Abstract
The evolution of eukaryotic organisms is often strongly influenced by microbial symbionts that confer novel traits to their hosts. Here we describe the intracellular Enterobacteriaceae symbiont of the invasive ant Cardiocondyla obscurior, ‘Candidatus Westeberhardia cardiocondylae'. Upon metamorphosis, Westeberhardia is found in gut-associated bacteriomes that deteriorate following eclosion. Only queens maintain Westeberhardia in the ovarian nurse cells from where the symbionts are transmitted to late-stage oocytes during nurse cell depletion. Functional analyses of the streamlined genome of Westeberhardia (533 kb, 23.41% GC content) indicate that neither vitamins nor essential amino acids are provided for the host. However, the genome encodes for an almost complete shikimate pathway leading to 4-hydroxyphenylpyruvate, which could be converted into tyrosine by the host. Taken together with increasing titers of Westeberhardia during pupal stage, this suggests a contribution of Westeberhardia to cuticle formation. Despite a widespread occurrence of Westeberhardia across host populations, one ant lineage was found to be naturally symbiont-free, pointing to the loss of an otherwise prevalent endosymbiont. This study yields insights into a novel intracellular mutualist that could play a role in the invasive success of C. obscurior.
Introduction
Interactions between organisms drive biological complexity (Maynard Smith and Szathmáry, 1997), shaping life as we know it. Symbioses with prokaryotes are considered to promote Eukaryote diversification (Brucker and Bordenstein, 2012), particularly in insects (Moya et al., 2008; Gil et al., 2010). Some bacterial symbionts provide novel ecological traits to their insect hosts, for example, defense against pathogens or parasitoids (Oliver et al., 2003; Kaltenpoth et al., 2005), enhanced stress tolerance (Russell and Moran, 2006) or nutrients (Douglas, 2009). Nutrient-providing symbionts are commonly found in hosts with restricted diets, for example, aphids feeding on phloem sap (Baumann, 2005), blood-feeding diptera (Wang et al., 2013) or grain weevils (Heddi et al., 1999). Symbionts can provide essential amino acids, vitamins or help in nitrogen recycling (Nakabachi et al., 2005; Feldhaar et al., 2007; Michalkova et al., 2014; Patino-Navarrete et al., 2014). Such bacteria are commonly harbored in bacteriocytes, specialized host cells that sometimes form special organ-like structures, the bacteriomes (Baumann, 2005) or are confined to the insect gut (Engel and Moran, 2013). Provisioning with nutrients can lead to increased fitness (Michalkova et al., 2014), which may enable invasive species to exploit novel habitats or food sources (Feldhaar, 2011).
Cardiocondyla obscurior (Wheeler, 1929) is an invasive ant that forms small multiqueen colonies in disturbed, arboreal habitats throughout the tropics. A peculiarity of the genus Cardiocondyla is the occurrence of wingless males that mate with closely related queens in their maternal nest (Oettler et al., 2010). New colonies are established via colony splitting (Heinze et al., 2006). This unique life history with frequent genetic bottlenecks and high levels of inbreeding makes it an interesting model for the study of rapid adaptation to novel environments (Schrader et al., 2014).
Here, we describe a so-far unknown intracellular symbiont of C. obscurior, for which we propose the name 'Candidatus Westeberhardia cardiocondylae' strain obscurior (from here on referred to as Westeberhardia). We analyzed its distribution within and across host populations, and compared infection of individual ants depending on morph and age. Furthermore, we localized Westeberhardia in the host and scrutinized its genome focusing on its metabolic functions. Westeberhardia has lost many metabolic capabilities, but retained most of the shikimate pathway and the ability to synthesize the tyrosine precursor 4-hydroxyphenylpyruvate. We suggest that its localization in gut-associated bacteriomes of pupae, and the increased titers during pupal development point to a role of Westeberhardia in cuticle formation.
Material and methods
Ant colonies
We reared C. obscurior colonies from Brazil (BR), Japan (JP) and Spain (SP) in the laboratory. The BR colonies originated from two collection sites ~70 km apart, a cacao plantation in Ilhéus (2009 and 2013) and a citrus plantation near Una (2012) (Brazilian Ministry of Science and Technology, permits 20324-1/40101-1). JP colonies were collected from two coral trees 100 m apart (lineages ‘OypB', ‘OypC') in the Oonoyama park in Naha, Okinawa (2011) and from additional trees of the same park (‘OypU', 2013). SP colonies were collected at a campsite in Los Realejos, Tenerife in 2012 and 2013. All colonies were housed in plaster nests under 12 h 28 °C light/12 h 23 °C dark cycles, with constant humidity and ad libitum provided honey and pieces of cockroaches. All animal treatment guidelines applicable to ants under international and German law have been followed.
Westeberhardia detection and phylogenomic analyses
During analyses of the C. obscurior genome (Schrader et al., 2014), we identified prokaryotic scaffolds and candidates for horizontal gene transfers (HGTs) (Wheeler et al., 2013). These were then further characterized by blasting (blastx) all annotated genes against a database of prokaryotic proteins. Besides Wolbachia, we identified six scaffolds of an unknown Enterobacteriaceae. Following de novo genome assembly and annotation (see below), we used translated CDS sequences for phylogenetic placement following Husník et al. (2011). Briefly, we performed Dayhoff6 recoding followed by a phylogenomic reconstruction with PhyloBayes v.3.3f (Lartillot et al., 2009), based on 64 single-copy protein clusters (Supplementary Information).
We detected one prokaryotic gene incorporated into the host genome. After manual correction of the HGT gene model, we used blastx analyses against NCBI's non-redundant database to identify homologs. RNAseq data were used to verify expression in seven larval and seven adult queens by mapping reads against the C. obscurior genome (Schrader et al., 2014). We generated count tables with htseq (Anders et al., 2015) against C. obscurior gene annotations (including the manually corrected gene) and calculated untransformed, size factor-normalized read counts.
Genome assembly, annotation and functional analyses
Paired-end Illumina reads from Schrader et al. (2014) were used for de novo assembly of the Westeberhardia genome. We removed Wolbachia sequences based on their blastx result, and then assembled remaining prokaryotic reads using SOAPdenovo2 (Luo et al., 2012). The resulting contigs were scaffolded using a custom-modified version of SSPACE v.2.0 (Boetzer et al., 2011). Raw reads were then mapped back to the contigs using MIRA 4.0.1 (Chevreux et al., 1999) and manually joined. Scaffold corroboration and visual inspection of contigs were performed in the Staden Package (Staden et al., 2000). Inconsistencies were broken and manually reassembled. Base-calling correction was carried out using POLISHER (Lapidus et al., 2008). No corrections were made to the consensus, which consisted of a single 532 684-bp contig (average coverage 204.5x).
The replication origin was predicted using originX (Worning et al., 2006). A first round of open reading frame prediction was performed using Prodigal v.2.5 (Hyatt et al., 2010) and the predicted open reading frames were annotated using the BASys server (Van Domselaar et al., 2005). tRNAs were predicted using the ‘COVE-only' algorithm of tRNAscan-SE v.1.3.1 (Lowe and Eddy, 1997), and checked with TFAM v.1.4 (Tåquist et al., 2007). tmRNAs and their tag peptides were predicted using ARAGORN v.1.2.36 (Laslett and Canback, 2004). The genome was searched against Rfam v.11 (Burge et al., 2013) using Infernal v.1.1 (Nawrocki and Eddy, 2013), and the resulting ncRNAs were manually integrated into the annotation following the INSDC conventions (http://www.insdc.org/files/feature_table.html). Ribosome-binding sites were predicted using RBSfinder (Suzek et al., 2001) and signal peptides were predicted using SignalP v.4.1 (Petersen et al., 2011). The resulting annotation was manually curated in Artemis (Rutherford et al., 2000).
