Abstract
Interactions between phytoplankton and bacteria play a central role in mediating biogeochemical cycling and food web structure in the ocean. However, deciphering the chemical drivers of these interspecies interactions remains challenging. Here, we report the isolation of 2-heptyl-4-quinolone (HHQ), released by Pseudoalteromonas piscicida, a marine gamma-proteobacteria previously reported to induce phytoplankton mortality through a hitherto unknown algicidal mechanism. HHQ functions as both an antibiotic and a bacterial signaling molecule in cell–cell communication in clinical infection models. Co-culture of the bloom-forming coccolithophore, Emiliania huxleyi with both live P. piscicida and cell-free filtrates caused a significant decrease in algal growth. Investigations of the P. piscicida exometabolome revealed HHQ, at nanomolar concentrations, induced mortality in three strains of E. huxleyi. Mortality of E. huxleyi in response to HHQ occurred slowly, implying static growth rather than a singular loss event (e.g., rapid cell lysis). In contrast, the marine chlorophyte, Dunaliella tertiolecta and diatom, Phaeodactylum tricornutum were unaffected by HHQ exposures. These results suggest that HHQ mediates the type of inter-domain interactions that cause shifts in phytoplankton population dynamics. These chemically mediated interactions, and other like it, ultimately influence large-scale oceanographic processes.
Keywords: infochemicals, algicidal compound, bacteria–phytoplankton interaction, HHQ, Pseudoalteromonas, Emiliania huxleyi, IC50, mortality
Introduction
In the marine environment, interactions between bacteria and eukaryotic phytoplankton are pervasive and drive oceanic biogeochemical cycles (Legendre and Rassoulzadegan, 1995), and can have consequences for both microbial communities (Bratbak and Thingstad, 1985) and structuring of marine food webs (Azam et al., 1983). Bacteria–phytoplankton interactions are complex, being both temporally variable (Danger et al., 2007) and species-specific (Fukami et al., 1997), and remain largely enigmatic. These interactions can be beneficiary, as bacteria and phytoplankton can support the growth of one another via the exchange or recycling of nutrients (Azam et al., 1983); however, the interaction can also be detrimental, resulting in phytoplankton morbidity or mortality (Mayali and Azam, 2004). The intricate relationships between these two kingdoms are often mediated via excreted compounds that can direct communication between the two organisms. Identifying these released compounds, and their influence on the population dynamics of both phytoplankton and bacteria, will enhance our understanding of the role of “infochemicals” on large-scale biogeochemical processes.
The marine genus Pseudoalteromonas constitutes 0.5–6.0% of bacterial species globally (Wietz et al., 2010), and has been found in seawater, marine sediments, and associated with marine eukaryotes (Bowman, 2007; Skovhus et al., 2007; Sneed et al., 2014). Secondary metabolites secreted by members of this genus fulfill a variety of ecological roles, including involvement in chemical protection, settlement, induction of metamorphosis in marine invertebrates, and commercially, as antifouling, antifungal agents (Bowman, 2007; Ross et al., 2014; Sneed et al., 2014). Moreover, Pseudoalteromonas species have been implicated in producing algal-lytic compounds that cause mortality in dinoflagellates (Skerratt et al., 2002; Kim et al., 2009), diatoms (Mitsutani et al., 2001), and raphidophytes (Lovejoy et al., 1998); however, in each of these cases the causative chemical compound is yet unidentified.
In order to gain a more mechanistic understanding of this chemically mediated phytoplankton mortality, we exposed the globally important coccolithophore, Emiliania huxleyi to cultures of Pseudoalteromonas piscicida. E. huxleyi plays a predominant role in oceanic carbon (Balch et al., 1992; Iglesias-Rodriguez et al., 2008) and sulfur (Simo, 2001) cycling; therefore, understanding the factors that mediate the population abundance and distribution of this algal species is important for predicting its role in marine nutrient cycling and global climate. Further, E. huxleyi has been shown to form complex relationships with bacteria. For example, it has been observed that the Roseobacter, Phaeobacter gallaeciensis maintains a mutualistic-turned-to-parasitic relationship with E. huxleyi dependent on the metabolic stage of the algae (Seyedsayamdost et al., 2011). While no direct relationship between a Pseudoalteromonas species and E. huxleyi has been established, Seymour et al. (2010) found that Pseudoalteromonas haloplanktis exhibited strong chemoattraction to dimethlysulfoniopropionate (DMSP), a sulfur compound abundantly produced by E. huxleyi. Additionally, bacterial OTUs, including Pseudoalteromonas, were positively correlated with coccolithophores and E. huxleyi abundance in samples from a temperate marine coastal site off Plymouth, UK (Gilbert et al., 2012). Thus, these two taxa do co-occur in the water column, and have opportunity to interact with one another.
Here, we report the isolation and identification of a potent algicidal compound excreted by P. piscicida with specificity for E. huxleyi. We found that the presence of P. piscicida resulted in mortality of E. huxleyi, and used bioassay-guided fractionation to identify the responsible chemical mediator(s) of this interaction. This research further reveals the importance of the bacterial exometabolome in inter-domain interactions, especially those that can have significant biogeochemical implications.
Materials and Methods
Culture Conditions and Crude Extract Preparation
A pure culture of P. piscicida was isolated from open ocean plastic debris in the North Atlantic (Mincer culture ID; A757; Whalen et al., 2015), cryopreserved in 10% sterile DMSO, and stored at -85°C prior to experiments. From these stocks, multiple ‘starter’ cultures were prepared by inoculating 8 mL of TSW media (1 g tryptone in 1 L of 75:25 seawater/MilliQ water) with 100 μL of cryopreserved culture, and then incubated at 23°C while shaking at 100 rpm for 3 days. After 3 days, 2 mL of ‘starter’ culture was used to inoculate seven, 1.5 L Fernbach flasks of TSY media (1 g tryptone, 1 g yeast extract, 75% seawater), the newly inoculated cultures were then grown at 100 rpm for 8 days at 23°C. On day 7, 20 mL (approximately weight = 7.8 g) of 1:1 mixture of sterile Amberlite® XAD-7 and XAD-16 resin that had been extensively washed in organic solvent, dried, and autoclaved was added to each 1.5 L culture. Twenty-four hours later (day 8), the resin was filtered from the bacterial culture through combusted stainless steel mesh under gentle vacuum (<5 in Hg), desalted by rinsing with MilliQ water, pooled, and allowed to dry overnight at room temperature. Bacterial metabolites were eluted from the resin first in 800 mL of (1:1) methanol (MeOH):dichloromethane (DCM), followed by 800 mL of methanol. This crude extract was dried under vacuum centrifugation and stored at -85°C until testing in phytoplankton growth assays.
Emiliania huxleyi (Plymouth Algal Culture Collection: DHB624 and National Center for Marine Algae: CCMP374, CCMP379), Dunaliella tertiolecta (CCMP1320), and Phaeodactylum tricornutum (CCMP2561) were used in these experiments. All phytoplankton cultures were grown in 0.2 μm sterile-filtered, autoclaved seawater (FSW), enriched with f/2 (P. tricornutum) or f/2 -Si (all other species) media (Guillard, 1975). All cultures were maintained on a 12:12 h light:dark at 18°C, salinity of approximately 20 psu, and light intensity of 85–100 μmol photon m-2 s-1. Hereafter, these conditions will be referred to as general culturing conditions. Phytoplankton cultures were transferred every 7–10 days to maintain exponential growth. Unless specified, cultures were not axenic. Axenic cultures of E. huxleyi 624 were prepared by adding 1 mL of a dense E. huxleyi culture into 4 mL of f/2 -Si containing an antibiotic cocktail of penicillin G (3 mM), dihydrostreptomycin sulfate (0.2 mM), and gentamicin (0.5 mM). The culture was incubated under the general culturing conditions described above for 48 h, after which a 500 μL aliquot of the culture was transferred to fresh, antibiotic-free f/2 -Si media. This culture was maintained in exponential growth, similar to the other phytoplankton cultures. The culture was tested with DAPI (4′, 6-diamidino-2-phenylindole) to ensure it remained axenic throughout the course of the experiments, and this culture was used for all experiments that called for axenic E. huxleyi. For all phytoplankton counts, a 200 μL aliquot of culture was run on a flow cytometer (Guava, Millipore). Cell abundance was determined by using species-specific settings determined based on chlorophyll a (692 nm) and side scatter (SSC) for each species examined.
