Abstract
Human campylobacteriosis is primarily associated with poultry but also cattle. In this study, 55 Campylobacter jejuni strains isolated from 382 dairy calves’ feces were differentiated by multilocus sequence typing and tested for antimicrobial resistance. The most prevalent sequence type (ST) was ST883 (20.0%), followed by ST48 (14.5%), and ST50 (9.1%). In contrast to ST48 and ST50, ST883 has rarely been described in cattle previously. Furthermore, risk factor analysis was performed for the presence of the most prevalent STs in these calves. Multiple regression analysis revealed that the type of farm (organic vs. conventional) and calf housing (place, and individual vs. group) were identified as significantly (p < 0.05) associated with the presence of ST883 in calves, and ST50 was associated with calf diarrhea. Antimicrobial resistance was detected in 58.2% of the isolates. Most of the resistant isolates (81.3%) were resistant to more than one antimicrobial. Most frequently, resistance to ciprofloxacin (49.1%), followed by nalidixic acid (42.8%), and tetracycline (14.5%) was observed. The results of the present study support the hypothesis that dairy calves may serve as a potential reservoir for C. jejuni and pose a risk for transmission, including antimicrobial resistant isolates to the environment and to humans.
Keywords: Campylobacter jejuni, dairy calf, MLST, antimicrobial resistance
Introduction
Campylobacter jejuni is one of the most common causes of bacterial gastro-enteritis in humans and is of significant public health concern worldwide. Human campylobacteriosis is primarily associated with poultry, followed by cattle (French et al., 2009; de Haan et al., 2010b; Mughini Gras et al., 2012). Main risk factors are consumption of contaminated food, particularly poultry meat, raw milk, and water, as well as close contact to animals (Schildt et al., 2006; Heuvelink et al., 2009; Mughini Gras et al., 2012).
To distinguish between different Campylobacter strains, various methods have been applied, whereby multilocus sequence typing (MLST) has been identified as one of the best methods for application in epidemiological studies (Dingle et al., 2001; Korczak et al., 2009). The most commonly identified C. jejuni clonal complexes (CC) in bovines are CC21 and CC61. Sequence types (ST) of CC21 are not only typical for bovines, but also for other ruminants (sheep), poultry, and humans. In contrast, CC61 STs have been described as cattle associated (Kärenlampi et al., 2007; French et al., 2009; Grove-White et al., 2010; Bianchini et al., 2014), but are also frequently identified in humans, but not in poultry, suggesting that cattle may be an important source of human infection by contamination of food and water (French et al., 2009; Grove-White et al., 2010; Bianchini et al., 2014). Furthermore, CC42, CC45, CC48, and CC403 are frequently detected in cattle. Most of these CCs (CC42, CC45, and CC48) are also frequently identified in humans (French et al., 2009). These findings underline the importance of cattle in the epidemiology of human campylobacteriosis.
Campylobacter has been classified by the European Union as a zoonotic pathogen to be screened for antimicrobial resistance (Council directive, 2003/99/EC). However, this screening is limited to chicken and turkey isolates and does not include isolates from cattle. Key reasons for this are missing extensive European wide information on the risk of cattle isolates for human disease and low rates in antimicrobial resistance reported for cattle isolates (Aarestrup et al., 1997).
The aim of the present study was to evaluate different genotypes of C. jejuni in feces of preweaned calves in Austrian dairy herds by use of MLST and to investigate their antimicrobial resistance.
Materials and Methods
Samples
Fecal samples were collected from preweaned calves on 100 dairy farms in two Austrian regions (Lower Austria and Styria) during 2009 and 2010. This study was part of a study designed to examine differences between calves and farms with and without diarrhea (Klein et al., 2013; Klein-Jobstl et al., 2014). For selection of farms, local veterinarians were asked to provide lists with dairy farms with a documented problem of calf diarrhea during the last year. A farm with diarrhea problems was defined as a farm with multiple treatments for calf diarrhea by the veterinarian. Out of these lists, farms were randomly chosen. Additionally, farms from the same geographical region and of similar structure, with no history of calf diarrhea problems and no diarrheic calf at the time of sampling, were examined. In herds with five or less preweaned calves (which was the case on 62 of all farms), all calves were tested. In herds with more than five preweaned calves, five animals were randomly chosen. On the assumption of an inter-herd prevalence of over 40% (Ellis-Iversen et al., 2009) five samples were required from each herd to detect one positive calf with 95% confidence (calculation by use of Win Episcope 2.01). Samples were taken directly from the rectum. Feces were placed in sterile plastic tubes and transported to the laboratory in coolers. Farm management characteristics were evaluated by a face to face interview by use of a questionnaire during the farm visit (Table 1; Klein et al., 2013). All sampled calves were examined clinically according to the clinical examination of ruminants (Radostits et al., 2007). Feces was evaluated as described by Larson et al. (1977), where score 3 and 4 were categorized as diarrheic. Furthermore, the calf rearing areas were inspected and hygiene was estimated by evaluation of calf housings (bedding and pen walls) and the calves themselves according to Lundborg et al. (2005).
Table 1.