Metabolic functions were automatically predicted and analyzed using Pathway Tools v.17.5 (Karp et al., 2010) against BioCyc and MetaCyc databases (Caspi et al., 2012), following manual curation. Functional information was retrieved from the EcoCyc (Keseler et al., 2013), KEEG (Ogata et al., 1999) and BRENDA (Scheer et al., 2011) databases.
Coverage analyses
For comparison of Westeberhardia infection between the sequenced reference colonies from BR (Ilhéus, 2009) and JP (OypB) (Schrader et al., 2014), we mapped genomic reads from pools of 30 BR and 26 JP males (140 million reads each) against the C. obscurior and Westeberhardia genomes, and compared coverage between BR and JP with samtools' depth algorithm (Li, 2011) and custom bash/perl/R scripts as described in Schrader et al. (2014).
Analyses of intraspecific infection dynamics by PCR and real-time qPCR
To assess Westeberhardia presence across host populations, we screened 42 C. obscurior samples collected worldwide and the sister species Cardiocondyla wroughtonii (Forel, 1890) by performing a diagnostic PCR assay on a 204-bp fragment of the nrdB (ribonucleoside-diphosphate reductase 1 subunit beta) gene of Westeberhardia (WEOB_403) (nrdB-for: 5′-GGAAGGAGTCCTAATGTTGCG-3′ nrdB-rev: 5′-ACCAGAAATATCTTTTGCACGTT-3′), using the ant housekeeping gene elongation factor 1-alpha 1 (Cobs_01649) (EF1-for: 5′-TCACTGGTACCTCGCAAGCCGA-3′ EF1-rev: 5′-AGCGTGCTCACGAGTTTGTCCG-3′, 104-bp fragment) as a control. We used DNA from a previous study (Oettler et al., 2010), samples from laboratory colonies and stored tissues from which we extracted DNA using a chloroform-based method (Sambrook and Russell, 2001) (Table 1). To verify infection with the same Westeberhardia species, we sequenced a 917-bp fragment of the 16S rRNA gene of Westeberhardia (WEOB_122) (WE16S-for: 5′-CATTTGAATATGTAGAATGGACC-3′ WE16S-rev: 5′-AACTTTTACAAGATCGCTTCTC-3′) from one individual each of the BR, JP and SP populations and of C. wroughtonii (see Supplementary Information for PCR details).
Table 1. Prevalence of Westeberhardia across populations of Cardiocondyla obscurior and the closely related species C. wroughtoniia.
| Sampling site (year) | Sample description | Morph | Westeberhardia (sample size) |
|---|---|---|---|
| C. obscurior | |||
| Brazil: Ilhéus (2004) | Laboratory colonies | W | Y (3) |
| Brazil: Ilhéus (2009) | Laboratory colonies | Q | Y (4) |
| Brazil: Una (2012) | Laboratory colonies | Q | Y (4) |
| Brazil: Ilhéus (2013) | Laboratory colonies | Q | Y (4) |
| Japan: Ishigaki (2002) | Oettler et al. (2010) | W | N (2) |
| Japan: Naha (2011) 'OypB' | Laboratory colonies | Q | N (4) |
| Japan: Naha (2011) 'OypC' | Laboratory colonies | Q | Y (4) |
| Japan: Naha (2013) 'OypU' | Laboratory colonies | Q | Y (4) |
| Spain: Tenerife (2012) | Laboratory colonies | Q | Y (4) |
| Egypt: Talkha (2003) | Oettler et al. (2010) | W | N (1) |
| Fiji (2007) | EtOH material | W | Y (1) |
| Malaysia: Ulu Gombak (2002) | Oettler et al. (2010) | W | Y (1) |
| Sri Lanka (2006) | Oettler et al. (2010) | W | N (1) |
| USA: Lake Alfred, Florida (2004) | Oettler et al. (2010) | W | Y (3) |
| C. cf. obscurior Singapore (2014) | EtOH material | W | Y (2) |
| C. wroughtonii | |||
| Japan: Nakijin (2013) | Laboratory colonies | W | Y (2) |
Abbreviations: EtOH, ethyl alcohol; Q, queen; N, no; W, worker; Y, yes.
Based on a diagnostic PCR screen using the nrdB gene.
We assessed inter- and intrapopulational Westeberhardia prevalence in laboratory colonies using PCR and quantitative PCR (qPCR) assays for workers and queens, respectively (Supplementary Information). For this purpose, we sampled 6–10 dealate queens and 9–10 workers from 6–8 colonies from three lineages from BR and JP, respectively, and from the SP population, resulting in a total of 538 analyzed workers and 517 queens. Worker and queen DNA was extracted using the hotshot method (Alasaad et al., 2008) and the NucleoSpinTissue XS Kit (Machery-Nagel, Düren, Germany), respectively. To control for age effects on infection (see below), we selected freshly eclosed workers when possible.
We quantified Westeberhardia of single individuals by determining normalized nrdB copy numbers with qPCR (Supplementary Information) across developmental stages (larvae and prepupae of unknown sex and caste, young and old female pupae) for JP (OypB) and BR (Una, 2012) and across morphs (queens, workers, winged males, wingless males), ages (queens=2, 14, 28 and 48 days; workers=2, 14 and 28 days), and fertilization state of queens (virgin, mated) for only the BR population (Una, 2012).
Fluorescence in situ hybridization
To localize Westeberhardia, we performed fluorescence in situ hybridization (FISH) as described previously (Kaltenpoth et al., 2012, 2014 and Supplementary Information) on abdominal sections of queen, worker and wingless male pupae from BR (Ilhéus, 2009) and adult queens from BR (Ilhéus, 2009) and JP (OypB) with the general eubacterial probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′) (Amann et al., 1990) and one of the Westeberhardia-specific probes Wcard1 (5′-ATCAGTTTCGAACGCCATTC-3′) and Wcard2 (5′-CGGAAGCCACAATTCAAGAT-3′), targeting the 16S rRNA gene. Probes were labeled with Cy3 or Cy5, and samples were counterstained with DAPI (4′,6-diamidino-2-phenylindole).
Test for reproductive manipulation and paternal inheritance
Several bacterial symbionts are known to be reproductive manipulators, with cytoplasmic incompatibility (CI) and parthenogenesis induction (PI) being the most common phenotypes (Cordaux et al., 2011). Although CI results in the incompatibility of crosses between infected males and uninfected queens, PI causes parthenogenetic production of diploid female offspring in infected females. We crossed uninfected JP queens (OypB) with infected BR males (Ilhéus, 2009), by placing sexual pupae together with 20 workers into new nests (n=10), which were observed twice a week for the presence of male and female brood.
To test for paternal inheritance of Westeberhardia, crosses of infected males and uninfected freshly eclosed virgin queens were initiated in a mating arena overnight. The following day, we dissected and macerated the spermathecae of queens (n=8) in dH2O and isolated DNA using the NucleoSpinTissue XS Kit (Machery-Nagel). We performed a diagnostic PCR assay with the nrdB gene and the housekeeping gene EF1 as a positive control. We further analyzed infection status of two worker pupae each emerging from four of the above crosses using the nrdB PCR assay.
Results
Microbial associates of C. obscurior
Blastx analyses of the C. obscurior hologenome (Schrader et al., 2014) retrieved 1.5 Mb of Wolbachia sequence and 543 172 bp in six scaffolds of an unknown γ-proteobacterium. A preliminary assembly of the Wolbachia sequences is hosted on antgenomes.org. The 16S sequence of the γ-proteobacterium showed 98.4% identity with an Enterobacteriaceae of a C. obscurior sample from Florida, USA (voucher RA0330, GenBank: GQ275143), detected during a survey of ant-associated bacteria (Russell et al., 2009).