Phytoplankton Growth Inhibition Assay
The growth rate of E. huxleyi (strain 624) was measured in response to both live P. piscicida cells (103 and 106 cell mL-1) and filtrate from P. piscicida cells equivalent to 103 and 106 cell mL-1 in TSY media (between <1 and 40 μL of filtrate per 1 mL of algal culture). Cells from strain 624 in their exponential growth phase were plated in 24- well plates at a final concentration of 105 cell mL-1, and exposed to either live P. piscicida cells or filtrate. These experiments were conducted with both xenic and axenic cultures of E. huxleyi. Each treatment was replicated in triplicate. Prior to inoculation into 24-well plates, the bacteria were enumerated on a flow cytometer and diluted appropriately. Filtrate from the P. piscicida culture was obtained by filtering 5 mL of live culture through a 0.2 μm sterile syringe tip filter. Prior to sampling for phytoplankton cell abundance, triplicate wells were gently mixed via pipet and 50–200 μL aliquots were taken from each replicate, pipetted into a 96-well plate, and run on the flow cytometer. Growth rate was calculated by using the exponential growth equation,
where A is the abundance and T is the time, over the first 72 h of the experiment. Significant differences in growth rates of E. huxleyi in response to chemical additions were determined by using a one-way analysis of variance (ANOVA) and Tukey’s HSD post hoc analysis. All statistical analyses were performed using MatLAB (v. 8.3). Those treatments where the growth rate was significantly different from the algae only control (p < 0.05) were considered to have activity.
Bioassay-Guided Fractionation of Exudates from P. piscicida
Assessment of semi-purified fractions via the growth inhibition assay was carried out using axenic E. huxleyi strain 624 to determine those algicidal compounds exuded by P. piscicida (Supplementary Figure S1). E. huxleyi (624) cells in exponential growth phase were plated at 105 cell mL-1 in to 24-well plates in triplicate and exposed to crude and semi-purified fractions dissolved in DMSO, with DMSO concentrations not exceeding 0.2% v/v. Appropriate controls were run with each growth inhibition assay, i.e., f/2 media only, DMSO without compounds, were performed in triplicate. Well plates were incubated under general culturing conditions, and enumerated on the flow cytometer, as described above. At each step of the chemical fractionation process described below, semi-purified fractions were tested in the growth inhibition assay using axenic E. huxleyi strain 624 and included the addition of paired controls (TSY media only, f/2, and DMSO). Statistical comparison of growth rates resulting from semi-purified fractions were compared to controls as described in Section “Phytoplankton Growth Inhibition Assay.”
The entire crude extract (1260 mg; described above) was applied to a silica gel column and eluted with a step-gradient of isooctane, (4:1) isooctane/ethylacetate (EtOAc), (3:2) isooctane/EtOAc, (2:3) isooctane/EtOAc, (1:4) isooctane/EtOAc, EtOAc, (1:1) EtOAc/MeOH, and 100% MeOH, yielding eight fractions. A total of 458 mg of material eluting with (1:1) EtOAc/MeOH from the silica column was separated further by solid-phase extraction (Supelclean ENVI-18) eluting with a step-gradient of (20:1) water/acetonitrile, (4:1) water/acetonitrile, (3:2) water/acetonitrile, (2:3) water/acetonitrile, (1:20) water/acetonitrile, and acetone. All solvents, except acetone, were acidified with 0.1% formic acid. The active fraction, totaling 9.73 mg, eluted with (2:3) water/acetonitrile, and was separated further by semipreparative HPLC using an Agilent 1200 series HPLC and a Phenomenex Kinetex 5 μm C18 100 Å (150 mm × 10 mm) column, heated to 30°C, and a gradient of 42–95% acetonitrile in water with a flow rate of 4 mL min-1. All solvents were acidified with 0.1% formic acid. Fractions were pooled according to UV adsorption characteristics at 276 nm. A single active fraction eluted in the 58% water/42% acetonitrile fraction. A total of 1 mg of the active compound, determined to be 2-heptyl-4-quinolone, was recovered from 10.5 L of culture after 8 days of growth, yielding a final concentration of 0.39 μM.
Structure Determination of Bacterial Compounds
NMR experiments were conducted using an Agilent NMR 500 MHz (Agilent Technologies, Santa Clara, CA, USA) with DMSO-d6 as the solvent (referenced to residual DMSO at δH 2.50 and δC 39.5) at 25°C. HHQ standard was acquired from Sigma Aldrich (St. Louis, MO, USA).
Accurate mass spectra for 2-heptyl-4-quinolone was acquired on an Agilent Technologies 6230 TOF with a Dual Agilent Jet Stream Electrospray Ionization source, equipped with an Agilent 1260 Infinity series HPLC with a Phenomenex Kinetex 2.6 μm, C18, 100 Å, LC column (150 mm × 2.1 mm) as the stationary phase with a flow rate of 0.2 mL min-1. The ion source was operated at 350°C and 3500 V with a nitrogen gas flow of 8 L min-1, nebulizer pressure of 40 psi and fragmentor voltage of 135 V. The chromatrography method was as follows: 0–10 min at (1:1) water/acetonitrile; then ramped to (1:20) water/acetonitrile over the next 7 min; held at (1:20) water/acetonitrile for 3 min; then returned to (1:1) water/acetonitrile and held for 3 min. All solvents were acidified with 0.1% formic acid. The instrument was equipped with an Agilent Mass Hunter Workstation version B0.4.00 software.
Bioinformatic Analysis of P. piscicida Genome
High purity P. piscicida genomic DNA was extracted, purified, and sequenced using the Ion PGMTM Hi-Q Sequencing Kit according to the methods of Agarwal et al. (2014). Genomic DNA sequences were assembled into contigs using the SPAdes Genome Assembler (v 3.6.0; Bankevich et al., 2012), and the draft genome was annotated using RAST (Aziz et al., 2008). Following RAST annotation, a homology search was conducted for P. piscicida homologs matching the Pseudomonas aeruginosa pqsABCDE operon, pqsR, and pqsH genes, previously determined to be involved in alkylquinoline synthesis (Dulcey et al., 2013; Drees and Fetzner, 2015) using SEED Viewer (Version 2.0; Overbeek et al., 2005).
Dose-Response Experiments
The dose-response relationship of 2-heptyl-4-quinolone was measured in E. huxleyi (strains 624, 374, 379), D. tertiolecta, and P. tricornutum in growth inhibition assays with pure compounds tested at various concentrations in triplicate. All the E. huxleyi strains were axenic, where as the strains of D. tertiolecta and P. tricornutum were xenic. Dose-response experiments were conducted similarly to the growth inhibition assays. Cells from exponentially growing cultures were grown in triplicate 24-well plates. Initial cell abundance was determined by carbon concentration (Menden-Deuer and Lessard, 2000), where each well had an initial carbon concentration of 1.2 × 106 pg C mL-1. Well plates were kept under general culturing conditions, and cell abundance was monitored daily using a flow cytometer (Guava, Millipore) as described above.
Phytoplankton growth rate (μ d-1) was plotted against the concentration of each pure compound in order to determine the concentration of compound resulting in 50% growth inhibition (IC50). Pure compound IC50 values were calculated and 95% confidence intervals were estimated using Prism 6.0 software (GraphPad) by fitting the log transformation of the response variable (I; inhibitor concentration) by non-linear regression to the equation (1); where the slope factor (Hill Slope) is equal to -1.0 and the “Top” and “Bottom” numerals are equal the plateaus of curve in units of growth rate.
Chemical Profiling and Phylogenetic Analysis of Pseudoalteromonas sp.
DNA sequencing and phylogenetic analysis of marine isolate B030a (GenBank No. KT804650) was performed as described in Whalen et al. (2015). Evolutionary history of Pseudoalteromonas isolates were inferred using the neighbor-joining methods and the optimal tree is shown for topology. Exuded metabolites from 37 Pseudoalteromonas isolates in our chemical library underwent untargeted metabolomic fingerprint analysis as described in Whalen et al. (2015). A list of chemical features [m/z – retention time (RT) pairs] was filtered to include only those features that had concentrations that were at least one order of magnitude higher in the Pseudoalteromonas sample than in the media blank. This filtered list of features was then searched for the corresponding ion and RT in (-)-HRESI mode matching the standard 2-heptyl-4-quinolone. Phylogenetic analysis was performed as described in Whalen et al. (2015) and a heat map was generated showing the relative abundances of 2-heptyl-4-quinolone in each Pseudoalteromonas isolate.
Results
Phytoplankton Response to Live Bacterial Cells and Crude Extract
The presence of live P. piscicida cells caused significant mortality in E. huxleyi. When cultures of E. huxleyi (strain 624) were exposed to “high” concentrations of P. piscicida cells (106 cells mL-1), the growth rate of both axenic and xenic E. huxleyi cultures decreased 14 ± 5 and 28 ± 9%, respectively (Figure 1A; axenic, p = 0.015; xenic, p = 0.001). When exposed to “low” concentrations of P. piscicida cells at 103 cells mL-1, only xenic E. huxleyi had a significantly depressed (6 ± 1%) growth rate (p = 0.0048); whereas, axenic E. huxleyi were unaffected compared to f/2 controls. Additionally, filtrate from P. piscicida cells at 106 cells mL-1 significantly reduced the growth of xenic E. huxleyi by 32 ± 6% (p < 0.001).
