of identity with the human VH domain. Nanobodies can be humanized without loss of affinity or antigen specificity [50,51], and no immune response was noticed even after repeated administrations in mice [52] or as a result of clinical Phase I/II testing of therapeutic nanobodies in humans (see http://www.ablynx.com). Also, the half-life of the nanobodies can be tailoured to vary from 30 min to 3 weeks to suit the specific application [53]. In addition, nanobodies are more resistant to extreme pH and temperature and resist attack by proteases to a greater extent than conventional antibodies [54]. Thus they are expected to be resistant to the acidic pH found in the stomach. As such, it might be possible to administer nanobodies orally to target gastrointestinal diseases. Finally, since nanobodies are encoded by single genes, they can efficiently be produced in large amounts by both prokaryotic and eukaryotic hosts, reducing their production costs.
In conclusion, we have generated the first nanobodies against mouse and human furin; these can be used as highly specific furin inhibitors that do not inhibit the activity of other PCs. This strict specificity may reduce potential side effects when used for therapeutic intervention. Furthermore, these nanobodies can protect cells from diphtheria-toxin-induced cytotoxicity, suggesting that nanobody-mediated furin inhibition might constitute an effective therapeutic strategy against a wide variety of diseases known to involve furin action.
Acknowledgments
FUNDING
This work was supported, in part, by the GOA (Geconcerteerde Onderzoeksacties) [grant number 12/016], KU Leuven R&D project [grant number 324000], Fonds voor Wetenschappelijk Onderzoek Vlaanderen [grant number G.A103.11] and the National Institutes of Health [grant number DA05084] (to I.L.). J.D. is a postdoctoral fellow of the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO).
Abbreviations
- AMC
7-amino-4-methylcoumarin
- D9R
nona-d-arginine
- EDC
N-ethyl-N′-(3-dimethylaminopropyl)carbodi-imide
- Fc
flow channel(s)
- GPC3
glypican 3
- HA
haemagglutinin
- HEK
human embryonic kidney
- LF
lethal factor
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide
- NHS
N-hydroxysuccinimide
- PA
protective antigen
- PC
proprotein convertase
- PCSK
PC subtilisin/kexin
- α1-PDX
α-1 antitrypsin Portland variant
- RU
response unit(s)
- TGFβ
transforming growth factor β
- TGN
trans-Golgi network.
Footnotes
AUTHOR CONTRIBUTION
Bart Roucourt and Guido David performed and analysed the Biacore experiments. Gholamreza Ghassabeh, Jörg Kinne and Serge Muyldermans generated the nanobodies. Iris Lindberg purified the human furin. Jingjing Zhu, Sandra Meulemans and Jeroen Declercq performed and analysed the other experiments. Alphons Vermorken and Wim Van de Ven proofread the paper before submission. Jeroen Declercq and John Creemers designed the study and wrote the paper.
