Abstract
Arsenic metabolism is proposed to be an ancient mechanism in microbial life. Different bacteria and archaea use detoxification processes to grow under high arsenic concentration. Some of them are also able to use arsenic as a bioenergetic substrate in either anaerobic arsenate respiration or chemolithotrophic growth on arsenite. However, among the archaea, bioenergetic arsenic metabolism has only been found in the Crenarchaeota phylum. Here we report the discovery of haloarchaea (Euryarchaeota phylum) biofilms forming under the extreme environmental conditions such as high salinity, pH and arsenic concentration at 4589 m above sea level inside a volcano crater in Diamante Lake, Argentina. Metagenomic analyses revealed a surprisingly high abundance of genes used for arsenite oxidation (aioBA) and respiratory arsenate reduction (arrCBA) suggesting that these haloarchaea use arsenic compounds as bioenergetics substrates. We showed that several haloarchaea species, not only from this study, have all genes required for these bioenergetic processes. The phylogenetic analysis of aioA showed that haloarchaea sequences cluster in a novel and monophyletic group, suggesting that the origin of arsenic metabolism in haloarchaea is ancient. Our results also suggest that arsenite chemolithotrophy likely emerged within the archaeal lineage. Our results give a broad new perspective on the haloarchaea metabolism and shed light on the evolutionary history of arsenic bioenergetics.
Introduction
Life forms use various bioenergetic strategies to obtain free energy from the environment and transform it into electrochemical gradients that generate ATP. The first microorganisms likely used inorganic compounds to fuel bioenergetic redox reactions. The most-accepted hypothesis is that energy obtained from the oxidation of H2 and the reduction of CO2 were the processes used by the common ancestor of Bacteria and Archaea known as last universal common ancestor (Martin et al., 2008). However, several other inorganic substrates seem to have been used in bioenergetic redox reactions in the last universal common ancestor based on the evolutionary analyses of key enzymes involved in these processes (Schoepp-Cothenet et al., 2012). Such is the case of arsenite (As(III)), which is proposed as a candidate electron donor, feeding bioenergetic chains in primordial life based on phylogenetic analysis of large subunit of arsenite oxidases (AioA) and paleogeochemical analyses (Lebrun et al., 2003; van Lis et al., 2013; Sforna et al., 2014). In fact, the arsenic seems to have been present at much higher concentrations in the ancient Earth's crust than it is today (Oremland et al., 2009). The characteristics and biochemical nature of the ancient arsenic metabolism is subject of an ongoing debate (Kulp et al., 2008; Schoepp-Cothenet et al., 2009). Members of both Bacteria and Archaea encode aio genes for As(III) oxidation, which could be used for chemolithotrophic growth (van Lis et al., 2013). Some Bacteria have also genes for dissimilatory arsenate (As(V)) reduction (arr genes), associated to anaerobic respiration using arsenate as terminal electron acceptor (Duval et al., 2008). It has been proposed that anaerobic arsenate respiration is a more recent metabolism than arsenite oxidation, emerged once oxygen became abundant at the atmosphere (van Lis et al., 2013). Although Archaea and Bacteria are able to grow anaerobically on arsenate, the arrA gene (the only well-characterized enzyme that catalyze respiratory arsenate reduction) has only been found in Bacteria.
Haloarchaea are the most studied Archaea (phylum: Euryarchaeota) and a very well studied group of microorganisms. They are free-living halophiles, aerobic and sometimes facultative anaerobic, heterotrophs that in some cases synthetize ATP yielding energy from light using bacteriorhodopsins (Falb et al., 2008). Oxygen is poorly soluble at high salt concentrations and anaerobic growth of haloarchaea has been observed using nitrate, dimethylsulfoxide, trimethylamine N-oxide and fumarate as electron acceptors, some of which are probably remnants of the ancestral haloarchaea metabolisms that prevailed when Earth's atmosphere was still anoxic (Oren, 2013). Here we report the discovery of haloarchaea forming biofilms in nature in a high altitude soda lake under high arsenic concentrations, which are most likely using arsenic as a bioenergetic substrate to sustain growth.
Materials and Methods
Sample processing
Biofilm samples were collected from Laguna Diamante, located inside the Galan Volcano crater and placed at 4589 m above sea level (coordinates: 26°00' 51.04"S, 67°01'46.42"O Figure 1). Galan Volcano is placed at Catamarca province in Argentina and reach 5912 m above sea level. It is one of the supervolcanoes of the World, which hosted several large-volume explosive eruptions from upper Miocene to Pleistocene (between 7 and 4 Ma ago), (Ruggieri et al., 2011; Soler, 2007). Galan Volcano has the largest uncovered caldera in the World with 40-km diameter. Owing to the high altitude and volcanic influence, Diamante Lake presents multiple extreme conditions that include: high UV radiation, salinity and arsenic, low O2 pressure and a hydrothermal vent input. Three independent samples were taken from Diamante Lake Red Biofilms and water that were collected in sterile plastic bags and flasks in February 2012. Samples for scanning electron microscopy (SEM) and for water chemical analyses were stored in the dark at 4 °C and processed within 1–2 weeks. Conductivity, temperature, pH, dissolved O2 and total dissolved solids were measured in situ with a multiparameter probe HANNA HI 9828. Nutrient, ions and general chemical analyses were performed in Estación Experimental Obispo Colombres, Tucumán, Argentina (http://www.eeaoc.org.ar/) a chemical IRAM-certified laboratory. Samples for DNA extraction were frozen in liquid nitrogen, stored in the dark and processed within a week. Water samples were delivered to the facility Estación Experimental Obispo Colombres, Tucumán, Argentina for chemical analyses. Total metagenomic DNA used for sequencing was extracted from a mixture of three triplicate using PowerBiofilm DNA Isolation Kit (MO BIO Laboratories, Inc.) from 0.5 g of collected biofilm and processed according to manufacturer's indications.