FIGURE 1.
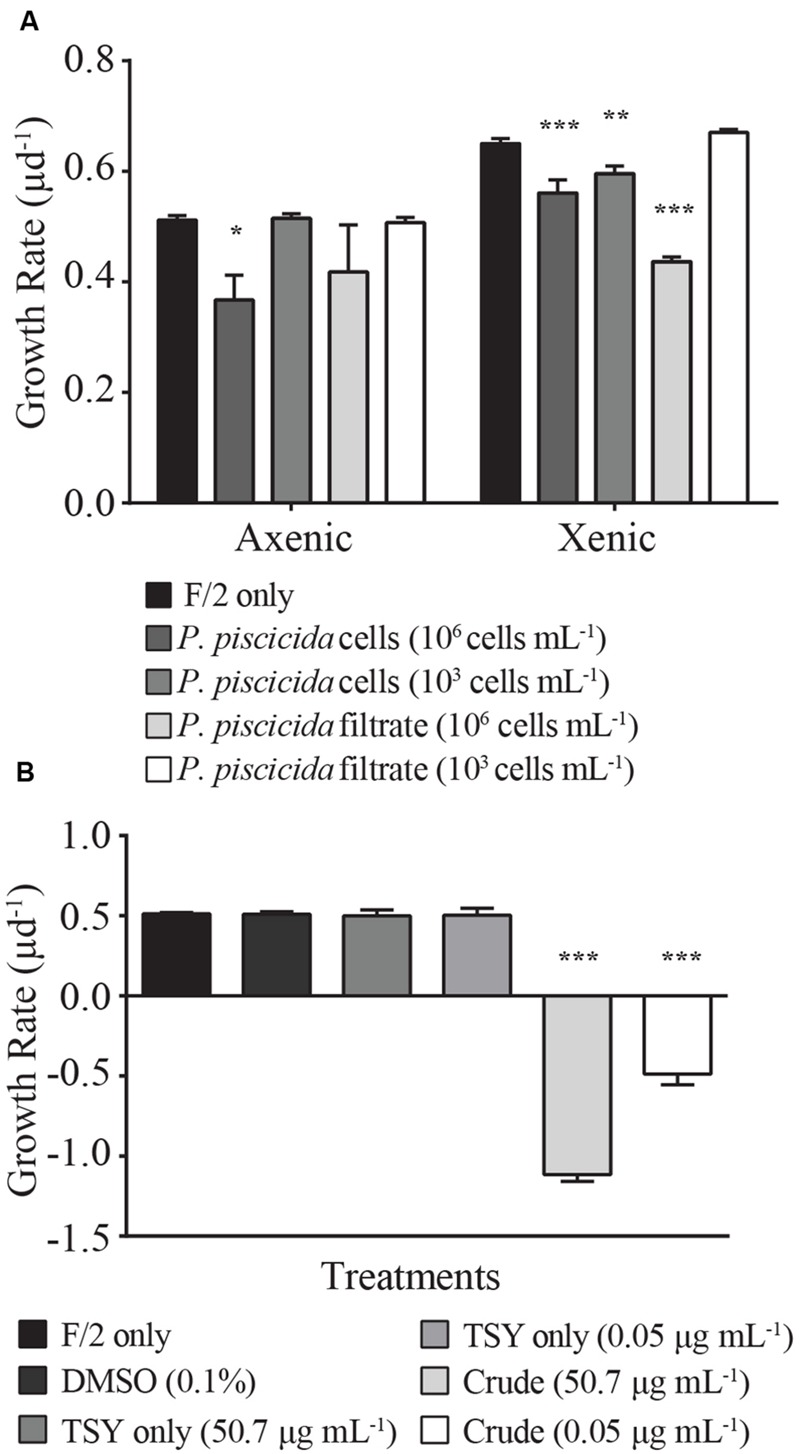
Growth rates (μ d-1) of Emiliania huxleyi (strain 624, either xenic or axenic) were measured in response to (A) live Pseudoalteromonas piscicida cells and filtrate. Growth rates (μ d-1) of axenic E. huxleyi (strain 624) were measured in response to (B) crude extracts of P. piscicida secreted metabolites and appropriate media (F/2 and TSY) and solvent (DMSO) controls. Compared to the control, the growth rate of E. huxleyi significantly decreased in response to high concentrations (106 cells mL-1) of P. piscicida and the crude extracts. Additionally, high concentrations of culture filtrate and low concentrations (103 cells mL-1) of P. piscicida resulted in a significant decrease in E. huxleyi growth but only in the xenic subculture. Significance is denoted as being less than 0.05 (∗), less than 0.01 (∗∗), or less than 0.001 (∗∗∗). For all treatments, n = 3. All error bars are one standard deviation of the mean.
Axenic E. huxleyi was exposed to crude extracts of exuded compounds produced by P. piscicida at a final concentration of 50.7 and 0.05 μg mL-1, concentrations approximately equivalent to the material exuded by 106 and 103 cells mL-1, respectively. Exposure to crude extracts resulted in significant mortality for E. huxleyi (Figure 1B; p < 0.001), with negative growth rates for E. huxleyi when exposed to either concentration of crude extract. Conversely, when exposed to crude extracts of TSY media, no significant difference in growth rate was observed relative to the DMSO and media only controls, indicating the causative agent of mortality was the result of a compound excreted by P. piscicida.
Compound Identification and Metabolomic Fingerprinting
The secreted algicidal compound produced by P. piscicida, was isolated using bioassay-guided fractionation. The purified active fraction was investigated with 1D-NMR and ESIMS experiments. 1H and 13C NMR showed the presence of a seven carbon straight alkyl chain (δH 0.85, 1.26, 1.32, 1.67, and 2.58 and δC 13.9, 22.0, 28.3, 28.4, 28.5, 31.1, and 33.2) and a 1,2 substituted benzene ring [δH 7.26 (d), 7.52 (t), 7.60 (t), and 8.03 (d); Figure 2A and Supplementary Figure S2]. These data along with a robust negative ESIMS ion at 242.155 m/z (Figure 2B) strongly suggested the known bacterial metabolite 2-heptyl-4-quinolone (HHQ, C16H21NO; Bredenbruch et al., 2005). Comparison of 1H and 13C NMR spectra of the active compound to an HHQ standard confirmed that the chemical structures were identical. Untargeted metabolomic profiling of Pseudoalteromonas crude extracts as detailed in Whalen et al. (2015), indicated HHQ was present in isolates A757, A754, A746, and B030a (Figures 2C,D). The production of HHQ is the most pronounced in a clade containing A757, A754, and A746 representing a distinct chemotype characterized by halogenation of the exometabolome (Whalen et al., 2015). 16S rDNA sequencing indicates A757, A754, and A756 are all P. piscicida and were isolated from plastic debris in the North Atlantic (Whalen et al., 2015). The HHQ-producing marine isolate B030a was isolated from macroalgae along the coast of Woods Hole, MA, USA, but is not in the same clade as A757, A754, and A746, rather this B030a falls within a large clade represented by diverse group of Pseudoalteromonas sp. (Figure 2C).
FIGURE 2.