REFERENCES
- 1.Taylor NA, Van De Ven WJ, Creemers JW. Curbing activation: proprotein convertases in homeostasis and pathology. FASEB J. 2003;17:1215–1227. doi: 10.1096/fj.02-0831rev. [DOI] [PubMed] [Google Scholar]
- 2.Creemers JW, Khatib AM. Knock-out mouse models of proprotein convertases: unique functions or redundancy? Front. Biosci. 2008;13:4960–4971. doi: 10.2741/3055. [DOI] [PubMed] [Google Scholar]
- 3.Roebroek AJ, Umans L, Pauli IG, Robertson EJ, van Leuven F, Van de Ven WJ, Constam DB. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development. 1998;125:4863–4876. doi: 10.1242/dev.125.24.4863. [DOI] [PubMed] [Google Scholar]
- 4.Roebroek AJ, Taylor NA, Louagie E, Pauli I, Smeijers L, Snellinx A, Lauwers A, Van de Ven WJ, Hartmann D, Creemers JW. Limited redundancy of the proprotein convertase furin in mouse liver. J. Biol. Chem. 2004;279:53442–53450. doi: 10.1074/jbc.M407152200. [DOI] [PubMed] [Google Scholar]
- 5.De Vos L, Declercq J, Rosas GG, Van Damme B, Roebroek A, Vermorken F, Ceuppens J, van de Ven W, Creemers J. MMTV-cre-mediated fur inactivation concomitant with PLAG1 proto-oncogene activation delays salivary gland tumorigenesis in mice. Int. J. Oncol. 2008;32:1073–1083. [PubMed] [Google Scholar]
- 6.Essalmani R, Susan-Resiga D, Chamberland A, Abifadel M, Creemers JW, Boileau C, Seidah NG, Prat A. In vivo evidence that furin from hepatocytes inactivates PCSK9. J. Biol. Chem. 2011;286:4257–4263. doi: 10.1074/jbc.M110.192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Louagie E, Taylor NA, Flamez D, Roebroek AJM, Bright NA, Meulemans S, Quintens R, Herrera PL, Schuit F, van de Ven WJM, Creemers JWM. Role of furin in granular acidification in the endocrine pancreas: identification of the V-ATPase subunit Ac45 as a candidate substrate. Proc. Natl. Acad. Sci. U.S.A. 2008;105:12319–12324. doi: 10.1073/pnas.0800340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pesu M, Watford WT, Wei L, Xu LL, Fuss I, Strober W, Andersson J, Shevach EM, Quezado M, Bouladoux N, et al. T-cell-expressed proprotein convertase furin is essential for maintenance of peripheral immune tolerance. Nature. 2008;455:U246–U273. doi: 10.1038/nature07210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassi DE, Mahloogi H, Al-Saleem L, Lopez De Cicco R, Ridge JA, Klein-Szanto AJ. Elevated furin expression in aggressive human head and neck tumors and tumor cell lines. Mol. Carcinog. 2001;31:224–232. doi: 10.1002/mc.1057. [DOI] [PubMed] [Google Scholar]
- 10.Cheng M, Watson PH, Paterson JA, Seidah N, Chretien M, Shiu RP. Pro-protein convertase gene expression in human breast cancer. Int. J. Cancer. 1997;71:966–971. doi: 10.1002/(sici)1097-0215(19970611)71:6<966::aid-ijc10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Page RE, Klein-Szanto AJ, Litwin S, Nicolas E, Al-Jumaily R, Alexander P, Godwin AK, Ross EA, Schilder RJ, Bassi DE. Increased expression of the pro-protein convertase furin predicts decreased survival in ovarian cancer. Cell. Oncol. 2007;29:289–299. doi: 10.1155/2007/930321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu ML, Gordon VM, Fitzgerald DJP, Leppla SH. Furin regulates both the activation of Pseudomonas exotoxin A and the quantity of the toxin receptor expressed on target cells. Infect. Immun. 1996;64:524–527. doi: 10.1128/iai.64.2.524-527.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuneoka M, Nakayama K, Hatsuzawa K, Komada M, Kitamura N, Mekada E. Evidence for involvement of furin in cleavage and activation of diphtheria-toxin. J. Biol. Chem. 1993;268:26461–26465. [PubMed] [Google Scholar]