Blastx analyses further revealed a 360-bp intronless gene of putative prokaryotic origin encoding a xanthine-guanine phosphoribosyltransferase (EC: 2.4.2.22), which is incorporated into the host genome and has its closest ortholog in Enterobacter cloacae (WP_023478997). Xanthine-guanine phosphoribosyltransferase has a central role in the synthesis of purine nucleotides through the salvage pathways, converting xanthine and guanine to XMP and GMP, respectively. The gene is present in genomic reads of C. obscurior from BR (Ilhéus, 2009) and JP (OypB). We used published RNAseq data from adult queens and queen-destined larvae (Schrader et al., 2014) to confirm in vivo transcription of the HGT and found a fivefold increased expression in larvae compared to adults (medianlarvae=1140.1, medianqueens=223.2, Mann–Whitney U-test: W=79, P<0.001).
Westeberhardia genome assembly, annotation and functional and phylogenomic analyses
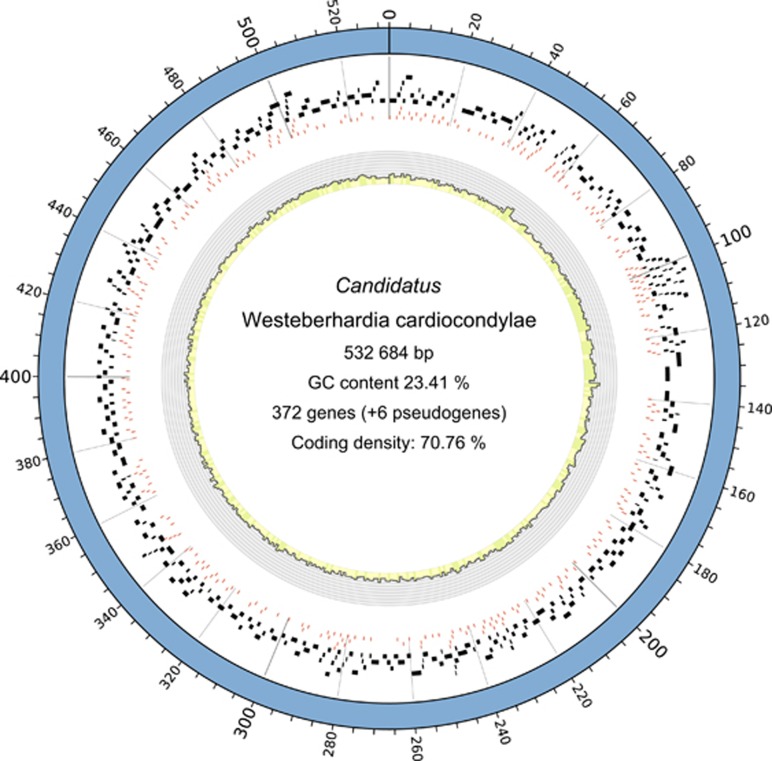
De novo assembly of the Westeberhardia genome produced a single scaffold representing a circular chromosome of 532 684 bp (23.41% GC content) with 372 protein-coding genes and six pseudogenes (Figure 1). Sequences are accessible through the European Nucleotide Archive (http://www.ebi.ac.uk/ena/data/view/LN774881) under study number PRJEB8217, chromosome accession number LN774881 and are hosted on antgenomes.org. The phylogenomic analysis placed Westeberhardia within a group of Enterobacteriaceae symbionts comprising both facultative and obligate insect endosymbionts, including Sodalis, Baumannia, Blochmannia and Wigglesworthia (Supplementary Figure S1).
Figure 1.
Genomic structure of Westeberhardia representational Circos plot (Krzywinski et al., 2009) illustrating genomic properties of Westeberhardia. Tile plots show the distribution of protein-coding genes (black bars) and ribosomal binding sites (red bars). The histogram in the inner circle shows GC content in percent for 1 kb windows.
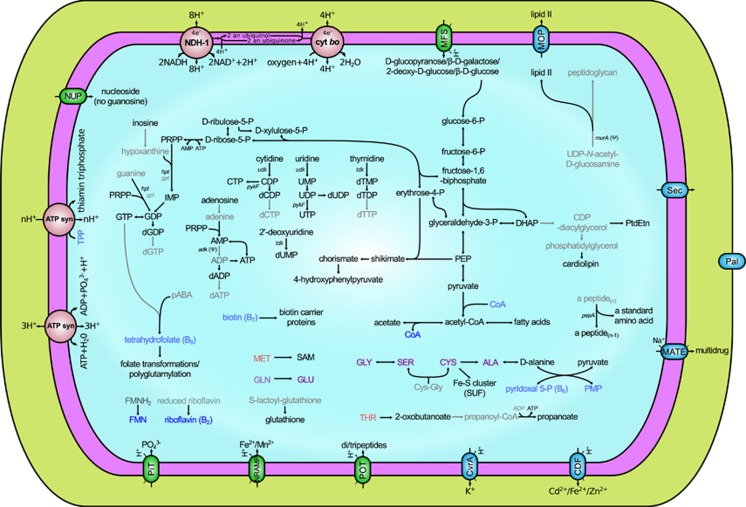
The genome codes for a simplified but functional informational machinery, with complete setups for DNA replication, transcription, translation and protein folding, but few genes involved in DNA repair (Figure 2). Westeberhardia has a limited metabolic repertoire but is capable of glycolysis, pentose phosphate pathway, fatty acid biosynthesis, nucleotide synthesis and ATP production through oxidative phosphorylation. The pathway for glycerophospholipid biosynthesis is impaired. Metabolite transport appears to be based on electrochemical potential-driven transporters and inorganic phosphate transporters, whereas ATP-binding cassette transporter as well as phosphotransferase system transporter genes are missing. Westeberhardia has lost the pathways for synthesis of all essential and most non-essential amino acids and cofactors, but has retained an incomplete shikimate pathway. Thus, it is able to synthesize chorismate, a central metabolite in the biosynthesis of many aromatic compounds (e.g. phenylalanine, tryptophan, p-hydroxybenzoate or enterobactin). However, it can only use chorismate for the biosynthesis of 4-hydroxyphenylpyruvate, a precursor of tyrosine (Hopkins and Kramer, 1992; Andersen, 2012). Although Westeberhardia cannot complete the last step in this pathway, the host genome codes for tyrosine aminotransferase (EC 2.6.1.5), converting 4-hydroxyphenylpyruvate to tyrosine (Cobs_01567). Further conversion of tyrosine to DOPA (3,4-dihyroxyphenylalanine), an important component of insect cuticles (Andersen, 2012), might occur through tyrosine 3-monooxygenase (EC 1.14.16.2) encoded in the host genome (Cobs_14710).
Figure 2.
Westeberhardia metabolic reconstruction. Intact pathways are shown in black lines and unclear pathways (missing a specific gene or having it pseudogenized) are shown in gray lines. Exporters are represented using green ovals, whereas exporters/importers are represented using blue ovals with the name of the family/superfamily they belong to, otherwise the protein or complex name is used. ATP synthase is shown twice to represent an additional metabolic capability. Essential and non-essential amino acids are shown in red and purple lettering, respectively. Cofactors and vitamins are represented in blue. Blurred compounds represent those for which biosynthesis or import cannot be accounted for based on the genomic data, according to MetaCyc. Relevant genes involved in the biosynthesis of nucleotides and peptidoglycan are indicated. A single frameshift is found in adk and murA; therefore, they might be young pseudogenes, or an RNA polymerase or ribosomal slippage would be required to produce a functional protein.
