Abstract
Newborn neurons enter an extended maturation stage, during which they acquire excitability characteristics crucial for development of presynaptic and postsynaptic connectivity. In contrast to earlier specification programs, little is known about the regulatory mechanisms that control neuronal maturation. The Pet-1 ETS (E26 transformation-specific) factor is continuously expressed in serotonin (5-HT) neurons and initially acts in postmitotic precursors to control acquisition of 5-HT transmitter identity. Using a combination of RNA sequencing, electrophysiology, and conditional targeting approaches, we determined gene expression patterns in maturing flow-sorted 5-HT neurons and the temporal requirements for Pet-1 in shaping these patterns for functional maturation of mouse 5-HT neurons. We report a profound disruption of postmitotic expression trajectories in Pet-1−/− neurons, which prevented postnatal maturation of 5-HT neuron passive and active intrinsic membrane properties, G-protein signaling, and synaptic responses to glutamatergic, lysophosphatidic, and adrenergic agonists. Unexpectedly, conditional targeting revealed a postnatal stage-specific switch in Pet-1 targets from 5-HT synthesis genes to transmitter receptor genes required for afferent modulation of 5-HT neuron excitability. 5-HT1a autoreceptor expression depended transiently on Pet-1, thus revealing an early postnatal sensitive period for control of 5-HT excitability genes. Chromatin immunoprecipitation followed by sequencing revealed that Pet-1 regulates 5-HT neuron maturation through direct gene activation and repression. Moreover, Pet-1 directly regulates the 5-HT neuron maturation factor Engrailed 1, which suggests Pet-1 orchestrates maturation through secondary postmitotic regulatory factors. The early postnatal switch in Pet-1 targets uncovers a distinct neonatal stage-specific function for Pet-1, during which it promotes maturation of 5-HT neuron excitability.
SIGNIFICANCE STATEMENT The regulatory mechanisms that control functional maturation of neurons are poorly understood. We show that in addition to inducing brain serotonin (5-HT) synthesis and reuptake, the Pet-1 ETS (E26 transformation-specific) factor subsequently globally coordinates postmitotic expression trajectories of genes necessary for maturation of 5-HT neuron excitability. Further, Pet-1 switches its transcriptional targets as 5-HT neurons mature from 5-HT synthesis genes to G-protein-coupled receptors, which are necessary for afferent synaptic modulation of 5-HT neuron excitability. Our findings uncover gene-specific switching of downstream targets as a previously unrecognized regulatory strategy through which continuously expressed transcription factors control acquisition of neuronal identity at different stages of development.
Keywords: adrenergic signaling, gene regulatory network, genomics, neuronal maturation, Pet-1, serotonin
Introduction
The development of mature neuronal identities is a crucial step in the formation of neural circuitry that enables behavioral output and plasticity (Fishell and Heintz, 2013). Neuronal maturation emerges progressively as earlier specification programs are suppressed and new gene expression trajectories commence to provide for acquisition of adult neuron-type morphological and functional characteristics. Tremendous progress has been made in elucidating the transcriptional mechanisms that control neural cell-type specification and differentiation (Shirasaki and Pfaff, 2002; Smidt and Burbach, 2007; Greig et al., 2013; Philippidou and Dasen, 2013). Much less is known, however, about the precisely controlled gene expression patterns required for subsequent postmitotic neuronal maturation and how they are transcriptionally controlled early in life to generate functionally mature neurons (Okaty et al., 2009). This constitutes a significant gap in understanding brain development as it is during the maturation stage that synaptic connectivity is shaped, which is crucial for the opening and closing of critical periods for experience-dependent plasticity (Hensch, 2005; de la Torre-Ubieta and Bonni, 2011; Le Magueresse and Monyer, 2013). Furthermore, genetic or environmental perturbation of gene expression trajectories underlying prenatal and postnatal neuronal maturation is thought to directly contribute to autism and other neurodevelopmental disorders (Levitt et al., 2004; Meredith et al., 2012; Tebbenkamp et al., 2014).
The regulatory mechanisms controlling the development of serotonin (5-HT) neurons are of particular interest, as 5-HT has wide-ranging modulatory effects on central neural circuitry. In addition, altered serotonergic gene expression early in life has been implicated in several neurodevelopmental disorders (Leonardo and Hen, 2008; Deneris and Wyler, 2012). Specification of mouse serotonergic progenitors and acquisition of 5-HT transmitter identity is controlled by a well-defined 5-HT gene regulatory network (GRN). The 5-HT GRN comprises several transcription factors (TFs), including Ascl1 and Foxa2, which act at the progenitor stage, and GATA-2, Lmx1b, and Pet-1, which act in postmitotic precursors to induce expression of a 5-HT gene battery [Tph2, Gch1, Gchfr, Slc6a4 (Sert), Slc22a3 (Oct3), Slc18a2 (VMAT2)] that enables initiation of 5-HT synthesis, reuptake, and vesicular transport (Deneris and Wyler, 2012). Newly differentiated 5-HT neurons then enter an extended fetal to early postnatal maturation stage, during which they migrate to form the various raphe nuclei, develop connections with numerous other neuron types, and acquire mature functional characteristics (Lidov and Molliver, 1982; Maejima et al., 2013). A critical event at this stage is the maturation of the excitability features needed to establish appropriate presynaptic and postsynaptic connectivity (Lidov and Molliver, 1982; Maejima et al., 2013). Indeed, intrinsic membrane properties, G-protein signaling, and neurotransmitter afferent synaptic responses of 5-HT neurons do not acquire adult characteristics until the early postnatal period (Rood et al., 2014). In contrast to well-studied earlier specification stages that determine 5-HT transmitter identity, little is known about the regulatory factors that control postnatal maturation of 5-HT neuron excitability.
Here, we used whole-genome approaches in fetal and postnatal 5-HT neurons to comprehensively investigate the regulatory strategies that shape maturation of postmitotic 5-HT neurons. We show that in addition to inducing brain 5-HT synthesis and reuptake, Pet-1 subsequently coordinates postmitotic expression trajectories of genes necessary for maturation of 5-HT neurons. The broad disruption of gene expression patterns in Pet-1−/− 5-HT neurons blocked functional maturation of 5-HT neuron passive and active membrane properties, multimodal afferent synaptic inputs, and G-protein signaling. To understand how Pet-1 controls 5-HT neuron maturation, we used conditional-targeting approaches and found that Pet-1 switches targets from those required in newborn 5-HT neurons for initiation of 5-HT synthesis to those required postnatally for extrinsic control of 5-HT neuron excitability. Further, we uncover an early postnatal sensitive period for control of 5-HT autoreceptor expression by Pet-1. Chromatin immunoprecipitation sequencing (ChIP-seq) analyses suggest Pet-1 controls maturation through direct activation and repression of downstream targets, including several TFs with known roles in neuronal maturation. Our findings reveal a previously unrecognized stage-specific function for Pet-1 that is critical for postnatal maturation of 5-HT neuron excitability.
Materials and Methods
Animals
Mice were maintained in ventilated cages on a 12 h light/dark cycle with access to food and water ad libitum with 2–5 mice per cage. All mice except the ePet-mycPet-1 mice were backcrossed ≥5 generations onto a C57BL/6J background. ePet-mycPet-1 mice were maintained on a mixed C57BL/6*129sv*SJL background. Embryonic age was defined by the number of days following the appearance of a vaginal plug, designated as embryonic day (E) 0.5. The date of birth was designated at postnatal day (P) 0. All procedures were approved by the Institutional Animal Care and Use Committees of Case Western Reserve University, Vanderbilt University, and Children Hospital of Philadelphia in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Mice were genotyped using the following primers: Pet-1: 5′-CGGTGGATGTGGAATGTGTGCG-3′, 5′-CGCACTTGGGGGGTCATTATCAC-3′, 5′-GCCTGATGTTCAAGGAAGACCTCGG-3′; floxed Pet-1: 5′-TAGGAGGGTCTGGTGTCTGG-3′, 5′-GCGTCCTTGTGTGTAGCAGA-3′; ePet-mycPet-1: 5′-GGGCCTATCCAAACTCAACTT-3′, 5′-GGGAGGTGTGGGAGGTTTT-3′; ePet-EYFP: 5′-TATATCATGGCCGACAAGCAG-3′, 5′-GAACTCCAGCAGGACCATGT-3′; eFev-βgal: 5′-CAAAGACAGGAGGAGGTTGGTAGC-3′, 5′-TTGGGTAACGCCAGGGTTTTCC-3′.
Flow cytometry and total RNA isolation
ePet-EYFP or Pet-1−/−;ePet-EYFP mice were used to collect YFP+ 5-HT neurons with flow cytometry. E11.5 and E15.5 hindbrain tissue was dissected between the mid-hindbrain boundary and rhombomere 4. For isolation of postnatal neurons, dorsal and median raphes from P1–P3 mice were dissected from midbrain. To dissociate cells, dissected embryonic tissue was treated with trypsin following a previously published protocol (Wylie et al., 2010; Wyler et al., 2015). Postnatal tissue was dissociated with a modified protocol: tissue was collected in SSS media [Hibernate A (Life Technologies), 2% B27, and 0.25% Glutamax (Invitrogen)] then washed in PBS and incubated on a shaker at 37°C for 40 min using a papain/DNase I solution (Papain Disassociation kit, Worthington; 20 U/ml papain, 1 mm l-cysteine, 0.5 mm EDTA, 0.05 mg/ml DNase I in Earle's balanced salt solution). Dissociated cells were washed in Leibovitz's L-15 (Life Technologies) media and resuspended in SSS media followed by trituration using a series of three fire-polished glass pipettes with decreasing bore width. Cells were passed through a 40 μm filter (BD Biosciences) and sorted on a Becton Dickinson FACS Aria digital cell sorter with an argon laser (200 mW at 488 nm) using an 85 μm nozzle. Cells were sorted directly into 500 μl of Trizol (Invitrogen). Each biological replicate was defined as an independent sorting experiment from pooled embryos on different days of different litters. Total RNA was isolated after addition of 10 μg of Glycoblue (Ambion) using chloroform extraction. RNA was purified and DNase I treated using the RNA Clean & Concentrator-5 kit (Zymo). RNA quality was analyzed using an Agilent 2100 bioanalyzer. All samples had a RNA integrity number ≥8.3.
RNA sequencing analysis
Purified total RNA was amplified with the SMARTer Ultra-low mRNA-Seq kit (Clontech) or Ovation RNA-Seq System V2 (NuGEN). Libraries were sequenced using paired-end reads for 50–100 cycles on the HiSeq 2500 system (Illumina). Sequence reads were mapped to the Mus musculus transcriptome [University of California, Santa Cruz, Genome Browser (UCSC) mm9] using annotation supplied by Illumina (ftp://igenome:G3nom3s4uussdftp.illumina.com/Mus_musculus/_UCSC/mm9/Mus_musculus_UCSC_mm9.tar.gz) using the tophat2 read mapper (Kim et al., 2013). Gene expression quantification and differential expression were analyzed using Cufflinks v2.2.1 (Trapnell et al., 2010). High Spearman rank correlations within biological replicate groups (>0.95) and between conditions (>0.85), as well as the number of high-quality mapped read pairs per sample, indicate the high-quality nature of the RNA sequencing (RNA-seq) data. Fragments per kilobase per million reads (FPKM) values were compared using the time-series option of Cufflinks. In the trajectory expression profiling and ePet-EYFP versus Pet-1−/−;ePet-EYFP comparisons, genes were called differentially expressed if fold-change was ≥1.5 and false discovery rate (FDR) was ≤5%. Hierarchical clustering of genes differentially expressed over development was performed in Matlab (Mathworks) using row-scaled expression values with average linkage and Euclidean distance. Gene ontology (GO) analysis was performed using WebGestalt (http://www.webgestalt.org), requiring ≥2 genes per category and hypergeometric test p value ≤0.01 (Zhang et al., 2005). Protein functional class annotation was performed using PantherDB version 9.0, which uses conserved protein families to categorize gene sets (Mi et al., 2013).