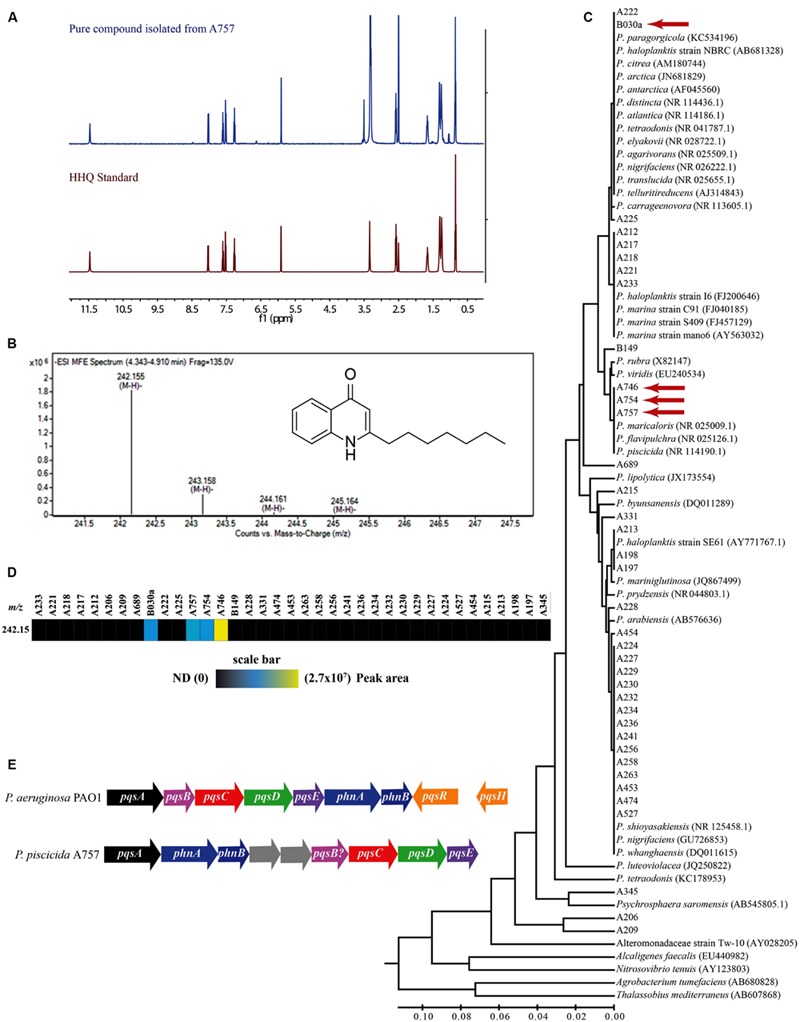
Composite figure showing HHQ structural elucidation from isolate A757, chemophylogenetic analysis of HHQ producing Pseudoalteromonas sp., and HHQ biosynthetic machinery in P. piscicida (A757). (A) Structural identification of 2-heptyl-4-quinolone (HHQ) from P. piscicida. 1H-NMR spectrum 500 MHz, DMSO-d6 of the pure compound isolated from P. piscicida (blue) and the authentic standard HHQ (red). (B) Mass spectrum [(-)-HRESI] of the active constituent from P. piscicida (A757; structure of HHQ in inset). (C) Phylogenetic tree and comparison of HHQ production in 37 isolates of Pseudoalteromonas sp. The evolutionary history was inferred using the UPGMA method (Sneath and Sokal, 1973). The optimal tree with the sum of branch length = 0.83629248 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) and are in the units of the number of base substitutions per site. The analysis involved 77 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 394 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013). Isolates identified to produce HHQ are indicated with a red arrow. (D) Heat map showing the relative abundance of HHQ produced by Pseudoalteromonas sp. profiled in Whalen et al. (2015). Colored bars indicate the peak area of HHQ for each marine isolate indicated by the column headings. (E) A comparison of the alkylquinoline biosynthetic pathway in Pseudomonas aeruginosa PAO1 and Pseudoalteromonas piscicida (A757; GenBank Accession numbers, KT879191–KT879199).
Identification of HHQ Biosynthetic Machinery in P. piscicida
The draft genome sequence of P. piscicida was determined to be 5.1 Mbp in size with an estimated 4543 coding sequences. The P. piscicida genome was mined using SEED Viewer for the identification of genes involved alkylquinoline synthesis (GenBank Accession numbers, KT879191–KT879199). In P. aeruginosa, quorum sensing is controlled by the transcriptional regulators LasR, RhIR, and MvfR (PqsR). The former two are controlled by homoserine lactones, while the signaling molecules that activate MvfR, a LysR-type transcriptional regulator, belong to a family of 4-hydroxy-2-alkylquinolines (HAQs), including 3,4-dihydroxy-2-heptylquinoline (PQS, 258.149 m/z [M-H]), its direct precursor 2-heptyl-4-quinolone (HHQ), and 4-hydroxy-2-heptylquinoline-N-oxide (HQNO; Dulcey et al., 2013). The presence of a single homolog to each of the P. aeruginosa pqsA, pqsC, pqsD, and pqsE genes were identified in the P. piscicida genome, with amino acid identities of 37.9, 28.5, 47.2, and 30.1%, respectively. However, in silico analysis indicated the operon arrangement was different to that of P. aeruginosa (Figure 2E). The coding sequence with the highest similarity to P. aeruginosa pqsB was positioned immediately upstream of the homolog pqsC in the P. piscicida genome with 20.3% amino acid identity. The position of this hypothetical protein within the operon containing other pqsABCDE homologs would suggest it is a possible homolog to pqsB. The genome of P. aeruginosa also encodes two anthranilate synthases, PhnA and PhnB, immediately downstream of the pqsABCDE operon, which supply anthranilic acid. The genome of P. piscicida contains two homologs of PhnA and PhnB (Figure 2E) with amino acid identities of 29.8 and 40.5%, respectively; however, their position is shifted in comparison to P. aeruginosa and downstream of the homolog to pqsA.
After finding the pqsABCDE homologs in our P. piscicida (A757) isolate, we were interested in determining if this operon exists in other Pseudoalteromonas sp. whose genome has been made publically available. Using the Department of Energy – Joint Genome Institute’s Integrated Microbial Genome (IMG) system, we searched 67 Pseudoalteromonas sp. available genomes that were categorized as finished, in permanent draft, or draft stage against two P. aeruginosa PAO1 genomes (637000218 and 2623620964) for pqsABCDE operon homologs. Using a minimum identity cutoff of 10% and e-value of 0.01, we were able to find representatives of many of the genes in the pqs operon (Supplementary Figure S3A). However, upon closer manual inspection of these genomes, only Pseudoalteromonas citrea NCIMB 1889 maintained the synteny of the pqs operon as seen in P. piscicida (A757; Supplementary Figure S3B).
Dose-Response Experiments with Various Phytoplankton Species
Dose-response experiments with axenic E. huxleyi (strain 624) determined the IC50 concentration for the isolated pure compound to be 88.3 ng mL-1, which was not significantly different (p = 0.92) from the IC50 obtained when exposed to the purchased HHQ standard (Figure 3A, Table 1; Sigma-Aldrich). For all three E. huxleyi strains, IC50 concentrations for the isolated pure compound ranged from 44.51 to 115.4 ng mL-1 (Figure 3B; Table 2). IC50 values were found to differ significantly (p = 0.003) when all three strains of E. huxleyi were compared. The naked strain, 374 was found to be the most sensitive to HHQ, having the lowest IC50 value, while the haploid strain, 379 was the least sensitive. A comparison of IC50 values obtained for all three strains of E. huxleyi with other phytoplankton species, D. tertiolecta and P. tricornutum, indicated E. huxleyi was significantly more susceptible to HHQ, despite both D. tertiolecta and P. tricornutum being vulnerable to the crude P. piscicida extract (Supplementary Figure S4).
FIGURE 3.
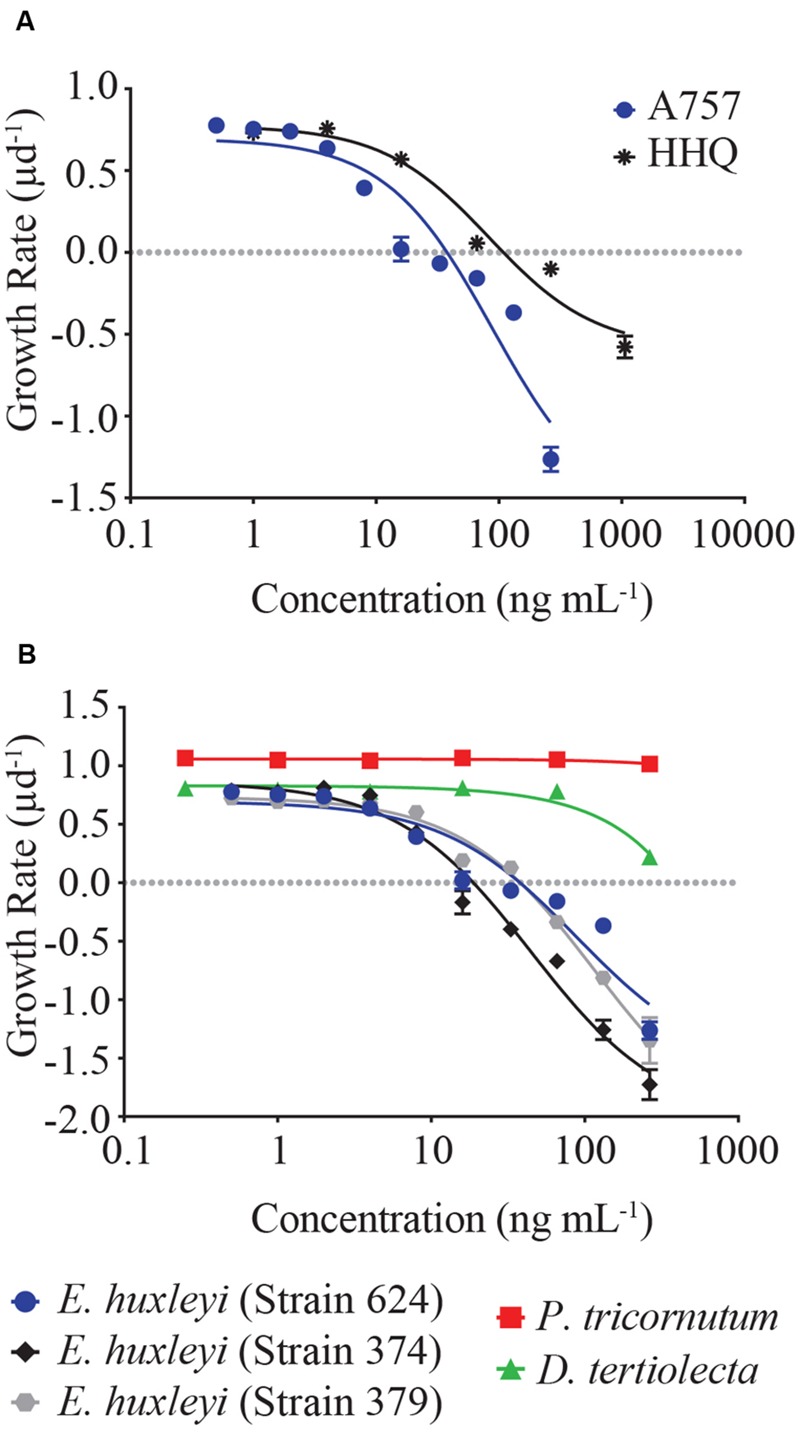
(A) Dose-response comparison of E. huxleyi (strain 624, axenic) growth rate in response to a gradient of extracted (labeled A757) and reference HHQ. (B) Dose-response curves of five different strains of phytoplankton in response to HHQ extracted from P. piscicida. Each symbol represents the mean of three independent replicates ± the standard deviation. IC50 values were calculated for each curve (see Table 1).