Intraspecific infection dynamics
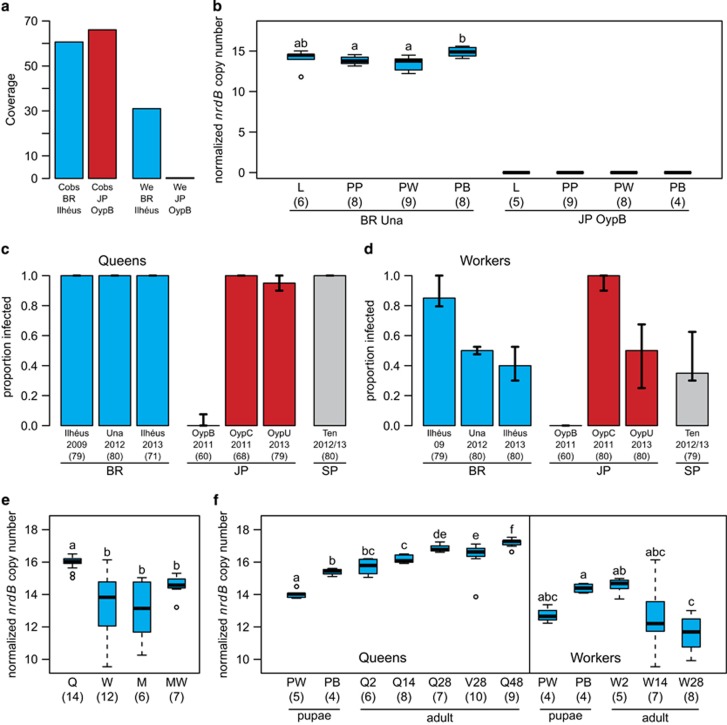
Coverage analysis of genomic reads showed that, in contrast to males from a BR lineage (Ilhéus, 2009), males from a JP lineage (OypB) are devoid of Westeberhardia (coverage BR: 30x; JP: 0.21; Figure 3a). qPCR of the Westeberhardia-specific nrdB gene in the female pupae, as well as in the larvae and prepupae of unknown sex and caste verified that Westeberhardia is completely absent in OypB (Figure 3b). Accordingly, Westeberhardia was not detected by FISH in sections of adult OypB queens.
Figure 3.
Intraspecific and temporal dynamics of Westeberhardia infection. (a) In genomic coverage data for pooled haploid males mapped against the Westeberhardia reference, Westeberhardia reads (We) were exclusively present in the Brazil, Ilhéus (2009) sample (BR, blue) and no reads could be detected in the OypB, Japan (JP, red) sample, whereas coverage of C. obscurior reads (Cobs) mapped against the C. obscurior reference is similar. (b) Real time-quantitative PCRs on DNA level for the nrdB gene confirm the absence of Westeberhardia in larvae (L), prepupae (PP) and female (queen and worker) pupae (PW=pupa white, PB=pupa brown) of the JP OypB population, whereas all those developmental stages are infected in the BR Una (2012) (letters indicate significances for within-population comparisons for BR). (c and d) Prevalence of Westeberhardia in queens (c) and workers (d) across different populations of C. obscurior from BR (blue), JP (red) and Tenerife, SP (gray), as revealed by qPCR (c) and diagnostic PCR (d), of the nrdB gene. For each lineage, 6–8 colonies and per colony 9–10 young workers and 6–10 queens were tested. Bars represent medians and whiskers denote quartiles. Note that while Westeberhardia infection status of workers varies between and within populations of C. obscurior, it is almost fixed in queens across all lineages except OypB. (e and f) Morph (e) and age (f) dependency of relative amounts of Westeberhardia in C. obscurior individuals from BR (Una, 2012) determined by real-time quantitative PCR. Normalized nrdB copy numbers are elevated in queens compared with all other morphs (Q=queens; W=workers; M=males winged; MW=males wingless) (e), increase with age in queens, but decrease with age in workers (numbers after Q/W show age in days, V=virgin queens, letters indicate significant differences for within-caste comparisons) (f). Sample sizes are given within parentheses.
Analyses of C. obscurior samples collected worldwide showed that Westeberhardia is present in 34 of 42 tested samples (81.0%), including all samples from BR, but absent in some JP populations and in material from Egypt and Sri Lanka (Table 1). The closely related species C. wroughtonii also contains Westeberhardia. A 917-bp 16S rDNA fragment of Westeberhardia is identical between the three C. obscurior populations from BR, JP and SP, and between C. obscurior and C. wroughtonii.
Westeberhardia infection of workers varies considerably within and among infected lineages, ranging from 42.5% to 96.3% (Figure 3d), whereas queen infection is almost fixed in all populations (88.6% to 100% Figure 3c and Supplementary Table 5). However, in the OypB lineage, only one of 60 workers and two of 60 queens were infected at low levels (indicated by weak bands on the agarose gel or late Cq values in the qPCR, respectively). These values are not significantly different from zero (one-sample t-tests: workers: t(59)=1, P=0.32; queens: t(59)=1.43, P=0.16), and could possibly be caused by contamination. Intriguingly, individuals from colonies collected in a tree merely 100 m away (OypC) show infection rates of 96.3% (workers) and 100% (queens).
In the BR (Una, 2012) population, relative densities of Westeberhardia increase significantly during pupal development (worker and queen pupae combined) from white (early) to brown (late) pupae (Npupa white=9, Npupa brown=8; t-test: t(14.7)=−4.3, P<0.001), but is not different between larvae, prepupa and early pupae (Figure 3b and Supplementary Table 1). Westeberhardia titers are higher in queen compared with worker pupae (t-test: t(13.3)=2.6, P=0.023). A comparison of 2- to 14-day-old adults of each morph (queens, workers, winged and wingless males) shows that Westeberhardia titers differ significantly across castes (Kruskal–Wallis: X2=24.2, d.f.=3, P<0.001; Figure 3e), with queens having significantly more Westeberhardia compared with the other morphs, which are not different from each other (pairwise Mann–Whitney U-tests with Benjamini–Hochberg correction for multiple testing (Benjamini and Hochberg, 1995); Supplementary Table 2). We calculated generalized linear models (with a Gaussian distribution and identity link function to model age dependency of Westeberhardia in adult females (Figure 3f). In workers, infection decreases with age (generalized linear model: d.f.=18, F=12.7, P=0.002; Supplementary Table 3) and is significantly more variable compared with that in queens (Fligner–Killeen test: X2=17.0, d.f.=1, P<0.001). In queens, Westeberhardia significantly increase with age from day 2 after eclosion to day 48 (generalized linear model: d.f.=38, F=29.4, P<0.001; Supplementary Table 4). Virgin and mated 28-day-old queens show no significant difference in infection (Wilcoxon's rank-sum test: W=49, P>0.05).
