Abstract
Objectives:
Today, genetic biomarkers have been demonstrated to play an important role in identifying at-risk subjects for familial or inherited cancers. We have identified a single-nucleotide polymorphism (SNP) that results in missplicing of the cholecystokinin (CCK) receptor gene and expressing a larger mutated receptor in pancreatic cancer. The purpose of this study was to evaluate the significance and specificity of this SNP as a potential biomarker in patients with pancreatic cancer compared with other gastrointestinal (GI) cancers that also have CCK receptors.
Methods:
DNA was isolated and genotyped for the CCK receptor SNP from frozen tumor tissue from banked specimens of patients with pancreas, gastric, or colon cancer and from human cancer cell lines. Genotype and allelic frequencies were compared between the cancer cohort and two normal control databases using Fisher's exact test and odds ratio (OR). The Kaplan–Meier method was used to estimate the survival for patients with the CCK-B receptor SNP compared with those with the wild-type genotype. Immunohistochemical staining of cancer cells was done to detect the mutated receptor.
Results:
Colon and gastric cancer patients had similar genotype frequencies for the CCK receptor SNP as that reported in the normal population. In contrast, the prevalence of the SNP in subjects with pancreatic cancer was twice that of controls and other GI cancers. Survival was adversely affected by the presence of the SNP only in those with pancreatic cancer. Immunoreactivity for the mutated receptor was positive in pancreatic cancer tissues with the SNP but absent in other GI cancers.
Conclusions:
A SNP of the CCK receptor is significantly increased in patients with pancreatic cancer but not in those with other GI malignancies. Therefore, this SNP may be a potential biomarker for pancreatic cancer.
INTRODUCTION
Pancreatic cancer carries a dismal prognosis1 with only ∼5–6% surviving 5 years.2 It is predicted that in the next decade, deaths from pancreatic cancer will surpass those from colon and breast cancer if the trend continues.3 One reason for this poor outcome is that no adequate screening tests or biomarkers have been identified to screen those at risk.4 Carbohydrate antigen 19-9 is the only serologic biomarker currently approved by the Food and Drug Administration (FDA) for the diagnosis of pancreatic cancer; however, this biomarker lacks specificity and is usually only elevated once the cancer becomes unresectable.5 Computerized tomography is not sensitive enough to detect lesions in the pancreas <2 cm that are deemed resectable.4 Another approach that has been used in an attempt to detect pancreatic cancer in “earlier” stages is endoscopic ultrasound and fine-needle biopsy,6 especially in subjects with a history of familial pancreatic cancer.7 Although screening family members of those with pancreatic cancer is an approach that may help improve earlier detection, unfortunately only ∼10% of the reported pancreatic cancer cases are familial.8 Hereditary conditions are associated with an increased risk of developing pancreatic cancer7 including Lynch II syndrome, Peutz–Jegher syndrome, familial atypical multiple mole melanoma, hereditary pancreatitis, Gardner's syndrome, and von Hippel–Lindau syndrome. Family members with these hereditary conditions are generally screened by either radiographic imaging or endoscopic ultrasound for pancreatic cancer.
In the past couple of decades, with our increased knowledge of the human genome, several mutations have been identified and associated with pancreatic cancer. In contrast to other malignancies, however, pancreatic cancer is extremely heterogeneous and contains numerous genetic alterations.9 Of the somatic mutations found (which are mutations within the cancer proper), point mutations in KRAS oncogene are the most common and occur in up to 90% of all pancreatic cancers.10
The majority of pancreatic cancers arise from histologic precancerous lesions called pancreatic intraepithelial neoplasia (PanIN). The genetics of the PanIN lesions have been studied, and researchers have found that KRAS gene mutations and telomere shortening tend to appear first (in PanIN-1 lesions), followed by the inactivation of p16/CDKN2A (usually in PanIN-2), and finally the inactivation of TP53 and MAD4/DPC4 (typically in PanIN-3 lesions).11 Many investigators have attempted to use KRAS mutations in pancreatic fluid secretions, stool samples, and fine-needle aspirates of pancreatic tissue to identify those at risk for cancer.12 Unfortunately, KRAS has been found to be nonspecific and elevated in noncancerous conditions such as chronic pancreatitis.
Approximately 15% of pancreatic cancers arise from cystic lesions in the pancreas called intraductal papillary mucinous neoplasms and mucinous cystic neoplasms. Analysis of the cystic fluid with mutational profiling13, 14 has been useful to identify potentially precancerous lesions. Whole-genome sequencing of DNA from intraductal papillary mucinous neoplasms revealed a high frequency of mutations in the guanine nucleotide-binding protein-α (GNAS).15 Recently, secretin-stimulated pancreatic juice evaluation has shown a correlation with KRAS and GNAS somatic mutations and risk of pancreatic cancer.16 Although these mutated genes may be useful in patients with morphological pancreatic abnormalities, they require invasive endoscopic procedures fluid or tissue analysis.
