Abstract
Plant species richness and the presence of certain influential species (sampling effect) drive the stability and functionality of ecosystems as well as primary production and biomass of consumers. However, little is known about these floristic effects on richness and community composition of soil biota in forest habitats owing to methodological constraints. We developed a DNA metabarcoding approach to identify the major eukaryote groups directly from soil with roughly species-level resolution. Using this method, we examined the effects of tree diversity and individual tree species on soil microbial biomass and taxonomic richness of soil biota in two experimental study systems in Finland and Estonia and accounted for edaphic variables and spatial autocorrelation. Our analyses revealed that the effects of tree diversity and individual species on soil biota are largely context dependent. Multiple regression and structural equation modelling suggested that biomass, soil pH, nutrients and tree species directly affect richness of different taxonomic groups. The community composition of most soil organisms was strongly correlated due to similar response to environmental predictors rather than causal relationships. On a local scale, soil resources and tree species have stronger effect on diversity of soil biota than tree species richness per se.
Introduction
Plant interactions and feedbacks with soil biota determine ecosystem functioning and primary productivity in terrestrial habitats (Wardle et al., 2004; van der Heijden et al., 2006; Bagchi et al., 2014; Wagg et al., 2014). Soil microorganisms and meiofauna (that is, microfauna and mesofauna) have key roles in nutrient cycling. In particular, fungi act as obligate root symbionts, decomposers or pathogens of other organisms. Soil meiofauna and protists consume living organisms and dead organic material and disperse these degradation products as well as fungal and bacterial propagules in soil (Wardle, 2002; Adl and Gupta, 2006).
Greater taxonomic and functional diversity of plants promotes ecosystem services and enhances stability (Cardinale et al., 2011; Gamfeldt et al., 2013). The plant diversity effects on these functions are more pronounced in stress conditions (Steudel et al., 2012) and become stronger with time (Reich et al., 2012). Through resource availability and niche differentiation, increase in plant biomass and species richness favours the accumulation of soil microbial and faunal biomass and abundance that accommodate greater number of species. Such bottom-up relationships among diversity of food-web organisms occur both aboveground and belowground and are reflected along the trophic cascades (Scherber et al., 2010; Eisenhauer et al., 2013; but see Porazinska et al., 2003). Recent studies on natural grassland plants showed that plant species richness is positively correlated with that of several major fungal groups on a local scale (Hiiesalu et al., 2014; Pellissier et al., 2014). Richness of free-living protists, meiofauna and saprotrophic fungi may similarly benefit from specialization on different sources of food or substrate for decomposition.
Top-down relationships may also regulate ecosystem functioning. In experimental systems, more diverse communities of arbuscular mycorrhizal fungi promote plant diversity, productivity and nutrient uptake (van der Heijden et al., 2006; Wagg et al., 2011) that could be related to the mediation of interspecific competition (van der Heijden et al., 2003) and differential benefits of phylogenetically distantly related fungi (Maherali and Klironomos, 2007). In ectomycorrhizal (EcM) symbiosis, fungal species provide differential benefits to their hosts (van der Heijden and Kuyper, 2003) and more diverse communities are more efficient in the uptake of organic phosphorus (Baxter and Dighton, 2005). Counterintuitively, greater diversity of pathogens may also enhance plant richness by specifically suppressing dominants (Bagchi et al., 2014) and reducing the yield in monospecific agroforestry systems (Cardinale et al., 2011).
In addition to taxonomic and functional richness, certain component species may determine the efficiency of ecosystem processes, a phenomenon termed as (taxonomic) sampling effect (Cardinale et al., 2006, 2011). Many of the pioneering ecological studies failed to separate sampling effect from the diversity effect per se, which requires optimizing the experimental design (Huston, 1997; Wardle, 1999; Tedersoo et al., 2014a). Sampling effect can be eliminated or accounted for by (i) comparing species performance in monocultures and polycultures (Wardle, 1999) or (ii) by using model selection or variation partitioning, incorporating component species as dummy variables (Healy et al., 2008; Wagg et al., 2011).
A vast majority of biodiversity studies have recorded short-term effects (but see Reich et al., 2012) and focussed on grassland ecosystems. However, ecosystems naturally dominated by woody plants cover nearly half of the land surface on the Earth. Compared with grassland plants, tree individuals may live for several centuries and they are more widely spaced, creating heterogeneous patches via stem flow and accumulation of root and leaf litter (Nadrowski et al., 2010). Tree species differ substantially in the quality of their litter that determine the chemical composition and microbial biomass directly or indirectly by stimulating earthworm activity (Frouz et al., 2013). Soil and litter quality affect degradation rates and community composition of saprotrophic and EcM fungi (Aponte et al., 2013; Prescott and Grayston, 2013) and meiofauna (Ayres et al., 2009). Tree species richness has usually a neutral (including unimodal relationships) or slightly positive effect on ecosystem processes (Nadrowski et al., 2010). Because of great differences in physiology and ecological properties, tree species drive many of the biochemical and ecological processes in soils (Nadrowski et al., 2010; Gamfeldt et al., 2013). The few available studies so far suggest that tree diversity has a neutral effect on richness of herbs and arthropods on a local scale, but the effects of tree species composition predominate (Vehviläinen et al., 2008; Ampoorter et al., 2014). Plant communities may also correlate with communities of soil organisms such as fungi (Bahram et al., 2012; Peay et al., 2013), but the statistical methods explicitly addressing community-wise relationships and their underlying mechanisms are poorly validated in ecological literature.
Nearly all previous biodiversity studies have used traditional morphology-based identification methods to determine the richness of consumers (but see Hiiesalu et al., 2014; Pellissier et al., 2014). This approach requires substantial taxonomic expertise and long processing time given the large number of samples and individuals (Scherber et al., 2010). Many microscopic taxa are comprised of cryptic species that potentially exhibit different ecological requirements, but they remain undetected owing to the paucity of taxonomically informative morphological character states. The alternative DNA-based tools have been developed and increasingly used for identification of bacteria, protists and fungi since two decades ago. More recently, the massively parallel DNA metabarcoding technology has been adopted for large-scale community-level identification of fungi (Jumpponen and Jones, 2009), protists (Chariton et al., 2010; Medinger et al., 2010) and animals (Porazinska et al., 2009). For meiofauna, metabarcoding studies have focussed on specific order to phylum-level groups such as nematodes or certain arthropods (Porazinska et al., 2009; Hajibabaei et al., 2011). The cytochrome I oxidase gene, the standard barcode for animals, has proven to be suboptimal for metabarcoding analyses owing to problems with primer coverage and large DNA insertions in certain taxa (for example, Creer et al., 2010; de Wit and Erseus, 2010; Deagle et al., 2014; Zhan et al., 2014). It has been outlined that multiple taxonomic groups should be addressed simultaneously for better documenting their relative abundance (Soininen et al., 2013) and understanding of ecological and biogeographic processes (Coleman, 2009; Soininen, 2014). For these reasons, the small subunit (SSU) of ribosomal DNA has been targeted following traditions in microbiology and kingdom-level phylogenetics (Bik et al., 2012). However, there are multiple primer mismatches and/or this marker is too conservative for species-level resolution in nearly all groups of protists, fungi, plants and animals (Pawlowski et al., 2012; Schoch et al., 2012; Tang et al., 2012; Bachy et al., 2013; Lindahl et al., 2013). As an alternative to these markers, the internal transcribed spacer 2 (ITS2) has been proposed as a common species-level metabarcoding marker in eukaryotes, although taxonomic groups differ somewhat in length and there is some intraindividual variation inherent to all nuclear markers (Coleman, 2009; Koetschan et al., 2010; Yao et al., 2010: Bengtsson-Palme et al., 2013; Wang et al., 2015). The ITS2 marker has been successfully used to target fungi (Clemmensen et al., 2013), various protist groups (for example, Arif et al., 2014) and plants (De Barba et al., 2014) in metabarcoding studies. Although a large number of animal ITS sequences have been deposited in public databases, this region has been hitherto overlooked in the DNA metabarcoding of meiofauna.
In this study, we first describe the development of a DNA metabarcoding method for identification of multiple eukaryotic organisms simultaneously at species-level resolution. We constructed multiple taxon-targeted primers for the ITS2 region in single PCR reactions to maintain ribosomal DNA-based proportions of organisms. By using the metabarcoding analysis of DNA extracted from pools of thousands of soil cores, we disentangled the relative roles of tree diversity, confounding sampling effects as well as spatial and edaphic variables on taxonomic richness and community composition of soil fungi, protists and meiofauna. We postulated the following alternative hypotheses: (1) tree diversity per se and taxonomic sampling effect influence microbial biomass and richness of soil biota; (2) the effect of these biotic variables is relatively stronger in biotrophic organisms compared with saprotrophs and trophically directly unrelated organisms; (3) vegetation has both direct effects, and indirect effects through altered soil chemistry and microbial biomass, on richness of soil biota; and (4) communities of soil biota shift in concordance mainly owing to similar responses to the environment.
Materials and methods
Experimental design and sampling
We selected two forest diversity experiments in Satakunta, Finland (61° N; 22° E) and Järvselja, Estonia (58° N; 27° E) to test our hypotheses. The Finnish experiment was established across three sites in a clear-cut boreal forest in 1999. We selected sites 1 and 3 for sampling, because these were the least damaged by moose (Ampoorter et al., 2014). At the time of sampling, trees had reached a height of 5–11 m and formed a closed canopy in most of the plots. The soils are podzols with a silty or sandy texture on granite bedrock. Seedlings of Pinus sylvestris L., Picea abies (L.) H. Karst., Larix sibirica Ledeb., Betula pendula Roth. and Alnus glutinosa (L.) Gaertn. were planted as monocultures or equal combinations of two, three or five species in 400-m2 square plots (Scherer-Lorenzen et al., 2006). To avoid edge effects, we restricted our sampling to 300 m2 in the centre of each plot by excluding the outermost row of trees.
Vegetation at the Estonian study system constitutes a remnant of a large-scale forest experiment established in early 1920 s on clear-cut forested land. The soils are formed on postglacial alluvial deposits and exhibit loamy or sandy texture. Certain forest quadrats were planted with trees, whereas others were left for natural regeneration. At the sapling stage, trees were selectively thinned and forest quadrats were deeply drained to stimulate tree growth and prevent waterlogging. In the second half of the twentieth century, intensity of management declined and forest development was subjected to natural succession that was affected by sporadic selective cutting and differential moisture regime owing to degradation of the ditch network. The combination of these treatments and processes resulted in the development of vegetation with different dominant trees (P. abies, P. sylvestris, B. pendula, A. glutinosa, Tilia cordata Mill. or Populus tremula L.) and a range in tree richness (2–11 species; other subdominant EcM trees Corylus avellana L., Quercus robur L., Salix caprea L. and arbuscular mycorrhizal hosts Ulmus glabra Huds., Fraxinus excelsior L., Acer platanoides L. and Sorbus aucuparia L.). Because the original quadrats were of unequal size and shape, we established round 2500-m2 plots in uniform patches of vegetation. For mature forests, greater plot size represents better the edaphic and floristic processes and the interacting biota (Bruelheide et al., 2014).