Variables surveyed on farm.
| Area of interest | Variable |
|---|---|
| Farm characteristics | Region; production (organic vs. conventional); number of cattle and cows on farm; other farm animals than cattle on farm; if yes, which other farm animals, contact to other farm animals; workers on farm; other animals (companion animals) with access to the cows‘ and calves’ stable; water source |
| Housing | Housing of cows; pasture; calving area; calf housing (location, type, bedding) |
| Calf feeding | Colostrum management; milk feeding; feeding of hay, and concentrates; water |
| Hygiene | Cleaning and disinfection of different areas and barns; feed hygiene; cleaning of feeding equipment; milking hygiene |
| Miscellaneous | Dry off regime; dry period length |
| Individually sampled calves | Age (days); housing; feeding; diseases; treatments; treatment with antimicrobials; feeding of non-saleable milk |
Results regarding risk factors for the presence of C. jejuni in calves were published elsewhere (Klein et al., 2013).
This study was discussed and approved by the institutional ethics committee of the University of Veterinary Medicine Vienna in accordance with Good Scientific Practice and national legislation.
Laboratory Procedures
All fecal samples were processed within one day, held chilled until processing. Samples were prepared for detection and isolation of thermophilic Campylobacter according to standards described by ISO-10272-(2002). Briefly, after enrichment in Bolton Broth (Oxoid, Basingstoke, England) at 42°C for 48 h under microaerophilic conditions (10% CO2, 5% O2, and 85% N2), the samples were plated on two selective agars, modified CCDA (charcoal cefoperazon deoxycholate; Oxoid, Basingstoke, England) and CampyFoodAgar (Bio Merieux, Marcy l‘Etoile, France) and incubated at 42°C for 48 h under microaerophilic conditions. Additionally, all fecal samples were directly streaked onto the two selective agars without prior enrichment. One morphological typical colony per sample was differentiated by aerobic incubation, PCR (Linton et al., 1997) and 16S-rRNA-gene sequencing on selected strains.
MLST
The MLST analysis was carried out as described by Dingle et al. (2001). Genomic DNA was extracted using a QIAamp DNA mini kit (Qiagen, Venlo, The Netherlands). The seven housekeeping loci defined by Dingle et al. (2001) as are aspA, glnA, gltA, glyA, pgm, tkt, and uncA were amplified using primers and protocols as described (Dingle et al., 2001). Sequencing was carried out by BigDye Terminator v3.1 cycle sequencing kit and an Applied Biosystems 310 ABI Prism genetic analyser. Sequence data were analyzed for MLST Types using the Campylobacter Multi Locus Sequence Typing website2 developed by Jolley and Maiden (2010) and funded by the Wellcome Trust.
Antimicrobial Resistance Testing
Antimicrobial resistance was determined using CLSI M45-A3 for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. Isolates were tested using disk susceptibility and the minimal inhibitory concentration was determined by antimicrobial dilution against ampicillin, amoxicillin/clavulanate, chloramphenicol, ciprofloxacin, colistin, erythromycin, gentamicin, nalidixic acid, neomycin, streptomycin, and tetracycline. As clinical breakpoints for C. jejuni are documented only for ciprofloxacin, erythromycin, and tetracycline (EUCAST) epidemiological cut-off-values, which have been determined by the European Committee on Antimicrobial Susceptibility Testing4, were applied on all C. jejuni isolates. For colistin and neomycin, no epidemiological cut-off-values have been determined. For these two antimicrobial substances cut-off were evaluated by comparing against values given in the literature and according to the distribution of our isolates were 16 mg/l for colistin and 4 mg/l for neomycin (El-Adawy et al., 2012; Ghimire et al., 2014).
Detection of gyrA Mutations
In all quinolone resistant isolates the quinolone resistance-determining region (QRDR) of the gyrA gene a 220Bp PCR product was ampliefied with primers GyrA-for 5′-gctatgcaaaatgatgaggc-3′ and GyrA-rev 5′-cagtataacgcatcgcagcgg-3′ to detect the responsible point mutation at codon 86. Genomic DNA used for amplification was extracted as described above (MLST).
Statistical Analysis
Data were statistically analyzed using PASW, version 20.0 (IBM Cooperation, New York, NY, USA).
The presence of each MLST ST was summarized in a binary variable. The presence of each ST was given as an individual variable, where presence of the given ST was categorized as one and not present as zero. Similarly, resistances against antimicrobials were categorized as either resistant (=1) or not resistant (=0). Different STs, resistances as well as farm characteristics or management factors were compared with the most prevalent STs and resistances against antimicrobials. Depending on the independent variable either Fisher’s exact test, Chi square test, binary logistic regression, or t-test was calculated. Correlation between C. jejuni MLST-types and resistance against antimicrobials were tested by Spearman correlation coefficient. The level of significance was set at a p-value of <0.05.
The presence of the most prevalent STs was associated with farm characteristics and management in a two-step process. First, the presence of the ST was compared to the different independent variables as described above by either Fisher’s exact test, Chi square test, binary logistic regression or t-test. All variables were tested for correlation among each other by Spearman correlation coefficient before entering the model. If a correlation between two variables >0.60 was given, one of the covariates was discarded taking biological plausibility into account. In a second step, variables with a p-value ≤0.20 were entered in a multiple logistic regression model. Confounding was monitored by the change in the coefficient of a variable after removing another variable (Dohoo et al., 2009). If the change of the estimates was ≥25% the removed variable was considered to have a potential confounding effect and was consequently forced into the model. Model fit was evaluated with the Hosmer–Lemeshow test for 10 groups.