Localization of Westeberhardia
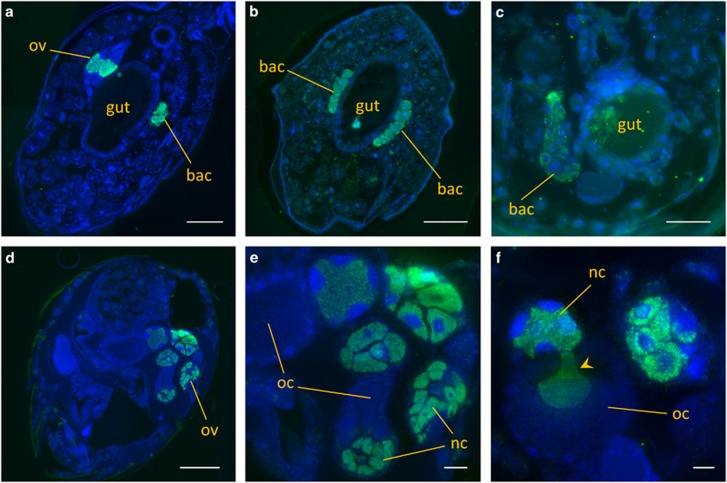
Westeberhardia is localized intracellularly in bacteriomes connected to the gut in queen, worker and wingless male pupae (Figures 4a–c). As in many other insect taxa with bacteriomes, bacteriocytes are densely packed with symbiont cells and exhibit enlarged host cell nuclei. In some sections, individual symbiont cells or symbiont-filled bacteriocytes were found in the gut lumen, suggesting beginning degradation of the bacteriome in later pupal stages. Concordantly, no bacteriomes were detected in adult queens (Figure 4d). However, both pupal and adult queens show high Westeberhardia abundances in the ovaries (Figures 4a, d–f). In particular, Westeberhardia is localized predominantly in the nurse cells. Several sections captured transgenerational infection events of the symbiont from the maternal nurse cells into late stages of the developing oocyte (Figure 4f).
Figure 4.
Localization of Westeberhardia in adults and pupae of C. obscurior (from Brazil, Ilhéus, 2009). Symbionts were stained in longitudinal sections through the abdomen with the Westeberhardia-specific probe Wcard2-Cy5 (green) and host cell nuclei were counterstained with DAPI (blue). (a–c) Localization of Westeberhardia in gut-associated bacteriomes (bac) in the pupae of a queen (a), a worker (b) and a male (c). Note the additional presence of symbionts in the queen ovaries (ov). (d) Section of the abdomen of an adult queen, with symbionts visible in the ovaries (ov). (e and f) Ovaries of an adult queen. Symbionts are mainly localized in the nurse cells (nc), but enter the developing oocyte (oc), probably during nurse cell depletion (arrowhead). Scale bars: 100 μm (a, b and d), 50 μm (c) and 20 μm (e and f).
Test for reproductive manipulation and paternal inheritance
Westeberhardia-free OypB queens mated to Westeberhardia-infected BR males produced viable diploid female and haploid male offspring. Furthermore, infected females generally also produce male offspring and unfertilized females exclusively produce males. In addition, Westeberhardia does not seem to be transmitted paternally, as revealed by the absence of the diagnostic nrdB gene in the spermatheca content of Westeberhardia-uninfected queens mated with infected males. Worker pupae emerging from the above crosses were also uninfected.
Description of ‘Ca. W. cardiocondylae'
In accordance with the guidelines of the International Committee of Systematic Bacteriology, unculturable bacteria should be classified as Candidatus (Murray and Stackebrandt, 1995). We propose the name ‘Ca. Westeberhardia cardiocondylae' strain obscurior for this newly discovered γ3-proteobacterium. The genus name Westeberhardia refers to Mary Jane West-Eberhard, expressing our admiration for her far-reaching advances in evolutionary developmental biology. The specific epithet, cardiocondylae, indicates that it is an endosymbiont of Cardiocondyla ants.
Discussion
The 16S sequence of an unknown Enterobacteriaceae isolated from C. obscurior was previously published (Russell et al., 2009), but the specificity and functionality of this association had not been addressed. Here, we describe it as ‘Ca. Westeberhardia cardiocondylae' strain obscurior and provide a first characterization of its relationship with C. obscurior. Phylogenomic analysis indicates that Westeberhardia is closely related to Blochmannia, the obligate endosymbiont of Camponotus ants (Feldhaar et al., 2007). Nevertheless, its phylogenetic placement has to be considered with caution, because of long-branch attraction. As already observed by Husník et al. (2011), the monophyly of the cluster formed by Sodalis, Baumannia, Blochmannia and Wigglesworthia, in which Westeberhardia appears, needs to be further tested.
Transmission of Westeberhardia
Maternal transmission of Westeberhardia occurs through a different process than described for other endosymbionts (Koga et al., 2012; Balmand et al., 2013). In adult queens, Westeberhardia is localized in ovarial syncytial nurse cells, which originate from the same germline stem cell as the oocyte and are responsible for provisioning of the oocyte with metabolites. During the process of nurse cell depletion, when large amounts of cytoplasmic material are channeled into the oocyte (Mahajan-Miklos and Cooley, 1994), cytoplasmic Westeberhardia are swept into the developing oocyte, ensuring complete vertical transmission (Figure 4f).
CI is a widespread phenotype induced by some bacterial endosymbionts (Gotoh et al., 2006; Werren et al., 2008), but has to our knowledge not been shown for any social insect. In social Hymenoptera, males develop from haploid, unfertilized eggs and females from diploid, fertilized eggs; consequently, CI would only affect diploid offspring. Westeberhardia does not appear to induce strong CI, if any, as uninfected queens mated to infected males produced diploid F1 females. Another common phenotype caused by reproductive manipulators, the induction of thelytokous PI, can also be excluded. This would lead to exclusive female offspring in infected queens, as C. obscurior does not show diploid male production (Schrempf et al., 2006). The regular occurrence of male offspring and the exclusive production of males by unfertilized females argue against PI. However, Wolbachia, also present in C. obscurior, could exert effects on host reproduction as well. In contrast to some other intracellular symbionts (Moran and Dunbar, 2006; Damiani et al., 2008; Watanabe et al., 2014), paternal transmission of Westeberhardia is unlikely as we did not detect Westeberhardia DNA in transferred sperm and/or seminal fluids stored in the spermatheca of uninfected queens mated to infected males.
Westeberhardia as a possible source of a HGT event
The bacterial gene gpt encoding the xanthine-guanine phosphoribosyltransferase (EC: 2.4.2.22), which was horizontally transferred into the host genome has its closest ortholog in Enterobacter, an Enterobacteriaceae. This indicates that Westeberhardia could be the origin of the HGT event. However, it could also be a relict of a former symbiont no longer present in C. obscurior (Husník et al., 2013). Homologs of gpt have been identified in most bacterial endosymbionts, including Buchnera, Moranella, Blochmannia, Sodalis and Wigglesworthia. The presence of the HGT in the OypB lineage suggests either an ancestral association between C. obscurior and Westeberhardia and a secondary loss of the symbiont in OypB, or the origin of the HGT from an unknown bacterium in the ancestor of both lineages. Westeberhardia is not capable of de novo synthesis of purines, but it is capable of producing all purine nucleotides from recovered bases and nucleosides. A functional genome annotation revealed the presence of hpt, a gene with similar function to gpt, in the Westeberhardia genome. However, RNAseq data show that infected hosts transcribe gpt. Therefore, Westeberhardia might be reliant on an effective salvage using the host-encoded gpt in some conditions (O'Reilly et al., 1984). Interestingly, gpt expression is higher in larval compared with adult queens, indicating that it is not correlated with Westeberhardia titers. Inhibiting gpt expression in Westeberhardia-infected and -uninfected individuals will help elucidate a putative effect of this gene on host and bacteria fitness.