In each plot, we determined the basal area (BA) of all tree species and coverage of understorey vascular plant species. In Finnish plots, we estimated the relative amount of birch coppice (cut in spring, 2010) in 10 abundance classes based on the number and size of stumps. For Estonian plots, we obtained additional information about productivity, volume and height of trees from the State Forest Management Centre (www.rmk.ee).
In summer 2011, we collected samples from 43 Finnish plots (spread over two sites 10 km distant, each roughly 4 ha) and 41 Estonian (spread evenly across 1000 ha) plots. In Finland, we randomly selected 40 trees per plot to equally represent the composition of planted species. At ca. 0.5 m distance from the trunk of each tree individual, we collected a single soil core by hammering a PVC tube (5 cm diameter) to 5 cm depth. In Estonia, we similarly sampled 40 soil cores per plot, but we collected each pair of cores 1–1.5 m distant from each of 20 randomly selected trees (>10 cm diameter at breast height) located at least 8 m distant from each other to account for spatial autocorrelation range in soil biota (Bahram et al., 2013). For Finnish and Estonian study areas, information about plant species composition and other metadata is given in Supplementary Table S1, and Supplementary Data S1 and S2.
In both study areas, the cores nearly always comprised both the organic layer and top mineral soil and included roots. Although deep soil may comprise some unique organisms adapted to anoxic conditions and low nutrients, our sampling was limited to topsoil, because >50% of microbial biomass and its biological activity occurs in the topmost organic soil (Serna-Chavez et al., 2013) and deeper sampling was impossible in Finnish soils owing to great abundance of rocks. All 40 soil cores per plot were pooled, thoroughly mixed and air-dried at 30–40 °C for 24 h. Drying was selected as an alternative to deep freezing or fresh extraction because of improved options for pulverization of large amounts of soil and necessity to standardize extractable material on a dry weight basis. Dried soil was stored air-tight in zip-lock plastic bags and ground into fine powder by heavy rubbing of the zip-lock bags followed by bead beating using 3-mm tungsten carbide balls in Mixer Mill MM400 (Retsch GmbH, Haan, Germany) at 30 Hz for 10 min for subsequent soil nutrient and molecular identification analyses.
Soil nutrients, phospholipid fatty acids and ergosterol
Concentrations of C, N, 13C and 15N were measured from 2 to 70 mg of soil using an elemental analyser (Eurovector, Milan, Italy) coupled with an isotope ratio mass spectrometer (MAT 253; Thermo Electron, Bremen, Germany) according to Tedersoo et al. (2012). Total phosphorus was extracted using ammonium lactate and determined using flow injection analysis. Potassium concentration was determined from the same extract by the flame photometric method (AOAC956.01). Exchangeable magnesium and calcium content were determined in ammonium acetate extract (pH=7.0).
Bacterial biomass was assessed using the phospholipid fatty acid analysis for which the samples were extracted with a mixture of chloroform–methanol–phosphate buffer (1:2:0.8; V/V) according to Bligh and Dyer (1959). Phospholipids were separated using solid-phase extraction cartridges LiChrolut Si60 (Merck, Whitehouse Station, NJ, USA), and the samples were subjected to mild alkaline methanolysis (Šnajdr et al., 2008). The samples were analysed with gas chromatography–mass spectrometry (450-GC, 240-MS; Varian, Palo Alto, CA, USA). Methylated fatty acids were identified according to their mass spectra using a mixture of chemical standards (Sigma, St Louis, MO, USA). Actinobacterial biomass was estimated as the sum of 10Me-16:0, 10Me-17:0 and 10Me-18:0. Total bacterial biomass was determined on the basis of i14:0, i15:0, a15:0, i16:0, i17:0, a17:0, 16:1ω7, 18:1ω7, cy17:0, cy19:0, 16:1ω5, 10Me-17:0, 10Me-18:0 and 10Me-16:0 (Šnajdr et al., 2011).
Fungal biomass was estimated based on ergosterol concentration. Total ergosterol was extracted and analysed as described previously (Šnajdr et al., 2008). Samples (0.5 g) were sonicated with 3 ml 10% KOH in methanol at 70 °C for 90 min. Distilled water (1 ml) was added and the samples were extracted three times with 2 ml cyclohexane, evaporated under nitrogen, redissolved in methanol and analysed isocratically using an high performance liquid chromatography system with methanol as a mobile phase at a flow rate of 1 ml min−1. Ergosterol was quantified by ultraviolet detection at 282 nm. Saprotroph and EcM fungal biomass was calculated based on the proportion of sequences corresponding to each functional group, assuming that these two groups have equal ergosterol-to-ribosomal DNA ratio on average (Štursova et al., 2014).
Molecular analyses
DNA was extracted from 2.0 g of soil per sample using the PowerMax Soil DNA Isolation Kit (MoBio, Carlsbad, CA, USA) according to the manufacurer's protocols. PCR was performed using a mixture of 11 forward primers (ITS3tagmix1-11 in equimolar concentration) analogous to ITS3 and a degenerate reverse primer ITS4ngs analogous to ITS4 (the original primers are described in White et al., 1990; Tedersoo et al., 2014b, 2015a). Both primers were shortened and modified to perfectly match >99.5% of all Fungi (except Tulasnellaceae and Microsporidia; Supplementary Table S2). The ITS3 primer mixes were designed to cover Cercozoa protists (amoebae from Rhizaria superkingdom), Ciliophora protists (Alveolata superkingdom), Chlorophyta (unicellular algae from Viridiplantae superkingdom), as well as soil animals (Acari, Nematoda, Collembola, Rotifera and Annelida (Metazoa). Based on a wide range of studies, these groups are the most abundant and species-rich eukaryote taxa in soil. For primer design, we downloaded available sequences from the International Nucleotide Sequence Databases Consortium (INSDC) and aligned the conserved 5.8 S and large subunit (LSU) regions by classes or phyla by using MAFFT ver. 7 (Katoh and Standley, 2013). The primer annealing sites for ITS3 and ITS4 were visually examined to distinguish true mismatches from low-quality sequences and mitochondrial LSU sequences. We further checked primer matching by running BLASTn searches against targeted organisms in INSDC, setting the number of comparisons greater than the number of species available for that taxonomic group. The ITS4ngs primer was tagged with one of the 110 identifier barcodes (10–12 bases) that were modified from those recommended by Roche (Basel, Switzerland) to differ by >3 bases, to start only with adenosine and to comprise the proportion of adenosine and thymidine between 0.3 and 0.7 to equalize their affinities in an adapter ligation step. The PCR cocktail comprised 0.6 μl DNA, 0.5 μl each of the primers (20 μM), 5 μl 5 × HOT FIREPol Blend Master Mix (Solis Biodyne, Tartu, Estonia) and 13.4 μl double-distilled water. PCR was carried out in four replicates in the following thermocycling conditions: an initial 15 min at 95 °C, followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min, and a final cycle of 10 min at 72 °C. PCR products (typically 350–400 bp) were pooled and their relative quantity was estimated by running 2 μl DNA on 1% agarose gel for 15 min. DNA samples yielding no visible band or a strong band were re-amplified using 35 and 25 cycles instead. We also used negative (for DNA extraction and PCR) and positive controls (Hydnoplicata whiteii specimen MURU5860) throughout the experiment. Amplicons were purified by use of exonuclease and Shrimp alkaline phosphatase enzymes (Fermantas, Kaunas, Lithuania) at 37 °C for 45 min and at 85 °C for 15 min. Purified amplicons were subjected to normalization of quantity by use of SequalPrep Normalization Plate Kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. Normalized amplicons were divided into two pools that were each subjected to 454 adaptor ligation, emulsion PCR and 454 pyrosequencing using the GS-FLX+ technology and Titanium chemistry as implemented in Beckman Coulter Genomics (Danvers, MA, USA).
Pyrosequencing resulted in 625 074 reads with a median length of 412 bases. By use of Acacia 1.52 (Bragg et al., 2012), these sequences were re-assigned to samples based on the barcodes, and quality trimmed (options: minimum average quality threshold=30, maximum k-mer distance=13; homopolymer significance threshold=−2) to exclude short and low-quality sequences. The resulting 564 305 sequences were subjected to removal of the flanking 5.8S and 28S rRNA genes for better resolution in clustering of ITS sequences and removal of chimeric sequences by use of the ITSx software (Bengtsson-Palme et al., 2013). We retained sequences of >99 bp in length to remove most of the partial sequences. We ran a second round of chimera checking using UCHIME (Edgar et al., 2011). After these quality-filtering steps, the 401 762 retained sequences were further clustered at 98.0% sequence similarity as implemented in CD-Hit 4.6.1 (Fu et al., 2012). Compared with the routinely used 97%, this threshold is a better proxy at species level in several groups of fungi (Kõljalg et al., 2013), protists (Litaker et al., 2007) and animals (Noge et al., 2005). Of 16 437 clusters, 6823 (41.5%) were represented by a single sequence. These singletons were removed from further analyses, because these comprise a high proportion of technical artefacts (Tedersoo et al., 2010). The longest sequence of each remaining 9614 clusters was selected as a representative for BLASTn sequence similarity search (word size=7; penalties: gap=−1; gap extension=−2; match=1) against the INSDC and UNITE (Abarenkov et al., 2010a) databases. In addition, we ran BLASTn searches against reference sequences of fungi in 99.0% similarity species hypotheses that include third-party taxonomic and metadata updates (Kõljalg et al., 2013) as implemented in the PlutoF workbench (Abarenkov et al., 2010b). For each query, we considered 10 best-matching references to annotate taxa as accurately as possible. If no taxonomy was reliably revealed, we ran manual BLASTn searches against INSDC with 500 best-matching sequences as output. We followed the regularly updated INSDC and Index Fungorum (www.indexfungorum.org) for higher-level taxonomy of eukaryotes and up to class-level taxonomy of fungi, respectively. We assigned each fungal genus, family or order to functional categories. If different lifestyles were present in specific genera, we chose the dominant group (>75% of species assigned to a specific category) or considered its ecology unknown (<75%). Taxa were considered to be EcM if they best matched to any sequences belonging to EcM lineages (Tedersoo and Smith, 2013) and exhibited sequence length/blast scores above predetermined lineage-specific thresholds. Targeted groups of protists and soil animals were too poorly represented in INSDC to allow reliable trophic categorization. We selected 98.0% sequence similarity to represent roughly species-level discrimination of operational taxonomic units (OTUs) in all taxa based on post-hoc determination of optimal metabarcoding thresholds (Põlme et al., 2014; Tedersoo et al., 2014c). Only for Collembola, we re-clustered all sequences at 94.0% sequence similarity, because this level distinguishes better among species (Anslan and Tedersoo, 2015).