ChIP-seq
YFP+ tissue between the mid-hindbrain boundary and rhombomere 4 of E12.5–E15.5 hindbrains from Pet-1−/−; ePet-mycPet-1; ePet-EYFP embryos was dissected and quickly flash frozen on dry ice. Chromatin was isolated and immunoprecipitated using a ChIP grade, goat anti-Myc antibody (ab9132, Abcam) with proprietary protocols (Active Motif). Reads were mapped to the Mus musculus genome (UCSC mm10) using the Burroughs-Wheeler Aligner and were filtered to select reads that map to a single location (Li and Durbin, 2009). Peak calling was performed with MACS (Model-Based Analysis for ChIP-Seq) v1.4.2 modified to accept a custom scaling factor of 0.8195945 derived from the Normalization ChIPseq software (Zhang et al., 2008; Liang and Keleş, 2012). MycPet-1 ChIP peaks were associated to genes using the GREAT (Genomic Regions Enrichment of Annotations Tool) web service v3.0 (mm10) with 5 kb intervals upstream and downstream of the transcription start site (TSS) and transcription terminal site (TTS) of each gene model along with peaks that overlap the gene body. ChIP peak signal coverage was visualized in the UCSC Genome Browser (Rhead et al., 2010).
Electrophysiology
Slice preparation and recording.
Brain slices from Pet-1−/− mice or their wild-type (+/+) littermates were prepared as previously described from P21 (P20–P24) or adult (>P60) mice (Crawford et al., 2011; Green et al., 2015). Brains were sectioned using a vibratome (Leica Microsystems) at ∼200 μm and maintained in ice-cold sucrose aCSF (see below; 248 mm sucrose substituted for NaCl) during sectioning as previously described (Beck et al., 2004; Calizo et al., 2011; Crawford et al., 2011). Once sectioned, slices were maintained in a holding chamber containing aCSF (in mm: 124 NaCl, 2.5 KCl, 1.25 NaH2PO4, 2.0 MgSO4, 2.5 CaCl2, 10 dextrose, 26 NaHCO3, and l-tryptophan) bubbled with 95%O2/5%CO2 mixture at 37°C for 1 h, then at room temperature until recording. L-tryptophan (2.5 μm; Sigma-Aldrich) was included in the holding chamber to maintain 5-HT synthesis, but was not present in aCSF when recording (Liu et al., 2005). During a recording session, slices were placed in a recording chamber (Warner Instruments) and bathed in continuous flow of aCSF heated to 32–34°C with an inline heater (Warner Instruments). Neurons were visualized using a Nikon E600 upright microscope (Nikon) and targeted under differential interference contrast. Recordings were made using glass electrodes (3–6 MΩ access resistance) filled with electrolyte (in mm: 130 K-gluconate, 5 NaCl, 10 disodium phosphocreatine, 1 MgCl2, 0.02 EGTA, 10 HEPES, 2 MgATP, and 0.5 Na2GTP, with 0.1% biocytin, pH 7.3). Whole-cell recordings were controlled using a Multiclamp 700B amplifier (Molecular Devices) and signals were collected and stored using a Digidata 1320 analog-to-digital converter and pClamp 9.0 or 10.0 software (Molecular Devices). Following recording, slices were stored in 4% paraformaldehyde at 4°C for immunohistochemical detection of Tph2 to verify 5-HT neuron identity. In some experiments, +/+ and Pet-1−/− possessed the eFEV-LacZ transgene, which was used to verify 5-HT neurons with anti-βgal immunostaining (Krueger and Deneris, 2008). Chemicals for buffers and electrolytes were purchased from Sigma-Aldrich. Statistical tests were performed using Statistica (StatSoft). Data were analyzed using one-factor and two-factor ANOVAs. Post hoc analyses were conducted using the Student–Newman–Keuls method. A probability level of p < 0.05 was considered significant in all analyses.
Passive and active membrane characteristics.
Passive and active membrane characteristics were recorded using current-clamp techniques as previously described (Calizo et al., 2011). To obtain data on neuronal membrane characteristics (e.g., resting membrane resistance, resting membrane potential, and membrane time constant), action potential (AP) characteristics (e.g., AP threshold, amplitude, duration, and afterhyperpolarization amplitude), and excitability (frequency vs current), each cell received 500 ms current injections starting at −100 pA and stepping to 180 pA in 20 pA steps. Current pulses were separated by 10 s and voltage responses were recorded in response to each current pulse.
Glutamatergic synaptic activity.
Baseline glutamate activity at adult stages, in the form of EPSCs, was examined by recording current in voltage-clamp mode for 2–5 min as previously described (Lemos et al., 2006; Crawford et al., 2011). Several characteristics of EPSCs were examined, including frequency, rise and decay time, amplitude, and charge. A minimum of 200 EPSC events was used to calculate average EPSC characteristics. Statistical comparisons of EPSC characteristics were calculated using Student's t tests.
Five-carboxamidotryptamine and GTPγS responses.
To investigate 5-HT1a autoreceptor function, the nonselective 5-HT1,7 agonist 5-carboxamidotryptamine (100 nm; Sigma-Aldrich) was added and current was recorded until a steady-state outward potassium current was obtained (a total of ∼5 min). This outward hyperpolarizing current has previously been characterized as mediated by the 5-HT1a subscripted receptor in the dorsal and median raphe. GTPγS is a nonhydrolyzable form of GTP that can be used to directly activate G-protein-coupled channels, including GIRK channels. In normal dorsal raphe (DR) 5-HT neurons, the inhibitory 5-HT1a subscripted autoreceptor activates GIRK channels, resulting in an outward potassium current. This response is mimicked by activation of G-protein-regulated channels by adding GTPγS to the electrode solution. To measure the effect of GTPγS activation, voltage-clamp techniques were used to record from 5-HT neurons from P21 Pet-1−/− and +/+ mice with either normal electrolyte or with electrolyte containing 10 μm GTPγS (15 mm; Roche Diagnostics) as previously described (Rood et al., 2014). Recordings of current were taken using voltage-clamp techniques immediately upon membrane rupture so that the outward current elicited by the GTPγS activation of the G-protein could be recorded as it dialyzed into the cell and achieved steady-state levels after ∼3–5 min. Statistical comparisons of GTPγS responses were made with a two-factor ANOVA using genotype and electrolyte solution as factors, followed by Student–Newman–Keuls t tests.
Multielectrode array recordings.
As there are differences in the electrophysiological properties of neurons in the medial and lateral wing subfields of the DR, placement on the array and the dimensions of the electrode grid were such that recordings were made only from ventromedial DR nucleus (DRN) neurons (vmDRN). Mid-DRN slices of 280 μm thickness were taken between −4.5 and −4.75 mm back from bregma. We used a 6 × 10 perforated array with electrodes that have a diameter of 30 μm and 100 μm spacing between electrodes. The slice was placed so the electrodes cover an area spreading 1200 μm down from the cerebral aqueduct in the ventral direction and 340 μm laterally on either side of the midline (for a 680 μm total recording width). Placement of the array in the vmDRN region assured that the vast majority of cells in our recordings were 5-HT neurons based on extensive immunohistochemical detection of Tph2, 5-HT, and genetic markers of 5-HT neurons in this region (Scott et al., 2005; Krueger and Deneris, 2008). Data files were saved as .mcd files and analyzed in off-line sorter. For analysis, a Besel filter with a 150 Hz frequency cutoff was applied to the raw data traces. The threshold for detection was set manually to a level that will include all legitimate spikes with the least amount of unipolar noise spikes included (between 13 and 35 μV). Once spikes were detected, they were sorted by a combination of a k-means scan method and manual verification. The manual verification was conducted after the k-means scan was run and divided spikes into groups based on such criteria as amplitude, power under the curve, and spike duration (for full list of criteria, see off-line sorter V3 manual under the k-means scan; Plexon). Once waveforms were sorted into groups and judged to be biologically relevant, each spike was validated by eye and spikes that did not fit the average waveform shape were invalidated. All unsorted spikes were visualized manually and any spikes that matched the average waveform shape in the relevant group were added to that group. Spikes were then sorted into two classes using mean spike width as well as coefficient of variance (COV; COV = SD/mean) of their firing pattern to categorize each cell. Spikes were sorted into one group that had a large waveform (≥0.2 ms starting from the initial depolarization to the end of recovery), as well as a low COV (<0.9 arbitrary units, calculated using Matlab). The other population was defined by a smaller waveform (<0.2 ms) and a more variable firing pattern (COV, >0.9). These cutoff values were determined by trial and error with most cells that had a waveform width >0.2 also displaying a COV <0.9 and vice versa.
Histology
Perfusion/sectioning.
Mice were anesthetized with Avertin (44 mm tribromoethanol, 2.5% tert-amyl alcohol), 20 ml/kg, and transcardially perfused with PBS followed by 4% paraformaldehyde (PFA; Electron Microscopy Sciences) in PBS for 20 min. Brains were extracted and fixed for an additional 2 h in 4% PFA/PBS and incubated in 30% sucrose in PBS at 4°C overnight. Sections were collected using a freezing sliding microtome and mounted on SuperFrost Plus slides (ThermoFisher Scientific). Slides were stored at −80°C until use. All histology was performed on sex-matched littermate controls. For all experiments, sections were taken from the entire rostral caudal axis of the DRN. Mice were between 2 and 4 months old unless stated otherwise.
Immunohistochemistry.
Twenty micrometer sections from 2-month-old female eFev-LacZ and Pet-1−/−; eFev-LacZ littermates (Krueger and Deneris, 2008) were washed for 15 min in 1× PBS, pH 7.4, with 0.3% Triton X-100 (ThermoFisher Scientific). Slides were blocked in 10% normal goat serum (NGS; Millipore) in PBS with 0.1% Triton (PBS-T). Slides were then incubated at 4°C overnight in 5% NGS PBS-T with rabbit anti-ADRA1B (1:500; NR-102, Protos) and chicken polyclonal anti-β-galactosidase (1:1000; ab9361, Abcam; validation at abcam.com). Slides were washed six times in PBS-T and incubated for 2 h at room temperature with Alexa Fluor-488 anti-chicken or anti-rabbit antibody, or Alexa Fluor-594 anti-chicken or anti-rabbit antibody (1:500; Invitrogen) in 5% NGS PBS-T. Slides were then washed six times in PBS-T and mounted with ProLong Antifade Reagents (ThermoFisher Scientific).
In situ hybridization.
RNA isolated from an adult C57BL/6 mouse was used to generate cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche). The cDNA template was amplified by PCR with T3 viral promoter sequences at the 5′ end of the reverse primer (Table 1). Probes were ∼500–600 nt and had <70% sequence identity to other paralogs [based on National Center for Biotechnology Information (NCBI) blastn (Basic Local Alignment Search Tool)]. PCR product was ligated into either the pCR2.1 Vector (TA Cloning Kit, Life Technologies) or pGEM-T Easy Vector (Promega) and transformed into One Shot TOP10 Chemically Competent Escherichia coli (Invitrogen). Each probe was verified by Sanger sequencing. T3 RNA polymerase (Roche) was used to generate antisense probes labeled with digoxigenin-11-UTP according to the manufacturer's instructions. To increase sensitivity for Lpar1, Slc22a3, Gria4, and Htr1a, two probes corresponding to different regions of the transcript were cohybridized to endogenous mRNA. Sections were treated for 10 min with 4% PFA in PBS (Electron Microscopy Sciences) and washed 3 × 3 min with 0.1% diethylpyrocarbonate (DEPC; Sigma-Aldrich) containing PBS. Sections were then incubated in 10 μg/ml proteinase K (Ambion) in 0.05 m Trizma buffer, 0.0156 m EDTA, pH 7.4, for 16 min. Sections were fixed for 5 min in 4% PFA and washed 3 × 3 min in DEPC-PBS. Sections were incubated for 10 min in 0.25% acetic acid anhydride (Sigma-Aldrich) v/v in 0.1 m triethanolamine (Sigma-Aldrich), pH 8.0. Slides were washed 3× in DEPC-PBS and prehybridized for 2 h in hybridization buffer [50% formamide (Roche), 5× SSC buffer (ThermoFisher Scientific), 5× Denhardt's solution (ThermoFisher Scientific), 250 μg/ml yeast RNA (Sigma-Aldrich), 500 μg/ml salmon sperm DNA (Sigma-Aldrich)]. Slides were incubated 8–16 h at 65°C with digoxigenin-11-UTP-labeled probe in hybridization buffer covered by Hybrislip coverslips (Life Technologies). Slides were washed twice for 1 h in 2× SSC, 50% formamide, at 65°C followed by 10 min in 1× SSC at 37°C. Slides were equilibrated at room temperature for 10 min in buffer B1 (0.1 m Trizma, 0.15 m NaCl, pH 7.5). Slides were blocked for 1 h in 10% heat-inactivated goat serum in buffer B1 and incubated at 4°C overnight in 1:2500 anti-digoxigenin-ap fab fragments (Roche) in 5% goat serum in buffer B1. Slides were washed 5 × 5 min in buffer B1 and incubated for 10 min in buffer B2 (0.1 m Trizma, 0.1 m NaCl, 50 mm MgCl2, and 2 μm levamisole). Slides were developed in a chromogen solution [340 mg/ml 4-nitro blue tetrazolium chloride (Roche), 180 mg/ml BCIP 4-toluidine salt (Roche) in buffer B2 for 6–24 h]. Slides were then fixed in 4% PFA for 10 min and serially dehydrated in 50, 60, 70, 80, 90, and 100% ethanols followed by d-limonene (MP Biochemicals) and mounted using VectaMount Permanent Mounting Media (Vector Laboratories). All histology was performed on sex-matched littermate controls.