Table 1.
Statistical comparison of the dose-response curves and IC50 values generated for Emiliania huxleyi (Strain 624) exposed to pure compound from A757 and the HHQ standard.
| Compound | IC50 (ng mL-1) | nM | R2 | 95% CI | p-value |
|---|---|---|---|---|---|
| A757 | 88.31 | 362 | 0.91 | 45.7–170.4 | 0.9218 |
| HHQ standard | 83.07 | 341 | 0.96 | 50.0–138.0 |
Table 2.
Inhibitory concentration (IC50) of HHQ isolated from Pseudoalteromonas piscicida in five different strains of phytoplankton.
| Species | IC50 (ng mL-1) | nM | R2 | 95% CI |
|---|---|---|---|---|
| Emiliania huxleyi (624) | 88.31 | 362 | 0.91 | 45.7–170.4 |
| E. huxleyi (374) | 44.51 | 182 | 0.98 | 33.3–59.4 |
| E. huxleyi (379) | 115.4 | 474 | 0.98 | 87.2–152.8 |
| Dunaliella tertiolecta | >100000 | – | – | – |
| Phaeodactylum tricornutum | >100000 | – | 0.93 | – |
In order to decipher when HHQ-induced mortality of E. huxleyi (strain 624) occurred, growth rates of the alga were examined across 24 h time increments (Figure 4). While the growth rate of E. huxleyi when exposed to HHQ concentrations was dose-dependent, daily growth rate in many of the concentrations remained relatively stable over time. For example, daily growth rate when exposed to 132 ng mL-1 of HHQ ranged from -0.19 to -0.32 d-1, over the experimental time period. This indicates that the total loss of E. huxleyi when exposed to high concentrations of HHQ occurs over time, rather than a singular loss event.
FIGURE 4.
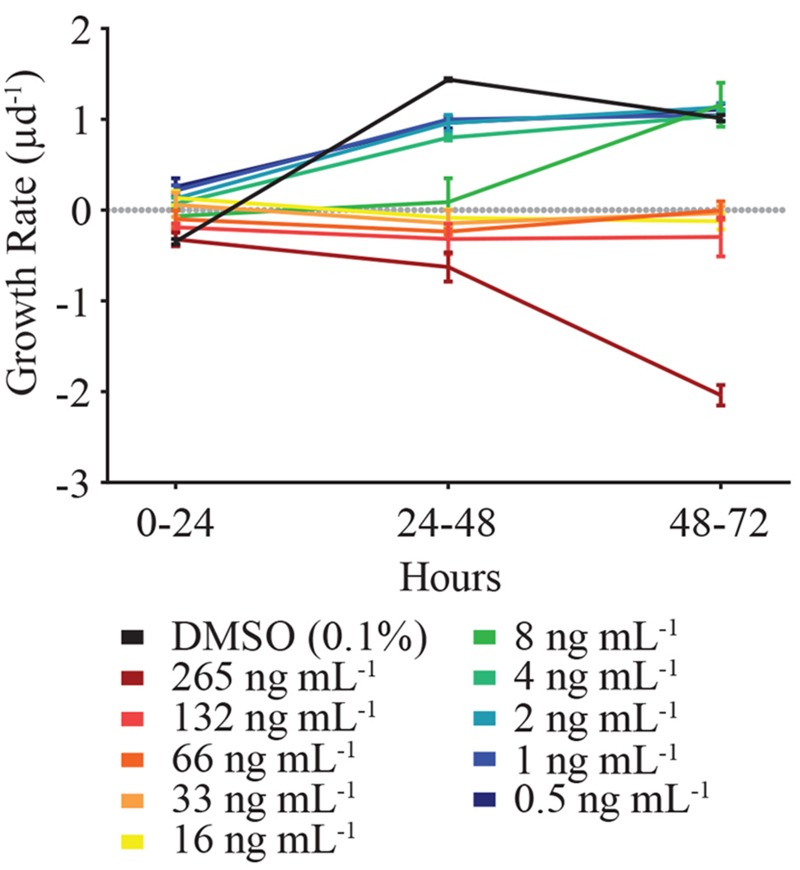
The incremental growth rate (μ d-1) of E. huxleyi (strain 624) over the experimental time period. At toxic concentrations (between 8 ng mL-1 and above), E. huxleyi growth rate remained relatively consistent among the time periods examined. Thus, the presence of HHQ results in static E. huxleyi growth rather than cell lysis. Error bars are one standard deviation of the mean.
Discussion
Bacterially Mediated Mortality of Marine Phytoplankton
Quorum sensing molecules mediate bacterial cell–cell communication allowing cells to respond collectively to changes in their environment. This study is the first to identify a quorum sensing precursor molecule that induces mortality in marine phytoplankton. Initial culturing experiments with high P. piscicida cell densities (106 cells mL-1) resulted in significant mortality in E. huxleyi cultures. This finding is concurrent with previous observations demonstrating high concentrations of Pseudoalteromonas sp. induced mortality in a wide range of phytoplankton species including raphidophytes, gymnodinoids, and diatoms (Lovejoy et al., 1998; Mitsutani et al., 2001). While the concentration of bacteria used in these experiments is high compared to natural abundances, bacterial behavior (e.g., swarming, biofilm formation) can create localized concentrations of bacteria on or around phytoplankton cells. Furthermore, in some instances, it is only when bacteria are attached to surfaces that they produce active metabolites, thereby generating microenvironments of high metabolite concentrations (Long et al., 2003).
Algicidal activity can be mediated either directly (bacterial cell – phytoplankton contact) or indirectly via chemical interactions (Figure 1). Filtrate from P. piscicida cells resulted in E. huxleyi mortality, indicating a possible chemical basis for the algicidal activity observed. Bio-assay guided fractionation revealed that HHQ, a compound released by P. piscicida, caused inhibition of E. huxleyi growth. This study is the first observation of HHQ production by a marine Pseudoalteromonas species. Previous work describes PQS and HHQ production as occurring during the stationary phase of growth, and both can function as antibiotics and signaling molecules in cell–cell communication, with roles in virulence, apoptosis, and viability of fungal and mammalian cells (Bredenbruch et al., 2005; Calfee et al., 2005; Reen et al., 2011). Unlike PQS, which has poor solubility in water; HHQ can passively diffuse into the external aqueous environment (Deziel et al., 2004). These diffusible signaling molecules, termed autoinducers, are a form of intercellular communication released by bacteria in a cell density-dependent manner, designed to engage cells in cooperative and coordinated behavior beneficial to the entire bacterial population. Once these autoinducers reach critical threshold, they cause the transcriptional activation of quorum-sensing-controlled genes that are key in biofilm formation, virulence factor production (Bredenbruch et al., 2005), secondary metabolite production, motility (Reen et al., 2011), and pathogenesis (Zaborin et al., 2009).