Statistical analyses
We chose to analyse richness and community composition in groups that were represented by >50 OTUs (Cercozoa, Ciliophora, Chlorophyta, Collembola, Nematoda as well as EcM, saprotrophic, plant pathogenic fungi). For richness analyses of soil biota, we calculated the residuals of OTU richness in relation to square root of the number of obtained sequences to account for differences in sequencing depth. We excluded two outlier samples from Finland and four outliers from Estonia that were dominated by a few species of moulds (relative abundance of sequences belonging to Trichocomaceae >5%, Mortierellaceae >20% or Mucoraceae >20%, that exceeds three times the mean+3 s.d.), which is indicative of substandard sample preservation (Tedersoo et al., 2014b). By use of vegan package of R, ‘individual'-based rarefied OTU accumulation curves were constructed separately for functional groups of fungi and major taxa of animals and protists in the two study systems (Supplementary Figure S1).
Concentrations of soil nutrients and vegetation measurements were logarithm-transformed prior to analyses to improve the distribution of residuals and reduce non-linearity. To estimate taxonomic sampling effects, the relative BA of each tree species was log-ratio transformed (Szava-Kovats et al., 2011). Besides species richness, we calculated both Shannon and Simpson indices of diversity for trees and understorey vascular plants (Magurran, 1988). To account for spatial autocorrelation that may arise from both spatially structured environmental factors and dispersal limitation, we calculated Principal Components of Neighbours Matrices (PCNM) spatial eigenvectors based on geographical coordinates of plots by using the vegan and packfor packages of R (R Core Development Team, 2014). These vectors represent spatial variation at different geographical scales over the study area and are used to control for spatial autocorrelation in ecological data sets (Legendre, 2008). To disentangle the effects of edaphic, floristic and spatial variables on residual richness of soil biota and biomass estimates, individual variables were subjected to multiple regression model selection based on the corrected Akaike information criterion. The components of best models were forward-selected to determine their adjusted coefficients of determination as implemented in the packfor package of R.
We used Structural Equation Models (SEM) to determine the direct and indirect paths between environmental predictors, biomass and richness of EcM fungi and saprotrophs. We also included bacterial biomass in these models for comparison. The initial SEM models were constructed based on the best models for these variables using Amos ver. 22 (SPSS, Chicago, IL, USA). We tested all direct and indirect relations among exogenous and endogenous variables, together with their error terms. Then the fit of models was maximized based on both chi-square test and root mean square error of approximation and Comparative Fit Index. We followed a backward stepwise elimination approach to remove non-significant links to maximize model fit.
The relative effects of edaphic, spatial and floristic variables on communities of soil organisms were determined based on Hellinger dissimilarity, exclusion of OTUs occurring in a single sample and a multivariate model selection procedure as implemented in DISTLM function of Permanova+ (Anderson, 2005). To obtain coefficients of determination (cumulative R2adjusted) and statistics (Fpseudo) for each variable, the components of best models were forward selected. We prepared Global Non-metric Multidimensional Scaling graphs in parallel, using the same options. Significant variables were fitted into the ordination space using the envfit function in the vegan package of R.
To test the correlation in community composition among soil biota, trees and herbs, we calculated the bidirectional Procrustes correlation coefficient using 4999 permutations as implemented in protest function in the vegan package of R. False discovery rate associated with multiple testing was reduced using the sharpened false discovery rate procedure (Benjamini and Hochberg, 2000). To test whether intimately associated communities display more similar beta diversity and to recover any sampling biases (size of matrices, connectance), we added the relatively small communities of trees, herbs and fungal subgroups belonging to mycoparasites, flagellates (Chytridiomycota) and white rot decomposers to the correlation matrices. The effects of association intimacy as well as the size of matrices and connectance on RProcrustes were tested using multiple regression analyses separately for Estonian and Finnish data sets. To further test whether the correlations among communities are potentially causal or related to the shared driving mechanisms of environmental variables, we calculated partial Procrustes association metrics accounting for the environmental and spatial predictors (Lisboa et al., 2014).
Variation partitioning analyses were conducted on standardized biomass and OTU richness data and Hellinger-transformed community data using the packfor and vegan packages of R. Because of the limits of variation partitioning, we generated four components of variance, including richness (tree and understorey richness and Shannon index), sampling effect (log ratio-transformed proportions of tree species), other floristic as well as edaphic characters (total BA, tree volume, productivity, herb and ericoid cover, soil pH and nutrients) and space (PCNM vectors). By using one-way analyses of variance, we further tested whether the groups of different mobility (mobile: actively moving organisms excluding amoeboid groups), body size (average cell diameter and organism diameter) and trophic strategies (biotrophic and non-biotrophic) exhibit significant differences in the relative importance of tree richness, individual tree species and space.
Results
Identification of soil biota
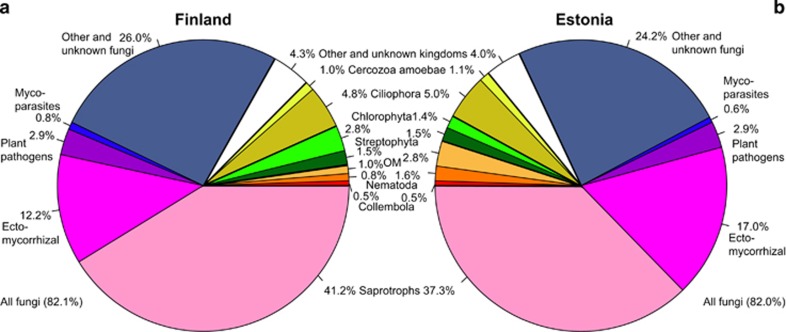
The metabarcoding approach enabled us to recover all targeted groups of eukaryotes from composite soil samples and identify them at different taxonomic levels (Supplementary Data S1). Across study systems, kingdom-level assignment of 1.0% of the recovered sequences and 3.9% OTUs remained unknown. Sequences and OTUs (98% similarity threshold) assigned to Fungi (81.1% of taxa), Alveolata (4.8%, mostly Ciliophora), Metazoa (4.8%, mostly Nematoda and Collembola) and Viridiplantae (4.0%, mostly Tracheophyta, Bryophyta and Chlorophyta) dominated among the soil eukaryote kingdoms both in Finland and Estonia (Figure 1). Saprotrophs, EcM mutualists and plant pathogens comprised 47.5, 17.7 and 3.5% of all fungal OTUs, respectively. The higher-level taxonomic distribution of soil organisms was remarkably similar in Finnish and Estonian samples (Figure 1). One of the negative controls produced two fungal sequences, whereas the two positive controls yielded six different fungal and oomycete sequences in addition to H. whiteii.
Figure 1.
Proportion of 98% sequence similarity-level OTUs belonging to the fungal, protist and animal groups in (a) Finland and (b) Estonia. OM, other Metazoa.
Richness and biomass
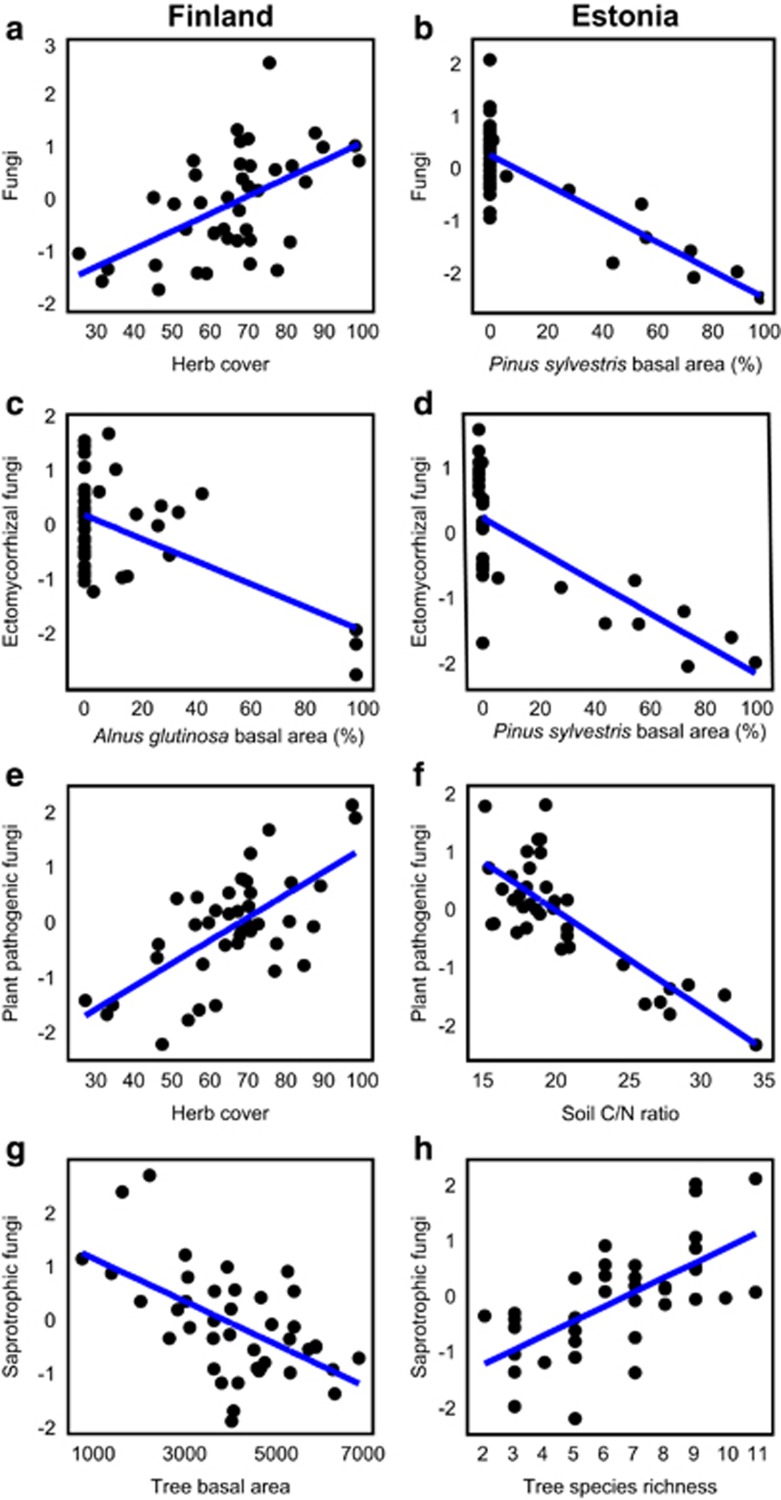
Tree species richness was positively correlated with richness of soil fungal groups in Estonia and EcM fungi in Finland, but it was poorly correlated with richness of protists and meiofauna (Supplementary Figure S2). Taxonomic sampling effect of plants and edaphic variables were usually among the best predictors of belowground richness depending on organisms and study systems (Figures 2 and 3; Supplementary Figure S3; Supplementary Table S3). Richness of all fungi was most strongly affected by herb cover (positive effect: F1,35=17.30; R2adj=0.289; P=0.001) and tree BA (negative effect: F1,35=17.30; R2adj,partial=0.108; P=0.007) in Finland. In Estonia, P. sylvestris BA (F1,32=63.29; R2adj=0.644; P<0.001) and tree species richness (F1,32=25.21; R2adj,partial=0.149; P<0.001), respectively, had a negative and positive effect on fungal richness. Richness of EcM fungi was negatively affected by A. glutinosa BA (F1,34=14.60; R2adj=0.289; P=0.003) but positively by increasing soil pH (F1,34=16.98; R2adj,partial=0.217; P=0.001) and tree diversity (Shannon index: F1,34=13.05; R2adj,partial=0.127; P=0.002) in Finland. In Estonia, BA of P. sylvestris (F1,32=40.28; R2adj=0.522; P<0.001) and BA of A. glutinosa (F1,32=11.35; R2adj,partial=0.109; P<0.003) had a strong negative effect on EcM fungal richness, explaining 63.1% of variation, but there was no significant tree diversity effect. Herb cover enhanced (F1,34=30.16; R2adj=0.422; P<0.001), but increasing C/N ratio reduced (F1,34=10.61; R2adj,partial=0.114; P=0.002), plant pathogen richness in Finland. Similarly, soil C/N ratio had a strong negative effect on plant pathogen richness in Estonia (F1,30=40.28; R2adj=0.627; P<0.001). Richness of saprotrophic fungi was negatively influenced by total BA of trees (F1,35=18.25; R2adj=0.301; P=0.001) and BA of P. abies (F1,35=9.94; R2adj,partial=0.117; P=0.004) in Finland. In Estonia, however, tree species richness per se had a strong positive effect on saprotroph richness (F1,32=25.60; R2adj=0.406; P<0.001).