Table 1.
Primers used to generate in situ probes
| Gene | Reverse primer containing T3 sequence | Forward primer | Probe size |
|---|---|---|---|
| Adra1b | AGAATTAACCCTCACTAAAGGGTTTGTCCCCTTGAGACCTTGTG | GAGGCTGCGCTTACACCTAC | 544 bp |
| Cited1 | AGAATTAACCCTCACTAAAGGGATCAGGTGAGGGGTAGGATG | ACTCTGAAGCGAGTGGCTAG | 472 bp |
| En1 | AGAATTAACCCTCACTAAAGGGGCAAGGTAGGGAGGAGG | CAAGAACGGCCTGGCGCTG | 539 bp |
| Gnaq | AGAATTAACCCTCACTAAAGGGGATGCCACCTGCCAGTAAAC | CGGGAAGGGCTTCTTAGTTTTGTAC | 524 bp |
| Gria1 | AGAATTAACCCTCACTAAAGGGCCCTCGTTGAGAAGCGTCTTC | GTTATCATTGGCAATGGACGAGC | 504 bp |
| Gria2 | AGAATTAACCCTCACTAAAGGGATTGAAGGTAGCTGTTCAGATAC | CTACATGTTACCATGTAGAATGCAAGTGG | 540 bp |
| Gria3 | AGAATTAACCCTCACTAAAGGGTTTAGGAATGCGTCTTAGTCTTTGC | TTTGGAGAACTCCCCATAGTGGAG | 516 bp |
| Gria4-1 | AGAATTAACCCTCACTAAAGGGCTTGTCTCAGCGGAAG | CACAGGAACTGCGATTAGACAGAG | 505 bp |
| Gria4-2 | AGAATTAACCCTCACTAAAGGGAACTGTATCATTGAACTCATTCATC | TGTGGGTTTTGTACTCACCC | 526 bp |
| Hcrtr1 | AGAATTAACCCTCACTAAAGGGAAGGGCTGAGCCCAAATTAGTTC | TCCTCTGCCAGACACAAGTCCTTG | 559 bp |
| Htr1a-1 | AGAATTAACCCTCACTAAAGGGCACCATAACCCAAAGTAGTTC | GGAAGAAGTGGAGGAAGAGTGTAG | 550 bp |
| Htr1a-2 | AGAATTAACCCTCACTAAAGGGCGAAGTTCAGATATAACGCAAAACC | CACGTAGAGGAGGTAAGGAGCAAAAG | 534 bp |
| Lpar1-1 | AGAATTAACCCTCACTAAAGGGTCATAGCATGACATTAGAGACCTTAGTCTCC | TATTGGAAGAATCTTGTGTATATAAAACTTTGCC | 499 bp |
| Lpar1-2 | AGAATTAACCCTCACTAAAGGGAAGCCTGTGTATACTCGATTG | GAGTTCAACTCTGCTATGAACCC | 538 bp |
| Nr3c1 | AGAATTAACCCTCACTAAAGGGTTACCGATCTTGATAGACCAGGCTGAC | AATCTGGGGAAAGGAAGGGGAC | 507 bp |
| Nxph4 | AGAATTAACCCTCACTAAAGGGCATACTGCGGCGTGGGCAATAC | TCGCGCGCTTTTAATTGCCAC | 550 bp |
| Pet-1 | AGAATTAACCCTCACTAAAGGGTTAATGGGGCTGAAAGGGATA | CCAGTGACCAATCCCATCCTC | 513 bp |
| Scn3b | AGAATTAACCCTCACTAAAGGGAGATGGTGAAGATTTTAAGTTGGCTG | GAGACAAAGATAGGTGTGTAGCTTC | 516 bp |
| Slc6a17 | AGAATTAACCCTCACTAAAGGGCCTCCAGATTGGAGATGTCCTTCATC | ATGATCGGGACCATGGCAG | 586 bp |
| Slc22a3-1 | AGAATTAACCCTCACTAAAGGGTGTATTCGGAGCTCAAGGGAAG | CAGACGGTGGAAGACGTAG | 535 bp |
| Slc22a3-2 | AGAATTAACCCTCACTAAAGGGATGGCTAGCTGGATGCCCTG | AGCTCCACGTCCCAGCTGAG | 577 bp |
| Tph2 | AGAATTAACCCTCACTAAAGGGATCCATCCCAACTGCTGTGT | CTACACGCAGAGCATTGAA | 658 bp |
Imaging
Slides were imaged on an Olympus Optical BX51 microscope using a SPOT RT color digital camera (Diagnostic Instruments). Images were converted to gray scale and brightness and contrast were adjusted using ImageJ (http://rsb.info.nih.gov/ij) across the entire image.
Prazosin binding assay
Prazosin binding was performed as previously described (Green et al., 2015). Homogenized midbrain membranes were incubated with [3H] prazosin [6 nm] in the presence or absence of prazosin [10 μm] to measure nonspecific and total binding, respectively. Reaction buffer included the α 1A receptor antagonist WB 4101 [10 μm]. Specific binding was calculated by subtracting nonspecific binding from total binding and expressed as bound ligand (femtomole) per milligrams protein. All experiments were performed in duplicate. Significance was calculated by a two-tailed t test for independent samples.
Viral injections
For P0 injection, pups were cryoanesthetized for the entire procedure. Pups were injected bilaterally with Stoelting standard stereotaxic instrument using coordinates from lambda (x = ±0.5 mm; y = −2 mm; z = −2.5 mm) with 0.5 μl AAV1.CMV.Pl.eGFP.WPRE.bGH (AAV1-GFP)/side or 0.5 μl AAV1.CMV.Pl.Cre.rBG (AAV-Cre)/side (University of Pennsylvania Viral Core). P22 pups and adults were anesthetized with isoflurane and bilaterally injected using the following coordinates: P22: x = ±0.5 mm; y = −1.0 mm; z = −3.5 mm; P60 and older: x = ±0.5 mm; y = 0.0 mm, z = −4.2 mm. Knock down of Pet-1 was verified by taking every third or fourth section and processing for Pet-1 expression by in situ hybridization (ISH).
RNA-seq and ChIP-seq NCBI Gene Expression Omnibus accession
All RNA-seq and ChIP-seq data are available at the NCBI Gene Expression Omnibus under accession code GSE74315.
Results
Gene expression trajectories in maturing 5-HT neurons
To study 5-HT neuron maturation, we first used RNA-seq in flow-sorted EYFP+ 5-HT neurons to temporally profile their genome-wide patterns of gene expression. EYFP+ 5-HT neurons were flow sorted at E11.5, when the vast majority of 5-HT neurons are born (Pattyn et al., 2003); E15.5, when 5-HT neurons are actively extending dendrites and axons; and postnatal days 1–3 (PN) when 5-HT neurons have coalesced into mature raphe nuclei and are acquiring mature functional properties. We isolated total RNA from three biological replicates at each time point and synthesized cDNA libraries suitable for mRNA-seq. We obtained an average of 37 million uniquely mapped paired-end reads to the mouse transcriptome for each biological replicate at each time point. Using time-series differential expression analysis, we identified 6126 genes whose expression changed ≥1.5-fold at ≤5% FDR from E11.5 to E15.5 and from E15.5 to PN. These data indicated that global changes in gene expression occur as newly born 5-HT neurons begin to establish synaptic connectivity and are acquiring mature functional characteristics.
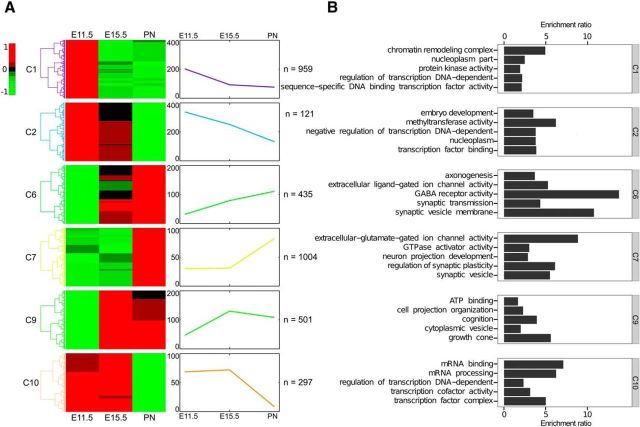
Unsupervised hierarchical clustering of genes with significantly altered expression across maturation revealed discrete groups that share highly similar temporal expression patterns characterized by either ascending or descending mean trajectories (Fig. 1A). GO analyses were used to predict shared function of expression clusters. Three ascending clusters, C6, C7, and C9, were significantly enriched for GO annotation terms associated with maturation of neuronal morphology and function: axonogenesis, growth cone, extracellular glutamate gated ion channel, synaptic vesicle membrane, and synaptic transmission (Fig. 1B). In contrast, clusters C1, C2, and C10 displayed descending trajectories and are enriched for GO terms associated with downregulation of earlier developmental programs involved in progenitor proliferation and specification (Fig. 1B). These data suggest that while earlier specification programs are downregulated, new programs commence to control morphogenesis and acquisition of mature functional properties.
Figure 1.
RNA-seq reveals temporal gene expression patterns in maturing 5-HT neurons. A, Total RNA from flow-sorted E11.5, E15.5, and P1–P3 (PN) YFP+ 5-HT neurons (n = 3 biological replicates/time point) was used for sequencing to determine expression patterns followed by hierarchical clustering of differentially expressed genes. Row-mean-normalized heat maps (left) and mean temporal expression levels (right) are shown for each cluster. Number of genes in each cluster is shown on the right of each trajectory. In total, 6126 genes were differentially expressed at ≥1.5-fold change and ≤5% FDR. B, GO enrichment analysis of gene expression clusters. GO terms enriched in clusters 6, 7, and 9 suggest increasing expression of genes related to neuronal maturation processes (hypergeometric test, p ≤ 0.01). GO terms enriched in clusters 1, 2, and 10 are associated with downregulation of earlier developmental programs involved in progenitor proliferation and specification (hypergeometric test, p ≤ 0.01).
Pet-1 broadly coordinates gene expression trajectories during maturation
To determine the regulatory strategies through which dynamic gene expression patterns are controlled in maturing 5-HT neurons, we focused on Pet-1 as it is continuously expressed in maturing 5-HT neurons and, unlike Lmx1b, Pet-1 is not required for their survival (Hendricks et al., 1999; Zhao et al., 2006; Krueger and Deneris, 2008; Kiyasova et al., 2011). Three biological replicates of flow-sorted +/+ and Pet-1−/− 5-HT neurons were collected at E15.5 and analyzed for differential expression by RNA-seq. As expected, expression of the known Pet-1-controlled 5-HT battery genes (Tph2, Slc6a4, Slc18a2, Htr1a, Gch1, Gchfr, Qdpr) were severely reduced in Pet-1−/− versus +/+ (data not shown) mice, thus validating our RNA-seq approach.
Loss of Pet-1 caused extensive changes in expression trajectories with hundreds of genes displaying substantially decreased as well as increased expression (Fig. 2A). Consistent with Pet-1 driving 5-HT neuron maturation, GO analysis showed enrichment for terms related to neuron growth and function (Fig. 2B). We manually annotated genes exhibiting significantly increased or decreased expression. A large number of TFs/chromatin modifiers, G-protein-coupled receptors (GPCRs), ion channels, transporters, and cell adhesion/axon guidance, peptide, synaptic, and broadly expressed genes showed significantly altered expression in Pet-1−/− 5-HT neurons (Fig. 2C). ISH verified Pet-1's broad regulatory scope (Fig. 2D). We also verified potential repression of some genes by Pet-1 and found that expression of the orexin receptor gene, Hcrtr1, was substantially upregulated in the Pet-1−/− DRN, but not in nonserotonergic sites of Hcrtr1 expression (Fig. 2D). These findings demonstrate that, in addition to controlling the 5-HT gene battery for acquisition of 5-HT transmitter identity, Pet-1 also positively and negatively regulates many disparate functional categories of genes as 5-HT neurons mature.