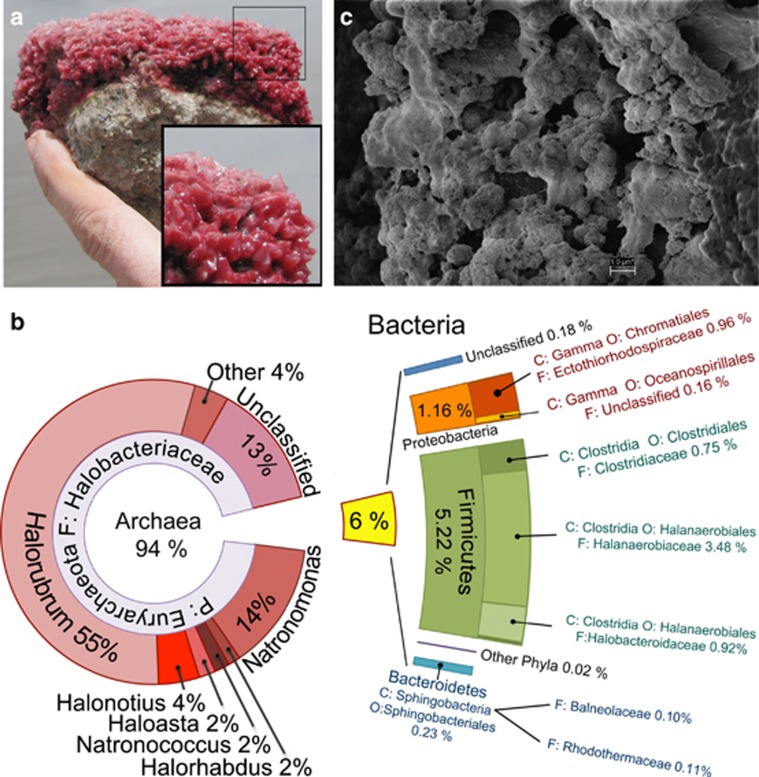
Figure 1.
General description of Diamante Lake Red Biofilms (DLRB). (a) Photography of DLRB obtained from the bottom of the lake. (b) Taxonomic composition of DLRB based on 692 16S rDNA sequences obtained by metagenomic shotgun sequencing of total DNA. Bacterial diversity was also assessed by 16S rRNA pyrotags. (c) Electron microscopy of DLRB showing round-shaped cells embedded in a net of extracellular polymeric substances (EPS).
Scanning electron microscopy
Samples for electron microscopy analyses were prepared scraping biofilm-associated crystals and fixed overnight at 4 °C in a Karnovsky fixative made with formaldehyde (8% v/v), glutar-aldehyde (16% v/v) and phosphate buffer (pH 7). Fixed samples were washed three times with phosphate buffer and CaCl2 for 10 min, and fixed with osmium tetroxide (2% v/v) overnight. Afterward, samples were washed twice with ethanol (30% v/v) for 10 min, dried at a critical point, and sputtered with gold. Specimens were observed under vacuum using a Zeiss Supra 55VP (Carl Zeiss NTS GmbH, Oberkochen, Germany) at the Centro Integral de Microscopía Electrónica (CIME, San Miguel de Tucumán, Argentina).
Isolation of Halorubrum strains from diamante lake
The medium used for the isolation of Halorubrum sp. AD156 and Halorubrum sp. BC156 was WSJ broth, a medium based on that used to isolate Salinibacter ruber (Anton et al., 2002; pH 8.5), containing: NaCl 252.16 g l−1; MgCl2. 6H2O 0.5 g l−1; MgSO4·7H2O 0.5 g l−1; KNO3 1.011 g l−1; KCl 5.84 g l−1; Peptone (Oxoid) 5 g l−1; Yeast extract (Oxoid) 1 g l−1; Trace element solution 1 ml (preparation: HCl, 25% 7.7 M 10 ml, FeCl2 1.5 g, ZnCl2 70 mg, MnCl2·4H2O 100 mg, H3BO3 6 mg, CoCl2·2H2O 190 mg, CuCl2·2H2O 2 mg, NiCl2·6H2O 24 mg, Na2MoO4·2H2O 36 mg), to a final volume of 1 l with distilled water. Solid media were prepared with 1.45% agar (Oxoid). Isolation was done from 5 g of red biofilm sampled from Diamante Lake, which was inoculated in fresh WSJ broth and left for enrichment during 5 days (37 ºC; 160 r.p.m.). After incubation, 100 μl of enriched media was plated on solid WSJ media and incubated aerobically at 37 °C for 12 days. Pure colonies were finally retrieved and PCR of the 16 S rRNA gene using the 344 F (5′-ACGGGGYGCAGCAGGCGCGA-3′) and 915 R (5′-GTGCTCCCCCGCCAATTCCT-3′) primers was performed to identify the taxonomy of isolates.
High-throughput DNA Sequencing
The total DNA sample obtained from DLRB was used to prepare a Metagenomic Shotgun Library. A Rapid library was constructed from 500 ng of DNA following Roche 454-FLX manual's instructions.
The V4 hypervariable region of the 16 S rDNA gene was amplified using F563-R802 universal primers that are reliable amplifying Bacteria but not Archaea. Primer sequences were obtained from RDP's Pyrosequencing Pipeline: http://pyro.cme.msu.edu/. A second short amplification round of PCR (10 cycles) was used to add barcodes and sequencing primers following the same procedure described in Rascovan et al. (2013). Three independent PCRs were performed and then pooled to reduce PCR bias.
Amplicon and metagenome shotgun libraries were sequenced on a Genome Sequencer FLX (454 Life Sciences, Branford, CT, USA) using Titanium Chemistry according to the manufacturer's instructions at INDEAR (Rosario, Argentina) Genome sequencing facility. A total of 1 315 139 454-FLX sequences were obtained from metagenomic shotgun sequencing and 2900 from 16 S rRNA amplicon sequencing. MG-RAST (Meyer et al., 2008) quality control tool was used to trim shotgun sequences by quality obtaining finally 1 053 206 high-quality sequences that were used for subsequent analysis.
Whole-genome shotgun sequencing was also done for the two strains isolated from Diamante Lake (named Halorubrum sp. AD156 and Halorubrum sp. BC156). Total DNA was obtained from the two strains using the regular DNA extraction protocols cited elsewhere. Illumina sequencing libraries were prepared from genomic DNA of each strain and sequenced in the Illumina Highseq 1500 following the manufacturer's instructions at INDEAR Genome sequencing facility.