An ideal biomarker would be one that can be easily found in a blood or urine sample without performing invasive procedures. Researchers have been performing large cohort genome-wide association studies on blood samples from patients with known pancreatic cancer in an effort to identify a germline (or inherited) genetic alteration such as a single-nucleotide polymorphism (SNP) that would predict risk for pancreatic cancer. In these studies some “hot spot” loci and SNPs have been identified, but the clinical correlation to specific phenotypic protein mutations remains undetermined.8, 17
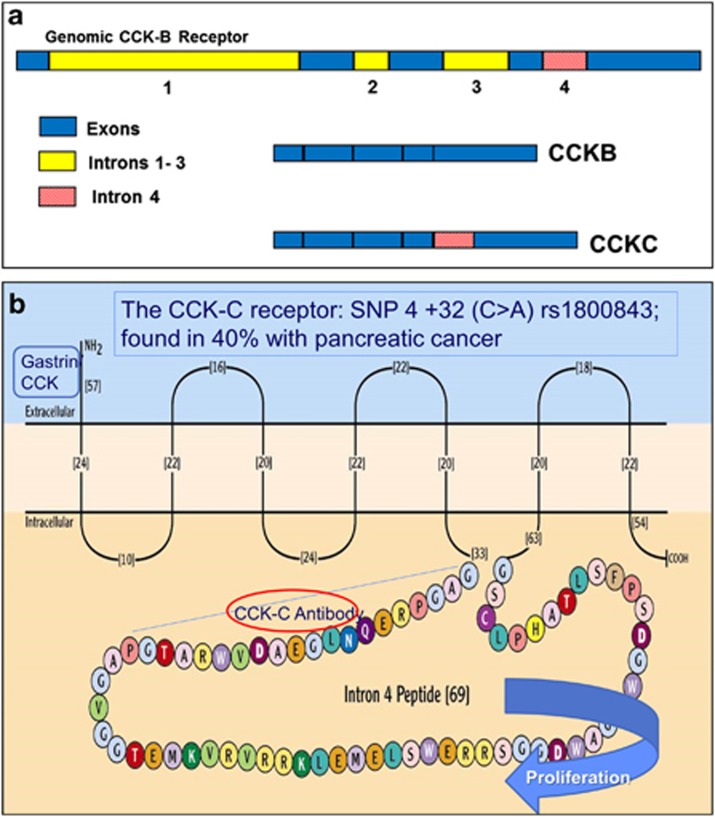
Using the candidate gene association study method, we approached the problem of identifying a biomarker for pancreatic cancer by examining a known mutated protein in pancreatic cancer tissues and identifying the genetic aberration associated with the abnormal cancer receptor phenotype. Growth of pancreatic cancer is stimulated by the actions of the gastrointestinal peptides gastrin18, 19 and /or cholecystokinin (CCK)20, 21 through the CCK-B receptor that is markedly overexpressed in pancreatic cancer.22, 23, 24 This receptor is a G protein–coupled receptor that is also overexpressed in other gastrointestinal (GI) malignancies including gastric cancer25, 26, 27 and colon cancer.26, 27, 28, 29, 30 We previously identified the mutated form of the CCK-B receptor in pancreatic cancer tissues31 that results from missplicing and retention of the fourth intron (Figure 1a), leading to an additional 69-amino acid insert to the third intracellular loop of this receptor, an area involved in cell proliferation (Figure 1b). As this mutated CCK receptor was found only in pancreas cancer and not in normal pancreas tissue, we termed it the “CCK-C receptor,” for cancer-associated receptor.32 Analysis of the DNA from pancreas cancer tissues revealed that the mutated CCK-B receptor was the result of a SNP (rs1800843) with a C>A transition at position 32 within the fourth intron.33 In a cohort of 761 pancreatic cancer patients, we found that the survival of those with the A-allele SNP genotype was significantly shortened, suggesting that the mutated receptor is associated with a more aggressive phenotype.34 Even when corrected for stage of disease, the SNP was determined to be the most important factor to predict survival from pancreatic cancer.34
Figure 1.
Diagram of the splice CCK-C receptor. (a) Schematic of the cholecystokinin-B (CCK-B) receptor gene. All four introns of the CCK-B receptor gene are spliced in normal tissues. The fourth intron of the CCK-B receptor is not spliced in pancreatic cancer cells that have the A-allele single-nucleotide polymorphism (SNP) mutation, creating the CCK-C receptor isoform. This isoform retains the fourth intron that when translated to protein adds an additional 69 amino acids to the CCK-B receptor, changing its phenotype. This larger mutant receptor is called the CCK-C receptor as it is only found in cancer. (b) Alternative splicing of the fourth intron of the CCK-BR gene adds 69 amino acids in the third intracellular loop, the area involved with GTP-protein signaling. The conserved gastrin or CCK binding site is at the extracellular domain of the N-terminal. An antibody was generated to the first 20 amino acids of the additional loop.
As CCK-B receptors have also been associated with normal GI mucosa as well as gastric cancer and colon cancer,23, 25, 30, 35 we studied the allelic frequency, survival, and expression of the mutated CCK receptor in these subjects to determine whether the germline SNP and CCK-C receptor we identified are specific for pancreatic cancer and may serve as a potential cancer biomarker for screening.
METHODS
Patient cohort
Surgical specimens were obtained from the tumor bank from the Penn State Cancer Institute from patients who underwent resection for potentially and curable pancreatic, gastric, or colon cancer. Control tissues were from subjects who had surgery for benign conditions of the pancreas including pancreatitis, pseudocysts, or benign pancreatic strictures and from deceased donors with normal pancreas tissues. All subjects or family members signed consent at the time of surgery to have their tissues deidentified from personal identifiers and banked for future research. Subjects with cancer also agreed for follow-up by clinic appointments or phone call for status evaluation and survival analysis. The protocol for tissue analysis and use of associated demographic information from the tumor bank or from procured pancreas tissues for transplantation was approved by the institutional research committee at the Pennsylvania State University.