Results
In total, 382 calves were sampled on 100 farms. Mean herd size was 40 ± 29 dairy cows (varying between 5 and 223 cows). Mean herd size did not differ between C. jejuni positive and negative farms (p = 0.67).
The median age of the sampled animals was 17 days [25 and 75% interquartile range (IR) 10–28]. C. jejuni positive calves were as young as 3 days and up to 67 days (median 18, IR 11-36). From 382 fecal samples, 55 (14.4%) were positive for C. jejuni. Another four samples (1.0%) were positive for C. coli. On farm level, on 30 of the 100 farms at least one animal shed C. jejuni, whereas only on five of these 30 farms all sampled animals were positive.
MLST of C. jejuni Isolates from Calves
The 55 C. jejuni isolates yielded 19 STs of which two were previously unreported. The isolates were assigned to eight clonal complexes (CC), dominated by three CCs (CC21, CC48, and CC206) that accounted for 74.5%. Half of the isolates (50.9%) belonged to CC21. The most prevalent STs were ST883 (20.0%), followed by ST48 (14.5%), and ST50 (9.1%; Table 2).
Table 2.
Campylobacter jejuni MLST types among 55 isolates from preweand dairy calves.
| CC | ST | N positive samples | N positive farms |
|---|---|---|---|
| 21 | 21 | 4 | 4 |
| 47 | 1 | 1 | |
| 50 | 5 | 4 | |
| 864 | 4 | 2 | |
| 883 | 11 | 6 | |
| 1943 | 3 | 1 | |
| 22 | 22 | 1 | 1 |
| 2497 | 3 | 2 | |
| 42 | 42 | 2 | 1 |
| 2580 | 1 | 1 | |
| 45 | 45 | 1 | 1 |
| 48 | 48 | 8 | 6 |
| 206 | 122 | 2 | 1 |
| 572 | 2 | 1 | |
| 6021 | 1 | 1 | |
| 353 | 356 | 1 | 1 |
| 354 | 4899 | 3 | 2 |
| Unknown | 2 | 2 |
On 24 of the 30 C. jejuni positive farms (80%) one ST was present. On five farms two and on one farm three different types were isolated.
MLST Types and Risk Factors
The presence of C. jejuni ST883 in calves compared to the presence of other STs in calves was significantly associated with season, the presence of calf diarrhea on farm, the type of farm (organic vs. conventional), workers on farm, the feeding of waste milk, the separation of the calf from its dam after birth, calf feeding, and calf housing (individual versus group, and within cows’ barn versus outside the barn). These variables were entered in the multiple logistic regression model. As farm had a confounding effect, this variable was forced into the model. In the final model, the type of farm and calf housing pointed out to be significantly associated with the presence of ST883 in calves (Table 3). On conventional farms, the risk for calves to shed ST883 was lower compared with organic farms. Housing calves in groups inside the cows’ barn was identified as a risk for shedding ST883.
Table 3.
Variables significantly associated with the presence of C. jejuni sequence type (ST) 883 in the final multiple logistic regression with farm forced into the model as a confounder.
| ST |
||||||
|---|---|---|---|---|---|---|
| Variable | ST883 | Others | OR1 | 95% CI2 | p | |
| Farm | 0.45 | |||||
| Type of farm | Organic | 6 | 6 | 1 | ||
| Conventional | 38 | 5 | 0.62 | 0.01-0.64 | 0.02 | |
| Calf housing | Individual | 40 | 4 | 1 | ||
| Group | 4 | 7 | 23.16 | 2.08-257.43 | 0.01 | |
| Within cows’ barn | 15 | 9 | 1 | |||
| Outside cows’ barn | 29 | 2 | 0.78 | 0.01-0.93 | 0.04 | |
1OR = odds ratio
2CI = confidence interval
Hosmer–Lemeshow for the model p = 0.77.
No significant associations were found with regard to the presence of ST48.
The presence of ST50 strains compared to other STs were associated with farm, type of farm (organic vs. conventional), farm size (number of cows on farm), the presence of poultry on farm, diarrhea, calf feeding, and antibiotic treatment in the calf. Farm was left in the multiple logistic model as it had a confounding effect. Finally, only one variable stayed significant in the final model. Calves suffering from diarrhea at the time of sampling had a higher risk to be ST50 positive than calves not suffering from diarrhea (OR 23.21, 95%CI 23.21-248.87, p = 0.01).
Antimicrobial Resistance in C. jejuni
Of the 55 C. jejuni strains, 32 (58.2%) were resistant to at least one of the tested antimicrobials. Strains were resistant to ampicillin, ciprofloxacin, nalidixic acid, neomycin, streptomycin, and tetracycline (Figure 1). Twenty-six of the isolates (47.3%) were resistant to at least two of the tested antimicrobials. Seven of these isolates were resistant against three to five antimicrobials. Most frequently, resistance to ciprofloxacin was observed (49.1%), followed by nalidixic acid with 42.8%, and tetracycline (14.5%). No resistance was observed against amoxicillin/clavulanate, chloramphenicol, colistin, erythromycin, and gentamicin.
FIGURE 1.