Bioinformatic analyses
Sequences for 16 S rRNA genes were identified from the metagenomic shotgun data set with Metaxa software (Bengtsson et al., 2011) using default parameters. The retrieved ribosomal RNA small subunit sequences were validated by BLASTN analysis against NCBI Database and the proportion of Archaea/Bacteria was determined by three methods: uclust and RDP classifier methods against greengenes database and best hits agains NCBI database. The three methods showed same results. Then 16 S rRNA sequences longer than 200 bp were used to determine the haloarchaea genera present in DLRB. Only best hits above 97% similarity to best hit against NCBI were considered for genera classification and below that threshold were considered unclassified haloarchaea. Amplicon sequences were demultiplexed and trimmed by quality using split_libraries.py script from QIIME software package (Caporaso et al., 2010). Sequences longer than 500 bp and shorter than 200 bp, with ambiguous base pairs, mean quality score below 27 and homopolymers longer than 6 bp were excluded from the analysis. Amplicon and 16 S rRNA shotgun sequences were taxonomically classified using the RDP classifier method with the Greengenes database using a confidence cutoff of 0.5. Since the set of primers used fail on amplifying archaea, the few sequences from haloarchaea that were obtained from amplicon data set (<7% of all amplicon sequences) were excluded from the analysis. The goal of this method was to determine the bacterial taxonomic diversity. The abundances of each bacterial taxon were then relativized to the total Bacteria abundance (6%).
DLRB metagenomic shotgun sequences were assembled using Newbler software using low coverage sequences (-utr parameter) and all other parameters by default. ORF prediction from single shotgun reads and from contigs was performed using FragGeneScan software (Rho et al., 2010). A first annotation step was performed by BLASTP of all predicted peptides against the full NCBI NR Database using an e-value cutoff of 1E-20. To identify the arsB, acr3, arsC, arsR, arrB, arrA, arrB, arrC/nrfD, arxA, aioA and aioB genes in the metagenomic data set, we generated custom curated databases for each gene using sequences obtained from NCBI and/or Uniprot. BLASTP searches of predicted peptides against each specific database were performed using an e-value threshold of 1E-20. Sequence hits for each gene were finally validated by checking the results obtained for the same sequences against the full NCBI Database.
Shotgun sequencing data from both Halorubrum strains was assembled using the A5 pipeline (Tritt et al., 2012). Coding sequences were predicted using FragGeneScan. Contigs with the genes of interest for this study (16 S rRNA, aioA and arrA) were identified by BLAST and full amino-acid and nucleotide sequences for these genes were retrieved for further analysis. All genes within these contigs were automatically annotated by BLAST against NCBI DB and then the arsenic metabolism related genes and other genes of interest were manually checked and curated. Other genomic analyses from these genomes were reserved for a future work, which will also be accompanied by a physiological characterization of the strains. Same procedures were followed with all genomes retrieved from NCBI database presented in Figure 4.
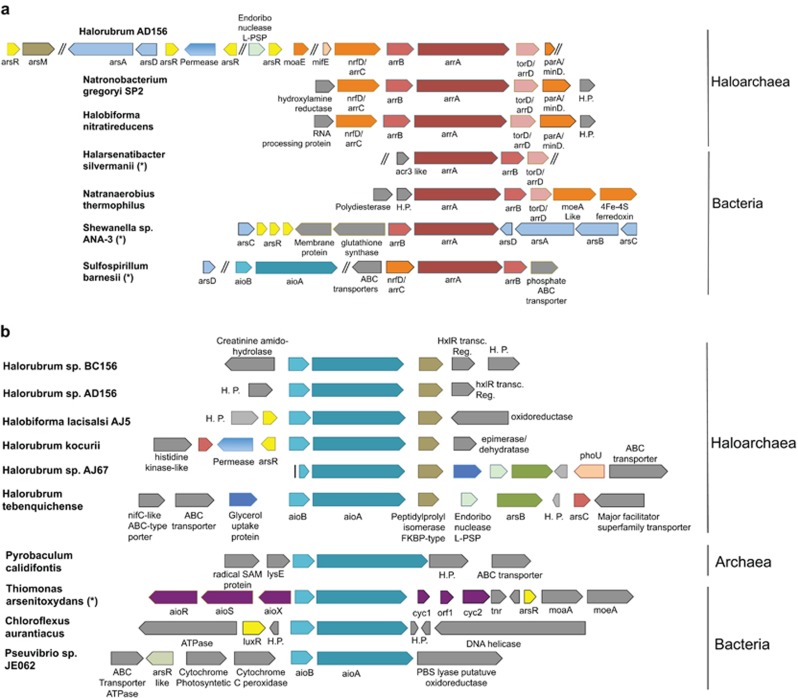
Figure 4.
Genomic organization of the arsenate reductases and arsenite oxidases loci in haloarchaea. The genomic context of the arrA (a) and aioA (b) genes in all haloarchaea genomes is shown. Genes are scaled to their relative amino-acid sequence sizes. Homologous or genes with similar function are presented using same colors. Genes that are not relevant for this work and hypothetical proteins (H.P.) are colored in gray. Double bars between genes indicate that some nonrelevant genes are present in that position. Species that are known arsenate-respiring bacteria or arsenite chemolithotrophs are marked with an asterisk.
Sequence similarity at the nucleotide and amino-acid level for the arrA and aioA genes between all haloarchaea (and best nonhaloarchaea hits in NCBI) was performed using the utility for sequence similarity computation in the T-COFFEE software (Notredame et al., 2000).
Phylogenetic analyses
As a positive control for our phylogenetic analyses, we used the 107 CISM/DMSO protein sequences alignment published by Duval et al. (2008). We identically reproduced their results using Neighbor joining (NJ), Poisson model, 500 bootstrap repetitions and uniform rates. Using the same alignment, we then estimated the best maximum likelihood (ML) model in MEGA (LG+G+I+F; Tamura et al., 2011). Using the ML method with these parameters we obtained a tree that shared 95.5% of the nodes with the NJ tree (calculated using the compare2trees software (Nye et al., 2006)). We then tested different de novo alignment software (Clustal, Muscle, T-Coffee) on the original data set and tested the NJ and ML methods. We obtained best results with T-Coffee, a more accurate de novo aligner than Clustal and Muscle (Wallace et al., 2005). By using the NJ method on the T-Coffee alignment, we obtained a tree that was 93.6% similar to the original and 94.7% using ML method (all differences were in internal nodes that did not affect the main tree topology). We then aligned the haloarchaea AioA and ArrA sequences (and the later discovered ArxA sequences that are available in NCBI database) with Clustal to the original alignment from Duval et al. and calculated the NJ and ML trees. In addition, we performed a de novo alignment using T-Coffee and performed a NJ and ML trees. All the trees were at least 86% similar to each other, and we selected the tree obtained by de novo alignment with T-Coffee and NJ method to present in this paper because it showed the best bootstrapping supports (Supplementary Figure 2). A network consensus tree calculated by SplitTree software (Huson and Bryant, 2006) with the trees obtained from all alignment and phylogenetic methods is also presented to show the nodes that presented discrepancies between methods (Supplementary Figure 3a).