Tumor DNA analysis
DNA was isolated from tissue or cancer cells using the DNeasy (Qiagen, Valencia, CA). Genotyping was done for the CCK-B receptor using a TaqMan SNP Genotyping Assay (ID C__22273290_10) (Applied Biosystems, Grand Island, NY). The SNP associated with the mutated receptor,33 rs1800843, was evaluated with the C-allele representing the wild-type receptor and the A-allele indicating the SNP transition.
Evaluation and detection of the mutated CCK receptor
Pancreatic and colon cancer cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and grown in vitro in the appropriate cell culture medium. Cultured log-phase cancer cells were grown on glass coverslips, fixed, and reacted with a monoclonal antibody selective for the mutated CCK-C receptor33 (1:100) overnight at 4 °C and an anti-murine IgG AlexaFluor 488 (Life Technologies, Frederick, MD) secondary antibody (1:2,000) at room temperature for 2 h. After antigen retrieval procedures, tissues were reacted with the CCK-C monoclonal antibody (1:200) and immunoreactivity was identified using Envision+labeled polymer/ horseradish peroxidase (Dako Cytomation, Carpinteria, CA). There is no crossreactivity of the CCK-C monoclonal antibody with the wild-type normal CCK-B receptor.33 Cellular immunofluorescence was examined on a Leica (Leica Camera Inc., Allendale, NJ) TCS SP2 AOBS confocal microscope. Nuclei were visualized with Hoechst 33342 (Pierce Thermo Scientific, Rockford, IL).
Statistical analysis
Genotypic and allelic frequencies in each population of cancer (pancreatic, gastric, and colon) vs. noncancer (benign) controls from both our local Pennsylvania tissue bank and controls from the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) 1000 Genome database normal control cohort were evaluated by Fisher's exact test using a two-sided, 0.05 significance level. The impact of A-allele SNP on the risk of pancreatic cancer over other GI cancers or no cancer controls was analyzed by logistic regression. The OR corresponding to each copy increase of the A-allele SNP is used to quantify its effect.
The Kaplan–Meier method was used to estimate the survival curve for patients with pancreatic, gastric, or colon cancer with the mutant SNP, an A-allele (A/A or A/C) compared with survival of those cancer patients with the wild-type C/C genotype by examining the 95% confidence interval for median survival of patients. The log-rank test was used to compare the difference between the two groups.
RESULTS
Patient demographics
Patients examined included N=37 with colon cancer, N=42 with gastric cancer, and N=51 with pancreatic cancer. The average age of the normal cohort (N=59) from Pennsylvania was 59±3.2 years, similar to that of the other gastrointestinal cancer controls. Genotype frequency was evaluated according to age at the time of cancer diagnosis in order to determine if those with the SNP may develop cancer earlier in life. In subjects with other GI cancers (gastric and colon) the mean age at cancer diagnosis for those with the wild-type receptor genotype (CC) was 61±2.5 years compared with 61.7±6.5 years for those with the mutated genotype (AA or AC). This difference was not statistically significant (P=0.83), suggesting that having the CCK receptor SNP does not appear to be associated with earlier occurrence or diagnosis of colon or gastric cancer. Although the age at the time of diagnosis of pancreatic cancer for those with the A-allele SNP (AA or AC) was earlier (64.9±4.9 years) compared with the age of those with the CC genotype (71±2.4 years), this difference was also not significant (P=0.17).
Gender did not influence the frequency of genotype in other GI cancers. Of those with either gastric or colon cancer with the wild-type CC genotype, 49% were female and 51% were male. There was equal distribution of gender type among those with the A-allele and other GI cancers; i.e., 50% were male and 50% were female. In the pancreatic cancer cohort, there was an increased percentage of males (64%) with the A-allele SNP compared with females with the A-allele (36%).
Frequency of the CCK- B SNP in GI cancers and normal controls
Ancestry genotyping was not performed but the self-described ancestry for race /ethnicity according to patients most closely resembled the Agilent population in the NCBI database. Therefore, the frequency of the rs1800843A allele in our cancer patient cohort was compared with the frequency of the A-allele in two normal control cohorts. The first normal cohort included 905 subjects and was selected from the 1000 Genome cohort of the NCBI website for this SNP in the normal Agilent population (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=1800843), and the second normal control cohort, as previously reported,33 included 59 subjects without cancer from a similar geographic, racial, and ethnic distribution as the cancer cohort (Central Pennsylvania). Genotype and allelic frequency for either the mutated CCK receptor SNP or the wild-type receptor is shown in Table 1. Genotyping distribution of the 37 colon cancer revealed AA=1, AC=8, and CC=28 and the 42 gastric cancer specimens revealed AA=2, AC=8, and CC=32. These GI cancers had similar genotype frequency to that reported for the normal subject cohort from central Pennsylvania population (AA=2, AC=10, and CC=47) and to the distribution of the NCBI database. In contrast, patients with pancreatic cancer had a significantly increased frequency of the SNP with the A-allele; nearly double that of the frequency in normal controls and colon or gastric cancer (Table 1). The frequency of the SNP and having AA or AC genotype was over 40% in those with pancreatic cancer compared with 26% of controls. The allelic frequency of the A-allele in normal controls (14.7%) is, however, comparable to the A-allelic frequency in both gastric (14%) and colon cancers (14%). In contrast, the A-allelic frequency is increased to 25% in pancreatic cancer subjects. The risk of pancreatic cancer was 2.74 times increased with one or two copies of the A-allele (P=0.0192) compared with the CC wild type.
