Abstract
One of the greatest challenges in microbial ecology remains to link the metabolic activity of individual cells to their taxonomic identity and localization within environmental samples. Here we combined mass-spectrometric imaging (MSI) through (matrix-assisted) laser desorption ionization time-of-flight MSI ([MA]LDI-TOF/MSI) with fluorescence in situ hybridization (FISH) to monitor antibiotic production in the defensive symbiosis between beewolf wasps and ‘Streptomyces philanthi' bacteria. Our results reveal similar distributions of the different symbiont-produced antibiotics across the surface of beewolf cocoons, which colocalize with the producing cell populations. Whereas FISH achieves single-cell resolution, MSI is currently limited to a step size of 20–50 μm in the combined approach because of the destructive effects of high laser intensities that are associated with tighter laser beam focus at higher lateral resolution. However, on the basis of the applicability of (MA)LDI-MSI to a broad range of small molecules, its combination with FISH provides a powerful tool for studying microbial interactions in situ, and further modifications of this technique could allow for linking metabolic profiling to gene expression.
Ecological analyses of microbial metabolites have thus far been hampered by the difficulty of localizing and quantifying these compounds in situ and tying their production to subpopulations or even single cells of individual microbial taxa. However, recent advances in mass-spectrometric imaging (MSI) techniques provide excellent tools to monitor metabolic processes and chemical communication in an ecological context (Svatoš, 2010, 2011). For example, matrix-assisted laser desorption/ionization mass spectrometric imaging (MALDI-MSI) has successfully been employed to observe antagonistic interactions between Streptomyces and Bacillus strains in vitro (Yang et al., 2009). The analysis of microbial interactions in situ, however, requires the combination of metabolic profiling with taxonomic identification and localization of the involved microorganisms. Previous studies employing microautoradiography or high-resolution secondary ion mass spectrometry in combination with in situ hybridization have provided insights into the metabolism of individually identified bacterial cells in environmental samples (Orphan et al., 2001; Kindaichi et al., 2004; Behrens et al., 2008; Musat et al., 2008). However, the need for isotopic labeling limits the application of these techniques to a subset of biological questions.
Here we combine MSI with fluorescence in situ hybridization (FISH) for simultaneous metabolite profiling and taxonomic identification of bacteria, using the defensive symbiosis between beewolf wasps and Streptomyces bacteria as a model. Beewolves of the genera Philanthus, Trachypus and Philanthinus (Hymenoptera, Crabronidae) cultivate ‘Streptomyces philanthi' in specialized antennal gland reservoirs (Kaltenpoth et al., 2006; Goettler et al., 2007; Kaltenpoth et al., 2014) and secrete the bacteria into their subterranean brood cells before oviposition (Kaltenpoth et al., 2010a). Later, the larva incorporates the symbionts into the cocoon silk, where the streptomycetes produce a cocktail of at least nine different antibiotics (Kroiss et al., 2010) and thereby protect the larva against pathogenic fungi and bacteria during the long (up to 9 months) and vulnerable phase of hibernation (Kaltenpoth et al., 2005; Koehler et al., 2013). Previous studies using MSI revealed that the antibiotics abound on the outer surface of the cocoon, while they are virtually absent from the inner surface (Kroiss et al., 2010).