For the ArrA and AioA phylogenetic analyses we also tested de novo alignments with Clustal and T-COFFEE and then NJ and ML phylogenetic methods using 1000 bootstrapping repetitions. We obtained the best bootstrapping supports using de novo alignment with T-COFFEE and ML likelihood method. LG+G+I was the best model in both analyses and 86% of the nodes showed over 70% bootstrapping support in AioA tree and 78% in ArrA. The NarB protein from Pseudomonas stutzeri was used as outgroup in the AioA phylogenetic analysis since the NarB nitrate reductase cluster was the closest to the AioA cluster in the CISM/DMSO tree. For the same reason an ArxA protein from Halomonas was used as outgroup in the ArrA tree. The topologies of the AioA trees obtained by all methods tested were at least 93% similar and >96% similar for ArrA trees. The network consensus trees calculated from all alignment and phylogenetic methods are also presented (Supplementary Figures 3b and c). Same procedures were used to build the AioB phylogenetic analysis presented in Supplementary Figure 4, where a Maximum likelihood analysis using the LG+G+I model showed the best results.
Results
Haloarchaea dominate Diamante Lake biofilms
We discovered red microbial formations attached to gaylussite crystals on the bottom of calcareous rocks (microbialites) that were submerged in Diamante Lake within the Puna region of South America (Figure 1a). Diamante Lake is an unexplored environment with a unique set of extreme conditions that include high pH (9 to 11), high arsenic concentrations (115 to 234 mg l−1), high salinity (270 g l−1, 217 mS cm−1), high UV radiation (84 W m−2 of UVA-B at noon), high day–night temperature range (−20 °C to +20 °C) and low O2 pressure (Table 1). It is situated inside the crater of the Galán volcano near a hydrothermal spring effluent at an altitude of 4570 meters above sea level. Environmental conditions in the area are thought to resemble some of the conditions that prevailed on ancient Earth and on Mars in the past (Cabrol et al., 2007, Sforna et al., 2014). Diamante Lake and the surrounding area is a good model to look for clues about ancient life forms and studying adaptations that permit to survive in extreme environments and other planets.
Table 1. Physicochemical properties of Diamante Lake water compared to other environments.
| Ions (g/l) | Diamante Lake | Socompa Lake | Mono Lakea | Searles Lakea | Sea water | Underground water |
|---|---|---|---|---|---|---|
| Sodium | 43.8–53.3 | 37.1 | 28 | 223.8 | 10 | 0.2 |
| Calcium | 0.001 | 2.4 | 0.004 | NA | 0.4 | 0.3 |
| Potassium | 8.8 | 5.3 | 1.8 | NA | 0.4 | 0.01 |
| Magnesium | 0.02 | 4.09 | 0.04 | NA | 1.2 | 0.08 |
| Chloride | 97–154 | 52.3 | 17.7 | 243.4 | 2 | 0.25 |
| Carbonate | 9.6 | 1.21 | 24.2 | 49.5 | 0.12 | 0.35 |
| Sulfate | 10 | 31.8 | 8.3 | 91 | 3 | 0.3 |
| Nitrate | 0.005 | 0.06 | 0.01 | NA | 0.001 | 0.3 |
| Phosphate | 0.25–0.7 | 0.025 | 0.06 | 1.9 | 0.00002 | 0.0001 |
| Total Arsenic | 0.12–0.23 | 0.02 | 0.01 | 0.29 | 1.00E-06 | 1.00E-06 |
| Others: | ||||||
| Conductivity (mS/Cm) | 192–217 | 115 | 80.8 | 230 | 45 | 0.2 |
| pH | 9–11 | 8.6 | 9.8 | 9.8 | 7.5–8.4 | Variable |
Abbreviation: NA, not available.
Mono Lake and Searless Lake data were obtained from http://www.monobasinresearch.org/onlinereports/deir1.htm and Kulp et al., (2008).
To assess the taxonomic composition and metabolic potential of the DLRB, we performed metagenomic shotgun sequencing of total extracted DNA. We recovered 692 16 S rRNA sequences, of which 94% were classified as Archaea and 6% as Bacteria (Figure 1b). All of the Archaea sequences were assigned to the Halobacteriaceae family known as haloarchaea. Almost 20 different haloarchaea genera were identified (Supplementary Table 1) where Halorubrum (55%), Natronomonas (14%), Halonotius (4%), Halohasta (2%), Natronococcus (2%) and Halorhabdus (1%) were the most abundant and 4% corresponded to other genera. We considered the 16 S rRNA sequences that showed <95% similarity to best hit against NCBI as unclassified haloarchaea (13%). We extended our characterization of the low-abundant Bacterial community by high-throughput sequencing of 16S rRNA amplicons. Most bacterial sequences were classified as anaerobic taxonomic groups (mainly Clostridia and Chromatiales), suggesting that at the microenvironment surrounding the microbial community, oxygen is probably limiting and the conditions microaerobic or transiently anaerobic (Figure 1b).
Electron microscopy showed a biofilm conformation with round cells embedded in a net of extracellular polymeric substances (Figure 1c). Therefore, DLRB represents a very unusual biofilm since it is composed almost exclusively of haloarchaea. Until now, haloarchaea biofilms had been only observed under artificial conditions in the lab (Frols et al., 2012).