- 14.Lea N, Lord JM, Roberts LM. Proteolytic cleavage of the A subunit is essential for maximal cytotoxicity of Escherichia coli O157: H7 Shiga-like toxin-1. Microbiology. 1999;145:999–1004. doi: 10.1099/13500872-145-5-999. [DOI] [PubMed] [Google Scholar]
- 15.Young JAT, Collier RJ. Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu. Rev. Biochem. 2007;76:243–265. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]
- 16.Abrami L, Fivaz M, Decroly E, Seidah NG, Jean F, Thomas G, Leppla SH, Buckley JT, van der Goot FG. The pore-forming toxin proaerolysin is activated by furin. J. Biol. Chem. 1998;273:32656–32661. doi: 10.1074/jbc.273.49.32656. [DOI] [PubMed] [Google Scholar]
- 17.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk HD, Garten W. Inhibition of furin-mediated cleavage activation of Hiv-1 hlycoprotein-Gp160. Nature. 1992;360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 18.Kido H, Okumura Y, Takahashi E, Pan HY, Wang S, Chida J, Le TQ, Yano M. Host envelope glycoprotein processing proteases are indispensable for entry into human cells by seasonal and highly pathogenic avian influenza viruses. J. Mol. Genet. Med. 2008;3:167–175. [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez-Reyes L, Ruiz-Arguello MB, Garcia-Barreno B, Calder L, Lopez JA, Albar JP, Skehel JJ, Wiley DC, Melero JA. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9859–9864. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maisner A, Mrkic B, Herrler G, Moll M, Billeter MA, Cattaneo R, Klenk HD. Recombinant measles virus requiring an exogenous protease for activation of infectivity. J. Gen. Virol. 2000;81:441–449. doi: 10.1099/0022-1317-81-2-441. [DOI] [PubMed] [Google Scholar]
- 21.Jean F, Thomas L, Molloy SS, Liu GP, Jarvis MA, Nelson JA, Thomas G. A protein-based therapeutic for human cytomegalovirus infection. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2864–2869. doi: 10.1073/pnas.050504297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scamuffa N, Siegfried G, Bontemps Y, Ma LM, Basak A, Cherel G, Calvo F, Seidah NG, Khatib AM. Selective inhibition of proprotein convertases represses the metastatic potential of human colorectal tumor cells. J. Clin. Invest. 2008;118:352–363. doi: 10.1172/JCI32040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarac MS, Cameron A, Lindberg I. The furin inhibitor hexa-D-arginine blocks the activation of Pseudomonas aeruginosa exotoxin A in vivo. Infect. Immun. 2002;70:7136–7139. doi: 10.1128/IAI.70.12.7136-7139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarac MS, Peinado JR, Leppla SH, Lindberg I. Protection against anthrax toxemia by hexa-D-arginine in vitro and in vivo. Infect. Immun. 2004;72:602–605. doi: 10.1128/IAI.72.1.602-605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiryaev SA, Remacle AG, Ratnikov BI, Nelson NA, Savinov AY, Wei G, Bottini M, Rega MF, Parent A, Desjardins R, et al. Targeting host cell furin proprotein convertases as a therapeutic strategy against bacterial toxins and viral pathogens. J. Biol. Chem. 2007;282:20847–20853. doi: 10.1074/jbc.M703847200. [DOI] [PubMed] [Google Scholar]
- 27.Becker GL, Sielaff F, Than ME, Lindberg I, Routhier S, Day R, Lu YH, Garten W, Steinmetzer T. Potent inhibitors of furin and furin-like proprotein convertases containing decarboxylated P1 arginine mimetics. J. Med. Chem. 2010;53:1067–1075. doi: 10.1021/jm9012455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cameron A, Appel J, Houghten RA, Lindberg I. Polyarginines are potent furin inhibitors. J. Biol. Chem. 2000;275:36741–36749. doi: 10.1074/jbc.M003848200. [DOI] [PubMed] [Google Scholar]