Table 1. Genotype and allelic frequency of the CCK receptor SNP in gastrointestinal cancers.
| Tissue type N=number of patients | A/A genotype (%) | A/C genotype (%) | C/C genotype (%) | A-allelic frequency (%) |
|---|---|---|---|---|
| Colon cancer N=37 | 2.7 | 21.6 | 75.7 | 14 |
| Gastric cancer N=42 | 4.8 | 19 | 76.2 | 14 |
| Pancreatic cancer33 N=51 | 9.8 | 31.4 | 58.8 | 25 |
| Normal controls N=905a and N=59b | 2.8 | 23.8 | 73.4 | 14.7 |
CCK, cholecystokinin; SNP, single-nucleotide polymorphism.
Normal controls without cancer from the National Center for Biotechnology Information (NCBI) 1000 Genome database Agilent population.
Normal controls from the Pennsylvania (PA) database.
Table 2 shows the specificity of the A-allele SNP in pancreatic cancer compared with each of the cohorts. The frequency of the A-allele is significantly increased in pancreatic cancer compared with other GI malignancies (OR 1.90; P=0.0349). Furthermore, having the A-allele SNP is also significantly increased in pancreatic cancer compared with both normal control noncancer cohorts (OR 2.21; P=0.0214 and OR 1.65; P=0.0389, respectively). There was no significant difference in the prevalence of the SNP among gastric and colon cancer subjects compared with the normal controls (P=0.643).
Table 2. Logistic regression analysis of the impact of A-allele SNP on the risk of pancreatic cancer over other GI cancers or no cancer controls.
| Condition | Comparator | Odds ratio and 95% confidence limits (for increase of one copy of A-allele SNP) | P-value |
|---|---|---|---|
| Pancreatic cancer | Other GI cancer (colon/gastric) | 1.90 (1.04–3.43) | 0.0349 |
| Pancreatic cancer | Pennsylvania no cancer control cohort | 2.21 (1.15–4.53) | 0.0214 |
| Pancreatic cancer | NCBI no cancer control cohort | 1.65 (1.01–2.63) | 0.0389 |
| Other GI cancer (colon/gastric) | Pennsylvania no cancer control cohort | 1.17 (0.60–2.27) | 0.643 |
GI, gastrointestinal; NCBI, national center for biotechnology information; SNP, single-nucleotide polymorphism.
Relationship of the CCK-B receptor SNP on patient survival
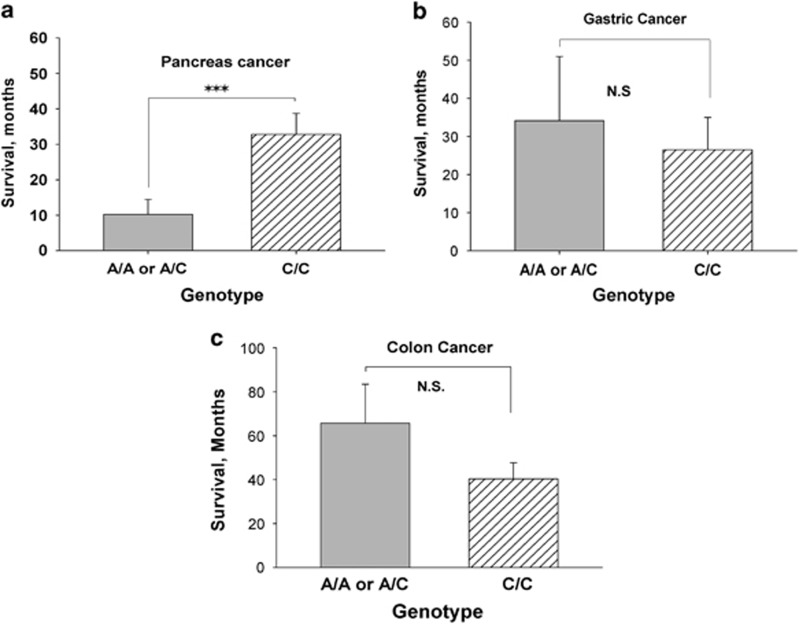
Time to death or last censored visit date was recorded for all cancer subjects. The role of the A-allele SNP expression on patient survival was evaluated in each respective cancer type to survival of those with the wild-type CCK receptor. As so few subjects were homozygote for the A-allele (AA), analysis was performed by combining subjects with either one (AC) or two A-alleles (AA) to that of the CC wild type. We previously demonstrated that protein expression of the CCK-C receptor occurs with expression of just one A-allele,33 and hence the phenotype of the protein expression is the same whether the subject is homozygous or heterozygous for the A-allele. Patients with pancreatic cancer who expressed the SNP and at least one A-allele had a significant shortened survival (P=0.001) compared with pancreatic cancer subjects without the transition or wild-type genotype (CC) (Figure 2a). In contrast, the presence of an A-allele did not affect survival in those with gastric cancer (P=0.352; Figure 2b) or those with colon cancer (P=0.811; Figure 2c). As stage of disease at diagnosis also influences survival, we conducted a multivariate analysis to correct for stage of disease. The hazard ratio for survival difference in those with the A-allele SNP compared with the CC genotype after correcting for stage of disease was significantly increased (hazard ratio=3.6; P=0.0004), indicting that having the A-allele SNP independently (independent of stage) predicts for a shorter survival from pancreatic cancer.