Figure 2.
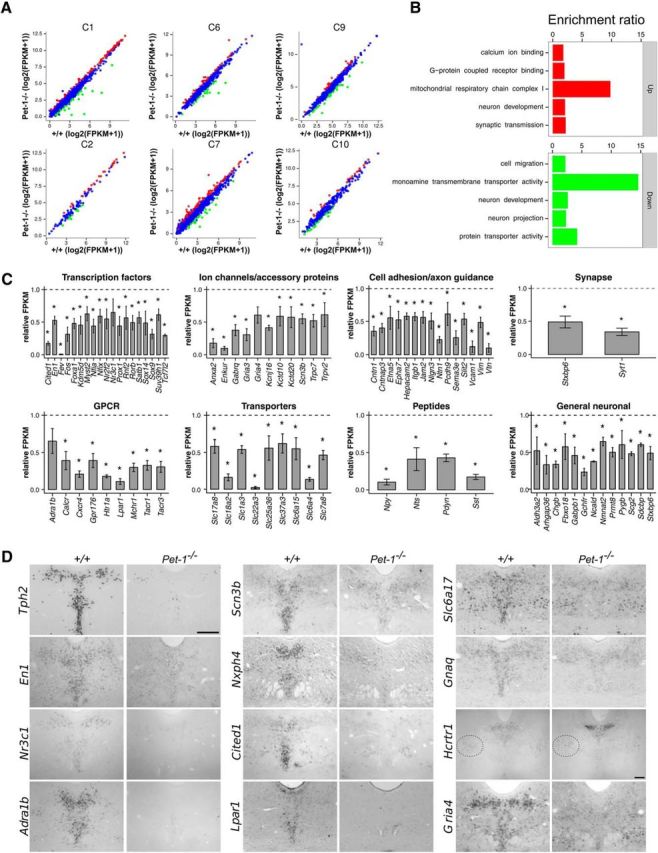
RNA-seq shows that Pet-1 globally controls the 5-HT transcriptome through positive and negative regulation of gene-expression trajectories. A, Scatterplots showing altered expression of genes in Pet-1−/− versus +/+ 5-HT neurons in each expression cluster (Fig. 1). B, GO enrichment analysis of genes upregulated (top) and downregulated (bottom) by Pet-1. C, Relative changes in expression (FPKM) for various categories of Pet-1-controlled genes. *, Benjamini–Hochberg q value ≤0.05; n = 3. Error bars are SEM. D, ISH verification of genes regulated by Pet-1. Scale bar, 300 μm. Dotted oval shows nonserotonergic site of Hcrtr1 expression.
Pet-1−/− 5-HT neuron passive and active membrane properties are permanently immature
To directly test whether Pet-1 was required for functional maturation of postmitotic 5-HT neurons, we used whole-cell patch-clamp electrophysiology to assess passive and active intrinsic membrane characteristics of Pet-1−/− 5-HT neurons. Previous studies demonstrated that 5-HT neuron passive and active membrane characteristics mature postnatally and do not exhibit maturity until ∼P21 (Rood et al., 2014). Thus, we directly compared functional membrane characteristics in Pet-1−/− and +/+ slices obtained from P21 mice and also in slices from adult mice to examine whether any differences persisted. Although not different at P21, resting membrane potential was significantly depolarized in slices obtained from Pet-1−/− adults compared with adult +/+ slices (Fig. 3B1), thus indicating an immature functional characteristic of Pet-1−/− 5-HT neurons as defined previously (Rood et al., 2014). In P21 slices, membrane resistance was increased in Pet-1−/− 5-HT neurons compared with membrane resistance in +/+ slices (Fig. 3B2), which further corresponds to an immature functional state (Rood et al., 2014). In addition, the membrane time constant, tau, was also increased in Pet-1−/− 5-HT neurons compared with tau in +/+ slices (Fig. 3B3).
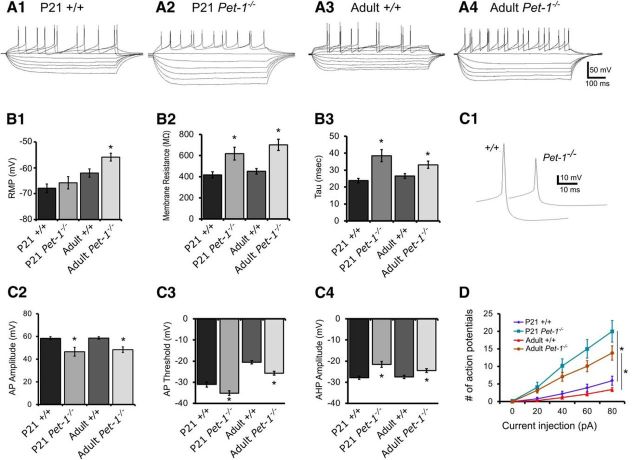
Figure 3.
Pet-1−/− 5-HT neuron passive and active membrane properties are permanently immature. A, Whole-cell recordings of membrane voltage responses to hyperpolarizing and depolarizing current injection from P21 and adult +/+ and Pet-1−/− 5-HT neurons in the DRN. B, Passive membrane properties of Pet-1−/− 5-HT neurons are functionally immature. B1, Resting membrane potential (RMP; ANOVA F(3,97) = 9.177, p < 0.0001; n = 26, 11, 37, 27; Student–Newman–Keuls post hoc test: adult Pet-1−/− vs adult +/+ 5-HT neurons was significantly different). B2, Membrane resistance (ANOVA F(3,97) = 11.92, p < 0.0001; n = 26, 11, 37, 27; Student–Newman–Keuls t test: P21 and adult Pet-1−/− 5-HT neurons were significantly different from P21 and adult +/+ 5-HT neurons, respectively). B3, Membrane time constant (tau; ANOVA F(3,97) = 9.574, p < 0.0001; n = 26, 11, 37, 24; Student–Newman–Keuls t test: P21 and adult Pet-1−/− 5-HT neurons were significantly different from P21 and adult +/+ 5-HT neurons, respectively, p < 0.05. C, Persistent immaturity of AP characteristics in P21 and adult Pet-1−/− 5-HT neurons. C1, Representative raw data traces of an AP recorded from a P21 +/+ and a P21 Pet-1−/− 5-HT neuron. C2, AP amplitudes in P21 and adult +/+ and Pet-1−/− 5-HT neurons (ANOVA F(3,95) = 10.25, p < 0.0001; n = 26, 11, 35, 27; Student–Newman–Keuls t test confirmed that AP amplitude in P21 and adult Pet-1−/− 5-HT neurons was significantly smaller than P21 and adult +/+ 5-HT neurons, respectively. C3, Pet-1−/− 5-HT neuron AP firing threshold was significantly more hyperpolarized in P21 and remained more hyperpolarized in adult Pet-1−/− 5-HT neurons (F(3,95) = 37.92, p < 0.0001; n = 26, 11, 35, 27; Student–Newman–Keuls t test confirmed that P21 and adult Pet-1−/− 5-HT neurons more hyperpolarized than P21 and adult +/+ 5-HT neurons, respectively, p < 0.05). C4, Afterhyperpolarization amplitudes were smaller in P21 and adult Pet-1−/− 5-HT neurons (F(3,95) = 8.153, p < 0.0001; n = 26, 11, 35, 27; Student–Newman–Keuls test confirmed that P21 and adult Pet-1−/− 5-HT neurons were smaller than P21 and adult +/+ 5-HT neurons, respectively. D, Excitability of Pet-1−/− 5-HT neurons. Increased numbers of APs were elicited with depolarizing current pulses in P21 and adult Pet-1−/− 5-HT neurons compared with P21 and adult +/+ 5-HT neurons (two-way ANOVA significant interaction; ANOVA F(12,296) = 13.13, p < 0.0001; n = 26, 11, 17, 24).
Several active membrane properties were also altered in Pet-1−/− mice (Fig. 3C) in a manner consistent with an immature functional state (Rood et al., 2014). AP amplitude was significantly decreased while AP firing threshold was more hyperpolarized and afterhyperpolarization amplitude was decreased in Pet-1−/− 5-HT neurons (Fig. 3C1–C4). These changes persisted into adulthood, indicating permanent immaturity. Most of these parameters combine to govern neuron excitability, thus changes in these characteristics would suggest changes in excitability. Depolarizing current pulse injection revealed significantly greater and permanent current-induced excitability of Pet-1−/− neurons (Fig. 3D). Together our findings reveal numerous altered passive and active membrane characteristics of Pet-1−/− neurons, indicative of an immature stage of 5-HT neurons (Rood et al., 2014), which therefore uncovers a crucial role for Pet-1 in programming their functional maturation.
Pet-1 controls maturation of glutamatergic and GPCR synaptic input to 5-HT neurons
GO analyses revealed significant enrichment in ascending clusters C6 and C7 for terms/genes associated with extracellular ligand-gated ion channel activity and glutamate-gated ion channel activity, respectively (Fig. 1B), which suggested that a key step in 5-HT maturation is expression of ionotropic receptors required for proper afferent control of 5-HT neuronal firing and transmitter release (Maejima et al., 2013). A major source of direct excitatory synaptic input to 5-HT neurons is glutamatergic afferents acting via AMPA/kainate receptors (Crawford et al., 2011). However, little is known about the specific glutamate receptor subtypes expressed in 5-HT neurons and how their expression is controlled.
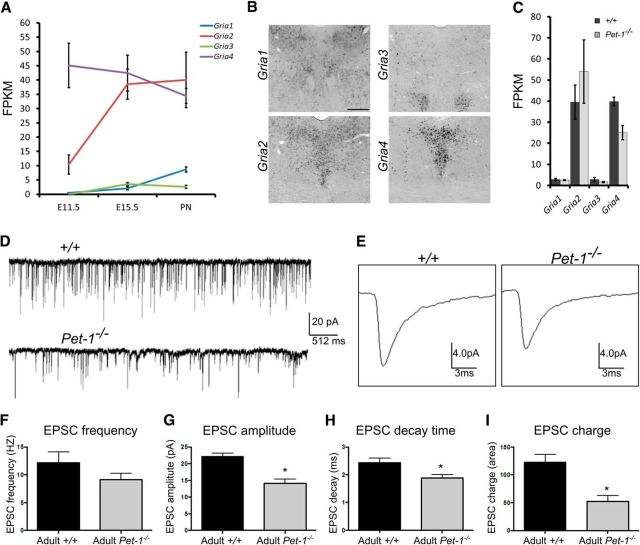
RNA-seq analyses indicated that Gria2 and Gria4 were the principal AMPA receptor (AMPAR) subunit genes expressed in maturing 5-HT neurons, while Gria1 and Gria3 were expressed at much lower levels throughout the E11 to P3 stage of maturation (Fig. 4A). ISH verified strong Gria2 and Gria4 expression with weak expression of Gria1 in the adult DRN; Gria3 expression was not detected in adult DRN 5-HT neurons (Fig. 4B). RNA-seq revealed that Pet-1 deficiency resulted in a specific decrease in Gria4 expression (Fig. 4C), which was verified by ISH (Fig. 2D).
Figure 4.
Pet-1 promotes maturation of AMPA excitatory synaptic input to 5-HT neurons by regulating Gria4. A, RNA-seq analysis of AMPAR subunit gene (Gria1–4) expression trajectories (n = 3). B, ISH for Gria1–Gria4 in control mice. C, FPKMs for Gria1–4 in +/+ and Pet-1−/− sorted E15.5 5-HT neurons (n = 3). D–E, Raw traces of EPSC synaptic activity (D) and averaged (from 200 individual events) single EPSC current events (E) recorded under voltage-clamp conditions. F–I, EPSC input onto adult Pet-1−/− 5-HT neurons is characteristic of immature EPSCs at early postnatal stages. F, EPSC frequency was not different; however, the variances differed significantly (Pet-1−/−: 9.132 ± 1.125, N = 17; +/+: 13.75 ± 2.458, N = 22; F = 6.179, p < 0.0005; n = 22, 17). EPSC events in Pet-1−/− 5-HT neurons on average had smaller amplitudes (G; t test, t = 5.102, df = 37, p < 0.0001; n = 22, 17), shorter decay time (H; t test, t = 2.461, df = 37, p = 0.0186; n = 22, 17), and smaller charge (I; t test, t = 3.755, df = 37, p = 0.0006; n = 22, 17). Error bars are SEM.