Arsenic metabolism genes in Diamante Lake biofilms
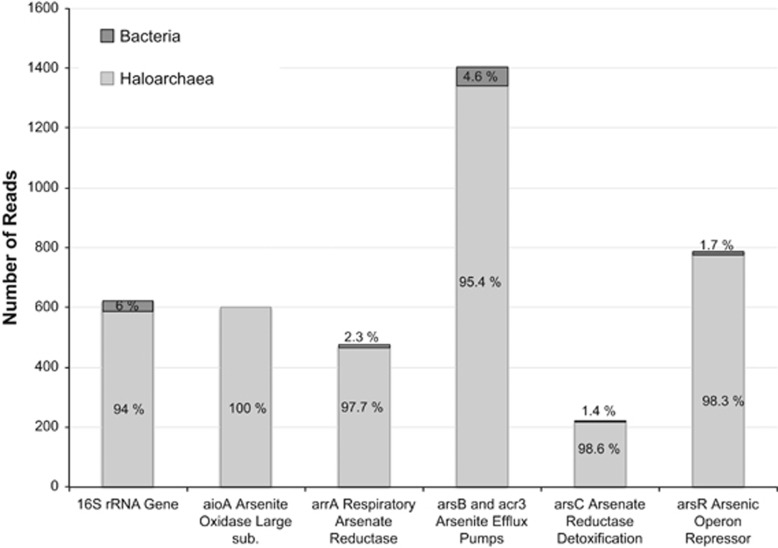
We were interested in determining the type of arsenic metabolisms present in DLRB owing to the extremely high arsenic concentration in Diamante Lake, higher than in many well-characterized arsenic-rich environments (Table 1), and the unexpected haloarchaea predominance in biofilms. Haloarchaea use the ars operon (arsABCRD) as a detoxifying mechanism to eliminate intracellular arsenic (Srivastava and Kowshik, 2013). We found that all genes of the ars operon were present in DLRB metagenome. The high abundance of arsB, arsC and arsR genes relative to the 16S rRNA gene suggest that DLRB haloarchaea use this arsenic detoxification mechanism to maintain nontoxic levels of intracellular arsenic, as it is expected for any microorganism growing under such arsenic concentrations (Figure 2). Differences in the gene sizes (that is, longer genes have a higher chance of being sequenced) and copy number (each of the ars operon genes are frequently found in other parts of the genomes) can explain the differences in the read counts between ars operon genes.
Figure 2.
Abundance of genes related to the arsenic metabolism in DLRB. Metagenomic shotgun sequences coding for arrA, aioA, arsB/acr3, arsC and arsR genes were identified by BLAST (e-value<1E-20) using custom databases of each gene. Results were validated based on BLAST hits on the full NCBI DB to confirm the identity of the sequences and then manually curated. Taxonomic assignments for Bacteria (red) and Haloarchaea (blue) were determined based on best BLAST hits on NCBI DB. Percentages are calculated relative to total counts for each gene. Read counts for 16S rRNA is shown as a reference gene that is present in all organisms in the biofilm.
Surprisingly, we also found high abundance of sequences for arsenite oxidases, large (aioA) and small (aioB) subunits, and respiratory arsenate reductases (arrA; Figure 2). These enzymes are frequently present in arsenic-rich environments, but they have never been described in haloarchaea. We found that aioA and arrA genes were present at almost the same abundance as the 16S rRNA gene indicating that most organisms in DLRB might encode at least one of the enzymes if not both. A BLAST search against NCBI Database showed that all aioA and 98% of the arrA sequences gave best hit against genes present in haloarchaea genomes, although these genes have never been reported in the literature. We found that four haloarchaea genomes (Halorubrum kocurii, Halorubrum sp. AJ67, Halobiforma lacisalsi and Halorubrum tebenquichense) have aioBA genes in their genomes, whereas two genomes (Halobiforma nitratireducens and Natronobacterium gregoryi) have arrA genes. We also obtained the full aioBA sequences from two isolates obtained from DLRB (Halorubrum sp. AD156 and Halorubrum sp. BC156), one of which had also an arrA gene, and additionally two aioA and two arrA full sequences from contigs obtained by assembling our metagenomic data. Preliminary results on the physiology of these isolates showed a remarkable high tolerance to arsenic concentrations (between 50 and 800 mM of As(V) and up to 1 mM of As(III)) and effective transformation of As(III) into As(V). We are currently working on the full characterization of the arsenic metabolism in the Halorubrum isolates.
We observed that the eight haloarchaea aioA sequences and the five arrA sequences showed a rather low amino-acid and nucleotide sequence similarity to each other (between ~60 and 95% for both genes) and a much lower sequence similarity to the best non-haloarchea hit in NCBI (~30 to 50%, Supplementary Figure 1). The divergence in the sequences of haloarchaea aioA and arrA genes and the wide geographical distribution of the species that harbor them could indicate a long evolutionary history of these enzymes in haloarchaea.
Phylogenetic analysis of arrA and aioA genes
Both aioA and arrA genes are part of the complex iron–sulfur molybdoenzyme (CISM) family (Rothery et al., 2008), also known as DMSO reductases. A phylogenetic analysis of all the haloarchaea AioA and ArrA within the CISM family showed not only that they clustered together with all known AioA and ArrA proteins, respectively, but also that both form a robust monophyletic branch within the group of each enzyme (Supplementary Figure 2). It worth noting that the haloarchaea ArrA proteins are clearly clustering together with all the other known ArrA enzymes and are not related to the recently described ArxA proteins (a clade of arsenite oxidases, proposed to be ancestor of the ArrA, which catalyze the oxidative reverse reaction; Zargar et al., 2012).
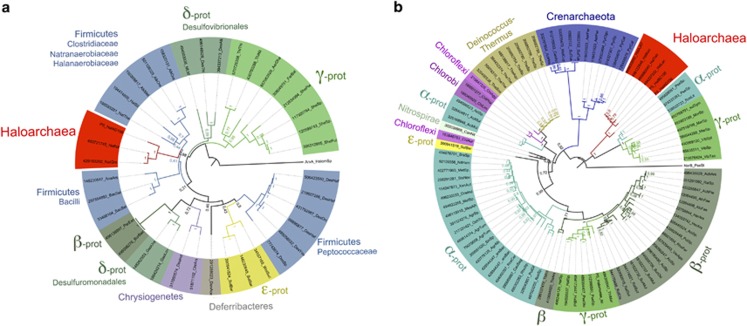
Haloarchaea ArrA sequences also formed a robust monophyletic branch in the phylogenetic analyses generated with all known ArrA sequences (Figure 3a). In all the analyses performed, using different phylogenetic methods, haloarchaea ArrA proteins were clustering within the Firmicutes lineage, between the Clostridia and Bacilli classes (Supplementary Figure 3C), suggesting that they could have been obtained by horizontal gene transfer from an ancient ancestor of modern Firmicutes. However, the low boostrapping support for the position of haloarchaea clade within the Firmicutes lineage makes this concept a speculation at this point. The ArrA phylogenetic tree does not resemble the evolutionary history of the bacterial lineages carrying ArrA enzymes and suggests that multiple horizontal gene transfer events might have taken place in the past, as it was also proposed previously (Duval et al., 2008). Nevertheless, our analyses indicate that most likely haloarchaea ArrA enzymes were obtained in one single (and ancient) event in the past, probably when arsenic-rich environments were more common scenarios on Earth.