We used (matrix-assisted) laser desorption ionization time-of-flight MSI ([MA]LDI-TOF/MSI) to visualize the abundance of two different antibiotics (piericidin A1 and B1) and subsequently localized the symbionts producing these compounds on beewolf cocoons using FISH. Pieces of beewolf cocoons were fixed to MALDI target plates without any pre-treatment using double-sided adhesive tape, with the outer cocoon surface facing upward. In order to allow for later alignment of ion-intensity maps and FISH images, the cocoon pieces were surrounded by thin paint markings (Edding751, 1–2 mm tip width, white), applied with the tip of a needle. This marker was chosen because it yielded characteristic signals in (MA)LDI-MS (measured at m/z 322.5±0.5) and also showed fluorescence at 640 nm, the excitation wavelength of the fluorescent dye used for FISH (that is, Cy5). MSI was carried out without any pretreatment of the samples (Hoelscher et al., 2009; Kroiss et al., 2010), or after application of 2,5-dihydroxybenzoic acid matrix by sublimation (Svatoš and Mock, 2013). A MALDI micro MX mass spectrometer (Waters, Milford, MA, USA) equipped with a nitrogen laser (337 nm) was used in the reflectron mode and positive polarity for data acquisition as previously reported (Kroiss et al., 2010). The step size in both x and y directions was set to 50 μm corresponding to 508 dots per inch resolution. Two-dimensional ion-intensity maps were reconstructed using the spectral data for the respective potassium adduct ions of piericidin A1 (PA1, m/z 454±0.5 [M+K]+) and piericidin B1 (PB1, m/z 468±0.5 [M+K]+) with the BioMAP software (Novartis Institutes for BioMedical Research, Basel, Switzerland). After (MA)LDI imaging, samples were subjected to FISH with the ‘S. philanthi'-specific probe SPT177-Cy5 (Kaltenpoth et al., 2005, 2006) as described previously (Kaltenpoth et al., 2010b). Fluorescence images were recorded on a Zeiss AxioImager Z.1 (Zeiss, Jena, Germany) using both the mosaic and z-stack options for obtaining high-resolution images with increased focusing depth. Overlays of (MA)LDI and FISH images were achieved in Adobe Photoshop CS5 Extended 12.0 by using the pen markings as a guide.
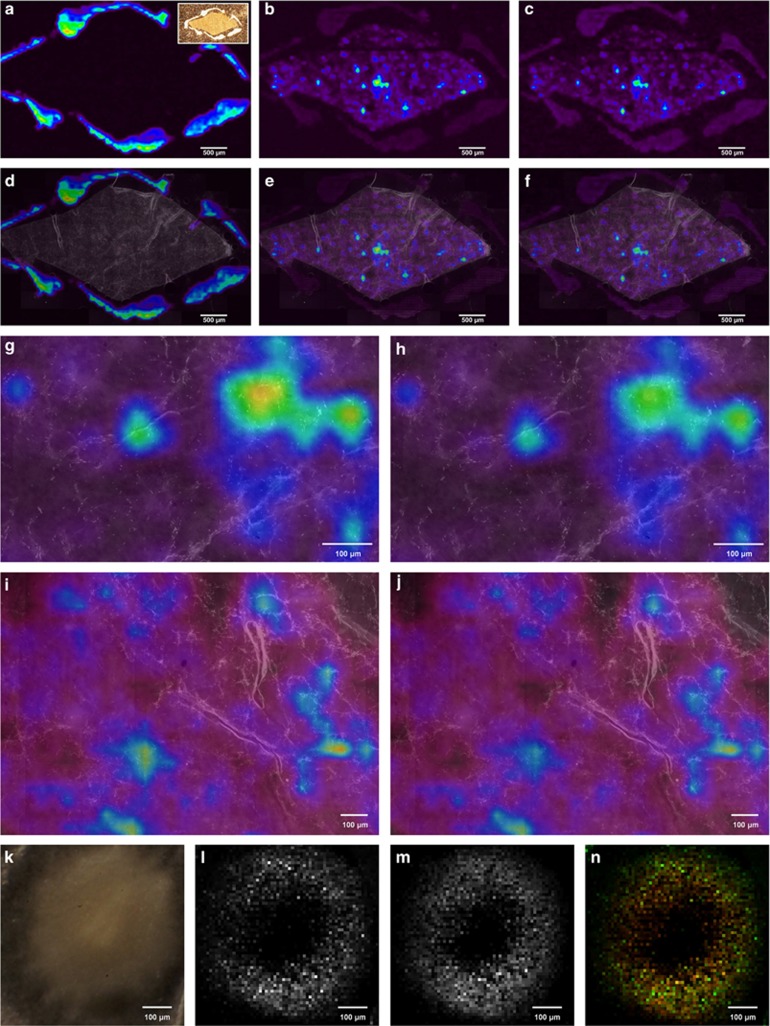
(MA)LDI-MSI revealed a patchy distribution of antibiotics across the outer cocoon surface of European beewolves. The two measured antibiotic substances showed very similar distributions (Figures 1a–j), suggesting that both compounds—as well as possibly the other seven antibiotics produced by the symbionts on the beewolf cocoon that could not be measured here because of their low concentrations—are produced by individual bacterial cells or subpopulations of cells. This is supported by MSI with a high-resolution atmospheric pressure scanning microprobe (AP-SMALDI-MSI) of PA1 and PB1 produced by ‘S. philanthi' on beewolf cocoons and in vitro, which confirmed the colocalization of both antibiotics (Figures 1k–n and Supplementary Figure S1, for experimental procedures see Supplementary Online Material). Thus, different symbiont subpopulations apparently do not specialize in the production of individual compounds, but instead produce a mixture of antibiotics.