Figure 2.
Linear regressions between the standardized residuals of richness of soil fungi and the strongest predictors as revealed from the best multiple regression models (Supplementary Table S2): (a, b) all fungi; (c, d) EcM fungi; (e, f) plant pathogens; (g, h) saprotrophs.
Figure 3.
Linear regressions between the standardized residuals of richness of soil protists or animals and the strongest predictors as revealed from the best multiple regression models (Supplementary Table S2): (a, b) Cercozoa; (c, d) Chlorophyta; (e, f) Ciliophora; (g, h) Collembola; (i, j) Nematoda.
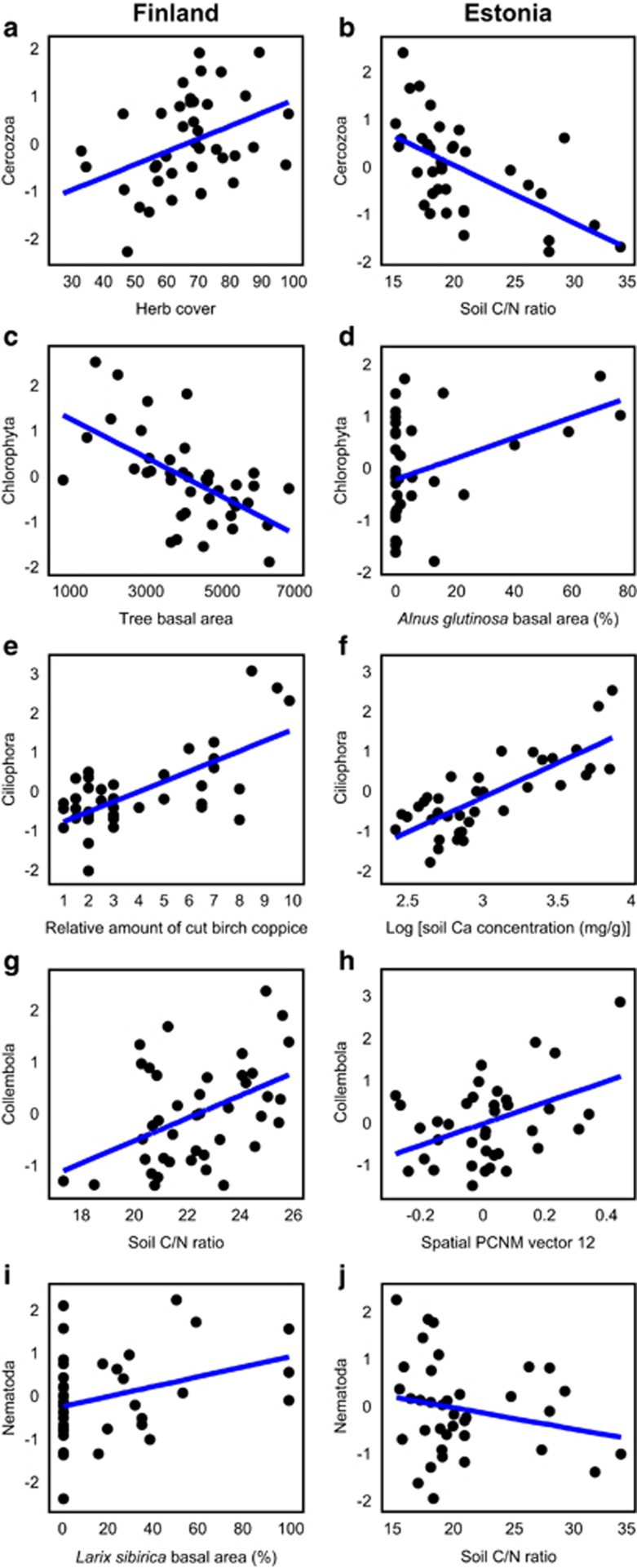
Richness of the Cercozoa amoebae was most strongly enhanced by herb cover (F1,34=8.72; R2adj=0.162; P=0.006) and soil pH (F1,34=9.46; R2adj,partial=0.149; P=0.010) in Finland but negatively affected by C/N ratio (F1,33=17.75; R2adj=0.318; P=0.001) and soil Ca concentration (F1,33=17.75; R2adj,partial=0.151; P=0.003) in Estonia. Chlorophyta richness was reduced by increasing total tree BA in Finland (F1,37=21.12; R2adj=0.335; P<0.001) but promoted by A. glutinosa BA in Estonia (F1,31=5.41; R2adj=0.109; P=0.022). Ciliophora richness was favoured by the relative amount of cut birch coppice (F1,35=32.57; R2adj=0.441; P<0.001) and understorey richness (F1,35=12.05; R2adj,partial=0123; P=0.004) in Finland but by soil Ca concentration in Estonia (F1,31=52.90; R2adj=0.590; P<0.001).
Of soil animals, the richness of Collembola was affected by soil C/N ratio (F1,37=9.93; R2adj=0.182; P=0.002) and spatial eigenvectors in Finland (F2,37=17.34; R2adj,partial=0.250; P=0.001) but driven only by spatial variables in Estonia (F2,34=15.17; R2adj,cumul=0.288; P=0.001). Nematoda richness responded positively to L. sibirica BA in Finland (F1,38=9.93; R2adj=0.072; P=0.047), whereas soil C/N ratio had a negative effect on roundworms in Estonia (F1,34=9.64; R2adj=0.196; P=0.003).
Although the Estonian and Finnish soils differed considerably in nutrient concentrations (Supplementary Figure S4), the biomass of microbial groups in both study systems was positively related to soil macronutrients (Figure 2). Model selection indicated that the total microbial biomass and bacterial biomass were positively influenced by soil P concentration both in Finland and Estonia (Supplementary Table S3). The biomass of Actinobacteria increased with soil P concentration in Finland (F1,38=45.55; R2adj=0.527; P<0.001), but it responded positively to increasing proportion of A. glutinosa BA (F1,33=14.43; R2adj=0.272; P=0.001) and soil Ca concentration (F1,33=8.84; R2adj,partial=0.133; P=0.011) in Estonia. Fungal biomass, including that of saprotrophs and EcM fungi, was largely determined by N concentration in both study systems. The ratio of saprotrophs to EcM symbionts increased with the relative BA of A. glutinosa in both systems (Supplementary Table S3). In addition, soil Ca concentration negatively affected the relative proportion of saprotrophs in Estonia (F1,33=14.85; R2adj,partial=0.254; P=0.001).
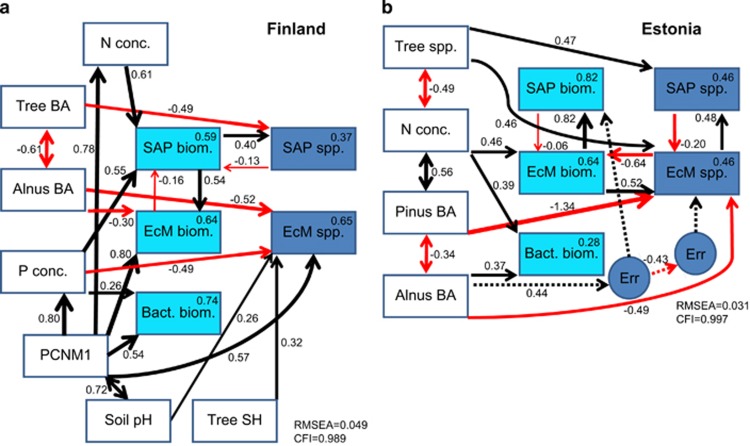
Structural equation modelling revealed that site (PCNM1) had a strong effect on soil pH, N and P concentrations in Finland (Figure 4a). All these variables as well as tree diversity (Shannon index) had a direct positive effect on EcM fungal richness, whereas A. glutinosa BA had a direct negative effect on both EcM fungal biomass and richness. Soil nitrogen concentration and total BA had, respectively, a direct positive and negative effect on the richness of saprotrophs. Saprotroph biomass positively influenced saprotroph richness and EcM fungal biomass, but the opposite paths were of minor importance (P>0.1). Similarly for the Estonian data set, SEM largely confirmed the results of model selection, emphasizing the direct positive effect of tree richness and EcM fungal biomass but negative effects of A. glutinosa and P. sylvestris BAs on EcM fungal richness (Figure 4b). According to SEM, EcM fungal biomass and richness had strong positive effects on saprotroph biomass and richness, respectively.
Figure 4.
Structural equation models demonstrating the direct and indirect effects of spatial, environmental and floristic variables on biomass and species richness of EcM and saprotrophic fungi in (a) Finland and (b) Estonia. Black and red arrows indicate positive and negative relationships, respectively. Dashed arrows indicate connections through error terms (Err). Numbers above arrows indicate standardized path coefficients. Numbers above variables indicate proportion of variation explained. SH, Shannon diversity index.