Exposure to HHQ resulted in a significant decrease in growth rate and eventual mortality of E. huxleyi (Figures 3 and 4). While this is the first observation of HHQ-induced mortality in a phytoplankton species, responses to other alkyl-quinolones have been documented in phytoplankton. For example, Long et al. (2003) found that exposure to 2-n-pentyl-4-quinolinol (PQ), also produced in the PQS pathway, resulted in mortality of several phytoplankton species. These results imply that HHQ and other compounds produced in the PQS pathway may have functions beyond quorum sensing and anti-microbial activity, and function as inter-domain cues that initiate eukaryotic cell death. While the concentration of HHQ in the marine environment has not been measured, acyl-homoserine lactones (AHLs), another class of autoinducers, have been measured at nanomolar concentrations in the aqueous phase or higher in biofilms (Wheeler et al., 2006) and have been implicated in the control of degradation of sinking detritus (Hmelo et al., 2011) and involved in phosphorus acquisition by epibonts of Trichodesmium sp. (Van Mooy et al., 2012). The IC50 observed for E. huxleyi in response to HHQ is within the nanomolar range (Tables 1 and 2), implying that if the concentration of HHQ parallels other quorum sensing molecules in the marine environment, especially within the context of a biofilm, E. huxleyi would be highly susceptible to this autoinducer.
It is worth noting that HHQ was not toxic to all phytoplankton. When D. tertiolecta and P. tricornutum were exposed to HHQ, no significant change in growth rate was observed, even at high concentrations (Figure 3). This specificity in phytoplankton susceptibility to bacterial cells has been observed previously in response to the presence of live Pseudoalteromonas sp. cells (Lovejoy et al., 1998), indicating differential species sensitivity resulting from variability in chemical resistance mechanisms or target-site insensitivity. Interestingly, when D. tertiolecta and P. tricornutum were exposed to extracts of P. piscicida exudate, both phytoplankton species exhibited significant mortality in comparison to controls, suggesting this bacterium may produce a cocktail of algicidal compounds specific to different phytoplankton species.
Phylogenetic Evaluation of HHQ Biosynthetic Pathways
Our survey of HHQ production in the Pseudoalteromonas sp. within our chemical library indicated that two clades contained representatives capable of producing HHQ at measureable levels in culture. Isolates A757, A754, A746, and B030a fall out by 16S rDNA into two clades that contain 19 different Pseudoalteromonas sp. HHQ-producing clades represent both open ocean and coastal species isolated from both abiotic and biotic surfaces, indicating HHQ-producing species can inhabit diverse marine environments. With the recent re-evaluation of the alkylquinoline biosynthetic pathway described from the pathogen P. aeruginosa in Dulcey et al. (2013) and Drees and Fetzner (2015), we identified homologs of the pqsABCDE operon responsible for HHQ synthesis in P. piscicida A757. PQS and HHQ are able to bind MvfR (PqsR) and induce the expression of the pqsABCDE operon, which controls biosynthesis of HAQs (Deziel et al., 2004; Dubern and Diggle, 2008). The presence of PqsA, anthranilate-CoA ligase, activates anthranilic acid into anthraniloyl-CoA which reacts with PqsD, a 3-oxoacyl synthase, to produce the intermediate, anthraniloyl-PqsD that is further modified by the synthases, PqsB, PqsC (Dulcey et al., 2013). The product of the gene pqsE is thought to be a thioesterase, hydrolyzing the biosynthetic intermediate 2-aminobenzoylacetyl-coenzyme A to form 2-aminobenzoylacetate, the precursor of HHQ (Drees and Fetzner, 2015).
We did not find PQS in our metabolomic analysis of Pseudoalteromonas sp. excretome, which is supported by the lack of a conclusive P. piscicida homolog to pqsH, the monooxygenase responsible for converting HHQ to PQS. The loss of the pqsH has been observed in two Burkholderia sp., which lacked PQS production, but produced HHQ (Diggle et al., 2006). The lack of an ion corresponding to PQS in P. piscicida A757 was confirmed by our untargeted metabolomic fingerprint analysis (Whalen et al., 2015). In P. aeruginosa, the LysR transcriptional regulator MvfR (PqsR) binds to the promoter of the pqsABCDE and is induced by HAQs. P. piscicida does contain a putative pqsR homolog identified as a LysR family transcriptional regulator with 23.2% similarity and located 2.1 Kbp away from psqE.
An analysis of the presence of this pathway in the publically available genomes of marine Pseudoalteromonads indicated that other species in this genus, including P. citrea, would have the biosynthetic capacity to produce HHQ. Interestingly, the mining of two additional P. piscicida publically available genomes (JGI: 2519899641 and 2541047159) indicated the lack of the pqs operon, suggesting the loss or reshuffling of these genes in subspecies. A recent study also indicated that the lack of PqsE homologs might not prohibit the synthesis of HHQ, rather the thioesterase function could be taken over to an extent by broad-specificity thioesterases, like TesB (Drees and Fetzner, 2015). This finding is significant to our study, as we generally lacked identifying homologs to pqsE in our genomic analysis.
In addition, other marine members of the gamma-proteobacteria have also been described to produce HHQ (Wratten et al., 1977; Debitus et al., 1998), including P. aeruginosa. Indeed, we confirmed the production of HHQ by comparison to the authentic standard via LC–MS from a marine isolate of P. aeruginosa (B323) in the Mincer Culture Collection isolated from water samples 430 km north of the British Virgin Islands (data not shown). These data indicate a more phylogenetically diverse grouping of marine members of the gamma-proteobacteria have the ability to produce algicidal compounds, including HHQ, consistent with earlier predictions of algicidal clades (Mayali and Azam, 2004); however, molecules responsible for the bioactivity have yet to be described. Additionally, Machan et al. (1992) reported HHQ inhibited the growth of Staphylococcus aureus and other gram-positive bacteria. While this finding was reported in clinical isolates of P. aeruginosa, an additional role of HHQ other than mediating cell–cell communication and phytoplankton abundance could include mediating Gram-positive bacterial populations, possibly associated with phytoplankton surfaces.
Mode of Mortality and Biogeochemical Implications
Exposure to HHQ resulted in static growth of E. huxleyi, which eventually led to mortality of the alga, rather than an immediate cell lysis event upon exposure (Figure 4). Static growth upon exposure to HHQ has been observed in bacteria, with growth inhibition observed in several Vibrio species and a marine sponge bacterial isolate (Reen et al., 2011). In contrast, other studies have observed phytoplankton that were exposed to Pseudoalteromonas sp. and lysed within several hours of exposure (Lovejoy et al., 1998), indicating multiple compounds are acting through different mechanisms of action. We did identify a second compound from A757 exudate whose activity results in immediate cell lysis; however, attempts to identify this compound are still ongoing. Cell lysis verses static growth is an important distinction, as it ultimately impacts the fate of algal cells and carbon cycling. With static growth, E. huxleyi cells would remain in the water column, which could increase aggregation and export. Conversely, cell lysis is a final fate for the alga, fueling the microbial loop and decreasing carbon flow to higher trophic levels.
These results suggest that pelagic bacteria are not passive receivers of dissolved organic material. For example, it has been observed that when co-cultured under low iron conditions, P. aeruginosa cells lysed Staphylococcus aureus for its useable iron. Similar relationships have been proposed for bacteria–phytoplankton interactions, with bacteria having the potential to control algal blooms (Doucette et al., 1998). When the concentration of bacteria is high enough to induce quorum sensing (and the production of HHQ) the bacterial population could use the nutrients regenerated from static and dead phytoplankton cells to maintain growth. Algal substrates can provide ecological niches for species-specific bacterial populations to thrive (Teeling et al., 2012). These surface-associated bacteria may be relatively impervious to changes in seawater chemistry (pCO2); however, their population dynamics are tightly coupled to phytoplankton bloom development (Allgaier et al., 2008). Even a small increase in the exponential growth rate of a bacterial population can have large consequences for overall population abundance. Our results, while preliminary, provide novel hypotheses for further inquiry into the role of antagonistic phytoplankton-bacterial interactions. Furthermore, identification of the chemical compounds responsible for phytoplankton mortality will provide an enhanced ability to project shifts in population dynamics of both organisms.
The ability of HHQ to modulate interspecies interactions and cross-kingdom behavior (i.e., fungi and animals) as seen here and other studies (Reen et al., 2011), implicates alkylquinolone-signaling molecules as having important ecological roles in regulating primary production and affecting phytoplankton successions. HHQ could potentially alter both bacterial and phytoplankton composition, and physiologic parameters. Elucidating the impacts of this pathway will enable us to better parameterize phytoplankton population dynamics, and ultimately, provide better understanding of planktonic food web structure and biogeochemical cycling.