Figure 2.
Survival according to genotype of the cholecystokinin (CCK) receptor. (a) Pancreatic cancer patients with the AA or AC genotype had a significantly shortened survival compared with those with the wild-type (CC) genotype of the CCK receptor (***P=0.0001); (b) CCK receptor genotype has no influence on survival of subjects with gastric cancer and; (c) CCK receptor genotype also does not have effect on survival of subjects with colon cancer. NS, not significant.
Expression of the mutated CCK receptor phenotype
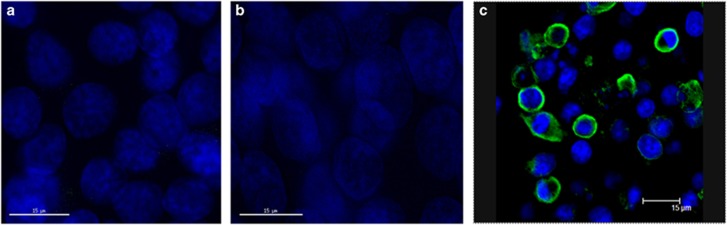
We previously raised a monoclonal antibody to the 69-amino acid segment of the mutated CCK receptor that reacts only to the variant receptor and with the normal CCK-B receptors.33 Two colon cancer cells lines (HCT-116 and HT-29) frequently used in the laboratory for in vitro and preclinical experiments were genotyped. The HCT-116 cells were homozygous for the A-allele (AA) and the HT-29 colon cancer cells were homozygous for the wild-type C-allele (CC). Immunohistochemical staining of HCT-116 cells (Figure 3a) and HT-29 cells (Figure 2b) failed to demonstrate any immunoreactivity to the CCK-C antibody regardless of the genotype of the CCK receptor. This finding indicates that even if the SNP was present in the colon cancer cell DNA, these cancer cells do not express the altered receptor, suggesting that the missplicing and abnormal receptor phenotype is specific for pancreatic cancer. In Figure 3c, COS cells that lack the CCK-C receptor were transiently transfected with the receptor and demonstrate positive immunoreactivity to the fluorescent antibody by confocal microscopy.
Figure 3.
Cancer cell immunoreactivity with the cholecystokinin-C (CCK-C) receptor antibody. (a) The CCK-C antibody fails to show reactivity in HCT-116 human colon cancer cells that are AA genotype. (b) The HT-29 human colon cancer controls are wild-type genotype CC and serve as the negative colon. Nuclei are stained blue with Hoechst. (c) COS cells that do not express the receptor were transiently transfected with the CCK-C receptor and immunohistochemistry reveals positive fluorescence by confocal microscopy.
Immunohistochemical staining with the CCK-C monoclonal antibody demonstrated reactivity in a pancreas cancer tissue of a subject with the A-allele SNP (Figure 4a). However, the immunoreactivity was negative from a normal (benign) human pancreas that was genotyped to be homozygote for A/A (Figure 4b) but without cancer. These results support that the phenotype of the mutated receptor is only found in the pancreas of A-allele SNP subjects once cancer is present.
Figure 4.
CCK-C monoclonal antibody only reacts in cancer tissues. (a) Immunohistochemical reaction of a human pancreatic cancer specimen from a patient with the A-allele single-nucleotide polymorphism (SNP) showing positive reactivity with a 1:200 dilution of a cholecystokinin-C (CCK-C) receptor monoclonal antibody. (b) Normal pancreas from a subject without cancer and with the A-allele SNP does not express the CCK-CR phenotype when reacted with the same CCK-CR antibody.
DISCUSSION
In this study we have shown that a SNP of the CCK-B receptor occurs with an increased prevalence in subjects with pancreatic adenocarcinoma compared with other GI cancers and may therefore be a novel genetic marker used to screen those at increased risk for familial pancreatic cancer. As normal gastrointestinal mucosa expresses CCK-B receptors,36 and also other cancers arising from GI epithelium also express CCK-B receptors,27, 37 it was important to evaluate these tissues as controls. This study demonstrated that a SNP (an inherited gene) that results in a mutated CCK-B receptor is associated with pancreatic cancer and not colon or gastric cancer. The frequency of the A-allele SNP in patients with colon or gastric cancer was the same as the prevalence of this SNP in the normal controls. Because the A-allele SNP was found more often in male subjects, this suggests that males with the CCK receptor A-allele SNP may be at greater risk of developing pancreatic cancer than females. As this anomaly is inherited, it is possible that evaluation for this SNP in blood DNA or a buccal mucosa DNA sample from patients with a family history of pancreatic cancer may increase the ability to identify those patients who require more intensive or invasive screening endoscopic procedures.
