Abstract
Although competition, niche partitioning, and spatial isolation have been used to describe the ecology and evolution of macro-organisms, it is less clear to what extent these principles account for the extraordinary levels of bacterial diversity observed in nature. Ecological interactions among bacteria are particularly challenging to address due to methodological limitations and uncertainties over how to recognize fundamental units of diversity and link them to the functional traits and evolutionary processes that led to their divergence. Here we show that two closely related marine actinomycete species can be differentiated based on competitive strategies. Using a direct challenge assay to investigate inhibitory interactions with members of the bacterial community, we observed a temporal difference in the onset of inhibition. The majority of inhibitory activity exhibited by Salinispora arenicola occurred early in its growth cycle and was linked to antibiotic production. In contrast, most inhibition by Salinispora tropica occurred later in the growth cycle and was more commonly linked to nutrient depletion or other sources. Comparative genomics support these differences, with S. arenicola containing nearly twice the number of secondary metabolite biosynthetic gene clusters as S. tropica, indicating a greater potential for secondary metabolite production. In contrast, S. tropica is enriched in gene clusters associated with the acquisition of growth-limiting nutrients such as iron. Coupled with differences in growth rates, the results reveal that S. arenicola uses interference competition at the expense of growth, whereas S. tropica preferentially employs a strategy of exploitation competition. The results support the ecological divergence of two co-occurring and closely related species of marine bacteria by providing evidence they have evolved fundamentally different strategies to compete in marine sediments.
Introduction
Molecular analyses reveal extraordinary levels of bacterial diversity in ocean environments (Sogin et al., 2006; Huber et al., 2007). This diversity creates a paradox in terms of how a limited range of resources can support unexpectedly large numbers of species, as has long been observed among phytoplankton (Hutchinson, 1961). The majority of bacterial diversity observed in nature falls into closely related groups of sequences that have been described as microdiverse sequence clusters (Acinas et al., 2004). The ecological implications of this microdiversity remain largely unknown, yet are crucial for understanding community structure and function (Koeppel et al., 2008). Among examples where phylogenetic diversity has been linked to ecological differentiation, it has been possible to identify functional traits that distinguish related groups of bacteria (Ferris et al., 2003; Sikorski and Nevo, 2005; Johnson et al., 2006) including links between sympatric speciation, competition-dispersal tradeoffs (Yawata et al., 2014) and resource partitioning (Hunt et al., 2008; Oakley et al., 2010). However, the delineation of fundamental units of diversity that maintains species-like properties and the ecological traits that make them distinct remains a major challenge in the field of microbial ecology (Fraser et al., 2009).
Resource competition is thought to be a major driver of evolutionary diversification (Svanbäck and Bolnick, 2007). Two competitive strategies by which organisms compete for resources are exploitation competition, which is characterized by rapid nutrient utilization, and interference competition, which occurs when one organism directly harms another. Each strategy involves tradeoffs in energy investment that are manifested as differences in growth rates and reproduction (Nicholson, 1954; Case and Gilpin, 1974; Little et al., 2008). Among bacteria, there is evidence that exploitation competition drives diversification in lab cultures (Hibbing et al., 2010), whereas interference competition is possibly best known in regards to suppressive soils (Weller et al., 2002) and has been proposed as a mechanism to explain the high levels of diversity observed in temporally constant and spatially homogenous environments (Czárán et al., 2002). Chemically mediated interference competition has been linked to improved fitness in biofilm-forming bacteria (Rao et al., 2005; Shank et al., 2011) and shown to affect community structure in hypersaline mats (Long et al., 2013) and freshwater sediments (Pérez-Gutiérrez et al., 2013), with theoretical models supporting the hypothesis that antibiotic production can improve fitness and stimulate biodiversity (Little et al., 2008; Hibbing et al., 2010). In the marine environment, it has been shown that interference competition is greater among Vibrio populations than within them (Cordero et al., 2012) and more common among particle-associated bacteria than those that are free living (Long and Azam, 2001). However, the extent to which bacteria employ interference vs exploitation competition in nature remains unknown due to a poor understanding of the spatiotemporal dynamics of microbial interactions and a lack of suitable methodologies for characterizing these processes. Furthermore, microbes experience other metabolic tradeoffs (Litchman et al., 2007; Flamholz et al., 2013) that complicate efforts to link phylogeny with specific functional traits. Although new genomic and spectral-imaging techniques have provided improved methods to address the ecology of both cultured and uncultured microbes (Watrous et al., 2012; Hugoni et al., 2013; Ottesen et al., 2013; Rinke et al., 2013), competitive interactions among groups of closely related bacteria remain largely unknown.