Figure 3.
Phylogenetic analyses of respiratory arsenate reductases (ArrA) and arsenite oxidases (AioA). Maximum likelihood trees build from 38 protein sequences for ArrA (a) and 93 for AioA (b). Haloarchaea branches are indicated in red. Bootstrapping support was calculated on 1000 repetitions. The outgroups were selected according the closest protein cluster in the CISM/DMSO family (the NarB nitrate reductase group in AioA tree and ArxA arsenite oxidase group for ArrA tree, see Supplementary Figure 2). The GI accession numbers and the first three letters of the genus and two or three letters of the species are indicated at each tip. The proteins original from this study are named PS.
The phylogenetic analyses of all known AioA enzymes showed that haloarchaea AioA formed a single, robust and monophyletic deep-branching cluster separated from all other known enzymes in this group (Figure 3b). The same result was obtained regardless of the phylogenetic method used (Supplementary Figure 3B) and indicated a common root between the haloarchaea (within the Euryarchaeota phylum) and the Crenarchaeota archaea lineages. Although phylogenetic analyses of short sequences are neither robust nor reliable, a phylogenetic tree of AioB rieske subunit (~180 amino acids) supports as well the ancient origin of aioBA within the Archaea domain (Supplementary Figure 4). Of note is a divergent AioA cluster containing some Alpha- and Gammaproteobacteria that is close to the branch of haloarchaea. All other AioA from the Proteobacteria phylum are located within the bacteria lineage clustering generally according to their taxonomic class. This group of Proteobacteria aioA has been previously considered as aio-like genes (Duval et al., 2008), although they have both aioA and aioB genes and other putative arsenic related enzymes within the same loci. The haloarchaea AioA cluster is the key missing piece of the puzzle that can now give a more rational explanation to the existence of such a divergent AioBA cluster in some Proteobacteria. We propose that this rare group of bacteria obtained their aioA sequences by horizontal gene transfer from haloarchaea or other related Euryarchaeota predecessor in an independent evolutionary event than all other Proteobacteria.
Genomic organization of arrA and aioA operons in Haloarchaea
Functional arsenic bioenergetic aioA and arrA genes are generally found together with other necessary accessory genes. The aioA gene of arsenite oxidizing microorganisms is always found together with the aioB gene encoding a protein from the Rieske family and frequently other genes from the ars operon or the arsenic metabolism are also found at the same genomic loci. On the other hand, the arrA gene of arsenate-respiring bacteria is found together with the arrB and often with the arrC/nrfD and arrD/torD genes (Duval et al., 2008; Slyemi and Bonnefoy, 2012).
We found that all the aioA and arrA accessory genes were found at a high and comparable abundance in DLRB (Supplementary Figure 5). We then analyzed in detail the genomic organization at the arrA and aioA loci of all publicly available haloarchaea genomes and from two isolates obtained from Diamante Lake. Whereas the strain Halorubrum sp. AD156 has both aioBA and arrA genes, the strain Halorubrum sp. BC156 has only the aioBA genes. We found that all genes required for anaerobic arsenate respiration (arrC, arrB, arrA and arrD) were present at the same genomic loci forming the same operon-like organization (Figure 4a) as occurs in arsenate-respiring bacteria. Moreover, in the Halorubrum sp. AD156 strain obtained directly from DLRB, other genes from the ars operon (arsA, arsD, and arsR) and the arsM gene (As(III)-S-adenosylmethionine methyltransferase), normally used to methylate arsenic as a detoxification mechanism) contiguous to the arsR gene were found close to the arrA operon. We also found that the aioA gene in haloarchaea is always accompanied by the aioB gene and a chaperon-coding gene from the peptidylprolyl isomerase FKBP type (Figure 4b). In some of the haloarchaea genomes other arsenic related genes (arsB, arsC and arsR) were also found in the close proximity of aioBA genes. These results indicate that haloarchaea arsenic bioenergetic genes are organized following a highly similar genomic organization as is found in the well-characterized arsenite oxidizing and arsenate-respiring bacterial models.
Discussion
Diamante Lake is an unusual and rare environment owing to its extreme conditions, resembling some of the ancient Earth lacustrine habitats. In this work we present the discovery of haloarchaea biofilms growing under such conditions, whose development seems to be conditioned by an arsenic-based bioenergetic metabolism. Our metagenomic results strongly suggest that the prevalent haloarchaeal part of the biofilm have all the genes necessary for anaerobic arsenate respiration and arsenite oxidation. The phylogenetic analyses, indicate that arsenic bioenergetics in haloarchea is ancient. This hypothesis is also reinforced by the relatively low sequence similarities among haloarchaea aioA and arrA genes and the finding of these enzymes in haloarchea isolated from very distant places on Earth, indicating that the acquisition of these enzymes was not a particular event that took place in Diamante Lake or a constrained area.
Our results suggest that chemolithotrophic use of arsenite could have emerged from a common ancestor of the two main lineages of archaea, the Crenarchaetoa and the Euryarchaeota. The haloarchaea originated about 3–2 Ga ago from methanogenic archaea (Gribaldo and Brochier-Armanet, 2006; Battistuzzi and Hedges, 2009). A recent work on the 2.7 billion-year-old anoxygenic Tumbiana lakes has pointed out at arsenic metabolism as the only pathway able to supply the energy necessary for microbial growth in that lacustrine habitat (Sforna et al., 2014). They also showed that conditions in Tumbiana Lake would have presented environmental conditions similar to those found in Mono Lake and Searles Lake, which are also similar to those found in Diamante Lake (Table 1). Interestingly, the same work also showed that methanogenesis was an important metabolic process in Tumbiana lake. We therefore hypothesize that arsenite oxidases were present in the ancestor of haloarchaea and methanogenic archaea, growing under similar conditions than those that prevailed in Tumbiana lake, and were eventually lost by modern methanogenic archaea and other Euryarchaeota lineages. Nevertheless, it is likely that new arsenite oxidizers from other families in the Euryarchaeota phylum will be found in arsenic-rich environments (most likely anaerobic). Our results indicate that haloarchaea have conserved arsenic metabolic mechanisms that are traces of the ancestors that prevailed in ancient Earth. The AioA phylogenetic analyses presented in this work challenge the current hypothesis of an origin of arsenite oxidases before the divergence of Archaea and Bacteria (Lebrun et al., 2003). In light of these results, we consider that further work is needed to better understand the antiquity and origin of the arsenic metabolism.