Dependency of Westeberhardia on host-provided metabolites
With a genome size reduction to 533 kb and a GC content of 23.41%, the Westeberhardia genome exhibits features of degenerative genome evolution following the transition to obligate symbiosis (Moya et al., 2008). In addition to reduced effective population sizes in host-associated bacteria compared with free-living relatives, small effective population size of C. obscurior (Schrader et al., 2014) and social insects in general (Romiguier et al., 2014) could lead to even faster genome degeneration. With a coding density of 70.76%, the genome is surprisingly loosely packed, compared with other endosymbionts with similar-length genomes (88% coding density on average) (McCutcheon and Moran, 2012). Furthermore, the occurrence of six pseudogenes indicates that genome erosion in Westeberhardia is still incomplete. It was previously shown that even in advanced mutualistic relationships, endosymbiont genome reduction continues (Gil et al., 2002). Nevertheless, despite the substantial genome reduction, Westeberhardia appears capable of DNA replication, transcription, translation and protein folding, suggesting that it is close to a minimal cell status (Gil et al., 2004). On the other hand, the lack of dnaA and any other alternative mechanism for replication initiation might be an indication that bacterial replication could fall under host control, as suggested for Wigglesworthia and Blochmannia (Akman et al., 2002; Gil et al., 2003), although no potential mechanism has been identified yet. It has also been suggested that, in the absence of recA, the maintenance of recBCD might indicate that this complex has a role in DNA replication initiation instead of recombination (Wu et al., 2006). The gene count with only 372 coding genes and the impairment of essential pathways such as cofactor and essential amino-acid biosynthesis indicate a metabolic dependency on extrinsic resources. In particular, a highly simplified cell envelope and the absence of most transporter genes point towards dependency on the host machinery. In this, Westeberhardia resembles Buchnera aphidicola BCc, which also lacks the ability to synthesize cell surface components (Pérez-Brocal et al., 2006). Intracellularity allows the host to control endosymbiont populations (Vigneron et al., 2012), which together with the lack of dnaA suggests that Westeberhardia populations are controlled by C. obscurior.
Potential for mutualism: Shikimate pathway
Westeberhardia has retained almost the complete shikimate pathway, which produces chorismate, the precursor for tryptophan, tyrosine and phenylalanine, but lacks the downstream enzymes necessary for the synthesis of these aromatic amino acids. However, it can produce 4-hydroxyphenylpyruvate, which can then be converted to tyrosine by the host. Tyrosine is a precursor for DOPA and thereby essential for cuticle formation in insects (Hopkins and Kramer, 1992; Andersen, 2012). Insects cannot synthesize aromatic amino acids and acquisition from diet and/or provisioning by endosymbionts is a common phenomenon. For example, B. aphidicola has evolved overproduction of phenylalanine and tryptophan (Lai et al., 1994; Jiménez et al., 2000). Similarly, B. floridanus can synthesize tyrosine, and increased tyrosine biosynthesis during the host's pupal stage (Zientz et al., 2006) coincides with elevated Blochmannia titers (Stoll et al., 2010; Ratzka et al., 2013). Accordingly, we found high densities of Westeberhardia in late C. obscurior pupae and young adults. Taken together with the detection of gut-associated bacteriomes in pupae, this suggests a role of Westeberhardia in cuticle synthesis during metamorphosis. Although the precise metabolites provided to the host are unclear at this stage, we hypothesize that the retention of the shikimate pathway is key to the mutualistic relationship between Westeberhardia and Cardiocondyla.
After eclosion, Westeberhardia declines slowly in workers but proliferates in queens with age. Although virgin queens exhibit significantly reduced egg laying rates compared with mated queens (Schrempf et al., 2005), we did not find an increase of Westeberhardia infection with reproductive output. Instead, it appears as if the mere availability of reproductive tissue allows proliferation of Westeberhardia. As a consequence of reproductive division of labor in a eusocial host, the major proportion of Westeberhardia encounters a dead end. Cardiocondyla workers completely lack ovaries (Heinze et al., 2006), thus likely impeding Westeberhardia proliferation in the absence of the appropriate microhabitat. In Camponotus floridanus ants, mid-gut connected bacteriomes populated by Blochmannia during the pupal stage become symbiont-free in adult queens and workers, whereas queens retain Blochmannia in their ovarioles (Sauer et al., 2000; Wolschin et al., 2004). Similarly, symbionts localized in gut-associated bacteriomes of cereal weevils are actively eliminated by the initiation of apoptosis after cuticle formation is finished, but ovary-associated symbionts are retained for vertical transmission (Vigneron et al., 2014). Probably due to slow degeneration of bacteriomes, Westeberhardia was still present in young adult males and workers. Bacteria detected in the gut lumen (from degrading bacteriomes) may be the source of continued bacterial infections found in adult workers and males. We have not ruled out that the symbiont continues to have a role in adult workers, although its general decline suggests that the role is not vital. It remains to be investigated if active degradation of bacterial populations in workers benefits individuals and, on a higher level, colony performance (Wenseleers et al., 2002).
Population differences cast doubts about the symbiosis status
We found a naturally occurring uninfected host lineage that continues to thrive in the laboratory, questioning the essentiality of Westeberhardia, at least under conditions including ad libitum protein provisioning. We verified the absence of Westeberhardia in freshly collected field colonies and established laboratory colonies with different methods and across different developmental stages. It remains elusive why Westeberhardia prevalence is so substantially different between colonies of two lineages (OypB, OypC) separated by such short distances (<100 m) in the field. This could indicate a facultative status of Westeberhardia as seen for many insect–microbe symbioses (e.g., Moran et al., 2005). However, Westeberhardia lacks the main characteristics shared by facultative symbionts, even those of a facultative symbiont in the transition to becoming obligate (i.e., large genomes with low coding density and abundance of pseudogenes, presence of repetitive sequences and transposable elements, high GC content (Manzano-Marín and Latorre, 2014)). The occurrence of Westeberhardia in C. wroughtonii and in different populations of C. obscurior indicates an ancestral infection and secondary loss of Westeberhardia in OypB. As the impact of facultative symbionts depends on the particular environmental conditions (Dale and Welburn, 2001; Hansen et al., 2007), a shift in diet or different gut microbiota could explain symbiont loss by natural selection. Owing to proximity of host lineages, an alternative, more parsimonious explanation is mutational loss of Westeberhardia and subsequent fixation through drift (Reuter et al., 2005). Future comparisons between infected and uninfected hosts with the same or different genetic backgrounds under varying environmental conditions will help to reveal potential effects of Westeberhardia on host fitness.
Conclusion
Our study describes a novel intracellular bacterium that maintains a mutualistic relationship with its host but can be lost in some conditions. Its genomic organization, metabolic capabilities, localization and prevalence during host development indicate a role of Westeberhardia in host cuticle formation, possibly facilitating an invasive lifestyle in nutrient poor arboreal environments. The putative monophyly with other insect endosymbionts, possibly facilitating cuticle buildup during development (Zientz et al., 2006; Vigneron et al., 2014), suggests a single origin of metamorphosis-based symbiosis.
Owing to novel traits emerging through host–symbiont associations, it is indispensable to evaluate possible fitness effects of symbionts on hosts, which are used as model organisms for broad biological questions. Although symbionts are situated along the boundary between biotic environmental factors and genomic composition of the host, it becomes obvious that selection pressures acting on the holobiont must be considered when studying adaptation: ‘Contrary to common belief, environmentally initiated novelties may have greater evolutionary potential than mutationally induced ones' (West-Eberhard, 2005).