The control of Gria4 by Pet-1 suggested that Pet-1 promotes maturation of functional AMPAR responses in 5-HT neurons. To test this idea, we performed whole-cell recordings in slices obtained from +/+ and Pet-1−/− slices (Fig. 4D–I). Although EPSC frequency was not altered, we found a significant decrease in the variability of EPSC frequency, indicating decreased glutamatergic input to Pet-1−/− neurons (Fig. 4F). Moreover, Pet-1−/− neurons exhibited an overall decrease in EPSC amplitude (Fig. 4G) and shortening of the EPSC decay time (Fig. 4H), leading to a reduction in the charge carried by each current (Fig. 4I). Together these findings suggest decreased postsynaptic AMPAR numbers and therefore Pet-1 is required for functional maturation of excitatory glutamatergic input onto 5-HT neurons through its specific control of the Gria4 expression trajectory.
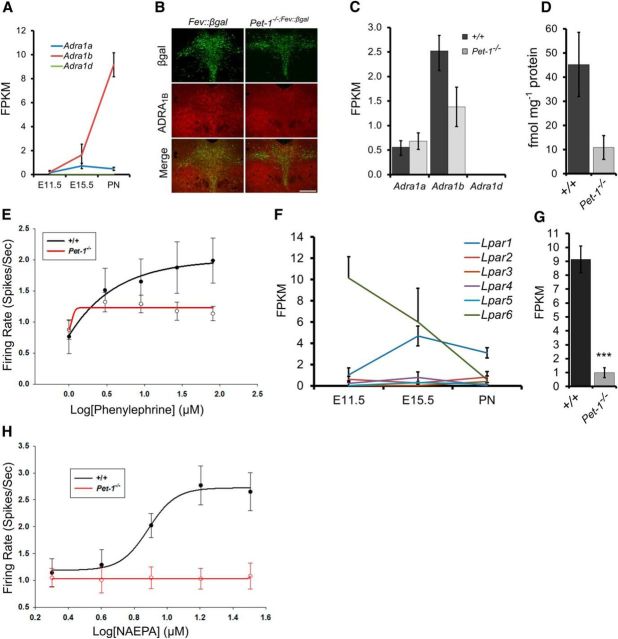
To determine whether Pet-1 plays a broader role in coordinating maturation of afferent synaptic responsivity in 5-HT neurons, we examined GPCR expression in our RNA-seq datasets. A critical synaptic input to the 5-HT system originates from noradrenergic neurons that drive tonic firing of 5-HT neurons through α1 adrenoceptors (Vandermaelen and Aghajanian, 1983). Adra1b expression was substantially increased during the maturation phase while Adra1a and Adra1d expression was weak or undetectable (Fig. 5A), which is consistent with previous studies (Day et al., 2004). Thus, late-fetal–early-postnatal upregulation of Adra1b appears to be a key event in the functional maturation of 5-HT neurons (Fig. 5A). Immunostaining (Fig. 5B), gene expression (Fig. 5C), and membrane ligand binding with the α1 adrenergic receptor antagonist [3H]-prazosin (Fig. 5D) revealed loss of the receptor in Pet-1−/− DRNs. Furthermore, multielectrode array (MEA) recordings demonstrated that Pet-1−/− 5-HT neurons lacked functional α1 adrenergic receptors as they failed to display an increase in excitability in response to increasing doses of phenylephrine, a selective α 1 adrenergic receptor (ADRA1) agonist (Fig. 5E).
Figure 5.
Pet-1 controls maturation of GPCR synaptic input to 5-HT neurons. A, RNA-seq analysis of α1 adrenergic receptor gene-expression trajectories in flow-sorted +/+ 5-HT neurons. B, coimmunostaining of β-galactosidase (eFev::lacZ) marked 5-HT neurons (green) and ADRA1B (red). C, RNA-seq analysis of Adra1 receptor gene expression in +/+ and Pet-1−/− neurons at E15.5. D, [3H]-prazosin binding in +/+ versus Pet-1−/− midbrain (t(5) = 2.43, p = 0.07; n = 3. E, MEA recordings of α1-selective agonist phenylephrine (PE) responses. +/+, n = 19 cells/6 mice; Pet-1−/−, n = 24 cells/9 mice. F, RNA-seq analysis of Lpar1–6 expression trajectories in flow-sorted +/+ 5-HT neuron. G, FPKMs for Lpar1 in +/+ and Pet-1−/− sorted E15.5 5-HT neurons (n = 3). H, MEA recording of LPA1 selective agonist, (Z)-N-[2-(phosphonooxy)ethyl]-9-octadecenamide (NAEPA). +/+, n = 27 cells/4 mice; Pet-1−/−, n = 9 cells/3 mice. Scale bar, 300 μm. ***p < 0.001.
Lysophosphatic acids (LPAs) are critical lipid signaling molecules in the nervous system. LPAs have been implicated in the regulation of neural development and cognition through six different GPCRs, LPA1–LPA6 (mouse gene names Lpar1–Lpar6; Yung et al., 2015). In contrast to other Lpar genes, Lpar1 expression increased from E11.5 to E15.5 with little change in expression level at birth (Fig. 5F). RNA-seq (Fig. 5G) indicated a loss of Lpar1 expression in Pet-1−/− 5-HT neurons and ISH verified that Pet-1 was required for Lpar1 expression in the DRN (Fig. 2D). MEA recordings indicated that in contrast to 5-HT neurons in +/+ slices, 5-HT neurons in Pet-1−/− slices did not exhibit a dose–response relationship with (Z)-N-[2-(phosphonooxy)ethyl]-9-octadecenamide, a selective LPA1 agonist (Fig. 5H).
We next investigated whether in addition to coordinating GPCR expression, Pet-1 controls downstream G-protein signaling, which also develops postnatally in 5-HT neurons (Rood et al., 2014). Voltage-clamp recordings were used to test activation of G-protein-coupled channels with the nonhydrolysable GTP analog, GTPγs (Rood et al., 2014). The magnitude of the response elicited by GTPγs in Pet-1−/− 5-HT neurons was significantly lower than the response observed in control 5-HT neurons (Fig. 6), which suggests immature G-protein to GIRK channel signaling in Pet-1−/− 5-HT neurons. Our collective electrophysiological findings presented in Figures 4, 5, and 6 demonstrate that Pet-1−/− 5-HT neurons fail to acquire mature ionotropic and GPCR synaptic pathways that provide for extrinsic control of 5-HT neuron excitability.
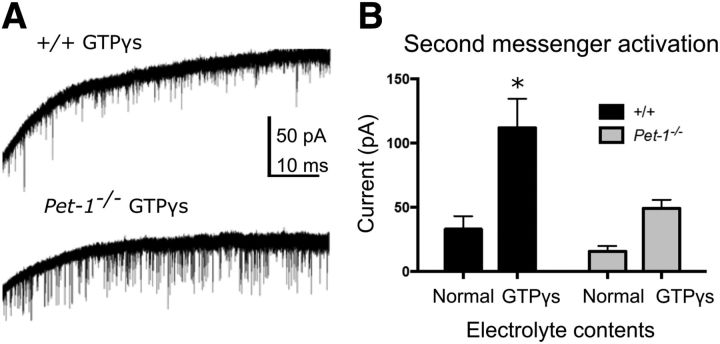
Figure 6.
Immature G-protein signaling in Pet-1−/− 5-HT neurons. A, Representative image of GTPγS elicited responses in +/+ and Pet-1−/− 5-HT neurons. B, Quantification of GTPγS elicited response. ANOVA aCSF content F(1,31) = 13.59 p = 0.0009; genotype F(1,31) = 6.798, p = 0.0139; n = 4, 9, 6, 16. Student–Newman–Keuls t tests indicated that +/+ normal versus Pet-1−/− normal was not significantly different, +/+ normal versus +/+ GTPγS electrolyte was significant (*p < 0.05), and Pet-1−/− normal vs Pet-1−/− GTPγS was not significant.
Stage-specific switching of Pet-1 targets
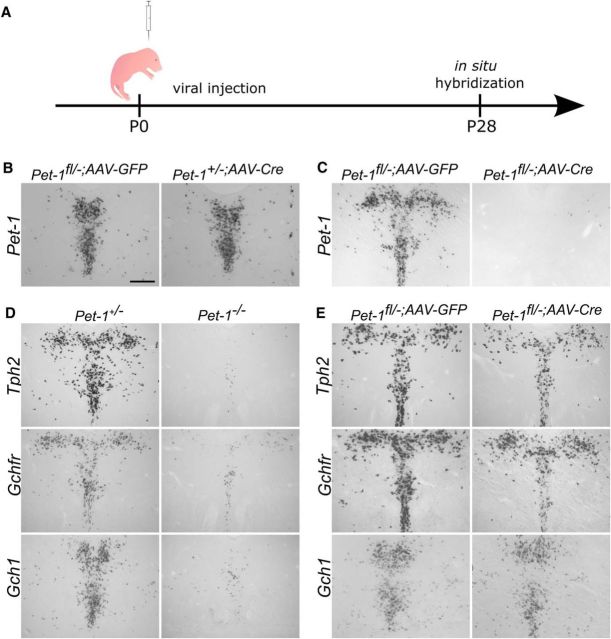
The continuous expression of Pet-1 led us to inquire whether its function is required specifically in the early postnatal period when excitability of 5-HT neurons is maturing. Thus, we developed an adeno-associated viral (AAV)-mediated Cre/loxP approach to target Pet-1 at different postnatal stages (Fig. 7A). Pet-1fl/− mice at P0 were stereotaxically injected with either AAV-Cre (Pet-1fl/−;AAV-Cre) or AAV-GFP (Pet-1fl/−;AAV-GFP). Four weeks following injection, Pet-1 expression was assayed by ISH throughout the DRN. Neither AAV-GFP injection into the Pet-1fl/− midbrain nor AAV-Cre injected into Pet-1+/− midbrain had any effect on the expression of Pet-1 (Fig. 7B). In contrast, AAV-Cre injections into the Pet-1fl/− brain eliminated >95% of Pet-1 expression throughout the entire DRN (Fig. 7C).
Figure 7.
5-HT synthesis genes lose sensitivity to Pet-1 as 5-HT neurons mature. A, Experimental scheme: Pet-1fl/− mice were injected with AAV-Cre or AAV-GFP to conditionally delete Pet-1 in the early postnatal period followed by ISH at P28. B, ISH for Pet-1fl/− and Pet-1+/− injected with AAV-GFP or AAV-Cre respectively. C, ISH for Pet-1 showing that AAV-Cre eliminates Pet-1 expression in the DRN. D, ISH reveals nearly complete elimination of expression of 5-HT synthesis genes Tph2, Gchfr, and Gch1 in the Pet-1−/− DRN. E, ISH in P0 AAV-injected mice reveals nearly total insensitivity of Tph2, Gchfr, and Gch1 expression to loss of Pet-1. Scale bar, 300 μm.
We next investigated the temporal requirements for Pet-1 in the control of 5-HT neuron transmitter identity. ISH assays verified that expression of 5-HT synthesis genes Tph2, Gch1, and Gchfr was nearly eliminated in Pet-1−/− mice (Fig. 7D). Unexpectedly, however, we found a dramatic change in the sensitivity of these genes to Pet-1 deficiency in the early postnatal period: Tph2 and Gchfr expression was only slightly reduced, while Gch1 expression was not altered (Fig. 7E). These findings reveal a surprising temporal change in the dependence of 5-HT synthesis genes on Pet-1 as 5-HT neurons mature. We then examined Htr1a as its expression is low at the onset of 5-HT synthesis in newborn 5-HT neurons and is subsequently upregulated as 5-HT neurons mature, which is consistent with the postnatal development of the 5-HT1a autoreceptor pathway (Liu et al., 2010; Rood et al., 2014). In striking contrast to 5-HT synthesis genes, neonatal targeting of Pet-1 resulted in severely decreased expression of Htr1a in the DRN (Fig. 8A). Similarly, neonatal targeting of Pet-1 nearly eliminated upregulation of Adra1b expression and substantially reduced Gria4 expression specifically in the DRN (Fig. 8A). We also examined whether Pet-1 function was required in the early postnatal period to repress some targets. Indeed, Hcrtr1 expression was increased after neonatal targeting of Pet-1 (Fig. 8B). These findings reveal a stage-specific switch in Pet-1 targets from 5-HT synthesis genes to GPCR excitability genes during early postnatal 5-HT neuron maturation.
Figure 8.