The presence of the arrA gene has been suggested as a reliable indicator of arsenate anaerobic respiration (Malasarn et al., 2004). To the best of our knowledge, in all organisms where the arrCBA genes (not the arxA) were found, the arsenate respiratory activity was observed when tested. The finding of arrA, arrB and arrC genes in the DLRB and the Halorubrum sp. AD156, together with the exclusive prevalence of anaerobic bacteria associated with the DLRB, the high arsenic concentration compared with other terminal electron acceptors and the already known capability of haloarchaea to grow under anaerobic conditions using different compounds as electron acceptors, strongly indicate that haloarchaea in Diamante Lake would be able to grow by facultative anaerobiosis on arsenate. The haloarchaea in DLRB together with the Halorubrum sp. AD156, Halobiforma nitratireducens and Natronobacterium gregoryi are the only known archaea that would use ArrA enzymes for anaerobic arsenate respiration. It has been reported that some archaea from the phylum Crenarchaeota are able to grow anaerobically by arsenate (and selenate) respiration (Huber et al., 2000). Some proteins with similarity to ArrA were proposed as candidates to perform this function (Cozen et al., 2009). However, we checked the two proposed candidate genes (PAE1265 and PAE2859) within the phylogeny presented in Supplementary Figure 2 and found that they clustered together with the tetrathionate reductases cluster (TtrA) and the polysulfide reductases (Psr/PshA), respectively. Considering the most likely acquisition of haloarchaea arrA by HTG from bacteria, our data supports the previous hypothesis proposing that arsenate respiration through arrA genes originated within the bacteria lineage (probably within the Gammaproteobacteria lineage) (Duval et al., 2008).
Ecological coupling of arsenic oxidation and reduction was proposed in Mono Lake red biofilms where different populations of bacteria perform either As(III) oxidation in the presence of light or anaerobic As(V) respiration in the dark (Hoeft et al., 2010). In contrast, in the DLRB, the same organisms would perform both functions. The environmental conditions in Diamante Lake are highly variable during the day and particularly across seasons. Red biofilms grow along the bottom of rocks during the summer, when the water level is at a minimum, mineral concentrations are highest (up to 234 mg l−1 of arsenic and conductivity of 217 mS cm−1) and levels of dissolved oxygen are low. During winter, when the water volume doubles (conductivity 90 mS cm−1), the biofilms disaggregates and haloarchaea adopt a free-living style. We hypothesize that DLRB employ different metabolic strategies in response to environmental changes and the heterogeneous conditions within the biofilm. As oxygen is probably a limiting factor, we propose that there is a respiratory and functional diversification to sustain biofilm growth and maintenance. Whereas some organisms might adopt anaerobic respiration on different substrates (mainly arsenate), others might grow aerobically using the limited oxygen available. At the same time, As(III) would contribute energetically to fuel biofilm growth. We propose that other limiting organic and/or light energy sources, together with the limiting oxygen as terminal electron acceptor also contribute to biofilm development. Thus, haloarchaea in DLRB would be performing a complete bioenergetic arsenic cycling, with As(III) oxidation coupled to O2 or nitrate reduction, which could also be complemented by light harvesting, and As(V) respiratory reduction coupled to organic carbon oxidation and, theoretically, also H2 and H2S oxidation (the presence of these compounds was not determined).
Our analyses of Diamante haloarchaea biofilms add novel support to the current hypothesis that arsenic metabolism is one of the main plausible metabolic strategies to meet the energetic demands of microbial mats of the Archaean era. Further characterization of the microbial communities of Diamante Lake will likely continue to shed light on the arsenic metabolism mechanisms, on the evolutionary history of haloarchaea and may give clues regarding the strategies that allow survival under extreme environmental conditions.
Acknowledgments
This work was supported by Préstamo BID PICT 2010 N°1788-ANPCyT, PICT-730 and Loreal-CONICET-UNESCO price for Women in Science. We thank Paola Scaffidi, Travis Dittmer, Reini F. Luco, Tom Misteli and Christelle Desnues for valuable discussions and comments on the manuscript and Catamarca province government for their valuable support.
Author contributions
MEF discovered the biofilms at Diamante Lake, organized the sampling campaigns and processed the samples, performed the physicochemical analyses and site characterization, designed the haloarchaea isolation strategy and revised the manuscript. NR conceived and organized the metagenomic and bioinformatic analyses and wrote the manuscript. JM isolated the two Halorubrum strains. MPV supervised the sequencing of the samples at the INDEAR facility. All authors read and approved the final version of the manuscript.
Data accession
The data reported in this paper are provided in the Supporting Online Material and archived at the following databases: [SRA: SRR1151110, MG-RAST: 4495434.3, MG-RAST: 4513627.3].