Figure 1.
(MA)LDI-FISH of antibiotics produced by symbiotic ‘Streptomyces philanthi' bacteria on a beewolf cocoon (Philanthus triangulum) and in vitro. Ion-intensity maps of (a) the paint marker for alignment of LDI and FISH pictures (m/z 322.5); inset: image of the cocoon piece surrounded by white paint markings on the LDI target plate, (b) piericidin A1 (PA1, m/z 454.5 [M+K]+) and (c) piericidin B1 (PB1, m/z 468.5 [M+K]+). (d–f) The same maps, overlayed with a FISH micrograph of the cocoon piece. Symbiont cells were labeled with the fluorescent oligonucleotide probe SPT177-Cy5. (g–h) Magnifications of (e, f), respectively, with individual bacterial cells visible. (i–j) MALDI-FISH of (i) PA1 (m/z 454.5 [M+K]+) and (j) PB1 (m/z 468.5 [M+K]+) on another cocoon piece. (k–n) AP-SMALDI imaging of antibiotics produced by ‘S. philanthi' in vitro. (k) Light microscopic image of an ‘S. philanthi' colony, (l) PA1 (m/z 416.27 [M+H]+), (m) PB1 (m/z 430.25 [M+H]+), (n) overlay of PA1 (green) and PB1 (red).
On the cocoon, FISH allowed for the visualization of individual symbiont cells, which were abundant across the entire cocoon surface and often occurred in highest densities along the outer cocoon threads (Supplementary Figure S2). The presence of the matrix had no influence on the efficiency of FISH after MSI (data not shown). The alignment of ion-intensity maps with FISH images revealed high concentrations of antibiotics around some subpopulations of cells, whereas other cell aggregations were surrounded by much lower amounts of antibiotics (Figures 1g–j), highlighting the possibility for cheating in the symbiosis. However, the current limitations in sensitivity and lateral resolution of MALDI-MSI do not permit the visualization of compounds on the single-cell level (~1 μm) and thereby may obscure fine-scale patterns of antibiotic production. This is supported by AP-SMALDI-MSI of beewolf cocoons at two different resolutions (step sizes 20 and 5 μm, Supplementary Figure S1): Whereas the high-resolution measurement revealed high concentrations of antibiotics along the outer cocoon threads, which agrees with the FISH experiments showing a similar pattern of symbiont cell densities (Supplementary Figure S2), this pattern was not as apparent at lower resolutions (Supplementary Figure S1). However, the high laser intensities required for a step size of 5 μm were destructive for the samples; therefore, subsequent FISH experiments could not be performed.