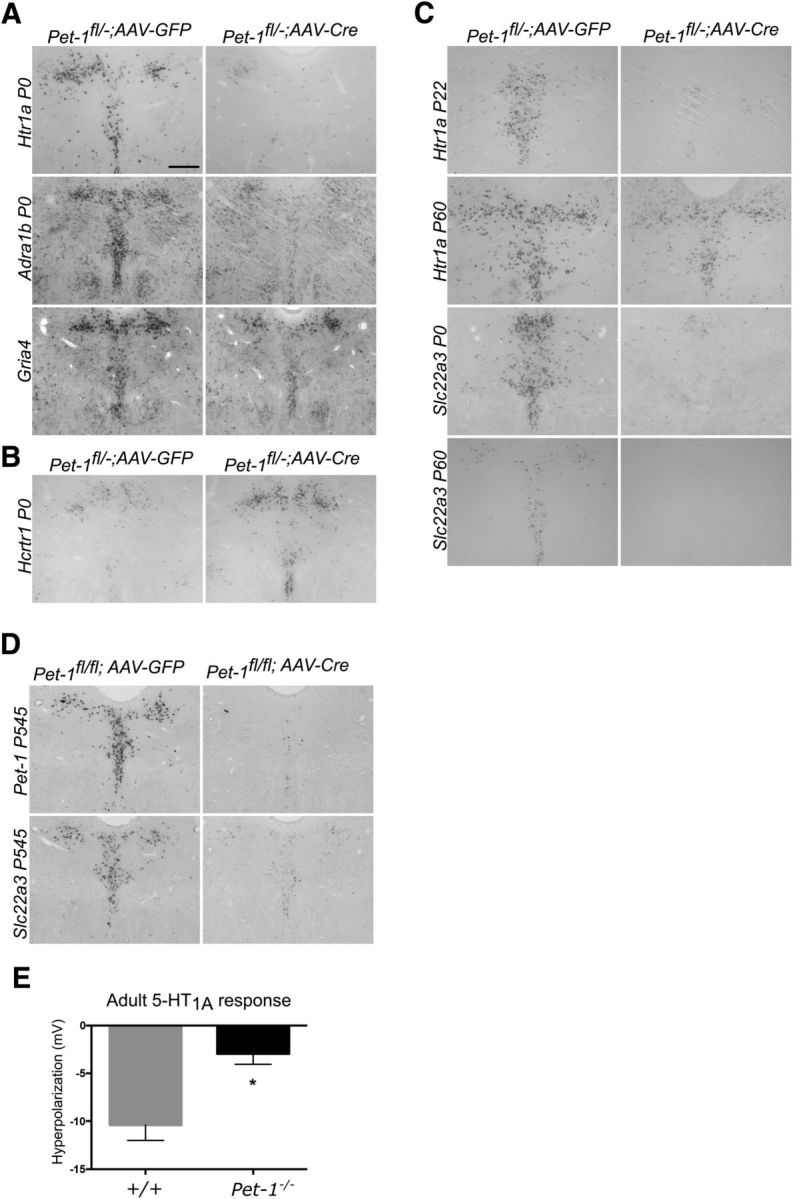
Early postnatal Pet-1 function is essential for control of multiple excitability genes. A, ISH of Htr1a, Adra1b, and, Gria4 expression in P0 injected mice. B, Neonatal targeting of Pet-1 results in increased Hcrtr1 expression in 5-HT neurons. C, Early postnatal transcriptional sensitive period for Htr1a control by Pet-1. Pet-1fl/− mice were injected with AAV-Cre or AAV-GFP at the indicated postnatal ages. ISH: Htr1a, P22 assayed at P43; Htr1a, P60 assayed at P180; Slc22a3, P0 assayed at P28; Slc22a3, P60 assayed at P90. D, ISH for Pet-1 and Slc22a3 of P545 injected mice assayed at P590. E, Permanent immaturity of 5-carboxamidotryptamine elicited responses in adult DRN slices. p = 0.0003, t(36) = 4.053. Pet-1−/−, n = 22; +/+, n = 16. Scale bar, 300 μm.
To further investigate stage-specific control of 5-HT excitability genes, we targeted Pet-1 at additional postnatal time points. AAV-Cre injection into Pet-1fl/− mice at P22 also led to a near complete loss of Htr1a expression (Fig. 8C). In contrast, when P60 Pet-1fl/− mice were injected with AAV-Cre, we found a markedly reduced sensitivity of Htr1a to loss of Pet-1 (Fig. 8C). The early life closing of Pet-1-dependent control of Htr1a was not due to a general loss of Pet-1 function as Slc22a3, the low-affinity high-capacity 5-HT transporter gene (Baganz et al., 2010), remained highly sensitive to loss of Pet-1 at P60 (Fig. 8C) and even in 18-month-old mice (Fig. 8D). Whole-cell recordings verified the Pet-1 dependence of 5-HT1a agonist responses in adult 5-HT neurons and further revealed that despite the eventual loss of Htr1a's sensitivity to Pet-1, compensatory restoration of autoreceptor function does not occur later in life (Fig. 8E). Together these findings uncover an early postnatal sensitive period for control of Htr1a expression by Pet-1.
Pet-1 directly controls the 5-HT neuron maturation-promoting factor, Engrailed 1
To investigate the regulatory mechanisms through which Pet-1 controls 5-HT neuron maturation, we performed ChIP-seq with the ePet-mycPet-1 mouse rescue line (Liu et al., 2010). We collected chromatin from 168 Pet-1-expressing rostral hindbrains of E12.5–E15.5 embryos to capture the early epoch of Pet-1 occupancy in 5-HT chromatin. With >27 million uniquely mapped sequencing reads from mycPet-1 immunoprecipitated DNA, we identified 4953 mycPet-1 ChIP peaks enriched over the input sample ≥2-fold (p ≤ 1.0e-05). As predicted for an ERG-type ETS domain TF (Wei et al., 2010), mycPet-1 ChIP peaks were enriched near TSSs, with 33% of peak regions located within 5 kb upstream of the TSS.
The MEME (Multiple Em for Motif Elicitation) suite was used to identify sequence motifs enriched within the mycPet-1 ChIP peaks (Bailey et al., 2009). Comprehensive examination in vitro has defined a position weight matrix (PWM) of Pet-1/FEV (Pet-1/Fifth Ewing Variant) high-affinity binding sites (Wei et al., 2010). Importantly, the most significant de novo enriched motif was an exact match to the Pet-1/FEV high-affinity binding site that provides for sequence-specific Pet-1/FEV transactivation or repression (Fyodorov et al., 1998; Maurer et al., 2003; Wang et al., 2013; Fig. 9A). We found that 82.8% of all mycPet-1 ChIP peaks contain ≥1 Pet-1/FEV high-affinity PWM hit (Fig. 9B; p ≤ 1.0e-03), which are highly phylogenetically conserved compared with PWM hits in random genomic regions (data not shown). The highly significant enrichment of high-affinity Pet-1/FEV motifs in mycPet-1 ChIP peaks provides strong independent validation of Pet-1 binding sites in vivo.
Figure 9.
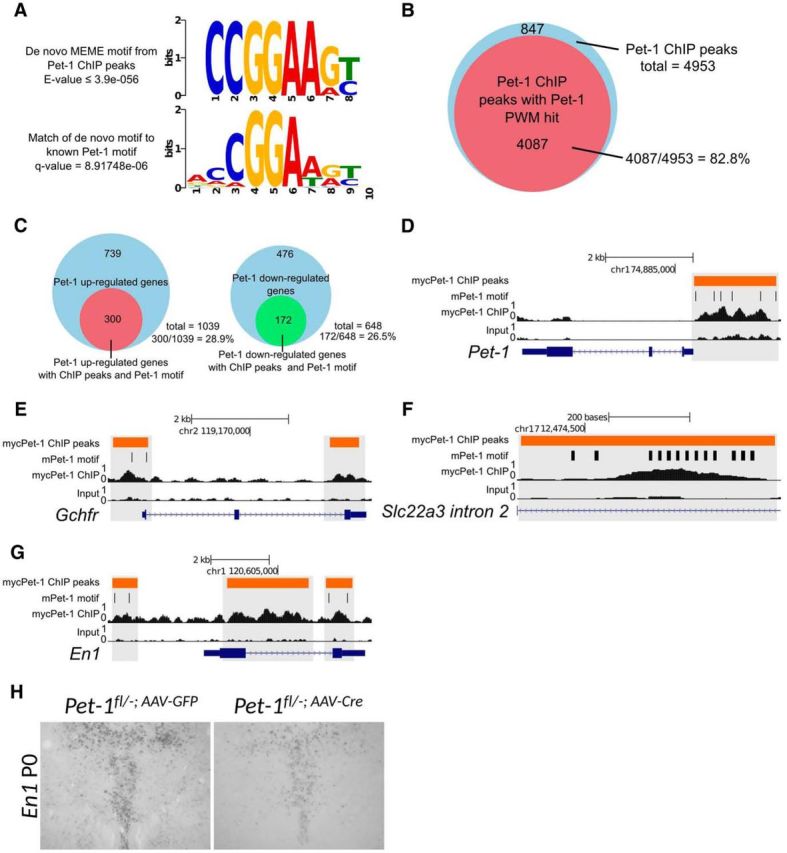
Pet-1 directly regulates the 5-HT neuron maturation factor Engrailed 1. A, De novo MEME motif analysis identifies the top significantly enriched motif in myc-Pet-1 peaks (top). TOMTOM Motif Comparison Tool identifies a highly significant match of the top enriched motif to Pet-1/FEV high-affinity binding site (bottom) defined in vitro (Wei et al., 2010). B, Fraction of mycPet-1 ChIP peaks with ≥1 match to the known Pet-1/FEV PWM motif. C, Fraction of Pet-1 upregulated (left) and downregulated (right) genes with mycPet-1 ChIP peaks within 5 kb from the TSS or TTS. D–G, Genome browser screen shots showing mycPet-1 enrichment over input control for Pet-1 (D), Gchfr (E), Slc22a3 (F), and En1 (G). Orange bars indicate area of significant peak enrichment. Black vertical lines indicate presence of Pet-1/FEV high-affinity motifs. H, P28 ISH for En1 of P0 Pet-1fl/− injected mice.
We identified a large number of Pet-1-regulated genes with ≥1 mycPet-1 peak containing the Pet-1/FEV high-affinity motif. Pet-1 itself and the 5-HT battery genes Slc22a3 and Gchfr had multiple mycPet-1 peaks with Pet-1/FEV high-affinity binding motifs (Fig. 9D–F). Interestingly, Slc22a3 possessed 13 Pet-1/FEV high-affinity motifs within a 626 bp region of mycPet-1 enrichment within intron 2 (Fig. 9F). Further, an additional 172 genes with decreased expression and 300 genes upregulated in Pet-1−/− 5-HT neurons were associated with ≥1 Pet-1/FEV motif containing mycPet-1 peak (Fig. 9C), suggesting that Pet-1 controls maturation of 5-HT neurons not only through direct gene activation, but also through direct repression.
Bioinformatic analyses with the Panther classification tool and GO term analyses revealed a significant enrichment for genes classified as nucleic acid binding or TF proteins in the intersected set of Pet-1-regulated genes with mycPet-1 occupancy near TSSs. Included were several TFs with well defined functions in the 5-HT neuron lineage (En1, Nkx2–2, Nr3c1) or in other postmitotic neuron types (Foxa1, Nr2f2; Pattyn et al., 2003; Espallergues et al., 2012; Fox and Deneris, 2012; Domanskyi et al., 2014; Jochems et al., 2015). For example, our previous studies showed that En1 is intrinsically required to control postmitotic 5-HT neuron identity, migration, and survival (Fox and Deneris, 2012). ChIP-seq revealed mycPet-1 peaks with Pet-1/FEV motifs upstream and downstream of the En1 TSS (Fig. 9G) and ISH analyses revealed that postmitotic expression of En1 critically depends on Pet-1 (Fig. 2D). Moreover, AAV-Cre targeting showed that Pet-1 was required in the early postnatal period to support continued postmitotic En1 expression (Fig. 9H). These results suggest that Pet-1 controls maturation of 5-HT neurons in part by directly regulating a known 5-HT neuron maturation factor and possibly several other potential 5-HT regulatory factors.