Limitations to this study include the fact that the sample size was small and included subjects from one site. Possibly, if a much larger cohort of colon and gastric cancer subjects were screened, the prevalence of the SNP may increase in these malignancies. Although the frequency of the CCK receptor SNP was very high in those with pancreatic cancer, the control subjects in this investigation showed an A-allelic frequency of ∼14% for the SNP. In addition, as the SNP is found in control subjects without cancer, although at a lower frequency, the CCK-BR SNP by itself most likely would not be an adequate biomarker for pancreatic cancer. Perhaps, the inclusion of the CCK-BR SNP in a panel of blood biomarkers would be more effective and would increase the sensitivity compared with using this SNP or other individual biomarkers alone. Long-term studies would help determine whether those in the “normal population” with the SNP may be at increased risk for development of pancreatic cancer.
Identification of germline or inherited genetic alterations such as SNPs has changed how we screen family members for cancer risk. Some classic example of germline mutations used as biomarkers for screening of familial cancers include the APC gene in familial adenomatous polyposis,38 BRCA-1 mutations in ovarian cancer,39 and RET mutations in medullary carcinoma of the thyroid.40 The germline mutation BRCA2 that has been associated with inherited breast cancer41 with a frequency of 35% has also been identified in pancreatic cancer.42 Although we often screen patients who have family members with pancreatic cancer for the BRCA2 mutation, BRCA2 alterations have only been reported to occur with a very low frequency (4.1–5.6%) in pancreatic cancer,43, 44 and hence it is found more often in breast cancer. Based upon what we know about BRCA2 and other germline mutations, we know that some germline mutations may increase cancer risk in other tissues but have the highest specificity for a particular organ site (i.e., BRCA2 with breast cancer). Our investigation showed that the CCK receptor germline A-allele SNP occurs in nearly 40% of those we evaluated with pancreatic cancer, making this genetic biomarker very significant. The frequency of the A-allele SNP in this study was similar to that we previously found in a larger cohort including 931 patients,34 suggesting that this value is reproducible and accurate. Although historically, evidence for a major gene influencing the risk of pancreatic cancer has been suggested based upon a hospital-based segregation and computational analysis,45 this “unknown” major gene has not been described. Herein, we report a germline inherited mutation of the CCK receptor that has an increased association with pancreatic cancer.
In order for a mutation to have clinical relevance, its presence or expression should influence the phenotype or survival of a cancer. In our study, survival of pancreatic cancer patients with the CCK receptor gene SNP was significantly shortened compared with those with the wild-genotype CCK-B receptor. However, the presence of this alteration did not influence survival of those with other GI cancers. We rationalize that the reason for the absence of survival difference in gastric and colon cancer patients having the CCK-B receptor SNP mutation is related to the lack of protein expression as demonstrated by our immunocytochemical experiments. Although the SNP is present in a small portion of the normal population and also in those with other GI malignancies, it appears that only in pancreatic cancer does the C>A transition result in translation of a unique protein receptor type. This unique difference supports the specificity of this germline mutation as a biomarker for pancreatic cancer.
The CCK-B receptor is a classic seven-transmembrane spanning G protein–coupled receptor.36, 46 The missplicing of the fourth intron that occurs results in translation of this intron into an additional 69 amino acids that are added to the third intracellular loop of the receptor.32 Although the fourth intron SNP describe herein does not alter the ligand binding site, the extended third intracellular loop of the receptor is the part of the receptor that interacts with G proteins and is crucial for intracellular signaling and cell proliferation.47 Therefore, it is conceivable that the change in the third intracellular loop of the CCK-B receptor may activate cell proliferation and shorten survival. Indeed, when this extended segment of the receptor is downregulated by RNA interference techniques in pancreas cancer cells in the lab,48 growth is markedly inhibited, implying the importance of this added protein segment to the third loop in cell proliferation. The expression of this mutated receptor in pancreatic cancer may be related to the aggressive nature of this disease and poor prognosis.
Currently, there are no adequate screening tests to evaluate patients who are not invasive. Imaging tests such as magnetic resonance and computerized tomography are useful for staging pancreas cancer, and endoscopic ultrasound is more sensitive to identify smaller lesions that could be amenable to fine-needle aspiration,49 but the ideal biomarker would be noninvasive, without ionizing radiation, and inexpensive.4 With the advancement in genetic technology, many have been searching for a gene associated with pancreatic cancer.8, 17
In conclusion, we have evaluated the specificity of an A-allele SNP of the CCK receptor for pancreatic cancer, a receptor known to stimulate growth of pancreatic cancer when activated. As this genetic alteration is found at a high frequency in this malignancy and is also associated with a protein phenotype, its presence may be very useful in screening those at risk for pancreatic cancer. The lack of protein expression and phenotypic changes (survival) in subjects with gastric or colon cancer with this SNP significantly increase its specificity for pancreatic cancer. Perhaps, adding the CCK-B receptor SNP to genetic screening will help identify those subjects who will need close surveillance in order to diagnose pancreatic cancer in the early or precancerous state. Through continued research together with improved scientific technologies, there is hope for better tests to improve the outcome of patients with pancreatic cancer.
Study Highlights
Acknowledgments
We appreciate the support and collection of specimens by Dan Beard of the Penn State Cancer Institute Tissue Bank. We appreciate the technical assistance of Calpurnia Jayakumar for performing confocal microscopy on transfected cells.
Footnotes
Guarantor of the article: Jill P. Smith, MD.