- 29.Misumi Y, Ohkubo K, Sohda M, Takami N, Oda K, Ikehara Y. Intracellular processing of complement pro-C3 and proalbumin is inhibited by rat α-1-protease inhibitor variant (Met352→Arg) in transfected cells. Biochem. Biophys. Res. Commun. 1990;171:236–242. doi: 10.1016/0006-291x(90)91382-3. [DOI] [PubMed] [Google Scholar]
- 30.Van Rompaey L, Ayoubi T, Van De Ven W, Marynen P. Inhibition of intracellular proteolytic processing of soluble proproteins by an engineered α2-macroglobulin containing a furin recognition sequence in the bait region. Biochem. J. 1997;326:507–514. doi: 10.1042/bj3260507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henrich S, Lindberg I, Bode W, Than ME. Proprotein convertase models based on the crystal structures of furin and kexin: explanation of their specificity. J. Mol. Biol. 2005;345:211–227. doi: 10.1016/j.jmb.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 32.Coppola JM, Hamilton CA, Bhojani MS, Larsen MJ, Ross BD, Rehemtulla A. Identification of inhibitors using a cell-based assay for monitoring Golgi-resident protease activity. Anal. Biochem. 2007;364:19–29. doi: 10.1016/j.ab.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiao GS, Cregar L, Wang JZ, Millis SZ, Tang C, O’Malley S, Johnson AT, Sareth S, Larson J, Thomas G. Synthetic small molecule furin inhibitors derived from 2,5-dideoxystreptamine. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19707–19712. doi: 10.1073/pnas.0606555104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conrath KE, Lauwereys M, Galleni M, Matagne A, Frere JM, Kinne J, Wyns L, Muyldermans S. β-lactamase inhibitors derived from single-domain antibody fragments elicited in the Camelidae. Antimicrob. Agents Chemother. 2001;45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauwereys M, Ghahroudi MA, Desmyter A, Kinne J, Holzer W, De Genst E, Wyns L, Muyldermans S. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 1998;17:3512–3520. doi: 10.1093/emboj/17.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen VK, Hamers R, Wyns L, Muyldermans S. Camel heavy-chain antibodies: diverse germline VHH and specific mechanisms enlarge the antigen-binding repertoire. EMBO J. 2000;19:921–930. doi: 10.1093/emboj/19.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne R, Muyldermans S, Wyns L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4586–4591. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Creemers JWM, Kormelink PJG, Roebroek AJM, Nakayama K, Vandeven WJM. Proprotein processing activity and cleavage site selectivity of the Kex2-like endoprotease Pace4. FEBS Lett. 1993;336:65–69. doi: 10.1016/0014-5793(93)81610-c. [DOI] [PubMed] [Google Scholar]
- 39.Creemers JWM, Pritchard LE, Gyte A, Le Rouzic P, Meulemans S, Wardlaw SL, Zhu XR, Steiner DF, Davies N, Armstrong D, et al. Agouti-related protein is posttranslationally cleaved by proprotein convertase 1 to generate agouti-related protein (AGRP)(83132): interaction between AGRP(83132) and melanocortin receptors cannot be influenced by syndecan-3. Endocrinology. 2006;147:1621–1631. doi: 10.1210/en.2005-1373. [DOI] [PubMed] [Google Scholar]
- 40.Ayoubi TAY, Meulemans SMP, Roebroek AJM, Van de Ven WJM. Production of recombinant proteins in Chinese hamster ovary cells overexpressing the subtilisin-like proprotein converting enzyme furin. Mol. Biol. Rep. 1996;23:87–95. doi: 10.1007/BF00424434. [DOI] [PubMed] [Google Scholar]