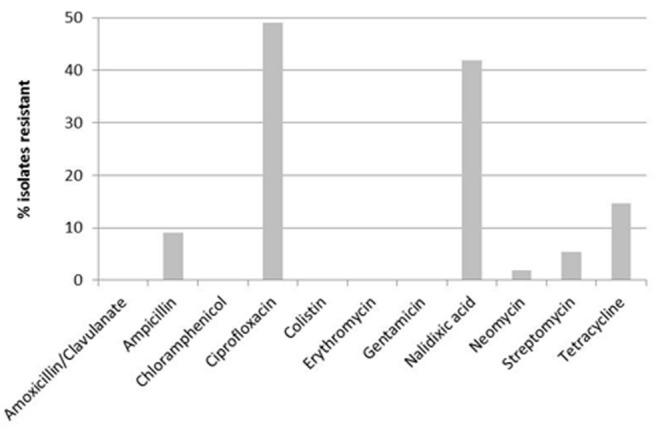
Antimicrobial resistance of Campylobacter jejuni (n = 55) isolated from preweaned dairy calves.
Associations could be observed in resistance against ciprofloxacin and nalidixic acid (p = 0.01), as well as against ciprofloxacin and ampicillin (p = 0.02). All isolates that were resistant against nalidixic acid or ampicillin were also resistant against ciprofloxacin.
Genetic Identification of Quinolone Resistant Isolates
As almost half of the isolates were identified to be quinolone resistant in all of these isolates the QRDR of the gyrA gene was sequenced to detect the responsible point mutation at codon 86 the most important mechanism in C. jejuni for quinolone resistance. In all but one of the 27 isolates the point mutation at codon 86 (ACA to ATA) resulting in a substitution of isoleucine for threonine in gyrase A has been detected. One sensitive isolate sequenced confirmed the original sequence (ACA) as seen in sensitive isolates. Another mutation detected in three of the isolates at location codon 110 did not change the amino acid composition of the enzyme. One isolate resistant to ciprofloxacin and nalidixic acid did not have any mutation in the sequenced region.
Antimicrobial Resistance and MLST-Types
Of the 11 STs that were detected repeatedly, in four STs (ST122, ST572, ST883, and ST4899) all isolates were ciprofloxacin resistant, whereas in ST883 this association was significant (p = 0.01). In ST42, ST864, ST1943, and ST2497, no resistance against ciprofloxacin were observed. Similar findings were obtained for nalidixic acid resistances, see Table 4.
Table 4.
Antimicrobial resistance of the 55 C. jejuni MLST types obtained from dairy preweaned calves.
| CC | ST | N isolates | N resistances | Antimicrobial resistance |
|---|---|---|---|---|
| 21 | 21 | 2 | 2 | CIP, NAL |
| 21 | 1 | 3 | AMP, CIP, NAL | |
| 21 | 1 | 1 | STREP | |
| 47 | 1 | 1 | TET | |
| 50 | 2 | 3 | CIP, NAL, TET | |
| 50 | 2 | 2 | CIP, NAL | |
| 50 | 1 | 0 | ||
| 864 | 2 | 1 | TET | |
| 864 | 2 | 0 | ||
| 883 | 9 | 2 | CIP, NAL | |
| 883 | 1 | 3 | CIP, NAL, STREP | |
| 883 | 1 | 3 | AMP, CIP, NAL | |
| 1943 | 3 | 0 | ||
| 22 | 22 | 1 | 0 | |
| 2497 | 3 | 0 | ||
| 42 | 42 | 2 | 0 | |
| 2580 | 1 | 0 | ||
| 45 | 45 | 1 | 0 | |
| 48 | 48 | 7 | 0 | |
| 48 | 1 | 2 | CIP, NAL | |
| 206 | 122 | 2 | 2 | CIP, NAL |
| 572 | 1 | 2 | AMP, CIP | |
| 572 | 1 | 3 | AMP, CIP, NAL | |
| 6021 | 1 | 5 | AMP, CIP, NEO, STREP, TET | |
| 353 | 356 | 1 | 0 | |
| 354 | 4899 | 2 | 2 | CIP, NAL |
| 4899 | 1 | 1 | CIP | |
| Unknown | 1 | 0 | ||
| Unknown | 1 | 1 | TET |
CIP, ciprofloxacin; NAL, nalidixic acid; TET, tetracycline; AMP, ampicillin; STREP, streptomycin; NEO, neomycin.
Testing for correlation between C. jejuni MLST-types and antimicrobial resistance revealed significant associations between ST48 and ciprofloxacin resistance (coefficient = –0.33; p = 0.01), between ST572 and resistance against ampicillin (coefficient = –0.61; p < 0.01), between ST864 and ciprofloxacin (coefficient = –0.28; p = 0.04) and tetracycline (coefficient = 0.28; p = 0.04) resistance, and between ST883 and ciprofloxacin and nalidixic acid, respectively (coefficient = 0.51; p < 0.01).
Only two of the 55 C. jejuni positive calves were previously treated with antimicrobials. Both of these calves shed antimicrobial resistant C. jejuni (ST50). Both strains were resistant to ciprofloxacin and nalidixic acid, one additionally against tetracycline.
There was no association between the evaluated use of antimicrobials on farm and resistances.
Discussion
To the knowledge of the authors studies examining C. jejuni MLST types in cattle have been limited to quinolone-resistant C. jejuni in Austria (Kovac et al., 2015) and studies in dairy calves are sparse.