Author Contributions
EH and KW designed experiments; KW performed the chemical isolation; EH performed phytoplankton growth assays; RD and DR solved the chemical structure; AE, MS, BM performed genome sequencing and assembly; EH and KW analyzed data; all authors participated in writing and editing manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank K. Bidle for provision of the E. huxleyi strains.
Footnotes
Funding. This research was support through funding from the Gordon and Betty Moore Foundation through Grant GBMF3301 to MJ and TM; NIH grant from the National Institute of Allergy and Infectious Disease (NIAID – 1R21Al119311-01) to TM and KW; the National Science Foundation (OCE – 1313747) and US National Institute of Environmental Health Science (P01-ES021921) through the Oceans and Human Health Program to BM. Additional financial support was provided to TM from the Flatley Discovery Lab. Instruments used for the various chemical analyses were supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (grant number 2 P20 GM103430). NMR data was acquired at a research facility supported in part by the National Science Foundation EPSCoR Cooperative Agreement #EPS-1004057.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00059
Flow chart detailing the bioassay guided fractionation process that was used to eventually identify HHQ as a causative compound influencing growth of E. huxleyi.
Structural identification of 2-heptyl-4-quinolone (HHQ) from P. piscicida. 13C-NMR spectrum (75 MHz, DMSO-d6) of the pure compound isolated from P. piscicida (blue) and the authentic standard HHQ (red).
Genome mining of pqs operon in Pseudoalteromonas sp. (A) The pqs operon from two species of Pseudomonas aeruginosa PAO1 were used to query 67 publically available Pseudoalteromonas sp. genomes. Joint Genome Institute (JGI) genome accession numbers are listed for each isolate in parentheses. Red boxes indicate the presence of a homolog and black boxes indicate no homolog identified with BLAST cutoffs set at ≥10% identity and a maximum e-value of 0.01. White asterisks indicate two homologs (pqsB and pqsE) from P. citrea that were initially identified as absent in the genome using the Integrated Microbial Genome (IMG) gene-genome comparison analysis, but upon manual inspection, were found to be present. (B) A comparison of the alkylquinoline biosynthetic pathway in Pseudoalteromonas piscicida (A757; GenBank Accession no., KT879191–KT879199) and P. citrea (NCIMB 1889). Shading of homologs in P. citrea indicates percent amino acid identity to P. piscicida (A757) genes.
Growth rate (μ d-1) for three species of phytoplankton exposed to the crude extract of the secreted metabolites of P. piscicida. Despite HHQ having little effect on D. tertiolecta and P. tricornutum, toxicity to the crude extract of P. piscicida is observed in all species, indicating that P. piscicida likely produces additional compounds that result in algal mortality. Error bars are one standard deviation from the mean.
References
- Agarwal V., El Gamal A. A., Yamanaka K., Poth D., Kersten R. D., Schorn M., et al. (2014). Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat. Chem. Biol. 10 640–647. 10.1038/nchembio.1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allgaier M., Riebesell U., Vogt M., Thyrhaug R., Grossart H.-P. (2008). Coupling of heterotrophic bacteria to phytoplankton bloom development at different pCO2 levels: a mesocosm study. Biogeosciences 5 1007–1022. 10.5194/bg-5-1007-2008 [DOI] [Google Scholar]
- Azam F., Fenchel T., Field J. G., Meyer-Reil L. A., Thingstad F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10 257–263. 10.3354/meps010257 [DOI] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W. M., Holligan P. M., Kilpatrick K. A. (1992). Calcification, photosynthesis and growth of the bloom-forming coccolithophore Emiliania huxleyi. Cont. Shelf Res. 12 1353–1374. 10.1016/0278-4343(92)90059-S [DOI] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. P. (2007). Bioactive compound synthetic capacity and ecological signifcance of marine bacterial genus Pseudoalteromonas. Mar. Drugs 5 220–241. 10.3390/md504220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratbak G., Thingstad T. F. (1985). Phytoplankton-bacteria interactions: an apparent paradox? Analysis of a model system with both competition and commensalism. Mar. Ecol. Prog. Ser. 25 23–30. [Google Scholar]
- Bredenbruch F., Nimtz M., Wray V., Moor M., Muller R., Haussler S. (2005). Biosynthetic pathway of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines. J. Bacteriol. 187 3630–3635. 10.1128/JB.187.11.3630-3635.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfee M. W., Shelton J. G., McCubrey J. A., Pesci E. C. (2005). Solubility and bioactivity of the Pseudomonas quinolone signal are increased by a Pseudomonas aeruginosa-produced surfactant. Infect. Immun. 73 878–882. 10.1128/IAI.73.2.878-882.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danger M., Oumarou C., Benest D., Lacroix G. (2007). Bacteria can control stoichiometry and nutrient limitation of phytoplankton. Funct. Ecol. 21 202–210. 10.1111/j.1365-2435.2006.01222.x [DOI] [Google Scholar]
- Debitus C., Guella G., Mancini I. I., Waikedre J., Guemas J. P., Nicolas J. L., et al. (1998). Quinolones from a bacterium and tyrosine metabolites from its host sponge, Suberea creba from the Coral Sea. J. Mar. Biotechnol. 6 136–141. [PubMed] [Google Scholar]
- Deziel E., Lepine F., Milot S., He J. X., Mindrinos M. N., Tompkins R. G., et al. (2004). Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. U.S.A. 101 1339–1344. 10.1073/pnas.0307694100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle S. P., Lumjiaktase P., Dipilato F., Winzer K., Kunakorn M., Barrett D. A., et al. (2006). Functional genetic analysis reveals a 2-alkyl-4-quinolone signalling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem. Biol. 13 701–710. 10.1016/j.chembiol.2006.05.006 [DOI] [PubMed] [Google Scholar]
- Doucette G. J., Kodama M., Franca S., Gallacher S. (1998). “Bacterial interactions with harmful algal bloom species: bloom ecology, toxigenesis, and cytology,” in The Physiological Ecology of Harmful Algal Blooms eds Anderson D. M., Cembella A. D., Hallegraeff G. M. (Berlin: Springer Verlag; ) 619–647. [Google Scholar]
- Drees S. L., Fetzner S. (2015). PqsE of Pseudomonas aeruginosa acts as pathway-specific thioesterase in the biosynthesis of alkylquinolone signaling molecules. Chem. Biol. 22 611–618. 10.1016/j.chembiol.2015.04.012 [DOI] [PubMed] [Google Scholar]
- Dubern J. F., Diggle S. P. (2008). Quorum sensing by a 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol. Biosyst. 4 882–888. 10.1039/b803796p [DOI] [PubMed] [Google Scholar]
- Dulcey C. E., Dekimpe V., Fauvelle D. A., Milot S., Groleau M. C., Doucet N., et al. (2013). The end of an old hypothesis: the Pseudomonas signalling molecules 4-hydroxy-2-alkylquinolines derive from fatty acids, not 3-ketofatty acids. Chem. Biol. 20 1481–1491. 10.1016/j.chembiol.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami K., Nishijima T., Ishida Y. (1997). Stimulative and inhibitory effects of bacteria on the growth of microalgae. Hydrobiologia 358 185–191. 10.1023/A:1003141104693 [DOI] [Google Scholar]
- Gilbert J. A., Steele J. A., Caporaso J. G., Steinbrück L., Reeder J., Temperton B., et al. (2012). Defining seasonal marine microbial community dynamics. ISME J. 6 298–308. 10.1038/ismej.2011.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard R. R. L. (1975). “Culture of phytoplankton for feeding marine invertebrates,” in Culture of Marine Invertebrate Animals eds Smith W. L., Chaney M. H. (New York, NY: Plenum Press; ) 29–60. [Google Scholar]
- Hmelo L. R., Mincer T. J., Van Mooy B. A. S. (2011). Possible influences of bacterial quorum sensing on the hydrolysis of sinking particulate organic carbon in marine environments. Environ. Microbiol. Rep. 3 682–688. 10.1111/j.1758-2229.2011.00281.x [DOI] [PubMed] [Google Scholar]
- Iglesias-Rodriguez M. D., Halloran P. R., Rickaby R. E. M., Hall I. R., Colmenero-Hidalgo E., Gittins J. R., et al. (2008). Phytoplankton calcification in a high-CO2 world. Science 320 336–340. 10.1126/science.1154122 [DOI] [PubMed] [Google Scholar]