The combination of (MA)LDI imaging and FISH provides a powerful tool for tying metabolite profiling to taxonomic identification in environmental samples. However, the laser intensity needs to be carefully adjusted for the desired application to achieve maximum sensitivity and resolution while at the same time conserving the structure of the sample. This problem can be circumvented by using desorption electrospray ionization (DESI) imaging, which we also successfully combined with FISH in preliminary experiments (data not shown). Still, the limitations of both (MA)LDI and DESI in lateral resolution and sensitivity currently prohibit single-cell resolution of metabolic profiling and restrict the technique to mapping the distribution of metabolites on the subpopulation level (Svatoš, 2011). Thus, the exploration of complex environmental samples is at present limited to microbial communities with distinct spatial structure. However, the major strength of MSI-FISH is its broad applicability to a wide range of small molecules as well as proteins (Table 1) (Svatoš, 2010). Therefore, (MA)LDI-FISH and DESI-FISH allow for addressing a multitude of questions in microbial ecology, ranging from interactions in mixed-species biofilms or cross-feeding associations to the chemical basis and dynamics of mutualistic and antagonistic encounters. As several signal enhancement techniques for FISH have been developed (for example, Schönhuber et al., 1997; Zwirglmaier, 2005), modifications of the approach described here could also be employed to tie metabolic profiling by (MA)LDI or DESI imaging to the presence (Moraru et al., 2010) or expression (Pernthaler and Amann, 2004) of particular genes of interest in microbial communities or eukaryotic tissues. Future studies should explore the possibility for using oligonucleotide labels and hybridization protocols that allow for simultaneous MSI of labeled cells and metabolites of interest, which would obviate the necessity for subsequent FISH and thereby circumvent the problems with high laser intensities. Alternatively, new MALDI matrices capable of dissipating the high ultraviolet-laser intensities and thus preventing DNA damage could be developed.
Table 1. Comparison of established methods for linking localization and taxonomic identification of microbes to the production of particular metabolites of interest in environmental samples.
| Metabolite imaging method | Bacterial visualization method | Metabolite imaging | Reference | ||||
|---|---|---|---|---|---|---|---|
| Equipment costs | Need for labeling | Versatility (compounds) | Lateral resolution | Sensitivity | |||
| Microautoradiography | FISH | $ | Yes | Low | High | Medium | Kindaichi et al., 2004 |
| nanoSIMS | FISH/HISH/EL-FISH | $$$$ | Yes | Low | High | High | Orphan et al., 2001; Behrens et al., 2008; Musat et al., 2008; Li et al., 2008 |
| (MA)LDI | FISH | $$$ | No | Very high | Medium | Medium | This study |
| DESI | FISH | $$ | No | Very high | Low | Low | This study |
Abbreviations: DESI, desorption electrospray ionization; EL-FISH, element labeling fluorescence in situ hybridization; FISH, fluorescence in situ hybridization; HISH, halogen in situ hybridization; (MA)LDI, (matrix-assisted) laser desorption/ionization; SIMS, secondary ion mass spectrometry.
Acknowledgments
We thank Benjamin Weiss for assistance with the FISH experiments and Taras Nechitaylo for help with the ‘S. philanthi' cultivation. We gratefully acknowledge funding from the Max Planck Society (MK and AS) and the German Science Foundation (DFG KA2846/2-1 to MK).