Campylobacter jejuni isolates detected in the calves of our study were dominated by CC21, a CC regularly associated with cattle (Manning et al., 2003; Kwan et al., 2008; Ragimbeau et al., 2008; de Haan et al., 2010b; Bianchini et al., 2014; Jonas et al., 2015). Ten of the 19 C. jejuni STs detected were previously described in cattle. ST 883, the most common ST in our study, has only been described in cattle sporadically (de Haan et al., 2010b; Bianchini et al., 2014), and was furthermore sporadically associated with poultry and human campylobacteriosis (Wirz et al., 2010; Kittl et al., 2013b). In the study by Kovac et al. (2015) where ciprofloxacin-resistant C. jejuni of 17 cattle from Austria were examined, only one was ST883. In contrast, ST48 and ST50, the second and third most prevalent STs in the present study, were commonly associated with bovines, including Austria (Kwan et al., 2008; Ragimbeau et al., 2008; Rapp et al., 2014; Kovac et al., 2015). This ST was also associated with humans and other species, e.g., poultry (Ragimbeau et al., 2008; de Haan et al., 2010a). In the PubMLST database so far 32 human isolates and 33 chicken isolates from Austria have been downloaded between 2008 and 2014. Interestingly CC21 the CC most prevalent in claves in our study was also dominant in human isolates in Austria (21% of all isolates) whereas the clonal complex CC464, CC353, and CC354 dominated in chicken isolates.
Different studies emphasized the role of cattle in human campylobacteriosis. Following poultry, bovines were frequently associated with human infections (Wilson et al., 2009; de Haan et al., 2010a; Mughini Gras et al., 2012). Risk factors that have been described are direct contact to cattle and to cattle feces, as well as consumption of raw milk (Eberhart-Phillips et al., 1997; Smith et al., 2004; Schildt et al., 2006). Direct contact and consumption of raw milk is given not only for farmers but also, e.g., during farm vacation which is popular in Austria with nearly 10,000 farms offering this service (Grüner Bericht, n.d.). Furthermore, contaminated food or water can play a role in human infection (Clark et al., 2005; Levesque et al., 2008; Mughini Gras et al., 2012). The three dominating CCs (CC21, CC48, and CC206) and STs (ST883, ST48, and ST50) of our study were also recovered from infected humans (Ragimbeau et al., 2008; Mullner et al., 2009; Sheppard et al., 2009; de Haan et al., 2010a). This result indicates that calves may be a potential source of human infection, but this cannot be proven by this study. Gilpin et al. (2008) found indistinguishable C. jejuni genotypes in dairy calves and humans, using Penner serotyping and pulsed field gel electrophoreses, and came to the same conclusion, that calves may be a source of human campylobacteriosis.
Early campylobacter infection in calves might be due to a high level of environmental contamination, as well as direct contact with feces and ingestion of milk (Bianchini et al., 2014). Contamination of calf housings represents a risk for early infection with C. jejuni ST883. Calves housed within the cows‘ barn were at higher risk to shed ST883 than calves housed in a special barn for calves and young animals or outside the barn. Additionally, grouping of animals was a risk factor for shedding ST883. These findings suggest that close contact to adult as well as to other young cattle lead to higher infection pressure and mutual infection between animals. Furthermore, grouping can be a stress factor and consequently lead to a higher rate of campylobacter shedding and infection (Rapp et al., 2014).
Furthermore, ST883 was associated with the type of farm. On conventional farms, the risk for preweaned calves to be ST883 positive was lower than on organic farms, a finding that can hardly be explained. Possibly a certain clone is circulating on these farms and has been established.
The presence of ST50 was associated with diarrhea at the time of sampling in calves. Although some authors (Al-Mashat and Taylor, 1983; Diker et al., 1990; Schulze, 1992) suggested a possible role of Campylobacter in calf diarrhea, in other studies no association between C jejuni and disease was given (De Rycke et al., 1986; Acha et al., 2004), as was also true for the calves of the present study (Klein et al., 2013) and more likely other pathogens generally associated with calf diarrhea may be the cause of disease.
More than half of the calves (55%) originated from farms where also poultry was kept. Most of the STs detected in the calves of our study were also described in poultry. In the present study, specific types like ST50, frequently detected in poultry (de Haan et al., 2010b; Griekspoor et al., 2010; Kovanen et al., 2014), were associated with the presence of poultry on farm, suggesting cross-contamination between the two species. In contrast, other STs (e.g., ST21, ST48, and ST883) also regularly detected in poultry (Wirz et al., 2010; Kittl et al., 2013b) appeared in calves independently of the presence of poultry.
Antimicrobial resistance, particularly multidrug resistance is of public health concern. In the present study, 58.2% of the C. jejuni isolates were resistant to at least one and 47.3% against at least two of the tested antimicrobials. Because only two of the 55 C. jejuni positive calves were previously treated with antibiotics, no valid conclusion can be drawn if resistance to STs in this study was associated with previous antibiotic treatment. Nevertheless, these two treated animals shed C. jejuni resistant to two and three of the tested antimicrobials, respectively. In the present study, antimicrobial resistance to quinolones was detected most often. This has also been described for isolates originating from other sources (Oporto et al., 2009; Kittl et al., 2013a), explained by the fact that a single mutation is sufficient to cause resistance (Wang et al., 1993). This was confirmed in our study as all but one resistant isolates harbored a point mutation in codon 86 of the gyrA gene. Quinolone resistance has been described to be associated with specific ST types as detected in our study (Kittl et al., 2013a; Kovac et al., 2015). A survey performed in Austria revealed that quinolones are frequently used by Austrian veterinarians for treatment of cattle (Mayrhofer et al., 2006). This might explain a high level of quinolone resistance in C. jejuni isolates from calves.