- Kim J.-D., Kim J.-Y., Park J.-K., Lee C.-G. (2009). Selective control of the Prorocentrum minimum harmful algal blooms by a novel algal-lytic bacterium Pseudoalteromonas haloplanktis AFMB-008041. Mar. Biotech. 11 463–472. 10.1007/s10126-008-9167-9 [DOI] [PubMed] [Google Scholar]
- Legendre L., Rassoulzadegan F. (1995). Plankton and nutrient dynamics in marine waters. Ophelia 41 153–172. 10.1080/00785236.1995.10422042 [DOI] [Google Scholar]
- Long R. A., Qureshi A., Faulkner D. J., Azam F. (2003). 2-n-Pentyl-4-quinolinol produced by a marine Alteromonas sp. and its potential ecological and biogeochemical roles. Appl. Environ. Microbiol. 69 568–576. 10.1128/AEM.69.1.568-576.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy C., Bowman J. P., Hallegraeff G. M. (1998). Algicidal effects of a novel marine Pseudoalteromonas isolate (class Proteobacteria, gamma subdivision) on harmful algal bloom species of the genera Chattonella, Gymnodinium, and Heterosigma. Appl. Environ. Microbiol. 64 2806–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machan Z. A., Taylor G. W., Pitt T. L., Cole P. J., Wilson P. (1992). 2-heptyl-4-hydroxyquinoline n-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J. Antimicrob. Chemother. 30 615–623. 10.1093/jac/30.5.615 [DOI] [PubMed] [Google Scholar]
- Mayali X., Azam F. (2004). Algicidal bacteria in the sea and their impact on algal blooms. J. Eukaryot. Microbiol. 51 139–144. 10.1111/j.1550-7408.2004.tb00538.x [DOI] [PubMed] [Google Scholar]
- Menden-Deuer S., Lessard E. J. (2000). Carbon to volume relationships for dinoflagellates, diatoms, and other protist plankton. Limnol. Oceanogr. 45 569–579. 10.4319/lo.2000.45.3.0569 [DOI] [Google Scholar]
- Mitsutani A., Yamasaki I., Kitaguchi H., Kato J., Ueno S., Ishida Y. (2001). Analysis of algicidal proteins of a diatom-lytic marine bacterium Pseudoalteromonas sp. strain A25 by two-dimensional electrophoresis. Phycologia 40 286–291. 10.2216/i0031-8884-40-3-286.1 [DOI] [Google Scholar]
- Overbeek R., Begley T., Butler R. M., Choudhuri J. V., Chuang H. Y., Cohoon M., et al. (2005). The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33 5691–5702. 10.1093/nar/gki866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reen F. J., Mooij M. J., Holcombe L. J., McSweeney C. M., McGlacken G. P., Morrissey J. P., et al. (2011). The Pseudomonas quinolone signal (PQS), and its precursor HHQ, modulate interspecies and interkingdom behaviour. FEMS Microbiol. Ecol. 77 413–428. 10.1111/j.1574-6941.2011.01121.x [DOI] [PubMed] [Google Scholar]
- Ross A. C., Gulland L. E., Dorrestein P. C., Moore B. S. (2014). Targeted capture and heterologous expression of the Pseudoalteromonas alterchormide gene cluster in Escherichia coli represents a promising natural products exploratory platform. ACS Synth. Biol. 4 414–420. 10.1021/sb500280q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost M. R., Carr G., Kolter R., Clardy J. (2011). Roseobacticides: small molecule modulators of an algal-bacterial symbiosis. J. Am. Chem. Soc. 133 18343–18349. 10.1021/ja207172s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour J. R., Simo R., Ahmed T., Stocker R. (2010). Chemoattraction to dimethyl-sulfoniopropionate throughout the marine microbial food web. Science 329 342–345. 10.1126/science.1188418 [DOI] [PubMed] [Google Scholar]
- Simo R. (2001). Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological, and evolutionary links. Trends Ecol. Evol. 16 287–294. 10.1016/S0169-5347(01)02152-8 [DOI] [PubMed] [Google Scholar]
- Skerratt J. H., Bowman J. P., Hallegraeff G., James S., Nichols P. D. (2002). Algicidal bacteria associated with blooms of a toxic dinoflagellate in a temperate Australian estuary. Mar. Ecol. Prog. Ser. 244 1–15. 10.3354/meps244001 [DOI] [Google Scholar]
- Skovhus T. L., Holmstrom C., Kjelleberg S., Dahllof I. (2007). Molecular investigation of the distribution, abundance, and diversity of the genus Pseudoalteromonas in marine samples. FEMS Microbiol. Ecol. 61 348–361. 10.1111/j.1574-6941.2007.00339.x [DOI] [PubMed] [Google Scholar]
- Sneath P. H. A., Sokal R. R. (1973). Numerical Taxonomy: The Principles and Practice of Numerical Classification. San Francisco, CA: W. H. Freeman. [Google Scholar]
- Sneed J. M., Sharp K. H., Ritchie K. B., Paul V. J. (2014). The chemical cue tetrabromopyrrole from a biofilm bacterium induces settlement of multiple Caribbean corals. Proc. Biol. Sci. 281:pii20133086 10.1098/rspb.2013.3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Nei M., Kumar S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Nat. Acad. Sci. U.S.A. 101 11030–11035. 10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling H., Fuchs B. M., Becher D., Klockow C., Kabisch A., Bennke C. M. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336 608–611. 10.1126/science.1218344 [DOI] [PubMed] [Google Scholar]
- Van Mooy B. A. S., Hmelo L. R., Sofen L. E., Campagna S. R., May A. L., Dyhrman S. T., et al. (2012). Quorum sensing control of phosphorus acquisition in Trichodesmium consortia. ISME J. 6 422–429. 10.1038/ismej.2011.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen K. E., Poulson-Ellestad K. L., Deering R. W., Rowely D. C., Mincer T. J. (2015). Enhancement of antibiotic activity against multidrug-resistant bacteria by the efflux pump inhibitor 3,4-dibromopyrrole-2,5-dione isolated from a Pseudoalteromonas sp. J. Nat. Prod. 78 402–412. 10.1021/np500775e [DOI] [PubMed] [Google Scholar]
- Wheeler G. L., Tait K., Taylor A., Brownlee C., Joint I. (2006). Acyl-homoserine lactones modulate the settlement rate of zoospores of the marine alga Ulva intestinalis via a novel chemokinetic mechanism. Plant Cell Environ. 29 608–618. 10.1111/j.1365-3040.2005.01440.x [DOI] [PubMed] [Google Scholar]
- Wietz M., Schramm A., Jorgensen B., Gram L. (2010). Latitudinal patterns in the abundance of major marine bacterioplankton groups. Aquat. Microb. Ecol. 61 179–189. 10.3354/ame01443 [DOI] [Google Scholar]
- Wratten S. J., Wolfe M. S., Andersen R. J., Faulkner D. J. (1977). Antibiotic metabolites from a marine pseudomonad. Antimicrob. Agents Chemother. 11 411–414. 10.1128/AAC.11.3.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborin A., Romanowski K., Gerdes S., Holbrook C., Lepine F., Long J., et al. (2009). Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc. Natl. Acad. Sci. U.S.A. 106 6327–6332. 10.1073/pnas.0813199106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow chart detailing the bioassay guided fractionation process that was used to eventually identify HHQ as a causative compound influencing growth of E. huxleyi.
Structural identification of 2-heptyl-4-quinolone (HHQ) from P. piscicida. 13C-NMR spectrum (75 MHz, DMSO-d6) of the pure compound isolated from P. piscicida (blue) and the authentic standard HHQ (red).
Genome mining of pqs operon in Pseudoalteromonas sp. (A) The pqs operon from two species of Pseudomonas aeruginosa PAO1 were used to query 67 publically available Pseudoalteromonas sp. genomes. Joint Genome Institute (JGI) genome accession numbers are listed for each isolate in parentheses. Red boxes indicate the presence of a homolog and black boxes indicate no homolog identified with BLAST cutoffs set at ≥10% identity and a maximum e-value of 0.01. White asterisks indicate two homologs (pqsB and pqsE) from P. citrea that were initially identified as absent in the genome using the Integrated Microbial Genome (IMG) gene-genome comparison analysis, but upon manual inspection, were found to be present. (B) A comparison of the alkylquinoline biosynthetic pathway in Pseudoalteromonas piscicida (A757; GenBank Accession no., KT879191–KT879199) and P. citrea (NCIMB 1889). Shading of homologs in P. citrea indicates percent amino acid identity to P. piscicida (A757) genes.
Growth rate (μ d-1) for three species of phytoplankton exposed to the crude extract of the secreted metabolites of P. piscicida. Despite HHQ having little effect on D. tertiolecta and P. tricornutum, toxicity to the crude extract of P. piscicida is observed in all species, indicating that P. piscicida likely produces additional compounds that result in algal mortality. Error bars are one standard deviation from the mean.