Specific author contributions: Study concept and design (Alsubai and Smith); acquisition of data (Alsubai, Matters, McGovern, Gilius, Liao, and Smith); analysis and interpretation of data (Alsubai, Matters, McGovern, Gilius, Liao, and Smith); drafting of the manuscript (Alsubai and Smith); critical revision of the manuscript for important intellectual content (all); statistical analysis (Liao); obtained funding (Smith); technical or material support (McGovern and Gilius); study supervision (Smith). All authors have approved the final draft submitted.
Financial support: This work was funded by a grant NIH R01 CA117926 (to J.P.S.) and support from the William Donner Foundation (to J.P.S.).
Potential competing interests: Smith and Matters are co-inventors on an issued US patent (8,821,872) for the CCK-C monoclonal antibody used to perform and identify the mutant CCK-C receptor in pancreatic cancer.
References
- 1Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014; 371: 1039–1049. [DOI] [PubMed] [Google Scholar]
- 2Siegel R, Ma J, Zou Z et al. Cancer statistics, 2014. CA Cancer J Clin 2014; 64: 9–29. [DOI] [PubMed] [Google Scholar]
- 3Rahib L, Smith BD, Aizenberg R et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014; 74: 2913–2921. [DOI] [PubMed] [Google Scholar]
- 4Kaur S, Baine MJ, Jain M et al. Early diagnosis of pancreatic cancer: challenges and new developments. Biomark Med 2012; 6: 597–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Poruk KE, Gay DZ, Brown K et al. The clinical utility of CA 19-9 in pancreatic adenocarcinoma: diagnostic and prognostic updates. Curr Mol Med 2013; 13: 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Canto MI, Goggins M, Hruban RH et al. Screening for early pancreatic neoplasia in high-risk individuals: a prospective controlled study. Clin Gastroenterol Hepatol 2006; 4: 766–781. [DOI] [PubMed] [Google Scholar]
- 7Klein AP. Identifying people at a high risk of developing pancreatic cancer. Nat Rev Cancer 2013; 13: 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Petersen GM, Amundadottir L, Fuchs CS et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet 2010; 42: 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Jones S, Zhang X, Parsons DW et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008; 321: 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol 2008; 3: 157–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Hruban RH, Maitra A, Goggins M. Update on pancreatic intraepithelial neoplasia. Int J Clin Exp Pathol 2008; 1: 306–316. [PMC free article] [PubMed] [Google Scholar]
- 12Siddiqui AA, Kowalski TE, Kedika R et al. EUS-guided pancreatic fluid aspiration for DNA analysis of KRAS and GNAS mutations for the evaluation of pancreatic cystic neoplasia: a pilot study. Gastrointest Endosc 2013; 77: 669–670. [DOI] [PubMed] [Google Scholar]
- 13Finkelstein SD, Bibbo M, Kowalski TE et al. Mutational analysis of cytocentrifugation supernatant fluid from pancreatic solid mass lesions. Diagn Cytopathol 2014; 42: 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Khalid A, Zahid M, Finkelstein SD et al. Pancreatic cyst fluid DNA analysis in evaluating pancreatic cysts: a report of the PANDA study. Gastrointest Endosc 2009; 69: 1095–1102. [DOI] [PubMed] [Google Scholar]
- 15Furukawa T, Kuboki Y, Tanji E et al. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci Rep 2011; 1: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Eshleman JR, Norris AL, Sadakari Y et al. KRAS and GNAS mutations in pancreatic juice collected from the duodenum of patients at high risk for neoplasia undergoing endoscopic ultrasound. Clin Gastroenterol Hepatol 2014; 13: 963–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Amundadottir L, Kraft P, Stolzenberg-Solomon RZ et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet 2009; 41: 986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Smith JP, Fantaskey AP, Liu G et al. Identification of gastrin as a growth peptide in human pancreatic cancer. Am J Physiol 1995; 268: R135–R141. [DOI] [PubMed] [Google Scholar]
- 19Smith JP, Shih A, Wu Y et al. Gastrin regulates growth of human pancreatic cancer in a tonic and autocrine fashion. Am J Physiol 1996; 270: R1078–R1084. [DOI] [PubMed] [Google Scholar]
- 20Smith JP, Solomon TE, Bagheri S et al. Cholecystokinin stimulates growth of human pancreatic adenocarcinoma SW-1990. Dig Dis Sci 1990; 35: 1377–1384. [DOI] [PubMed] [Google Scholar]
- 21Smith JP, Kramer ST, Solomon TE. CCK stimulates growth of six human pancreatic cancer cell lines in serum-free medium. Regul Pept 1991; 32: 341–349. [DOI] [PubMed] [Google Scholar]
- 22Smith JP, Rickabaugh CA, McLaughlin PJ et al. Cholecystokinin receptors and PANC-1 human pancreatic cancer cells. Am J Physiol 1993; 265: G149–G155. [DOI] [PubMed] [Google Scholar]