Acknowledgments
We are most grateful to Masaki Suefuji, Tina Wanke and Jacques HC Delabie for collecting the JP, SP and BR colonies. We thank Helena Lowack for help in the laboratory, the ‘Molecular Ecology' class of 2014 (University Regensburg) and Franziska Scherl for help with the population screens. We also thank Benjamin Weiss for help with the sectioning of ant specimens for FISH; Yannick Wurm for hosting Wobs and WEobs on antgenomes.org; and Horacio Frydman, Abdelaziz Heddi, Filip Husník, Eva Schultner and Mary Jane West-Eberhard for helpful discussions. This study was funded by a grant from the German Science Foundation (DFG He1623/31) (to JO and JH). AL received Grant BFU2012-39816-C02-01 from the Spanish Ministry of Economy and Competitivity cofinanced by FEDER funds, and AM was funded by grant PrometeoII/2014/065 from Valencian Community (Spain). AMM was supported by a Marie Curie Grant (FP7-PEOPLE-2010_ITN SYMBIOMICS 264774) and a fellowship from the Mexican Federal Government (CONACYT 327211/381508). MK and LF were funded by the Max Planck Society, and an additional grant from the German Science Foundation (to MK) (DFG KA2846/2-1). DW and JW were supported by US NSF DEB0821936; JW was further supported by US NSF DEB1257053; and DW was further funded by a start-up grant from Massey University.
Author contributions
Designed the study: AK, LS, JO; wrote the manuscript: AK, LS, JH, JO; performed the experiments: AK, JO; analyzed the data: AK, LS; prokaryote sequence annotation and HGT detection: DW, JW; Westeberhardia genome assembly, functional annotation and phylogenomic analysis: AL, AM, AMM, RG; Westeberhardia localization (FISH): LF, MK. All authors read, reviewed and accepted the final version of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M et al. (2002). Genome sequence of the endocellular obligate symbiont of tsetse flies Wigglesworthia glossinidia. Nat Genet 32: 402–407. [DOI] [PubMed] [Google Scholar]
- Alasaad S, Rossi L, Maione S, Sartore S, Soriguer RC, Pérez JM et al. (2008). HotSHOT Plus ThermalSHOCK, a new and efficient technique for preparation of PCR-quality mite genomic DNA. Parasitol Res 103: 1455–1457. [DOI] [PubMed] [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56: 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SO. (2012) Cuticular sclerotization and tanning. In: Gilbert LI (ed) Insect Molecular Biology and Biochemistry. Academic Press: Waltham, MA, USA, pp 167–192. [Google Scholar]
- Balmand S, Lohs C, Aksoy S, Heddi A. (2013). Tissue distribution and transmission routes for the tsetse fly endosymbionts. J Invertebr Pathol 112: S116–S122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P. (2005). Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol 59: 155–189. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate: a pratical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300. [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. (2011). Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27: 578–579. [DOI] [PubMed] [Google Scholar]
- Brucker RM, Bordenstein SR. (2012). Speciation by symbiosis. Trends Ecol Evol 27: 443–451. [DOI] [PubMed] [Google Scholar]
- Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP et al. (2013). Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 41: D226–D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM et al. (2012). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 40: D742–D753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, Wetter T, Suhai S. (1999). Genome sequence assembly using trace signals and additional sequence information. Proc German Conf Bioinf 99: 45–56. [Google Scholar]
- Cordaux R, Bouchon D, Grève P. (2011). The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends Genet 27: 332–341. [DOI] [PubMed] [Google Scholar]
- Dale C, Welburn SC. (2001). The endosymbionts of tsetse flies: manipulating host-parasite interactions. Int J Parasitol 31: 628–631. [DOI] [PubMed] [Google Scholar]
- Damiani C, Ricci I, Crotti E, Rossi P, Rizzi A, Scuppa P et al. (2008). Paternal transmission of symbiotic bacteria in malaria vectors. Curr Biol 18: R1087–R1088. [DOI] [PubMed] [Google Scholar]
- Douglas AE. (2009). The microbial dimension in insect nutritional ecology. Funct Ecol 23: 38–47. [Google Scholar]
- Engel P, Moran NA. (2013). The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev 37: 699–735. [DOI] [PubMed] [Google Scholar]
- Feldhaar H. (2011). Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol Entomol 36: 533–543. [Google Scholar]
- Feldhaar H, Straka J, Krischke M, Berthold K, Stoll S, Mueller MJ et al. (2007). Nutritional upgrading for omnivorous carpenter ants by the endosymbiont Blochmannia. BMC Biol 5: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, Latorre A, Moya A. (2010). Evolution of prokaryote–animal symbiosis from a genomics perspective. Microbiol Monogr 19: 207–233. [Google Scholar]
- Gil R, Sabater-Munoz B, Latorre A, Silva FJ, Moya A. (2002). Extreme genome reduction in Buchnera spp.: toward the minimal genome needed for symbiotic life. Proc Natl Acad Sci USA 99: 4454–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, Silva FJ, Peretó J, Moya A. (2004). Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev 68: 518–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, Silva FJ, Zientz E, Delmotte F, González-Candelas F, Latorre A et al. (2003). The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc Natl Acad Sci USA 100: 9388–9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh T, Noda H, Ito S. (2006). Cardinium symbionts cause cytoplasmic incompatibility in spider mites. Heredity 98: 13–20. [DOI] [PubMed] [Google Scholar]
- Hansen AK, Jeong G, Paine TD, Stouthamer R. (2007). Frequency of secondary symbiont infection in an invasive psyllid relates to parasitism pressure on a geographic scale in California. Appl Environ Microbiol 73: 7531–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddi A, Grenier AM, Khatchadourian C, Charles H, Nardon P. (1999). Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc Natl Acad Sci USA 96: 6814–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze J, Cremer S, Eckl N, Schrempf A. (2006). Stealthy invaders: the biology of Cardiocondyla tramp ants. Insect Soc 53: 1–7. [Google Scholar]
- Hopkins TL, Kramer KJ. (1992). Insect cuticle sclerotization. Annu Rev Entomol 37: 273–302. [Google Scholar]
- Husník F, Chrudimský T, Hypša V. (2011). Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol 9: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husník F, Nikoh N, Koga R, Ross L, Duncan RP, Fujie M et al. (2013). Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153: 1567–1578. [DOI] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez N, González-Candelas F, Silva FJ. (2000). Prephenate dehydratase from the aphid endosymbiont (Buchnera displays changes in the regulatory domain that suggest its desensitization to inhibition by phenylalanine. J Bacteriol 182: 2967–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenpoth M, Gottler W, Herzner G, Strohm E. (2005). Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol 15: 475–479. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth M, Yildirim E, Gürbüz MF, Herzner G, Strohm E. (2012). Refining the roots of the beewolf—Streptomyces symbiosis: antennal symbionts in the rare genus Philanthinus (Hymenoptera, Crabronidae). Appl Environ Microbiol 78: 822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenpoth M, Roeser-Mueller K, Koehler S, Peterson A, Nechitaylo TY, Stubblefield JW et al. (2014). Partner choice and fidelity stabilize coevolution in a Cretaceous—age defensive symbiosis. Proc Natl Acad Sci USA 111: 6359–6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Paley SM, Krummenacker M, Latendresse M, Dale JM, Lee TJ et al. (2010). Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinform 11: 40–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martínez C et al. (2013). EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res 41: D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Meng XY, Tsuchida T, Fukatsu T. (2012). Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc Natl Acad Sci USA 109: E1230–E1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CY, Baumann L, Baumann P. (1994). Amplification of trpEG: adaptation of Buchnera aphidicola to an endosymbiotic association with aphids. Proc Natl Acad Sci USA 91: 3819–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus A, LaButti K, Foster B, Lowry S, Trong S, Goltsman E. (2008). POLISHER: An effective tool for using ultra short reads in microbial genome assembly and finishing. AGBT, Marco Island, FL, USA. [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. (2009). PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25: 2286–2288. [DOI] [PubMed] [Google Scholar]