The variation partitioning analysis was generally consistent with model selection (Supplementary Table S4; Supplementary Figure S3). There were no general differences in the relative strength of tree diversity or individual species effects on organisms with different mobility, body size or biotrophic associations (P>0.2). Variation partitioning analysis ascribed much of the environmental variation to a shared effect among space, soil and vegetation (Supplementary Table S4), confirming the SEM results that much of the floristic effects are spatially and edaphically structured. Across all data sets, the effect of space was significantly greater in Finland than in Estonia (F1,42=7.90; P=0.007). This could be ascribed to the arrangement of plots in two distinct blocks (sites) in Finland as opposed to a more uniform plot distribution in Estonia. Furthermore, the Estonian experiment was subjected to natural regeneration that may have been influenced spatially structured soil parameters.
Community composition
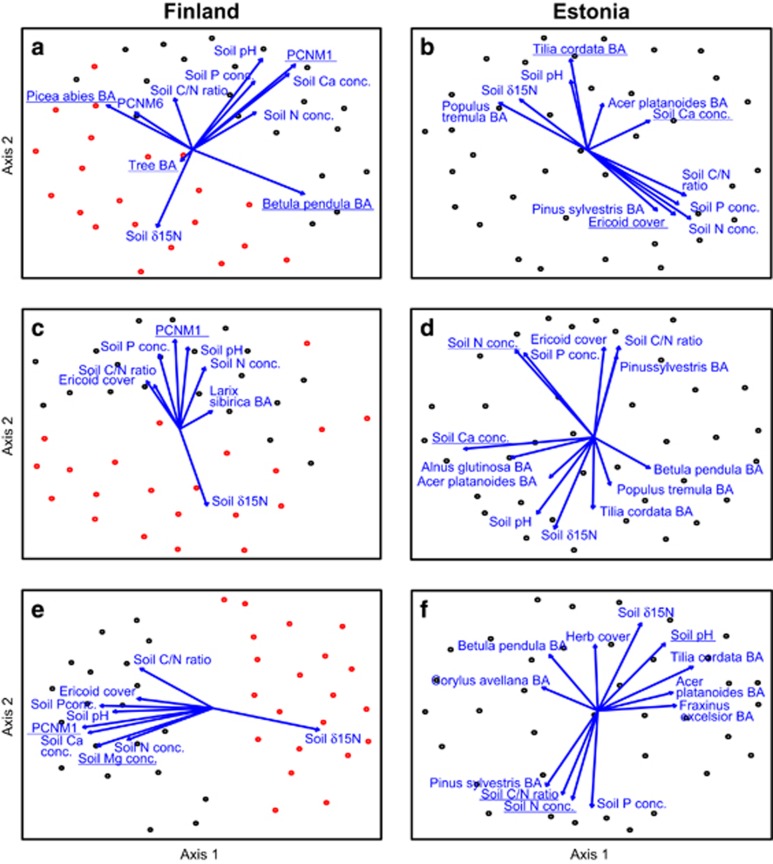
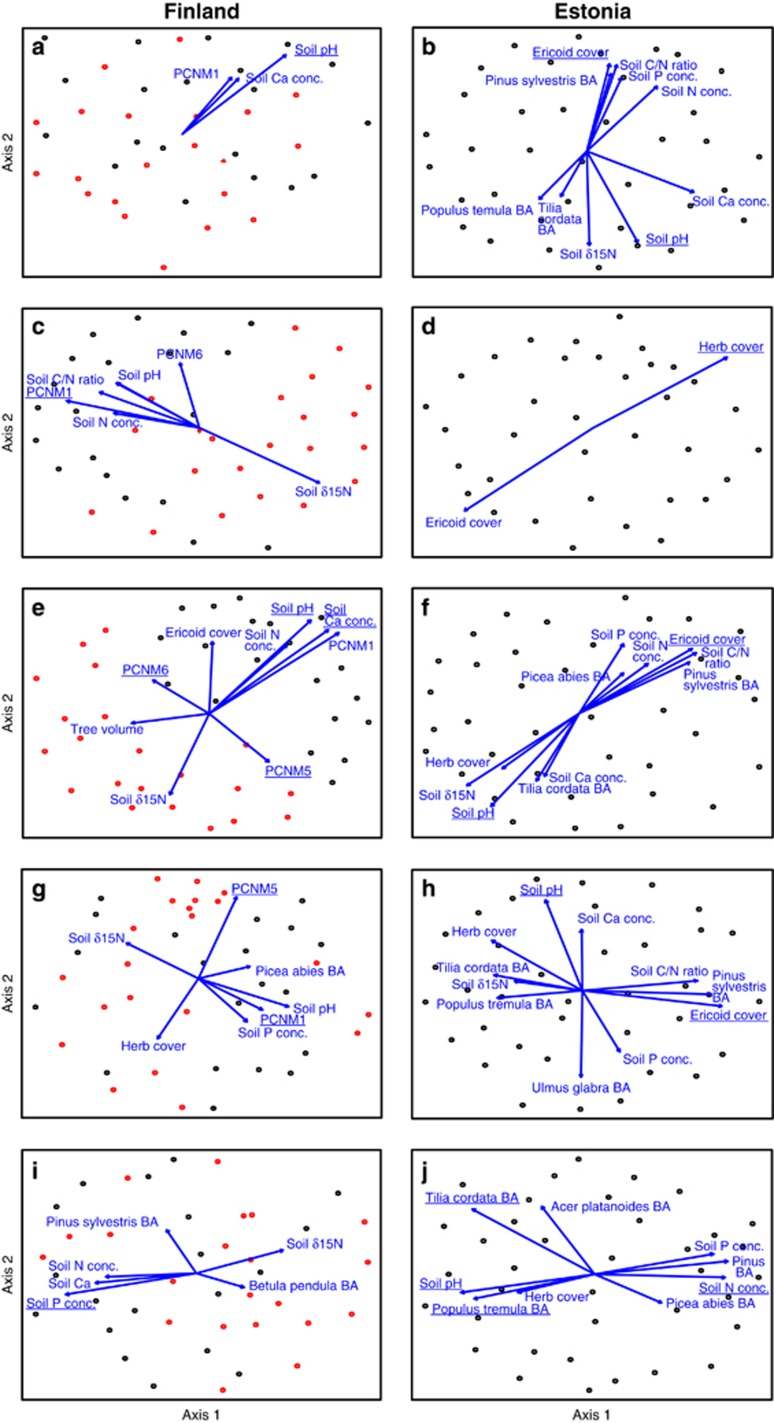
Communities of soil biota were generally driven by spatial vectors and soil variables in Finland and Estonia, respectively (Supplementary Table S5; Figures 5 and 6). According to the best multivariate model, the total fungal community was driven by spatial variation in Finland (F1,37=8.09; R2adj=0.151; P<0.001) but by soil C/N ratio (F1,33=7.08; R2adj=0.144; P<0.001) and Ca concentration (F1,33=3.90; R2adj,partial=0.086; P=0.001) in Estonia. The community composition of EcM fungi was mostly affected by spatial structure (F1,36=4.31; R2adj=0.076; P=0.001) and BA of B. pendula (F1,36=4.37; R2adj,partial=0.073; P=0.001) in Finland. In Estonia, the EcM fungal community was mostly affected by the cover of Ericaceae (F1,33=5.44; R2adj,partial=0.110; P<0.001). The community structure of plant pathogens was driven by spatial variation (F1,38=4.10; R2adj,partial=0.072; P=0.001) in Finland. Pathogen community was most strongly influenced by soil N (F1,34=5.37; R2adj=0.108; P<0.001) and Ca (F1,34=3.87; R2adj,partial=0.068; P=0.001) concentration in Estonia. Finnish saprotroph communities were primarily affected by spatial distance (F1,38=11.47; R2adj=0.207; P<0.001), whereas Estonian saprotroph communites were influenced by soil C/N ratio (F1,33=8.96; R2adj=0.181; P<0.001), N concentration (F1,33=4.20; R2adj,partial=0.069; P=0.001) and pH (F1,33=4.12; R2adj,partial=0.063; P=0.001).
Figure 5.
Non-metric Multidimensional Scaling graphs of communities of soil fungal functional groups: (a, b) EcM fungi; (c, d) plant pathogens; and (e, f) saprotrophs. Left panes, Finland (different shades depict sites); right panes, Estonia. Only those variables with significant fit (P<0.05) are indicated. Variables included in the best community models are underlined.
Figure 6.
Non-metric Multidimensional Scaling graphs of communities of soil protists and animals: (a, b) Cercozoa; (c, d) Chlorophyta; (e, f) Ciliophora; (g, h) Collembola; (i, j) Nematoda. Left panes, Finland (different shades depict sites); right panes, Estonia. Only those variables with significant fit (P<0.05) are indicated. Variables included in the best community models are underlined.
In protists, community composition of Cercozoa was strongly influenced by soil pH both in Finland (F1,39=3.33; R2adj=0.055; P=0.001) and in Estonia (F1,34=4.88; R2adj=0.097; P=0.001), whereas the community structure of Chlorophyta was mainly affected by space in Finland (F1,39=4.01; R2adj=0.071; P=0.001) and by herb cover in Estonia (F1,35=2.71; R2adj=0.076; P=0.001). Community structure of Ciliophora responded most strongly to soil Ca concentration (F1,36=7.83; R2adj=0.146; P=0.001) in Finland but to Ericaceae cover in Estonia (F1,34=5.05; R2adj,partial=0.101; P<0.001).
Of soil animals, the community of Collembola was affected by spatial structure in Finland (F2,38=5.53; R2adj,cumul=0.083; P=0.001) but by soil pH in Estonia (F1,34=3.27; R2adj,partial=0.059; P=0.001). Nematode communities were weakly affected by soil P concentration in Finland (F1,39=2.66; R2adj=0.040; P=0.005) but by soil N concentration in Estonia (F1,32=4.78; R2adj=0.095; P=0.001).
Procrustes analysis revealed that communities of nearly all soil organisms and those of trees and herbs significantly correspond to each other in both study systems (Supplementary Table S6). Further analysis revealed that RProcrustes was strongly linearly related to the logarithm of the size of the community matrix (Finland: t=10.6; R2adj=0.634; P<0.001; Estonia: t=7.9; R2adj=0.446; P<0.001), indicating that the Procrustes statistic may have inherent biases related to sampling depth and richness. Partial Procrustes tests revealed that strong and highly significant correlations between communities were lost after accounting for environmental and spatial predictors (Supplementary Table S6). Consistent with our past hypothesis, these results suggest that the same spatial or environmental variables drive the community composition of different organisms convergently with no causal relationships among these groups.