Conclusion
The results of the present study support the hypothesis that cattle including dairy calves may be a reservoir for C. jejuni and represent a risk for transmission of these bacteria to the environment and to humans. Cattle have not been recognized as an important source for antimicrobial resistant Campylobacter sp. or other bacteria, yet. Nevertheless, high resistance rates found in this and other recent studies point out that screening for antimicrobial resistance in cattle is necessary to better understand the epidemiology of resistance and its spread.
Author Contributions
DK-J designed the study, took all samples, performed statistical analysis, and drafted the manuscript. DS performed laboratory work and analysis. MI advised statistical analysis and interpretation, and reviewed the manuscript. MD supported the statistical analysis and reviewed the manuscript. FH designed the study together with DK-J and supervised the study, supervised, and performed laboratory work and analysis, and provided valuable references and suggestions during the preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was financed by the University of Veterinary Medicine Vienna by a start-up project (Profillinie 2).
Footnotes
References
- Aarestrup F. M., Nielsen E. M., Madsen M., Engberg J. (1997). Antimicrobial susceptibility patterns of thermophilic Campylobacter spp. from humans, pigs, cattle, and broilers in Denmark. Antimicrob. Agents Chemother. 412244–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acha S. J., Kuhn I., Jonsson P., Mbazima G., Katouli M., Mollby R. (2004). Studies on calf diarrhoea in Mozambique: prevalence of bacterial pathogens. Acta Vet. Scand. 45 27–36. 10.1186/1751-0147-45-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mashat R. R., Taylor D. J. (1983). Production of enteritis in calves by the oral inoculation of pure cultures of Campylobacter fetus subspecies intestinalis. Vet. Rec. 112 54–58. 10.1136/vr.112.7.141 [DOI] [PubMed] [Google Scholar]
- Bianchini V., Borella L., Benedetti V., Parisi A., Miccolupo A., Santoro E., et al. (2014). Prevalence in bulk tank milk and epidemiology of Campylobacter jejuni in dairy herds in Northern Italy. Appl. Environ. Microbiol. 80 1832–1837. 10.1128/AEM.03784-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C. G., Bryden L., Cuff W. R., Johnson P. L., Jamieson F., Ciebin B., et al. (2005). Use of the oxford multilocus sequence typing protocol and sequencing of the flagellin short variable region to characterize isolates from a large outbreak of waterborne Campylobacter sp. strains in Walkerton, Ontario, Canada. J. Clin. Microbiol. 43 2080–2091. 10.1128/JCM.43.5.2080-2091.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Council directive (2003/99/EC). DIRECTIVE 2003/99/EC of the European Parliament and of the Council on the Monitoring of Zoonoses and Zoonotic Agents, Amending Council Decision 90/424/EEC and Repealing Council Directive 92/117/EEC. Available at: http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex:32003L0099. [Google Scholar]
- de Haan C. P., Kivisto R., Hakkinen M., Rautelin H., Hanninen M. L. (2010a). Decreasing trend of overlapping multilocus sequence types between human and chicken Campylobacter jejuni isolates over a decade in Finland. Appl. Environ. Microbiol. 76 5228–5236. 10.1128/AEM.00581-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C. P., Kivisto R. I., Hakkinen M., Corander J., Hanninen M. L. (2010b). Multilocus sequence types of Finnish bovine Campylobacter jejuni isolates and their attribution to human infections. BMC Microbiol. 10:200 10.1186/1471-2180-10-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rycke J., Bernard S., Laporte J., Naciri M., Popoff M. R., Rodolakis A. (1986). Prevalence of various enteropathogens in the feces of diarrheic and healthy calves. Ann. Rech. Vet. 17 159–168. [PubMed] [Google Scholar]
- Diker K. S., Diker S., Ozlem M. B. (1990). Bovine diarrhea associated with Campylobacter hyointestinalis. Zentralbl. Veterinarmed. B. 37 158–160. [DOI] [PubMed] [Google Scholar]
- Dingle K. E., Colles F. M., Wareing D. R., Ure R., Fox A. J., Bolton F. E., et al. (2001). Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39 14–23. 10.1128/JCM.39.1.14-23.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohoo I., Martin W., Stryhn H. (2009). Veterinary Epidemiologic Research. Charlottettown, PE: Atlantic Veterinary College. [Google Scholar]
- Eberhart-Phillips J., Walker N., Garrett N., Bell D., Sinclair D., Rainger W., et al. (1997). Campylobacteriosis in New Zealand: results of a case-control study. J. Epidemiol. Commun. Health 51 686–691. 10.1136/jech.51.6.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Adawy H., Hotzel H., Dupre S., Tomaso H., Neubauer H., Hafez H. M. (2012). Determination of antimicrobial sensitivities of Campylobacter jejuni isolated from commercial turkey farms in Germany. Avian Dis. 56 685–692. 10.1637/10135-031912-Reg.1 [DOI] [PubMed] [Google Scholar]