- Laslett D, Canback B. (2004). ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32: 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Cooley L. (1994). Intercellular cytoplasm transport during Drosophila oogenesis. Dev Biol 165: 336–351. [DOI] [PubMed] [Google Scholar]
- Manzano-Marín A, Latorre A. (2014). Settling down: the genome of Serratia symbiotica from the aphid Cinara tujafilina zooms in on the process of accommodation to a cooperative intracellular life. Genome Biol Evol 6: 1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Szathmáry E. (1997) The Major Transitions in Evolution. Oxford University Press: Oxford, UK. [Google Scholar]
- McCutcheon JP, Moran NA. (2012). Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10: 13–26. [DOI] [PubMed] [Google Scholar]
- Michalkova V, Benoit JB, Weiss BL, Attardo GM, Aksoy S. (2014). Vitamin B6 generated by obligate symbionts is critical for maintaining proline homeostasis and fecundity in tsetse flies. Appl Environ Microbiol 80: 5844–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Dunbar HE. (2006). Sexual acquisition of beneficial symbionts in aphids. Proc Natl Acad Sci USA 103: 12803–12806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Russell JA, Koga R, Fukatsu T. (2005). Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl Environ Microbiol 71: 3302–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Peretó J, Gil R, Latorre A. (2008). Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet 9: 218–229. [DOI] [PubMed] [Google Scholar]
- Murray RG, Stackebrandt E. (1995). Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int J Syst Bacteriol 45: 186–187. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Shigenobu S, Sakazume N, Shiraki T, Hayashizaki Y, Carninci P et al. (2005). Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci USA 102: 5477–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. (2013). Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29: 2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly C, Turner PD, Smith-Keary PF, McConnell DJ. (1984). Molecular cloning of genes involved in purine biosynthetic and salvage pathways of Salmonella typhimurium. Mol Gen Genet 196: 152–157. [DOI] [PubMed] [Google Scholar]
- Oettler J, Suefuji M, Heinze J. (2010). The evolution of alternative reproductive tactics in male Cardiocondyla ants. Evolution 64: 3310–3317. [DOI] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. (1999). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KM, Russell JA, Moran NA, Hunter MS. (2003). Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci USA 100: 1803–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino-Navarrete R, Piulachs M-D, Belles X, Moya A, Latorre A, Peretó J. (2014). The cockroach Blattella germanica obtains nitrogen from uric acid through a metabolic pathway shared with its bacterial endosymbiont. Biol Lett 10: 20140407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, Heijne von G, Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Meth 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Pérez-Brocal V, Gil R, Ramos S, Lamelas A, Postigo M, Michelena JM et al. (2006). A small microbial genome: the end of a long symbiotic relationship? Science 314: 312. [DOI] [PubMed] [Google Scholar]
- Ratzka C, Gross R, Feldhaar H. (2013). Gene expression analysis of the endosymbiont-bearing midgut tissue during ontogeny of the carpenter ant Camponotus floridanus. J Insect Physiol 59: 611–623. [DOI] [PubMed] [Google Scholar]
- Reuter M, Pedersen JS, Keller L. (2005). Loss of Wolbachia infection during colonisation in the invasive Argentine ant Linepithema humile. Heredity 94: 364–369. [DOI] [PubMed] [Google Scholar]
- Romiguier J, Lourenco J, Gayral P, Faivre N, Weinert LA, Ravel S et al. (2014). Population genomics of eusocial insects: the costs of a vertebrate-like effective population size. J Evol Biol 27: 593–603. [DOI] [PubMed] [Google Scholar]
- Russell JA, Moran NA. (2006). Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc Biol Sci USA 273: 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JA, Moreau CS, Goldman-Huertas B, Fujiwara M, Lohman DJ, Pierce NE. (2009). Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc Natl Acad Sci USA 106: 21236–21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A et al. (2000). Artemis: sequence visualization and annotation. Bioinform Appl Note 16: 944–945. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press: New York, NY, USA. [Google Scholar]
- Sauer C, Stackebrandt E, Gadau J, Hölldobler B, Gross R. (2000). Systematic relationships and cospeciation of bacterial endosymbionts and their carpenter ant host species: proposal of the new taxon Candidatus Blochmannia gen. nov. Int J Syst Evol Microbiol 50: 1877–1886. [DOI] [PubMed] [Google Scholar]
- Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M et al. (2011). BRENDA, the enzyme information system in 2011. Nucleic Acids Res 39: D670–D676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader L, Kim JW, Ence D, Zimin A, Klein A, Wyschetzki K et al. (2014). Transposable element islands facilitate adaptation to novel environments in an invasive species. Nat Comms 5: 5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrempf A, Aron S, Heinze J. (2006). Sex determination and inbreeding depression in an ant with regular sib-mating. Heredity 97: 75–80. [DOI] [PubMed] [Google Scholar]
- Schrempf A, Heinze J, Cremer S. (2005). Sexual cooperation: mating increases longevity in ant queens. Curr Biol 15: 267–270. [DOI] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. (2000). The Staden package, 1998. Methods Mol Biol 132: 115–130. [DOI] [PubMed] [Google Scholar]
- Stoll S, Feldhaar H, Fraunholz MJ, Gross R. (2010). Bacteriocyte dynamics during development of a holometabolous insect, the carpenter ant Camponotus floridanus. BMC Microbiol 10: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek BE, Ermolaeva MD, Schreiber M, Salzberg SL. (2001). A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics 17: 1123–1130. [DOI] [PubMed] [Google Scholar]
- Tåquist H, Cui Y, Ardell DH. (2007). TFAM 1.0: an online tRNA function classifier. Nucleic Acids Res 35: W350–W353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Domselaar GH, Stothard P, Shrivastava S, Cruz JA, Guo A, Dong X et al. (2005). BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res 33: W455–W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron A, Charif D, Vincent-Monégat C, Vallier A, Gavory F, Wincker P et al. (2012). Host gene response to endosymbiont and pathogen in the cereal weevil Sitophilus oryzae. BMC Microbiol 12: S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron A, Masson F, Vallier A, Balmand S, Rey M, Vincent-Monégat C et al. (2014). Insects recycle endosymbionts when the benefit is over. Curr Biol 24: 2267–2273. [DOI] [PubMed] [Google Scholar]
- Wang J, Weiss BL, Aksoy S. (2013). Tsetse fly microbiota: form and function. Front Cell Infect Microbiol 3: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Yukuhiro F, Matsuura Y, Fukatsu T, Noda H. (2014). Intrasperm vertical symbiont transmission. Proc Natl Acad Sci USA 111: 7433–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenseleers T, Sundstrom L, Billen J. (2002). Deleterious Wolbachia in the ant Formica truncorum. Proc Biol Sci 269: 623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. (2008). Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6: 741–751. [DOI] [PubMed] [Google Scholar]
- West-Eberhard MJ. (2005). Developmental plasticity and the origin of species differences. Proc Natl Acad Sci USA 102: 6543–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D, Redding AJ, Werren JH. (2013). Characterization of an ancient lepidopteran lateral gene transfer. PLoS One 8: e59262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolschin F, Hölldobler B, Gross R, Zientz E. (2004). Replication of the endosymbiotic bacterium Blochmannia floridanus is correlated with the developmental and reproductive stages of its ant host. Appl Environ Microbiol 70: 4096–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worning P, Jensen LJ, Hallin PF, Staerfeldt H-H, Ussery DW. (2006). Origin of replication in circular prokaryotic chromosomes. Environ Microbiol 8: 353–361. [DOI] [PubMed] [Google Scholar]
- Wu D, Daugherty SC, Van Aken SE, Pai GH, Watkins KL, Khouri H et al. (2006). Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol 4: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientz E, Beyaert I, Gross R, Feldhaar H. (2006). Relevance of the endosymbiosis of Blochmannia floridanus and carpenter ants at different stages of the life cycle of the host. Appl Environ Microbiol 72: 6027–6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.