Discussion
Taxonomic richness and biomass
Diversity and identity of trees exhibited context-dependent effects on taxonomic richness of soil biota, depending on the study system and taxonomic group, which only partly supports our first hypothesis. Nonetheless, tree diversity was an important driver of the richness of EcM fungi in Finland and saprotrophic fungi in Estonia, suggesting that richness of both mutualistic and free-living organisms may benefit from greater producer diversity in certain conditions. Consistent with previous research on understorey richness (Ampoorter et al., 2014) and ecosystem services (Nadrowski et al., 2010; Gamfeldt et al., 2013), neutral effects of tree diversity prevailed in plant–soil biota relationships in both study systems. The low tree diversity impact contrasts with implications from grasslands, in which richness effects on functioning increase with ecosystem's age (Hooper et al., 2012; Reich et al., 2012).
The effects of individual tree species were usually stronger than diversity effects on richness and biomass of soil biota, corroborating the relatively strong sampling effects on ecosystem services (Cardinale et al., 2006; Nadrowski et al., 2010; Gamfeldt et al., 2013). Our results indicate that the magnitude and directionality of individual species effects are system specific. For example, the increasing proportion of P. sylvestris strongly suppressed richness of all fungi and in particular that of saprotrophs in Estonia, but it had a slight but significant positive effect on these groups in Finland. The richness of Cercozoa and Chlorophyta responded to different tree species in the two study systems. By contrast, the relative abundance of A. glutinosa consistently suppressed EcM fungal richness in both study systems that corroborates with the low mycobiont range in Alnus spp. worldwide (Põlme et al., 2013).
Taken together, negative and positive sampling effects were nearly equally represented, suggesting that both stimulating and suppressive species effects are common in tree–soil biota richness relationships. The positive effects are probably related to the abundance of a particularly suitable substrate or facilitation, whereas the negative effects may stem from low palatability, poor compatibility with mutualistic partners or strong defence mechanisms (such as allelochemicals) against soil biota. Variation partitioning further revealed that tree diversity and sampling effects did not differ between biotrophic (pathogens, mutualists) and free-living organisms, providing no support to our second hypothesis.
Both SEM and model selection revealed that soil pH and nutrient concentration were generally the strongest direct or indirect predictors for richness of soil biota in spite of great differences in vegetation. Consistent with the third hypothesis, soil N and P concentrations determined the biomass of bacteria and fungi, especially that of saprotrophs; biomass was the strongest direct predictor of saprotrophic fungal richness in Finland and EcM fungal richness in Estonia. The positive biomass effects are in agreement with studies in grasslands, in which the abundance of individuals or their biomass determines taxonomic richness of particular groups of meiofauna both aboveground and belowground (Scherber et al., 2010; Borer et al., 2012). Soil nutrient concentration or lower C/N ratio had a strong positive effect on richness of Ciliophora in Finland and that of plant pathogenic fungi, Cercozoa amoebae, Ciliophora and Nematoda in Estonia. Apart from nutrients, richness of EcM fungi and Cercozoa responded positively to increasing soil pH in Finland, which is consistent with the substantial pH effect on phylogenetic composition of soil microbes (Rousk et al., 2010).
Community composition
Plant biodiversity experiments carried out so far have seldom addressed community composition of the responding biota. Understanding whether species composition of organisms in one trophic level affects the community structure of organisms in a linked trophic level enables ecologists to further shed light into biological processes shaping the communities of interacting organisms and into the stability of the interaction networks (Nuismer et al., 2013). In our study systems, community composition of most groups of soil biota were related to edaphic variables, especially soil pH and Mg concentration in Finland but to soil pH, C/N ratio, Ca and N concentration in Estonia. Individual tree species had a minor effect at the community level, except that of T. cordata on EcM fungi and Nematoda, and ericoid plant cover on EcM fungi, Cercozoa, Ciliophora and Collembola in Estonia. Such sampling effects were fewer in Finland, but it could be due to the absence of T. cordata and paucity of Ericaceae in the Finnish plots. These two taxa transform the soil environment into extremities in terms of pH and C/N ratio (Read et al., 2004; Frouz et al., 2013).
Our results indicate that different environmental variables drive biomass, richness and community composition of soil organisms. Only site effect and soil pH were among the statistically significant shared determinants of both richness and community composition for groups of soil biota in Finland, whereas soil pH and soil Ca concentration sometimes determined both richness and community composition in Estonia. These results suggest that soil pH is universally related to both environmental filtering and niche differentiation that underlie richness and community development, respectively (Pärtel, 2002; Lauber et al., 2009; Tedersoo et al., 2014b).
Methodological advances
This is the first study to address organisms from multiple eukaryotic kingdoms simultaneously using a molecular marker with species-level resolution. The mixture of 11 forward primers designed to perfectly match Fungi, Viridiplantae, Ciliophora, Cercozoa, Straminipila and selected groups of Metazoa enabled us to recover the identity of all these target organisms in a metabarcoding analysis of a single PCR template. Our approach allows addition of further primer variants to capture additional taxonomic groups. Alternatively, different groups can be targeted in separate PCR reactions or using different markers followed by estimation of biomass or individuals by subsampling or quantitative PCR (Fierer et al., 2005; de Barba et al., 2014; Lentendu et al., 2014). Multiple pairs of MID-tagged primers and preparing several metabarcoding libraries per sample dramatically enhance requirements for time and analytical costs. Our approach of simultaneous identification of multiple organism groups from soil offers a relatively cheap and powerful alternative. The drawback of this method is that it renders cost-effective MID-tagging only the more conserved primer, but updated technologies exist for adding unique identifiers to templates before amplification (Lunderg et al., 2013). With further increase in sequence length (>700 bp), the full ITS region could be targeted as there are more universal eukaryote primer sites in the end of SSU (Tedersoo et al., 2015a). Adding further degenerate positions to the ITS4ngs primer (5′-TCCTSSGCTTANTDATATGC-3′) would render it universal to nearly all eukaryotes.
Previous studies have used ribosomal DNA SSU genes to target taxonomic composition of various protist kingdoms in soil and water (Moon-van der Staay et al., 2001; Bates et al., 2013). However, SSU offers poor species-level resolution in most eukaryote groups (Pawlowski et al., 2012; Schoch et al., 2012; Tang et al., 2012), and the ‘universal' SSU primers exhibit several mismatches to many large and important eukaryote kingdoms (Pawlowski et al., 2012; Tedersoo et al., 2015a, 2015b). Although SSU and cytochrome I oxidase have additional problems with multiple introns in several taxonomic groups, there is also considerable variation in the length of ITS sequences (for example, Acari, many Insecta; Wang et al., 2015). These biological phenomena cause exclusion of a small fraction of taxa from any metabarcoding data sets using a single marker (de Barba et al., 2014).
Ecologists may argue that our strategy of metabarcoding composite soil samples is better suited to detect immobile organisms. Other studies of soil animals have typically relied on identification of organisms obtained from specifically extracted pools of individuals (Porazinska et al., 2009; Hajibabaei et al., 2011). That approach is more labour intensive and enables to address only specific group(s) of soil biota, usually resulting in capturing the most mobile subset of the target species that respond to light or bait. Nonetheless, inclusion of only the active community members rather than eggs, dormant stages and pieces of cuticle may pose an advantage of that approach in certain cases. Alternatively, inactive community members can be discriminated against by targeting RNA instead of DNA (Baldrian et al., 2012).
Based on our analysis of DNA and microbial biomass, the amount of cells and ribosomal DNA molecules of soil animals is in minority compared with that of bacteria and fungi, which is in agreement with previous studies targeting SSU and LSU (Baldwin et al., 2013; Ramirez et al., 2014; Tedersoo et al., 2015a). The fungal dominance could be related to the actual differences in the number of cells and/or copies of ribosomal DNA (Medinger et al., 2010; Vetrovsky and Baldrian, 2013). One gram of organic forest soil may comprise >103 m of hyphae that roughly translates into 108 cells (Leake et al., 2004). The same amount of soil harbours thousands of nematodes that altogether comprise 106–107 cells (Wardle, 2002). The estimated cell numbers of protists generally fall into the same orders of magnitude (Adl and Gupta, 2006). Deeper sequencing using the Illumina (San Diego, CA, USA) or forthcoming ultra-high-throughput platforms are likely to provide more accurate richness estimates by exhaustively capturing taxonomic groups that are common but exhibit a relatively low biomass or marker gene content (Smith and Peay, 2014).
Conclusions
Compared with the effects of individual species and soil parameters, tree diversity per se has generally relatively low influence on taxonomic richness of soil biota. Our results outline that biodiversity effects are contex dependent and that experiments and field studies should be replicated to secure representativeness and understand system specificity (Vehviläinen et al., 2008; Bruelheide et al., 2014). Corresponding changes in beta diversity among vegetation and soil biota are largely explained by the convergent effect of environmental predictors, indicating that these variables must be accounted for in addressing community-wise relationships.
Acknowledgments
The data are available as Supplementary Data S1 and S2. Raw sequence data have been deposited in Short Read Archive under accession no. SRP043706. We thank I Jussila, M Mett and V Veldre for assistance during sampling, M Haugas for isotope sample preparation and L Neuenkamp for discussion about Procrustes analysis. This project received support from the Estonian Science Foundation grants 9286, 171PUT, EMP265, RMK and FIBIR.