- Ellis-Iversen J., Cook A. J., Smith R. P., Pritchard G. C., Nielen M. (2009). Temporal patterns and risk factors for Escherichia coli O157 and Campylobacter spp, in young cattle. J. Food Prot. 72 490–496. [DOI] [PubMed] [Google Scholar]
- French N. P., Midwinter A., Holland B., Collins-Emerson J., Pattison R., Colles F., et al. (2009). Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children’s playgrounds. Appl. Environ. Microbiol. 75 779–783. 10.1128/AEM.01979-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghimire L., Singh D. K., Basnet H. B., Bhattarai R. K., Dhakal S., Sharma B. (2014). Prevalence, antibiogram and risk factors of thermophilic Campylobacter spp. in dressed porcine carcass of Chitwan, Nepal. BMC Microbiol. 14:85 10.1186/1471-2180-14-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin B. J., Thorrold B., Scholes P., Longhurst R. D., Devane M., Nicol C., et al. (2008). Comparison of Campylobacter jejuni genotypes from dairy cattle and human sources from the Matamata-Piako District of New Zealand. J. Appl. Microbiol. 105 1354–1360. 10.1111/j.1365-2672.2008.03863.x [DOI] [PubMed] [Google Scholar]
- Griekspoor P., Engvall E. O., Olsen B., Waldenstrom J. (2010). Multilocus sequence typing of Campylobacter jejuni from broilers. Vet. Microbiol. 140 180–185. 10.1016/j.vetmic.2009.07.022 [DOI] [PubMed] [Google Scholar]
- Grove-White D. H., Leatherbarrow A. J., Cripps P. J., Diggle P. J., French N. P. (2010). Temporal and farm-management-associated variation in the faecal-pat prevalence of Campylobacter jejuni in ruminants. Epidemiol. Infect. 138 549–558. 10.1017/S0950268809991051 [DOI] [PubMed] [Google Scholar]
- Grüner Bericht. (n.d.). Available at: http://www.gruenerbericht.at/cm3/download/summary/82-gruener-bericht-oesterreich/1392-gruener-bericht-2014.html. [Google Scholar]
- Heuvelink A. E., van Heerwaarden C., Zwartkruis-Nahuis A., Tilburg J. J., Bos M. H., Heilmann F. G., et al. (2009). Two outbreaks of campylobacteriosis associated with the consumption of raw cows’ milk. Int. J. Food Microbiol. 134 70–74. 10.1016/j.ijfoodmicro.2008.12.026 [DOI] [PubMed] [Google Scholar]
- Jolley K. A., Maiden M. C. (2010). BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas R., Kittl S., Overesch G., Kuhnert P. (2015). Genotypes and antibiotic resistance of bovine Campylobacter and their contribution to human campylobacteriosis. Epidemiol. Infect. 143 2373–2380. 10.1017/S0950268814003410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärenlampi R., Rautelin H., Schonberg-Norio D., Paulin L., Hanninen M. L. (2007). Longitudinal study of Finnish Campylobacter jejuni and C. coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl. Environ. Microbiol. 73 148–155. 10.1128/AEM.01488-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittl S., Heckel G., Korczak B. M., Kuhnert P. (2013a). Source attribution of human Campylobacter isolates by MLST and fla-typing and association of genotypes with quinolone resistance. PLoS ONE 8:e81796 10.1371/journal.pone.0081796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittl S., Korczak B. M., Niederer L., Baumgartner A., Buettner S., Overesch G., et al. (2013b). Comparison of genotypes and antibiotic resistances of Campylobacter jejuni and Campylobacter coli on chicken retail meat and at slaughter. Appl. Environ. Microbiol. 79 3875–3878. 10.1128/AEM.00493-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D., Alispahic M., Sofka D., Iwersen M., Drillich M., Hilbert F. (2013). Prevalence and risk factors for shedding of thermophilic Campylobacter in calves with and without diarrhea in Austrian dairy herds. J. Dairy Sci. 96 1203–1210. 10.3168/jds.2012-5987 [DOI] [PubMed] [Google Scholar]
- Klein-Jobstl D., Iwersen M., Drillich M. (2014). Farm characteristics and calf management practices on dairy farms with and without diarrhea: a case-control study to investigate risk factors for calf diarrhea. J. Dairy Sci. 97 5110–5119. 10.3168/jds.2013-7695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korczak B. M., Zurfluh M., Emler S., Kuhn-Oertli J., Kuhnert P. (2009). Multiplex strategy for multilocus sequence typing, fla typing, and genetic determination of antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolates collected in Switzerland. J. Clin. Microbiol. 47 1996–2007. 10.1128/JCM.00237-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovac J., Cadez N., Stessl B., Stingl K., Gruntar I., Ocepek M., et al. (2015). High genetic similarity of ciprofloxacin-resistant Campylobacter jejuni in central Europe. Front. Microbiol. 6:1169 10.3389/fmicb.2015.01169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovanen S. M., Kivisto R. I., Rossi M., Hanninen M. L. (2014). A combination of MLST and CRISPR typing reveals dominant Campylobacter jejuni types in organically farmed laying hens. J. Appl. Microbiol. 117 249–257. 10.1111/jam.12503 [DOI] [PubMed] [Google Scholar]