- 23Smith JP, Liu G, Soundararajan V et al. Identification and characterization of CCK-B/gastrin receptors in human pancreatic cancer cell lines. Am J Physiol 1994; 266: R277–R283. [DOI] [PubMed] [Google Scholar]
- 24Smith JP, Solomon TE. Cholecystokinin and pancreatic cancer: the chicken or the egg? Am J Physiol Gastrointest Liver Physiol 2014; 306: G91–G101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Smith JP, Shih AH, Wotring MG et al. Characterization of CCK-B/gastrin-like receptors in human gastric carcinoma. Int J Oncol 1998; 12: 411–419. [PubMed] [Google Scholar]
- 26Wang TC, Dockray GJ. Lessons from genetically engineered animal models. I. Physiological studies with gastrin in transgenic mice. Am J Physiol 1999; 277: G6–G11. [DOI] [PubMed] [Google Scholar]
- 27Watson S, Durrant L, Morris D. Gastrin: growth enhancing effects on human gastric and colonic tumor cells. Br J Cancer 1989; 59: 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Singh P, Walker JP, Townsend CM Jr et al. Role of gastrin and gastrin receptors on the growth of a transplantable mouse colon carcinoma (MC-26) in BALB/c mice. Cancer Res 1986; 46: 1612–1616. [PubMed] [Google Scholar]
- 29Smith JP, Solomon TE. Effects of gastrin, proglumide, and somatostatin on growth of human colon cancer. Gastroenterology 1988; 95: 1541–1548. [DOI] [PubMed] [Google Scholar]
- 30Smith JP, Stock EA, Wotring MG et al. Characterization of the CCK-B/gastrin-like receptor in human colon cancer. Am J Physiol 1996; 271: R797–R805. [DOI] [PubMed] [Google Scholar]
- 31Smith JP, Verderame MF, McLaughlin PJ et al. Characterization of the CCK-C (cancer) receptor in human pancreatic cancer. Gastroenterology 1999; 116: A1164. [Google Scholar]
- 32Smith JP, Verderame MF, McLaughlin P et al. Characterization of the CCK-C (cancer) receptor in human pancreatic cancer. Int J Mol Med 2002; 10: 689–694. [PubMed] [Google Scholar]
- 33Smith JP, Harms JF, Matters GL et al. A single nucleotide polymorphism of the cholecystokinin-B receptor predicts risk for pancreatic cancer. Cancer Biol Ther 2012; 13: 164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Smith JP, Whitcomb DC, Matters GL et al. Distribution of cholecystokinin-B receptor genotype between patients with pancreatic cancer and controls and its impact on survival. Pancreas 2015; 44: 236–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35McWilliams DF, Watson SA, Crosbee DM et al. Coexpression of gastrin and gastrin receptors (CCK-B and delta CCK-B) in gastrointestinal tumour cell lines. Gut 1998; 42: 795–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Wank SA, Pisegna JR, de Weerth A. Cholecystokinin receptor family. Molecular cloning, structure, and functional expression in rat, guinea pig, and human. Ann NY Acad Sci 1994; 713: 49–66. [DOI] [PubMed] [Google Scholar]
- 37Watson SA, Clarke PA, Smith AM et al. Expression of CCKB/gastrin receptor isoforms in gastro-intestinal tumour cells. Int J Cancer 1998; 77: 572–577. [DOI] [PubMed] [Google Scholar]
- 38Beroud C, Soussi T. APC gene: database of germline and somatic mutations in human tumors and cell lines. Nucleic Acids Res 1996; 24: 121–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Berchuck A, Heron KA, Carney ME et al. Frequency of germline and somatic BRCA1 mutations in ovarian cancer. Clin Cancer Res 1998; 4: 2433–2437. [PubMed] [Google Scholar]
- 40Marsh DJ, Andrew SD, Eng C et al. Germline and somatic mutations in an oncogene: RET mutations in inherited medullary thyroid carcinoma. Cancer Res 1996; 56: 1241–1243. [PubMed] [Google Scholar]
- 41Couch FJ, Farid LM, DeShano ML et al. BRCA2 germline mutations in male breast cancer cases and breast cancer families. Nat Genet 1996; 13: 123–125. [DOI] [PubMed] [Google Scholar]
- 42Hahn SA, Greenhalf B, Ellis I et al. BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst 2003; 95: 214–221. [DOI] [PubMed] [Google Scholar]
- 43Axilbund JE, Wiley EA. Genetic testing by cancer site: pancreas. Cancer J 2012; 18: 350–354. [DOI] [PubMed] [Google Scholar]
- 44Ferrone CR, Levine DA, Tang LH et al. BRCA germline mutations in Jewish patients with pancreatic adenocarcinoma. J Clin Oncol 2009; 27: 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Klein AP, Beaty TH, Bailey-Wilson JE et al. Evidence for a major gene influencing risk of pancreatic cancer. Genet Epidemiol 2002; 23: 133–149. [DOI] [PubMed] [Google Scholar]
- 46Dufresne M, Seva C, Fourmy D. Cholecystokinin and gastrin receptors. Physiol Rev 2006; 86: 805–847. [DOI] [PubMed] [Google Scholar]
- 47Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer 2007; 7: 79–94. [DOI] [PubMed] [Google Scholar]
- 48Smith JP, Stanley WB, Verderame MF et al. Functional significance of the CCK-C receptor in human pancreatic cancer. Pancreas 2004; 29: 271–277. [DOI] [PubMed] [Google Scholar]
- 49Cote GA, Smith J, Sherman S et al. Technologies for imaging the normal and diseased pancreas. Gastroenterology 2013; 144: 1262–1271. [DOI] [PubMed] [Google Scholar]