Author contributions
LT and JK designed the experiments; LT, MB, SP, SA and IH performed sampling; HH, TC, FB and KP did part of the molecular work; LT, MB, KA and SP analysed the data; LT wrote the manuscript with input from other authors.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abarenkov K, Nilsson RH, Larsson K-H, Alexander IJ, Eberhardt U, Erland S et al. (2010. a). The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytol 186: 281–285. [DOI] [PubMed] [Google Scholar]
- Abarenkov K, Tedersoo L, Nilsson RH, Vellak K, Saar I, Veldre V et al. (2010. b). PlutoF – a web based workbench for ecological and taxonomic research with an online implementation for fungal ITS sequences. Evol Bioinform Online 6: 189–196. [Google Scholar]
- Adl SM, Gupta VVSR. (2006). Protists in soil ecology and forest nutrient cycling. Can J For Res 36: 1805–1817. [Google Scholar]
- Ampoorter E, Baeten L, Koricheva J, Vanhellemont M, Verheyen K. (2014). Do diverse overstoreys induce diverse understoreys? Lessons learnt from an experimental -observational platform in Finland. Forest Ecol Manag 318: 206–215. [Google Scholar]
- Anderson MJ. (2005) PERMANOVA: a FORTRAN Computer Program for Permutational Multivariate Analysis of Variance. University of Auckland: Auckland, New Zealand. [Google Scholar]
- Anslan S, Tedersoo L. (2015). Performance of cytochrome c oxidase subunit I (COI), ribosomal DNA Large Subunit (LSU) and Internal Transcribed Spacer 2 (ITS2) in DNA barcoding of Collembola. Eur J Soil Biol 69: 1–7. [Google Scholar]
- Aponte C, García LV, Marañón T. (2013). Tree species effects on nutrient cycling and soil biota: a feedback mechanism favouring species coexistence. Forest Ecol Manag 309: 36–46. [Google Scholar]
- Arif C, Daniels C, Bayer T, Banguera-Hinestroza E, Barbrook A, Howe CJ et al. (2014). Assessing Symbiodinium diversity in scleractinian corals via next-generation-based genotyping of the ITS2 rDNA region. Mol Ecol 23: 4418–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres E, Steltzer H, Berg S, Wallenstein MD, Simmons BL, Wall DH. (2009). Tree species traits influence soil physical, chemical, and biological properties in high elevation forests. PLoS One 4: e5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachy C, Dolan JR, Lopez-Garcia P, Deschamps P, Moreira D. (2013). Accuracy of protist diversity assessments: morphology compared with cloning and direct pyrosequencing of 18 S rRNA genes and ITS regions using the conspicuous tintinnid ciliates as a case study. ISME J 7: 244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi R, Gallery RE, Gripenberg S, Gurr SJ, Narayan L, Addis CE et al. (2014). Pathogens and insect herbivores drive rainforest plant diversity and composition. Nature 506: 85–88. [DOI] [PubMed] [Google Scholar]
- Bahram M, Kõljalg U, Courty P-E, Diedhiou A, Kjøller R, Põlme S et al. (2013). The distance-decay of similarity in communities of ectomycorrhizal fungi in different ecosystems and scales. J Ecol 101: 1335–1344. [Google Scholar]
- Bahram M, Põlme S, Kõljalg U, Zarre S, Tedersoo L. (2012). Regional and local patterns of ectomycorrhizal fungal diversity and community structure along an altitudinal gradient in the Hyrcanian forests of northern Iran. New Phytol 193: 465–473. [DOI] [PubMed] [Google Scholar]
- Baldrian P, Kolarik M, Stursova M, Kopecky J, Valaskova V, Vetrovsky T et al. (2012). Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J 6: 248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin DS, Colloff MJ, Rees GN, Chariton AA, Watson GO, Court LN et al. (2013). Impacts of inundation and drought on eukaryote biodiversity in semi-arid floodplain soils. Mol Ecol 22: 1746–1758. [DOI] [PubMed] [Google Scholar]
- Bates ST, Clemente JC, Flores GE, Walters WA, Wegener Parfrey L et al. (2013). Global biogeography of highly diverse protistan communities in soil. ISME J 7: 652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter JW, Dighton J. (2005). Phosphorus source alters host plant response to ectomycorrhizal diversity. Mycorrhiza 15: 513–523. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A et al. (2013). Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Meth Ecol Evol 4: 914–919. [Google Scholar]
- Benjamini Y, Hochberg Y. (2000). On the adaptive control of the false discovery rate in multiple testing with independent statistics. J Educ Behav Stat 25: 60–83. [Google Scholar]
- Bik HM, Porazinska D, Creer S, Caporaso JG, Knight R, Thomas WK. (2012). Sequencing our way towards understanding global eukaryotic biodiversity. Trends Ecol Evol 27: 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. (1959). A rapid method of total lipid extraction and purification. Can J Biochem Phys 37: 911–917. [DOI] [PubMed] [Google Scholar]
- Borer ET, Seabloom EW, Tilman D. (2012). Plant diversity contrls arthropod biomass and temporal stability. Ecol Lett 15: 1457–1464. [DOI] [PubMed] [Google Scholar]
- Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. (2012). Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat Methods 9: 425–426. [DOI] [PubMed] [Google Scholar]
- Bruelheide H, Nadrowski K, Assmann T, Bauhus J, Both S, Buscot F et al. (2014). Designing forest biodiversity experiments: general considerations. illustrated by a new large experiment in subtropical China. Methods Ecol Evol 5: 74–89. [Google Scholar]
- Cardinale BJ, Matulich KL, Hooper DU, Byrnes JE, Duffy E, Gamfeldt L et al. (2011). The functional role of producer diversity in ecosystems. Am J Bot 98: 572–592. [DOI] [PubMed] [Google Scholar]
- Cardinale BJ, Srivastava DS, Duffy JE, Wright JP, Downing AL, Sankaran M et al. (2006). Effects of biodiversity on the functioning of trophic groups and ecosystems. Nature 443: 989–992. [DOI] [PubMed] [Google Scholar]
- Chariton AA, Court LN, Hartley DM, Coloff MJ, Hardy CM. (2010). Ecological assessment of estuarine sediments by pyrosequencing eukaryotic ribosomal DNA. Front Ecol 8: 233–238. [Google Scholar]
- Clemmensen KE, Bahr A, Ovaskainen O, Dahlberg A, Ekblad A, Wallander H et al. (2013). Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science 339: 1615–1618. [DOI] [PubMed] [Google Scholar]
- Coleman AW. (2009). Is there a molecular key to the level of 'biological species' in eukaryotes? A DNA guide. Mol Phyl Evol 50: 197–203. [DOI] [PubMed] [Google Scholar]
- Creer S, Fonseca VG, Porazinska DL, Giblin-Davis RM, Sung W, Power DM et al. (2010). Ultrasequencing of the meiofaunal biosphere: practice, pitfalls and promises. Mol Ecol 19: S4–S20. [DOI] [PubMed] [Google Scholar]
- Deagle BE, Jarman SN, Coissac E, Pompanon F, Taberlet P. (2014). DNA metabarcoding and the cytochrome c oxidase subunit I marker: not a perfect match. Biol Lett 10: 20140562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Barba M, Boyer MF, Rioux MD, Coissac E, Taberlet P. (2014). DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: application to omnivorous diet. Mol Ecol Res 14: 306–323. [DOI] [PubMed] [Google Scholar]
- de Wit P, Erseus C. (2010). Genetic variation and phylogeny of Scandinavian species of Grania (Annelida, Clitellata, Enchytraediae), with discovery of a cryptic species. J Zool Syst Evol Res 48: 285–293. [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhauer N, Dobies T, Cesarz S, Hobbie SE, Worm K, Reich PB et al. (2013). Plant diversity effects on soil food webs are stronger than those of elevated CO2 and N deposition in a long-term grassland experiment. Proc Natl Acad Sci USA 17: 6889–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson JA, Vilgalys R, Jackson RB. (2005). Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol 71: 4117–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frouz J, Liveckova M, Albrechtova J, Chronakova A, Cajthaml T, Pizl V et al. (2013). Is the effect of trees on soil properties mediated by soil fauna? A case study from post-mining sites. For Ecol Manage 309: 87–95. [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 23: 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamfeldt L, Snäll T, Bagchi R, Jonsson M, Gustafsson L, Kjellander P et al. (2013). Higher levels of multiple ecosystem services are found in forests with more tree species. Nat Commun 4: 1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajibabaei M, Shokralla S, Zhou X, Singer GC, Baird DJ. (2011). Environmental barcoding: a next-generation sequencing approach for biomonitoring applications using river benthos. PLoS One 6: e17497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy C, Gotelli NJ, Potvin C. (2008). Partitioning the effects of biodiversity and environmental heterogeneity for productivity and mortality in a tropical tree plantation. J Ecol 96: 903–913. [Google Scholar]
- Hiiesalu I, Pärtel M, Davison J, Gerhold P, Metsis M, Moora M et al. (2014). Species richness of arbuscular mycorrhizal fungi: associations with grassland plant richness and biomass. New Phytol 203: 233–234. [DOI] [PubMed] [Google Scholar]
- Hooper DU, Adair EC, Cardinale BJ, Byrnes JEK, Hungate BA, Matulich KA et al. (2012). A global synthesis reveals biodiversity loss as a major driver of ecosystem change. Nature 486: 105–108. [DOI] [PubMed] [Google Scholar]
- Huston MA. (1997). Hidden treatments in ecological experiments: re-evaluating the ecosystem function of biodiversity. Oecologia 110: 449–460. [DOI] [PubMed] [Google Scholar]
- Jumpponen A, Jones KL. (2009). Massively parallel 454 sequencing indicates hyperdiverse fungal communities in temperate Quercus macrocarpa phyllosphere. New Phytol 184: 438–448. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koetschan C, Förster F, Keller A, Schleicher T, Ruderisch B, Schwarz R et al. (2010). The ITS2 Database III - sequences and structures for phylogeny. Nucleic Acid Res 38: D275–D279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M et al. (2013). Towards a unified paradigm for sequence-based identification of Fungi. Mol Ecol 22: 5271–5277. [DOI] [PubMed] [Google Scholar]
- Lauber C, Hamady M, Knight R, Fierer N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75: 5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake J, Johnson D, Donnelly D, Muckle G, Boddy L, Read DJ. (2004). Networks of power and influence: the role of mycorrhizal mycelium in controlling plant communities and agroecosystem functioning. Can J Bot 82: 1016–1045. [Google Scholar]
- Legendre P. (2008). Studying beta diversity: ecological variation partitioning by multiple regression and canonical analysis. J Plant Ecol 1: 3–8. [Google Scholar]
- Lentendu G, Wubet T, Chatzinotas A, Wilhelm C, Buscot F, Schlegel M. (2014). Effects of long-term differential fertilization on eukaryotic microbial communities in an arable soil: a multiple barcoding approach. Mol Ecol 23: 3341–3355. [DOI] [PubMed] [Google Scholar]
- Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R et al. (2013). Fungal community analysis by high-throughput sequencing of amplified markers – a user's guide. New Phytol 199: 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisboa FJG, Peres-Neto PR, Chaer GM, EdC Jesus, Mitchell RJ et al. (2014). Much beyond Mantel: bringing procrustes association metric to the plant and soil ecologist's toolbox. PLoS One 9: e101238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litaker RW, Vandersea MW, Kibler SR, Reece KS, Stokes NA, Lutzoni FM et al. (2007). Recognizing dinoflagellate species using ITS rDNA sequences. J Phycol 43: 344–355. [Google Scholar]
- Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. (2013). Practical innovations for high-throughput amplicon sequencing. Nat Methods 10: 999–1002. [DOI] [PubMed] [Google Scholar]
- Magurran AE. (1988) Ecological Diversity and its Measurement. Princeton University Press: Princeton, NJ, USA. [Google Scholar]
- Maherali H, Klironomos JN. (2007). Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316: 1746–1748. [DOI] [PubMed] [Google Scholar]
- Medinger R, Nolte V, Pandey RM, Jost S, Ottenwälder B, Schlötterer C et al. (2010). Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol Ecol 19: 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon-van der Staay SY, De Vachter R, Vaulot D. (2001). Oceanic 18 S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409: 607–610. [DOI] [PubMed] [Google Scholar]
- Nadrowski K, Wirth C, Scherer-Lorenzen M. (2010). Is forest diversity driving ecosystem function and service? Curr Opin Environ Sustain 2: 75–79. [Google Scholar]
- Noge K, Mori N, Tanaka C, Nishida R, Tsuda M, Kuwahara Y. (2005). Identification of astigmatid mites using the second internal transcribed spacer (ITS2) region and its application for phylogenetic study. Exp Appl Acarol 35: 29–46. [DOI] [PubMed] [Google Scholar]
- Nuismer SL, Jordano P, Bascompte J. (2013). Coevolution and the architecture of mutualistic networks. Evolution 67: 338–354. [DOI] [PubMed] [Google Scholar]
- Pärtel M. (2002). Local plant diversity patterns and evolutionary history at the regional scale. Ecology 83: 2361–2366. [Google Scholar]
- Pawlowski J, Audic S, Adl S. (2012). CBOL Protist Working Group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol 10: e1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peay KG, Baraloto C, Fine PVA. (2013). Strong coupling of plant and fungal community structure across western Amazonian rainforests. ISME J 7: 1852–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellissier L, Niculita-Hirzel H, Dubuis A, Pagni M, Guex N, Ndiribe C et al. (2014). Soil fungal communities in grasslands are environmentally structured at a regional scale in the Alps. Mol Ecol 23: 4274–4290. [DOI] [PubMed] [Google Scholar]
- Põlme S, Bahram M, Kõljalg U, Tedersoo L. (2014). Global biogeography of Alnus-associated Frankia Actinobacteria. New Phytol 204: 979–988. [DOI] [PubMed] [Google Scholar]
- Põlme S, Bahram M, Yamanaka T, Nara K, Dai YC, Grebenc T et al. (2013). Biogeography of ectomycorrhizal fungi associated with alders (Alnus spp.) in relation to biotic and abiotic variables at the global scale. New Phytol 198: 1239–1249. [DOI] [PubMed] [Google Scholar]
- Porazinska DL, Bardgett RD, Blaauw MB, Hunt HW, Parsons AN, Seastedt TR et al. (2003). Relationships at the aboveground-belowground interface: plants, soil biota and soil processes. Ecol Monogr 73: 377–395. [Google Scholar]
- Porazinska DL, Giblin-Davis RM, Faller L, Farmerie W, Kanzaki N, Morris K et al. (2009). Evaluating high-throughput sequencing as a method for metagenomic analysis of nematode diversity. Mol Ecol Res 9: 1439–1450. [DOI] [PubMed] [Google Scholar]
- Prescott CE, Grayston SJ. (2013). Tree species influence on microbial communities in litter and soil: current knowledge and research needs. For Ecol Manage 309: 19–27. [Google Scholar]
- R Core Development Team. (2014). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. Available from http://www.R-project.org/.