- 41.Veugelers M, De Cat B, Muyldermans SY, Reekmans G, Delande N, Frints S, Legius E, Fryns JP, Schrander-Stumpel C, Weidle B, et al. Mutational analysis of the GPC3/GPC4 glypican gene cluster on Xq26 in patients with Simpson-Golabi-Behmel syndrome: identification of loss-of-function mutations in the GPC3 gene. Hum. Mol. Genet. 2000;9:1321–1328. doi: 10.1093/hmg/9.9.1321. [DOI] [PubMed] [Google Scholar]
- 42.van de Loo JWHP, Creemers JWM, Bright NA, Young BD, Roebroek AJM, Van de Ven WJM. Biosynthesis, distinct post-translational modifications, and functional characterization of lymphoma proprotein convertase. J. Biol. Chem. 1997;272:27116–27123. doi: 10.1074/jbc.272.43.27116. [DOI] [PubMed] [Google Scholar]
- 43.Anderson ED, Thomas L, Hayflick JS, Thomas G. Inhibition of Hiv-1 Gp160-dependent membrane-fusion by a furin-directed α1-antitrypsin variant. J. Biol. Chem. 1993;268:24887–24891. [PubMed] [Google Scholar]
- 44.Molloy SS, Thomas L, Vanslyke JK, Stenberg PE, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO. J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjannet S, Savaria D, Laslop A, Munzer JS, Chretien M, Marcinkiewicz M, Seidah NG. α1-antitrypsin portland inhibits processing of precursors mediated by proprotein convertases primarily within the constitutive secretory pathway. J. Biol. Chem. 1997;272:26210–26218. doi: 10.1074/jbc.272.42.26210. [DOI] [PubMed] [Google Scholar]
- 46.Fugere M, Appel J, Houghten RA, Lindberg I, Day R. Short polybasic peptide sequences are potent inhibitors of PC5/6 and PC7: use of positional scanning-synthetic peptide combinatorial libraries as a tool for the optimization of inhibitory sequences. Mol. Pharmacol. 2007;71:323–332. doi: 10.1124/mol.106.027946. [DOI] [PubMed] [Google Scholar]
- 47.Roebroek AJM, Taylor NA, Louagie E, Pauli I, Smeijers L, Snellinx A, Lauwers A, Van de Ven WJM, Hartmann D, Creemers JWM. Limited redundancy of the proprotein convertase furin in mouse liver. J. Biol. Chem. 2004;279:53442–53450. doi: 10.1074/jbc.M407152200. [DOI] [PubMed] [Google Scholar]
- 48.Senzer N, Barve M, Kuhn J, Melnyk A, Beitsch P, Lazar M, Lifshitz S, Magee M, Oh J, Mill SW, et al. Phase I trial of “bi-shRNAi(furin)/GMCSF DNA/autologous tumor cell” vaccine (FANG) in advanced cancer. Mol. Ther. 2012;20:679–686. doi: 10.1038/mt.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Revets H, De Baetselier P, Muyldermans S. Nanobodies as novel agents for cancer therapy. Expert Opin. Biol. Ther. 2005;5:111–124. doi: 10.1517/14712598.5.1.111. [DOI] [PubMed] [Google Scholar]
- 50.Vincke C, Loris R, Saerens D, Martinez-Rodriguez S, Muyldermans S, Conrath K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J. Biol. Chem. 2009;284:3273–3284. doi: 10.1074/jbc.M806889200. [DOI] [PubMed] [Google Scholar]
- 51.De Groeve K, Deschacht N, De Koninck C, Caveliers V, Lahoutte T, Devoogdt N, Muyldermans S, De Baetselier P, Raes G. Nanobodies as tools for in vivo imaging of specific immune cell types. J. Nucl. Med. 2010;51:782–789. doi: 10.2967/jnumed.109.070078. [DOI] [PubMed] [Google Scholar]
- 52.Hmila I, Saerens D, Ben Abderrazek R, Vincke C, Abidi N, Benlasfar Z, Govaert J, El Ayeb M, Bouhaouala-Zahar B, Muyldermans S. A bispecific nanobody to provide full protection against lethal scorpion envenoming. FASEB J. 2010;24:3479–3489. doi: 10.1096/fj.09-148213. [DOI] [PubMed] [Google Scholar]
- 53.Jevsevar S, Kunstelj M, Porekar VG. PEGylation of therapeutic proteins. Biotechnol. J. 2010;5:113–128. doi: 10.1002/biot.200900218. [DOI] [PubMed] [Google Scholar]
- 54.Deffar K, Shi HL, Li L, Wang XZ, Zhu XJ. Nanobodies: the new concept in antibody engineering. Afr. J. Biotechnol. 2009;8:2645–2652. [Google Scholar]