Discussion
The regulatory strategies through which continuously expressed TFs control postmitotic neuronal development are poorly understood. Using a combination of RNA sequencing of flow-sorted 5-HT neurons, electrophysiological studies, and conditional targeting approaches, we investigated Pet-1, a regulatory factor continuously expressed in 5-HT neurons, at different stages across fetal to early postnatal life. We report that in addition to its well known role in initiating brain 5-HT synthesis in newly born 5-HT neurons (Hendricks et al., 2003), Pet-1 subsequently plays a much broader role in coordinating global postmitotic expression trajectories of genes necessary for functional maturation of 5-HT neurons. RNA sequencing revealed that loss of Pet-1 led to altered expression of hundreds of genes encoding various TFs, GPCRs, ion channels, and transporters, among others. Whole-cell recordings indicated that several passive and active membrane properties of Pet-1−/− 5-HT neurons as well as G-protein to GIRK channel signaling were highly characteristic of immature neonatal 5-HT neurons (Rood et al., 2014). Further, Pet-1 was necessary for coordinating maturation of glutamatergic, adrenergic, serotonergic, and lipid excitatory synaptic input to 5-HT neurons through control of Gria4, Adra1b, Htr1a, and Lpar1 expression trajectories, respectively. This previously unrecognized extensive role for Pet-1 in postnatal 5-HT neuron maturation led us to probe the temporal requirements for Pet-1. Thus, we developed an AAV-Cre targeting approach to eliminate Pet-1 expression at different postnatal stages. Unexpectedly, we found that as 5-HT neurons mature, 5-HT synthesis genes Tph2, Gch1, and Gchfr lost sensitivity to Pet-1 but 5-HT excitability genes Htr1a, Adra1b, and Hcrtr1 critically depended on Pet-1 in early postnatal life. These distinct gene-specific temporal dependencies on Pet-1 reveal a previously unrecognized stage-specific regulatory strategy in which continuously expressed Pet-1 switches transcriptional targets to control maturation of 5-HT neuron excitability.
Previous studies showed that Pet-1 acts to induce expression of 5-HT synthesis genes Tph2, Gch1, and Gchfr at the serotonergic precursor stage and thereby initiate brain 5-HT synthesis (Hendricks et al., 2003; Wyler et al., 2015). Activation of these genes in Pet-1−/− precursors is severely reduced, which results in a deficiency of 5-HT in newly born 5-HT neurons. Interestingly, through conditional postnatal targeting of Pet-1, we found that the dependence of 5-HT synthesis genes on Pet-1 greatly diminishes in magnitude as 5-HT neurons progress from fetal to postnatal stages of development. Although Pet-1's control of Tph2 substantially diminishes by the early postnatal period, Tph2 is partially dependent on Pet-1 in adulthood and therefore its sensitivity to Pet-1 may fluctuate throughout life (Liu et al., 2010). In contrast to 5-HT synthesis genes, several GPCR genes were highly dependent on Pet-1 expression in the early postnatal period. Expression of Adra1b and Htr1a was nearly eliminated after neonatal targeting of Pet-1 in 5-HT neurons of the DRN; Hcrtr1 was substantially upregulated, suggesting ongoing repression by Pet-1 during maturation. The critical dependence of Adra1b and Htr1a on Pet-1 coincides with their postnatal upregulation and the stage at which 5-HT neurons begin to develop appropriate GPCR pathways needed for responses to diverse afferent synaptic input (Rood et al., 2014). These findings suggest that as postmitotic development proceeds there is a switch in Pet-1 targets from those required in newborn 5-HT neurons for initiation of 5-HT synthesis to those required postnatally for maturation of extrinsically controlled 5-HT neuron excitability. Further conditional targeting studies indicate that Htr1a expression becomes nearly independent of Pet-1 in young adulthood while other genes, such as Slc22a3, remain completely dependent on Pet-1 throughout life. These findings reveal a transcriptional sensitive period for Pet-1-dependent control of Htr1a expression. Although several TFs are required in postmitotic neurons for maintenance of gene expression (Kadkhodaei et al., 2013; Deneris and Hobert, 2014; Laguna et al., 2015), to our knowledge early postnatal transcriptional sensitive periods have not been described. How discrete gene-specific regulatory sensitive periods open and close is unclear, but may involve highly dynamic changes in chromatin accessibility that occurs at cis-regulatory elements as neurons mature (Ding et al., 2013; Frank et al., 2015).
The existence of a regulatory sensitive period for Htr1a is potentially significant in view of abundant evidence supporting the early postnatal period in rodents as a critical period for serotonergic control of behaviors related to depression, anxiety, and fear (Leonardo and Hen, 2008). Alterations in 5-HT signaling during the early postnatal period, but not in adulthood, can alter emotional behaviors later in life (Rebello et al., 2014). Moreover, suppression of Htr1a expression during the P14–P30 stage (but not in adulthood) resulted in increased anxiety-like behaviors and decreased social behaviors later in life, suggesting a neurodevelopmental critical period for 5-HT1a function (Donaldson et al., 2014). The transcriptional sensitive period we have uncovered for Htr1a coincides with the critical period for Htr1a function, which highlights the necessity of precise transcriptional control of Htr1a autoreceptor expression during the early postnatal maturation stage. These findings raise the possibility that the regulatory sensitive period for Htr1a represents a time-restricted window when Htr1a function is particularly susceptible to alterations in Pet-1-driven postnatal regulatory programs (Meredith et al., 2012). Disruption of the transcriptional controls on Htr1a within this window may precipitate life-long adverse consequences on emotional health.
The results presented here provide insight into vertebrate terminal selector-like function. Terminal selectors were originally described in Caenorhabditis elegans as TFs that are continuously expressed in postmitotic neurons and function to initiate expression of neuron-type identity features during development and then maintain those features later in life through direct binding to conserved cis-regulatory motifs (Hobert, 2008). Several vertebrate TFs that control acquisition of transmitter identity are continuously expressed in specific neuron types (Holmberg and Perlmann, 2012; Kadkhodaei et al., 2013; Deneris and Hobert, 2014; Allan and Thor, 2015). However, in most cases there has yet to be an in-depth analysis of their terminal selector properties.
Pet-1's functional characteristics in 5-HT neurons fulfill the defining criteria of a terminal selector-type TF (Cheng et al., 2003; Liu et al., 2010). In addition to developmental stage-specific switching of Pet-1 targets discussed above, our results further illuminate Pet-1's terminal selector characteristics. Pet-1 (FEV) possesses a strong autonomous transcriptional repressor domain in a conserved alanine-rich C-terminal region and can function as a transcriptional repressor through binding to ETS high-affinity binding sites at least in cell line reporter assays (Fyodorov et al., 1998; Maurer et al., 2003). Here, we present evidence in support of a prominent role for Pet-1-mediated repression in regulating 5-HT neuron maturation in vivo. Indeed, RNA-seq analyses identified a greater number of significantly upregulated genes than downregulated ones in Pet-1−/− 5-HT neurons. ChIP-seq revealed that a large number of upregulated genes in Pet-1−/− 5-HT neurons possessed ≥1 Pet-1/FEV high-affinity binding sites within mycPet-1 peaks situated near transcriptional start sites. Previous ChIP-seq studies of ETS factors reported that Pet1/FEV-like ETS factors occupied regions enriched for high-affinity ETS sequence motifs, suggesting high-affinity FEV ETS binding sites mediate ETS factor function in vivo (Wei et al., 2010). The homeodomain factor Nkx2-2 was significantly derepressed in Pet-1−/− 5-HT neurons and two mycPet-1 peaks were detected within 5 kb of the Nkx2-2 TSS. Nkx2-2 is expressed in the ventricular zone, where it is required for specification of 5-HT progenitors (Pattyn et al., 2003). These findings suggest that Pet-1 helps to suppress earlier progenitor specification programs by repressing Nkx2-2 as 5-HT precursors become postmitotic. Further, neonatal targeting of Pet-1 resulted in a dramatic upregulation of Hcrtr1 expression, suggesting ongoing Pet-1-mediated repression may set appropriate level of orexin input to 5-HT neurons.
Our results suggest that Pet-1 promotes 5-HT neuron maturation through direct regulation of secondary regulatory factors. This notion is well illustrated with evidence in support of En1 as a direct Pet-1 target. We showed previously that En1 is intrinsically required for maturation and survival of 5-HT neurons in the DRN (Fox and Deneris, 2012). RNA-seq and ISH analyses revealed that Pet-1 was essential for sustained expression of En1 in 5-HT neurons. ChIP-seq revealed multiple mycPet-1 peaks within and upstream of the En1 locus, with most possessing conserved high-affinity Pet-1/FEV binding sites. Thus, these findings indicate that similar to certain C. elegans terminal selectors (Etchberger et al., 2007), Pet-1 is a regulatory intermediary that directly controls secondary maturation factors. Pet-1 and En1 might function in a feedforward manner analogous to direct control of the Otx-type TF, ceh-36, by the dedicated maintenance zinc-finger factor, che-1, which subsequently operate together to control several identity features of ASE chemosensory neurons in C. elegans (Etchberger et al., 2007).
In summary, we present new insights into how continuously expressed terminal selector regulatory factors control postmitotic neuronal development. Our findings show that continuously expressed Pet-1 acts as postnatal maturation-promoting factor of 5-HT neuron excitability through a stage-specific switch in its transcriptional targets and through direct control of secondary maturation regulatory factors. The discovery of a previously unrecognized early postnatal sensitive period for Pet-1-dependent control of the 5-HT1a autoreceptor opens a new direction for study of stage-specific transcriptional control of 5-HT signaling critical periods.
Footnotes
This research was supported by National Institutes of Health Grants P50 MH096972 and RO1 MH062723 to E.D., R21 MH099488 to S.G.B., and T32 NS067431 to S.C.W. We thank Randy Blakely, Pat Levitt, Ron Emeson, and other members of the Vanderbilt Conte Center for their many helpful comments throughout the course of this project. We thank Heather Broihier for valuable comments. We thank Meredith Sorenson Whitney for her help with stereotaxic injections. We thank Katherine Lobur for assistance with mouse genotyping and breeding.
The authors declare no competing financial interests.