The authors declare no conflicts of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Behrens S, Loesekann T, Pett-Ridge J, Weber PK, Ng W-O, Stevenson BS et al. (2008). Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl Environ Microbiol 74: 3143–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goettler W, Kaltenpoth M, Herzner G, Strohm E. (2007). Morphology and ultrastructure of a bacteria cultivation organ: the antennal glands of female European beewolves, Philanthus triangulum (Hymenoptera, Crabronidae). Arthropod Struct Dev 36: 1–9. [DOI] [PubMed] [Google Scholar]
- Hoelscher D, Shroff R, Knop K, Gottschaldt M, Crecelius A, Schneider B et al. (2009). Matrix-free UV-laser desorption/ionization (LDI) mass spectrometric imaging at the single-cell level: distribution of secondary metabolites of Arabidopsis thaliana and Hypericum species. Plant J 60: 907–918. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth M, Gottler W, Herzner G, Strohm E. (2005). Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol 15: 475–479. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth M, Goettler W, Dale C, Stubblefield JW, Herzner G, Roeser-Mueller K et al. (2006). 'Candidatus Streptomyces philanthi', an endosymbiotic streptomycete in the antennae of Philanthus digger wasps. Int J Syst Evol Microbiol 56: 1403–1411. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth M, Goettler W, Koehler S, Strohm E. (2010. a). Life cycle and population dynamics of a protective insect symbiont reveal severe bottlenecks during vertical transmission. Evol Ecol 24: 463–477. [Google Scholar]
- Kaltenpoth M, Schmitt T, Polidori C, Koedam D, Strohm E. (2010. b). Symbiotic streptomycetes in antennal glands of the South American digger wasp genus Trachypus (Hymenoptera, Crabronidae). Physiol Entomol 35: 196–200. [Google Scholar]
- Kaltenpoth M, Roeser-Mueller K, Koehler S, Peterson A, Nechitaylo T, Stubblefield JW et al. (2014). Partner choice and fidelity stabilize co-evolution in a Cretaceous-age defensive symbiosis. Proc Natl Acad Sci USA 111: 6359–6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindaichi T, Ito T, Okabe S. (2004). Ecophysiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by microautoradiography-fluorescence in situ hybridization. Appl Environ Microbiol 70: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler S, Doubský J, Kaltenpoth M. (2013). Dynamics of symbiont-mediated antibiotic production reveal efficient long-term protection for beewolf offspring. Front Zool 10: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroiss J, Kaltenpoth M, Schneider B, Schwinger M-G, Hertweck C, Maddula RK et al. (2010). Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol 6: 261–263. [DOI] [PubMed] [Google Scholar]
- Li T, Wu T-D, Mazeas L, Toffin L, Guerquin-Kern J-L, Leblon G et al. (2008). Simultaneous analysis of microbial identity and function using NanoSIMS. Environ Microbiol 10: 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraru C, Lam P, Fuchs BM, Kuypers MMM, Amann R. (2010). GeneFISH - an in situ technique for linking gene presence and cell identity in environmental microorganisms. Environ Microbiol 12: 3057–3073. [DOI] [PubMed] [Google Scholar]
- Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F et al. (2008). A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci USA 105: 17861–17866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. (2001). Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293: 484–487. [DOI] [PubMed] [Google Scholar]
- Pernthaler A, Amann R. (2004). Simultaneous fluorescence in situ hybridization of mRNA and rRNA in environmental bacteria. Appl Environ Microbiol 70: 5426–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönhuber W, Fuchs B, Juretschko S, Amann R. (1997). Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl Environ Microbiol 63: 3268–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svatoš A. (2010). Mass spectrometric imaging of small molecules. Trends Biotechnol 28: 425–434. [DOI] [PubMed] [Google Scholar]
- Svatoš A. (2011). Single-cell metabolomics comes of age: new developments in mass spectrometry profiling and imaging. Anal Chem 83: 5037–5044. [DOI] [PubMed] [Google Scholar]
- Svatoš A, Mock H-P. (2013) MALDI mass spectrometric imaging of plants. In: Weckwerth W, Kahl G (eds). The Handbook of Plant Metabolomics. Wiley-VCH: Weinheim, Germany, pp 93–110. [Google Scholar]
- Yang YL, Xu YQ, Straight P, Dorrestein PC. (2009). Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol 5: 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwirglmaier K. (2005). Fluorescence in situ hybridisation (FISH) - the next generation. FEMS Microbiol Lett 246: 151–158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.