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anton J, Oren A, Benlloch S, Rodriguez-Valera F, Amann R, Rossello-Mora R. (2002). Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int J Syst Evol Microbiol 52: 485–491. [DOI] [PubMed] [Google Scholar]
- Battistuzzi F, Hedges S. (2009) Archaebacteria. Oxford University Press: New York. [Google Scholar]
- Bengtsson J, Eriksson KM, Hartmann M, Wang Z, Shenoy BD, Grelet GA et al. (2011). Metaxa: a software tool for automated detection and discrimination among ribosomal small subunit (12 S/16 S/18 S) sequences of archaea, bacteria, eukaryotes, mitochondria, and chloroplasts in metagenomes and environmental sequencing datasets. Antonie van Leeuwenhoek 100: 471–475. [DOI] [PubMed] [Google Scholar]
- Cabrol NA, McKay CP, Grin EA, Kiss KT, Acs E, Tóth B et al. (2007). Signatures of habitats and life in Earth's high-altitude lakes: clues to Noachian aqueous environments on Mars. In Mary Chapman (ed) The Geology of Mars. Cambridge University Press: Cambridge, UK, pp 349–370. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozen AE, Weirauch MT, Pollard KS, Bernick DL, Stuart JM, Lowe TM. (2009). Transcriptional map of respiratory versatility in the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. J Bacteriol 191: 782–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval S, Ducluzeau AL, Nitschke W, Schoepp-Cothenet B. (2008). Enzyme phylogenies as markers for the oxidation state of the environment: the case of respiratory arsenate reductase and related enzymes. BMC Evol Biol 8: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falb M, Muller K, Konigsmaier L, Oberwinkler T, Horn P, von Gronau S et al. (2008). Metabolism of halophilic archaea. Extremophiles 12: 177–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frols S, Dyall-Smith M, Pfeifer F. (2012). Biofilm formation by haloarchaea. Environ Biol 14: 3159–3174. [DOI] [PubMed] [Google Scholar]
- Gribaldo S, Brochier-Armanet C. (2006). The origin and evolution of Archaea: a state of the art. Philos Trans R Soc Lond B Biol Sci 361: 1007–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft SE, Kulp TR, Han S, Lanoil B, Oremland RS. (2010). Coupled arsenotrophy in a hot spring photosynthetic biofilm at Mono Lake, California. Appli Environ Microbiol 76: 4633–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R, Sacher M, Vollmann A, Huber H, Rose D. (2000). Respiration of arsenate and selenate by hyperthermophilic Archaea. Syst Appl Microbiol 23: 305–314. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. (2006). Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23: 254–267. [DOI] [PubMed] [Google Scholar]
- Kulp TR, Hoeft SE, Asao M, Madigan MT, Hollibaugh JT, Fisher JC et al. (2008). Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake, California. Science 321: 967–970. [DOI] [PubMed] [Google Scholar]
- Lebrun E, Brugna M, Baymann F, Muller D, Lievremont D, Lett MC et al. (2003). Arsenite oxidase, an ancient bioenergetic enzyme. Mol Biol Evol 20: 686–693. [DOI] [PubMed] [Google Scholar]
- Malasarn D, Saltikov CW, Campbell KM, Santini JM, Hering JG, Newman DK. (2004). arrA is a reliable marker for As(V) respiration. Science 306: 455. [DOI] [PubMed] [Google Scholar]
- Martin W, Baross J, Kelley D, Russell MJ. (2008). Hydrothermal vents and the origin of life. Nat Rev Microbiol 6: 805–814. [DOI] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M et al. (2008). The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. (2000). T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217. [DOI] [PubMed] [Google Scholar]
- Nye TM, Lio P, Gilks WR. (2006). A novel algorithm and web-based tool for comparing two alternative phylogenetic trees. Bioinformatics 22: 117–119. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Saltikov CW, Wolfe-Simon F, Stolz JF. (2009). Arsenic in the Evolution of Earth and Extraterrestrial Ecosystems. Geomicrobiol J 26: 522–536. [Google Scholar]
- Oren ALife at high salt concentrations Rosenberg E, DeLong E, Lory S, Stackebrandt E, Thompson F. (2013) The Prokaryotes. Springer: Berlin Heidelberg, 421–440. [Google Scholar]
- Rascovan N, Carbonetto B, Revale S, Reinert M, Alvarez R, Godeas A et al. (2013). The PAMPA datasets: a metagenomic survey of microbial communities in Argentinean pampean soils. Microbiome 1: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho M, Tang H, Ye Y. (2010). FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res 38: e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothery RA, Workun GJ, Weiner JH. (2008). The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim Biophys Acta 1778: 1897–1929. [DOI] [PubMed] [Google Scholar]
- Ruggieri F, Saavedra J, Fernandez-Turiel JL, Gimeno D, Garcia-Valles M. (2011). Environmental geochemistry of recent volcanic ashes. J Hazard Mater 183: 353–365. [DOI] [PubMed] [Google Scholar]
- Schoepp-Cothenet B, Duval S, Santini JM, Nitschke W. (2009). Comment on ‘Arsenic (III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake, California'. Science 323: 583 author reply 583. [DOI] [PubMed] [Google Scholar]
- Schoepp-Cothenet B, van Lis R, Philippot P, Magalon A, Russell MJ, Nitschke W. (2012). The ineluctable requirement for the trans-iron elements molybdenum and/or tungsten in the origin of life. Sci Rep 2: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sforna MC, Philippot P, Somogyi A, van Zuilen MA, Medjoubi K, Schoepp-Cothenet B et al. (2014). Evidence for arsenic metabolism and cycling by microorganisms 2.7 billion years ago. Nature Geosci 7: 811–815. [Google Scholar]
- Slyemi D, Bonnefoy V. (2012). How prokaryotes deal with arsenic. Environ Microbiol Rep 4: 571–586. [DOI] [PubMed] [Google Scholar]
- Soler MM, Caffe PJ, Coira BL, Onoe AT, Mahlburg Kay S. (2007). Geology of the Vilama caldera: A new interpretation of a large-scale explosive event in the Central Andean plateau during the Upper Miocene. J Volcanol Geoth Res 164: 27–53. [Google Scholar]
- Srivastava P, Kowshik M. (2013). Mechanisms of metal resistance and homeostasis in haloarchaea. Archaea 2013: 732864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritt A, Eisen JA, Facciotti MT, Darling AE. (2012). An integrated pipeline for de novo assembly of microbial genomes. PLoS One 7: e42304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lis R, Nitschke W, Duval S, Schoepp-Cothenet B. (2013). Arsenics as bioenergetic substrates. Biochim Biophys Acta 1827: 176–188. [DOI] [PubMed] [Google Scholar]
- Wallace IM, Blackshields G, Higgins DG. (2005). Multiple sequence alignments. Curr Opin Struct Biol 15: 261–266. [DOI] [PubMed] [Google Scholar]
- Zargar K, Conrad A, Bernick DL, Lowe TM, Stolc V, Hoeft S et al. (2012). ArxA, a new clade of arsenite oxidase within the DMSO reductase family of molybdenum oxidoreductases. Environ Microbiol 14: 1635–1645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.