References
- Allan DW, Thor S. Transcriptional selectors, masters, and combinatorial codes: regulatory principles of neural subtype specification. Wiley Interdiscip Rev Dev Biol. 2015;4:505–528. doi: 10.1002/wdev.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baganz N, Horton R, Martin K, Holmes A, Daws LC. Repeated swim impairs serotonin clearance via a corticosterone-sensitive mechanism: organic cation transporter 3, the smoking gun. J Neurosci. 2010;30:15185–15195. doi: 10.1523/JNEUROSCI.2740-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck SG, Pan YZ, Akanwa AC, Kirby LG. Median and dorsal raphe neurons are not electrophysiologically identical. J Neurophysiol. 2004;91:994–1005. doi: 10.1152/jn.00744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calizo LH, Akanwa A, Ma X, Pan YZ, Lemos JC, Craige C, Heemstra LA, Beck SG. Raphe serotonin neurons are not homogenous: electrophysiological, morphological and neurochemical evidence. Neuropharmacology. 2011;61:524–543. doi: 10.1016/j.neuropharm.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Chen CL, Luo P, Tan M, Qiu M, Johnson R, Ma Q. Lmx1b, Pet-1, and Nkx2.2 coordinately specify serotonergic neurotransmitter phenotype. J Neurosci. 2003;23:9961–9967. doi: 10.1523/JNEUROSCI.23-31-09961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford LK, Craige CP, Beck SG. Glutamatergic input is selectively increased in dorsal raphe subfield 5-HT neurons: role of morphology, topography and selective innervation. Eur J Neurosci. 2011;34:1794–1806. doi: 10.1111/j.1460-9568.2011.07882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day HE, Greenwood BN, Hammack SE, Watkins LR, Fleshner M, Maier SF, Campeau S. Differential expression of 5HT-1A, alpha 1b adrenergic, CRF-R1, and CRF-R2 receptor mRNA in serotonergic, gamma-aminobutyric acidergic, and catecholaminergic cells of the rat dorsal raphe nucleus. J Comp Neurol. 2004;474:364–378. doi: 10.1002/cne.20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Bonni A. Transcriptional regulation of neuronal polarity and morphogenesis in the mammalian brain. Neuron. 2011;72:22–40. doi: 10.1016/j.neuron.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneris ES, Hobert O. Maintenance of postmitotic neuronal cell identity. Nat Neurosci. 2014;17:899–907. doi: 10.1038/nn.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneris ES, Wyler SC. Serotonergic transcriptional networks and potential importance to mental health. Nat Neurosci. 2012;15:519–527. doi: 10.1038/nn.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Wang W, Selvakumar T, Xi HS, Zhu H, Chow CW, Horton JD, Gronostajski RM, Kilpatrick DL. Temporal regulation of nuclear factor one occupancy by calcineurin/NFAT governs a voltage-sensitive developmental switch in late maturing neurons. J Neurosci. 2013;33:2860–2872. doi: 10.1523/JNEUROSCI.3533-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanskyi A, Alter H, Vogt MA, Gass P, Vinnikov IA. Transcription factors Foxa1 and Foxa2 are required for adult dopamine neurons maintenance. Front Cell Neurosci. 2014;8:275. doi: 10.3389/fncel.2014.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson ZR, Piel DA, Santos TL, Richardson-Jones J, Leonardo ED, Beck SG, Champagne FA, Hen R. Developmental effects of serotonin 1A autoreceptors on anxiety and social behavior. Neuropsychopharmacology. 2014;39:291–302. doi: 10.1038/npp.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espallergues J, Teegarden SL, Veerakumar A, Boulden J, Challis C, Jochems J, Chan M, Petersen T, Deneris E, Matthias P, Hahn CG, Lucki I, Beck SG, Berton O. HDAC6 regulates glucocorticoid receptor signaling in serotonin pathways with critical impact on stress resilience. J Neurosci. 2012;32:4400–4416. doi: 10.1523/JNEUROSCI.5634-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, Holt RA, Moerman DG, Hobert O. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G, Heintz N. The neuron identity problem: form meets function. Neuron. 2013;80:602–612. doi: 10.1016/j.neuron.2013.10.035. [DOI] [PubMed] [Google Scholar]
- Fox SR, Deneris ES. Engrailed is required in maturing serotonin neurons to regulate the cytoarchitecture and survival of the dorsal raphe nucleus. J Neurosci. 2012;32:7832–7842. doi: 10.1523/JNEUROSCI.5829-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CL, Liu F, Wijayatunge R, Song L, Biegler MT, Yang MG, Vockley CM, Safi A, Gersbach CA, Crawford GE, West AE. Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat Neurosci. 2015;18:647–656. doi: 10.1038/nn.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov D, Nelson T, Deneris E. Pet-1, a novel ETS domain factor that can activate neuronal nAchR gene transcription. J Neurobiol. 1998;34:151–163. doi: 10.1002/(SICI)1097-4695(19980205)34:2%3C151::AID-NEU5%3E3.3.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Green NH, Jackson CR, Iwamoto H, Tackenberg MC, McMahon DG. Photoperiod programs dorsal raphe serotonergic neurons and affective behaviors. Curr Biol. 2015;25:1389–1394. doi: 10.1016/j.cub.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks T, Francis N, Fyodorov D, Deneris ES. The ETS domain factor Pet-1 is an early and precise marker of central 5-HT neurons and interacts with a conserved element in serotonergic genes. J Neurosci. 1999;19:10348–10356. doi: 10.1523/JNEUROSCI.19-23-10348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron. 2003;37:233–247. doi: 10.1016/S0896-6273(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Hensch TK. Critical period plasticity in local cortical circuits. Nat Rev Neurosci. 2005;6:877–888. doi: 10.1038/nrn1787. [DOI] [PubMed] [Google Scholar]
- Hobert O. Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc Natl Acad Sci U S A. 2008;105:20067–20071. doi: 10.1073/pnas.0806070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg J, Perlmann T. Maintaining differentiated cellular identity. Nat Rev Genet. 2012;13:429–439. doi: 10.1038/nrg3209. [DOI] [PubMed] [Google Scholar]
- Jochems J, Teegarden SL, Chen Y, Boulden J, Challis C, Ben-Dor GA, Kim SF, Berton O. Enhancement of stress resilience through histone deacetylase 6-mediated regulation of glucocorticoid receptor chaperone dynamics. Biol Psychiatry. 2015;77:345–355. doi: 10.1016/j.biopsych.2014.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadkhodaei B, Alvarsson A, Schintu N, Ramsköld D, Volakakis N, Joodmardi E, Yoshitake T, Kehr J, Decressac M, Björklund A, Sandberg R, Svenningsson P, Perlmann T. Transcription factor Nurr1 maintains fiber integrity and nuclear-encoded mitochondrial gene expression in dopamine neurons. Proc Natl Acad Sci U S A. 2013;110:2360–2365. doi: 10.1073/pnas.1221077110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyasova V, Fernandez SP, Laine J, Stankovski L, Muzerelle A, Doly S, Gaspar P. A genetically defined morphologically and functionally unique subset of 5-HT neurons in the mouse raphe nuclei. J Neurosci. 2011;31:2756–2768. doi: 10.1523/JNEUROSCI.4080-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger KC, Deneris ES. Serotonergic transcription of human FEV reveals direct GATA factor interactions and fate of Pet-1-deficient serotonin neuron precursors. J Neurosci. 2008;28:12748–12758. doi: 10.1523/JNEUROSCI.4349-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguna A, Schintu N, Nobre A, Alvarsson A, Volakakis N, Jacobsen JK, Gómez-Galán M, Sopova E, Joodmardi E, Yoshitake T, Deng Q, Kehr J, Ericson J, Svenningsson P, Shupliakov O, Perlmann T. Dopaminergic control of autophagic-lysosomal function implicates Lmx1b in Parkinson's disease. Nat Neurosci. 2015;18:826–835. doi: 10.1038/nn.4004. [DOI] [PubMed] [Google Scholar]
- Le Magueresse C, Monyer H. GABAergic interneurons shape the functional maturation of the cortex. Neuron. 2013;77:388–405. doi: 10.1016/j.neuron.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Lemos JC, Pan YZ, Ma X, Lamy C, Akanwa AC, Beck SG. Selective 5-HT receptor inhibition of glutamatergic and GABAergic synaptic activity in the rat dorsal and median raphe. Eur J Neurosci. 2006;24:3415–3430. doi: 10.1111/j.1460-9568.2006.05222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo ED, Hen R. Anxiety as a developmental disorder. Neuropsychopharmacology. 2008;33:134–140. doi: 10.1038/sj.npp.1301569. [DOI] [PubMed] [Google Scholar]
- Levitt P, Eagleson KL, Powell EM. Regulation of neocortical interneuron development and the implications for neurodevelopmental disorders. Trends Neurosci. 2004;27:400–406. doi: 10.1016/j.tins.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Keleş S. Normalization of ChIP-seq data with control. BMC Bioinformatics. 2012;13:199. doi: 10.1186/1471-2105-13-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidov HG, Molliver ME. An immunohistochemical study of serotonin neuron development in the rat: ascending pathways and terminal fields. Brain Res Bull. 1982;8:389–430. doi: 10.1016/0361-9230(82)90077-6. [DOI] [PubMed] [Google Scholar]
- Liu C, Maejima T, Wyler SC, Casadesus G, Herlitze S, Deneris ES. Pet-1 is required across different stages of life to regulate serotonergic function. Nat Neurosci. 2010;13:1190–1198. doi: 10.1038/nn.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu RJ, Lambe EK, Aghajanian GK. Somatodendritic autoreceptor regulation of serotonergic neurons: dependence on L-tryptophan and tryptophan hydroxylase-activating kinases. Eur J Neurosci. 2005;21:945–958. doi: 10.1111/j.1460-9568.2005.03930.x. [DOI] [PubMed] [Google Scholar]
- Maejima T, Masseck OA, Mark MD, Herlitze S. Modulation of firing and synaptic transmission of serotonergic neurons by intrinsic G protein-coupled receptors and ion channels. Front Integr Neurosci. 2013;7:40. doi: 10.3389/fnint.2013.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer P, T'Sas F, Coutte L, Callens N, Brenner C, Van Lint C, de Launoit Y, Baert JL. FEV acts as a transcriptional repressor through its DNA-binding ETS domain and alanine-rich domain. Oncogene. 2003;22:3319–3329. doi: 10.1038/sj.onc.1206572. [DOI] [PubMed] [Google Scholar]
- Meredith RM, Dawitz J, Kramvis I. Sensitive time-windows for susceptibility in neurodevelopmental disorders. Trends Neurosci. 2012;35:335–344. doi: 10.1016/j.tins.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okaty BW, Miller MN, Sugino K, Hempel CM, Nelson SB. Transcriptional and electrophysiological maturation of neocortical fast-spiking GABAergic interneurons. J Neurosci. 2009;29:7040–7052. doi: 10.1523/JNEUROSCI.0105-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Vallstedt A, Dias JM, Samad OA, Krumlauf R, Rijli FM, Brunet JF, Ericson J. Coordinated temporal and spatial control of motor neuron and serotonergic neuron generation from a common pool of CNS progenitors. Genes Dev. 2003;17:729–737. doi: 10.1101/gad.255803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P, Dasen JS. Hox genes: choreographers in neural development, architects of circuit organization. Neuron. 2013;80:12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebello TJ, Yu Q, Goodfellow NM, Caffrey Cagliostro MK, Teissier A, Morelli E, Demireva EY, Chemiakine A, Rosoklija GB, Dwork AJ, Lambe EK, Gingrich JA, Ansorge MS. Postnatal day 2 to 11 constitutes a 5-HT-sensitive period impacting adult mPFC function. J Neurosci. 2014;34:12379–12393. doi: 10.1523/JNEUROSCI.1020-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, Pohl A, Pheasant M, Meyer LR, Learned K, Hsu F, Hillman-Jackson J, Harte RA, Giardine B, Dreszer TR, Clawson H, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood BD, Calizo LH, Piel D, Spangler ZP, Campbell K, Beck SG. Dorsal raphe serotonin neurons in mice: immature hyperexcitability transitions to adult state during first three postnatal weeks suggesting sensitive period for environmental perturbation. J Neurosci. 2014;34:4809–4821. doi: 10.1523/JNEUROSCI.1498-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MM, Wylie CJ, Lerch JK, Murphy R, Lobur K, Herlitze S, Jiang W, Conlon RA, Strowbridge BW, Deneris ES. A genetic approach to access serotonin neurons for in vivo and in vitro studies. Proc Natl Acad Sci U S A. 2005;102:16472–16477. doi: 10.1073/pnas.0504510102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Burbach JP. How to make a mesodiencephalic dopaminergic neuron. Nat Rev Neurosci. 2007;8:21–32. doi: 10.1038/nrn2039. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbenkamp AT, Willsey AJ, State MW, Sestan N. The developmental transcriptome of the human brain: implications for neurodevelopmental disorders. Curr Opin Neurol. 2014;27:149–156. doi: 10.1097/WCO.0000000000000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermaelen CP, Aghajanian GK. Electrophysiological and pharmacological characterization of serotonergic dorsal raphe neurons recorded extracellularly and intracellularly in rat brain slices. Brain Res. 1983;289:109–119. doi: 10.1016/0006-8993(83)90011-2. [DOI] [PubMed] [Google Scholar]
- Wang L, Liu T, Xu L, Gao Y, Wei Y, Duan C, Chen GQ, Lin S, Patient R, Zhang B, Hong D, Liu F. Fev regulates hematopoietic stem cell development via ERK signaling. Blood. 2013;122:367–375. doi: 10.1182/blood-2012-10-462655. [DOI] [PubMed] [Google Scholar]
- Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, Yan J, Talukder S, Turunen M, Taipale M, Stunnenberg HG, Ukkonen E, Hughes TR, Bulyk ML, Taipale J. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyler SC, Donovan LJ, Yeager M, Deneris E. Pet-1 controls tetrahydrobiopterin pathway and Slc22a3 transporter genes in serotonin neurons. ACS Chem Neurosci. 2015;6:1198–1205. doi: 10.1021/cn500331z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie CJ, Hendricks TJ, Zhang B, Wang L, Lu P, Leahy P, Fox S, Maeno H, Deneris ES. Distinct transcriptomes define rostral and caudal serotonin neurons. J Neurosci. 2010;30:670–684. doi: 10.1523/JNEUROSCI.4656-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung YC, Stoddard NC, Mirendil H, Chun J. Lysophosphatidic acid signaling in the nervous system. Neuron. 2015;85:669–682. doi: 10.1016/j.neuron.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZQ, Scott M, Chiechio S, Wang JS, Renner KJ, Gereau RW, 4th, Johnson RL, Deneris ES, Chen ZF. Lmx1b is required for maintenance of central serotonergic neurons and mice lacking central serotonergic system exhibit normal locomotor activity. J Neurosci. 2006;26:12781–12788. doi: 10.1523/JNEUROSCI.4143-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]