- Ramirez KS, Leff JW, Barberan A, Bates ST, Betley J, Crowther TW et al. (2014). Biogeographic patterns in below-ground diversity in New York City's Central Park are similar to those observed globally. Proc R Soc B 281: 20141988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read D, Leake JR, Perez-Moreno J. (2004). Mycorrhizal fungi as drivers of ecosystem processes in heathland and boreal forest biomes. Can J Bot 82: 1243–1263. [Google Scholar]
- Reich PB, Tilman D, Isbell F, Mueller K, Hobbie SE, Flynn DFB et al. (2012). Impacts of biodiversity loss escalate through time as redundancy fades. Science 336: 589–592. [DOI] [PubMed] [Google Scholar]
- Rousk J, Baath E, Brokes PC, Lauber CL, Lozupone C, Caporaso JG et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4: 1340–1351. [DOI] [PubMed] [Google Scholar]
- Scherber C, Eisenhauer N, Weisser WW, Schmid B, Voigt W, Fischer M et al. (2010). Bottom-up levels of plant diversity on multitrophic interactions in a biodiversity experiment. Nature 468: 553–556. [DOI] [PubMed] [Google Scholar]
- Scherer-Lorenzen M, Potvin C, Koricheva J, Schmid B, Hector A, Bornik Z et al. (2006). The design of experimental tree plantations for functional biodiversity research. Ecol Stud 176: 348–375. [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna-Chavez H, Fierer N, van Bodegom PM. (2013). Global drivers and patterns of microbial abundance in soil. Glob Ecol Biogeogr 10: 1162–1172. [Google Scholar]
- Smith DP, Peay KG. (2014). Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS One 9: e90234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šnajdr J, Cajthaml T, Valášková V, Merhautová V, Petranková M, Spetz P et al. (2011). Transformation of Quercus petraea litter: successive changes in litter chemistry are reflected in differential enzyme activity and changes in the microbial community composition. FEMS Microbiol Ecol 75: 291–303. [DOI] [PubMed] [Google Scholar]
- Šnajdr J, Valášková V, Merhautová V, Herinková J, Cajthaml T, Baldrian P. (2008). Spatial variability of enzyme activities and microbial biomass in the upper layers of Quercus petraea forest soil. Soil Biol Biochem 40: 2068–2075. [Google Scholar]
- Soininen EM, Zinger L, Gielly L, Bellemain E, Bråthen KA, Brochmann C et al. (2013). Shedding new light on the diet of Norwegian lemmings: DNA metabarcoding of stomach content. Polar Biol 36: 1069–1076. [Google Scholar]
- Soininen J. (2014). A quantitative analysis of species sorting across organisms and ecosystems. Ecology 95: 3284–3292. [Google Scholar]
- Steudel B, Hector A, Friedl T, Löfke C, Lorenz M, Wesche M et al. (2012). Biodiversity effects on ecosystem functioning change along environmental stress gradients. Ecol Lett 15: 1397–1405. [DOI] [PubMed] [Google Scholar]
- Štursova M, Šnajdr J, Cajthaml T, Barta J, Šantruckova H, Baldrian P. (2014). When the forest dies: the response of forest soil fungi to a bark beetle-induced tree dieback. ISME J 8: 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szava-Kovats RC, Zobel M, Pärtel M. (2011). The local-regional species richness relationship: new perspectives on the null-hypothesis. Oikos 121: 321–326. [Google Scholar]
- Tang CQ, Leasi F, Obertegger U, Kieneke A, Cbarraclough TG, Fontaneto D. (2012). The widely used small subunit 18 S rDNA molecule greatly underestimates true diversity in biodiversity surveys of the meiofauna. Proc Natl Acad Sci USA 109: 16208–16212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo L, Anslan S, Bahram M, Põlme S, Riit T, Liiv I et al. (2015. a). Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 10: 1–43. [Google Scholar]
- Tedersoo L, Bahram M, Dickie IA. (2014. a). Does host plant richness explain diversity of ectomycorrhizal fungi? Re-evaluation of Gao et al. (2013) data sets reveals sampling effects. Mol Ecol 23: 992–995. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Põlme S, Anslan S, Riit T, Kõljalg U et al. (2015. b). Analytical biases in microbial diversity studies. Science (in press). [DOI] [PubMed]
- Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R et al. (2014. b). Global diversity and geography of soil fungi. Science 346: 1078. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Ryberg M, Otsing E, Kõljalg U, Abarenkov K. (2014. c). Global biogeography of the ectomycorrhizal /sebacina lineage (Fungi, Sebacinales) as revealed from comparative phylogenetics analyses. Mol Ecol 23: 4168–4183. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Naadel T, Bahram M, Pritsch K, Buegger F, Leal M et al. (2012). Enzymatic activities and stable isotope patterns of ectomycorrhizal fungi in relation to phylogeny and exploration types in an afrotropical rain forest. New Phytol 195: 832–843. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Nilsson RH, Abarenkov K, Jairus T, Sadam A, Saar I et al. (2010). 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol 188: 291–301. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Smith ME. (2013). Lineages of ectomycorrhizal fungi revisited: foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biol Rev 27: 83–99. [Google Scholar]
- van der Heijden EW, Kuyper TW. (2003). Ecological strategies of ectomycorrhizal fungi of Salix repens: root manipulation versus root replacement. Oikos 103: 668–680. [Google Scholar]
- van der Heijden MGA, Streitwolf-Engel R, Riedl R, Siegrist S, Neudecker A, Ineichen K et al. (2006). The mycorrhizal contribution to plant productivity, plant nutrition and soil structure in experimental grassland. New Phytol 172: 739–752. [DOI] [PubMed] [Google Scholar]
- van der Heijden MGA, Wiemken A, Sanders IR. (2003). Different arbuscular mycorrhizal fungi alter coexistence and resource distribution between co-occurring plant. New Phytol 157: 569–578. [DOI] [PubMed] [Google Scholar]
- Vehviläinen H, Koricheva J, Ruohomäki K. (2008). Effects of stand tree species composition and diversity on abundance of predaory arthropods. Oikos 117: 935–943. [Google Scholar]
- Vetrovsky T, Baldrian P. (2013). The variability of the 16 S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8: e57923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagg C, Bender SF, Widmer F, van der Heijden MGA. (2014). Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc Natl Acad Sci USA 111: 5266–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagg C, Jansa J, Schmid B, van der Heijden MGA. (2011). Belowground biodiversity effects of plant symbionts support aboveground productivity. Ecol Lett 14: 1001–1009. [DOI] [PubMed] [Google Scholar]
- Wang X-C, Liu C, Huang L, Bengtsson-Palme J, Chen H, Zhang J-H et al. (2015). ITS1: a DNA barcode better than ITS2 in eukaryotes? Mol Ecol Res (in press). [DOI] [PubMed]
- Wardle DA, Bardgett RD, Klironomos JN, Setälä H, van der Putten WH, Wall DH. (2004). Ecological linkages between aboveground and belowground biota. Science 304: 1629–1633. [DOI] [PubMed] [Google Scholar]
- Wardle DA. (1999). Is ‘sampling effect' a problem for experiments investigating biodiversity-ecosystem function relationships? Oikos 87: 403–407. [Google Scholar]
- Wardle DA. (2002) Communities and Ecosystems: Linking the Aboveground and Belowground Components. Princeton University Press: New York, NY, USA. [Google Scholar]
- White TJ, Bruns TD, Lee S, Taylor J. (1990). Amplification and direct sequencing of fungal ribosomal RNAgenes for phylogenetics. In: Innis MA, Gelfand DH (eds). PCR Protocols: A Guide to Methods and Applications. Academic Press: London, UK, pp 315–322. [Google Scholar]
- Yao H, Song J, Liu C, Luo K, Han J, Li Y et al. (2010). Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One 5: e13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan A, Bailey SA, Heath DJ, MacIsaac HJ. (2014). Performance comparison of genetic markers for high-throughput sequencing-based biodiversity assessment in complex communities. Mol Ecol Res 14: 1049–1059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.