- Kwan P. S., Birtles A., Bolton F. J., French N. P., Robinson S. E., Newbold L. S., et al. (2008). Longitudinal study of the molecular epidemiology of Campylobacter jejuni in cattle on dairy farms. Appl. Environ. Microbiol. 74 3626–3633. 10.1128/AEM.01669-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson L. L., Owen F. G., Albright R. D., Appleman R. D., Lamb R. C., Miller L. D. (1977). Guidelines toward more uniformity in measuring and reporting calf experimental data. J. Dairy Sci. 60 989–991. 10.3168/jds.S0022-0302(77)83975-1 [DOI] [Google Scholar]
- Levesque S., Frost E., Arbeit R. D., Michaud S. (2008). Multilocus sequence typing of Campylobacter jejuni isolates from humans, chickens, raw milk, and environmental water in Quebec, Canada. J. Clin. Microbiol. 46 3404–3411. 10.1128/JCM.00042-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton D., Lawson A. J., Owen R. J., Stanley J. (1997). PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35 2568–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundborg G. K., Svensson E. C., Oltenacu P. A. (2005). Herd-level risk factors for infectious diseases in Swedish dairy calves aged 0-90 days. Prev. Vet. Med. 68 123–143. 10.1016/j.prevetmed.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Manning G., Dowson C. G., Bagnall M. C., Ahmed I. H., West M., Newell D. G. (2003). Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69 6370–6379. 10.1128/AEM.69.11.6370-6379.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrhofer S., Paulsen P., Smulders F. J., Hilbert F. (2006). Antimicrobial resistance in commensal Escherichia coli isolated from muscle foods as related to the veterinary use of antimicrobial agents in food-producing animals in Austria. Microb. Drug Resist. 12 278–283. 10.1089/mdr.2006.12.278 [DOI] [PubMed] [Google Scholar]
- Mughini Gras L., Smid J. H., Wagenaar J. A., de Boer A. G., Havelaar A. H., Friesema I. H., et al. (2012). Risk factors for campylobacteriosis of chicken, ruminant, and environmental origin: a combined case-control and source attribution analysis. PLoS ONE 7:e42599 10.1371/journal.pone.0042599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullner P., Spencer S. E., Wilson D. J., Jones G., Noble A. D., Midwinter A. C., et al. (2009). Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect. Genet. Evol. 9 1311–1319. 10.1016/j.meegid.2009.09.003 [DOI] [PubMed] [Google Scholar]
- Oporto B., Juste R. A., Hurtado A. (2009). Phenotypic and genotypic antimicrobial resistance profiles of Campylobacter jejuni isolated from cattle, sheep, and free-range poultry faeces. Int. J. Microbiol. 2009:456573 10.1155/2009/456573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radostits O. M., Gay C. C., Hinchcliff K. W., Constable P. D. (eds). (2007). “Examination of the patient,” in Veterinary Medicine. A Textbook of the Diseases of Cattle, Horses, Sheep, Pigs, and Goats 10th Edn (Philadelphia, PA: Saunders/Elsevier Limited; ) 8–21. [Google Scholar]
- Ragimbeau C., Schneider F., Losch S., Even J., Mossong J. (2008). Multilocus sequence typing, pulsed-field gel electrophoresis, and fla short variable region typing of clonal complexes of Campylobacter jejuni strains of human, bovine, and poultry origins in Luxembourg. Appl. Environ. Microbiol. 74 7715–7722. 10.1128/AEM.00865-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp D., Ross C. M., Cave V., Muirhead R. W. (2014). Prevalence, concentration and genotypes of Campylobacter jejuni in faeces from dairy herds managed in farm systems with or without housing. J. Appl. Microbiol. 116 1035–1043. 10.1111/jam.12425 [DOI] [PubMed] [Google Scholar]
- Schildt M., Savolainen S., Hanninen M. L. (2006). Long-lasting Campylobacter jejuni contamination of milk associated with gastrointestinal illness in a farming family. Epidemiol. Infect. 134 401–405. 10.1017/S0950268805005029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze F. (1992). [Campylobacter as the cause of diarrhea in calves]. Dtsch. Tierarztl. Wochenschr. 99 458–461. [PubMed] [Google Scholar]
- Sheppard S. K., Dallas J. F., MacRae M., McCarthy N. D., Sproston E. L., Gormley F. J., et al. (2009). Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int. J. Food Microbiol. 134 96–103. 10.1016/j.ijfoodmicro.2009.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. E., Stenzel S. A., Bender J. B., Wagstrom E., Soderlund D., Leano F. T., et al. (2004). Outbreaks of enteric infections caused by multiple pathogens associated with calves at a farm day camp. Pediatr. Infect. Dis. J. 23 1098–1104. [PubMed] [Google Scholar]
- Wang Y., Huang W. M., Taylor D. E. (1993). Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob. Agents Chemother. 37 457–463. 10.1128/AAC.37.3.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. J., Gabriel E., Leatherbarrow A. J., Cheesbrough J., Gee S., Bolton E., et al. (2009). Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol. Biol. Evol. 26 385–397. 10.1093/molbev/msn264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirz S. E., Overesch G., Kuhnert P., Korczak B. M. (2010). Genotype and antibiotic resistance analyses of Campylobacter isolates from ceca and carcasses of slaughtered broiler flocks. Appl. Environ. Microbiol. 76 6377–6386. 10.1128/AEM.00813-10 [DOI] [PMC free article] [PubMed] [Google Scholar]