As a major source of biologically active natural products (Berdy, 2005), actinomycetes represent a particularly interesting subject for chemical ecology studies. They are common inhabitants of complex environments such as terrestrial soils and marine sediments, where it has long been hypothesized that the secondary metabolites they produce mediate interactions with competing microbes (Williams et al., 1989; Jarvis, 1995). The actinomycete genus Salinispora is readily cultured from marine sediments (Jensen et al., 2005; Mincer et al., 2005) and has proven to be a useful model to address bacterial biogeography (Jensen and Mafnas, 2006), species concepts (Jensen, 2010), and the evolution of secondary metabolism (Freel et al., 2011; Ziemert et al., 2014). It is comprised of three closely related species, Salinispora arenicola, Salinispora tropica and Salinispora pacifica (Maldonado et al., 2005; Ahmed et al., 2013), which share 99% 16 S ribosomal RNA gene sequence identity (Jensen and Mafnas, 2006), thus placing them within a microdiverse sequence cluster (Acinas et al., 2004). The cells form branching filaments that develop into a mycelium and produce dormant, non-motile spores that are broadly distributed over large geographic areas (Jensen et al., 2005). Existing evidence suggests that S. tropica and S. pacifica are geographically isolated (Mincer et al., 2005; Freel et al., 2012), whereas the co-occurrence of both species with the more cosmopolitan and abundant species S. arenicola has been used as evidence for ecological divergence (Jensen and Mafnas, 2006). Although patterns of secondary metabolite production (Jensen et al., 2007) and biosynthetic gene cluster distribution (Penn et al., 2009; Ziemert et al., 2014) have been linked to Salinispora species-level divergence, functional support for sympatry has remained elusive.
In this study, we assessed the effects of S. arenicola and S. tropica on the growth of a diverse collection of co-occurring environmental bacteria in an effort to determine the extent to which secondary metabolites mediate competitive interactions. Direct challenge assays revealed distinct temporal patterns in the onset of growth inhibition, with S. arenicola employing interference competition mediated by antibiotic production and the relatively fast growing S. tropica preferentially employing exploitation competition. Although these two mechanisms of competition are well known to occur among plants, the results provide evidence that competitive strategies represent functional traits that can be used to distinguish between closely related yet ecologically distinct populations of bacteria.
Materials and methods
Sediment collection and processing
Sediment samples were collected via SCUBA at depths from 3 to 16 m in July 2012 during a research cruise aboard the R/V Walton Smith (U Miami). Individual sediment samples (5–10 g per sample) were collected from the sediment surface to depths of ca. −3 cm using sterile Whirl-Pak bags (Nasco, Ft. Atkinson, WI, USA). Locations included sites off Miami and the Dry Tortugas in the United States and Cancún, Cozumel, Akumal, and Banco Chinchorro in the Mexican Caribbean. All samples were processed immediately aboard ship using two methods: drying and stamping for selective actinomycete cultivation (Mincer et al., 2002) and serial dilution and plating for the general cultivation of marine bacteria. For the latter, ca. 1 g of sediment was serially diluted 1:1, 1:10 and 1:100 in sterile seawater, vortex mixed, and 50 μl of the supernatant inoculated onto agar media and spread with a sterile glass rod. Three types of media were used for both methods: (1) 25% marine agar (9.3 g Marine Broth Difco 2216, 16-g agar, 750-ml 0.2-μm filtered seawater, 250-ml deionized water), (2) seawater-agar (16-g agar, 1-l 0.2-μm filtered seawater) and (3) 25% A1 (2.5-g starch, 1-g yeast extract, 0.5-g peptone, 16-g agar, 750-ml 0.2-μm filtered seawater, 250-ml deionized water).
Strain isolation
Salinispora strains were recognized based on colony morphology (Mincer et al., 2002) and repeatedly transferred onto new agar media until pure cultures were obtained as evidenced by uniform colony morphology. The collection of sediment-derived bacteria used in the direct challenge assays were purified in a similar manner and selected to represent a diverse range of colony morphologies and pigmentation. All strains were maintained on medium A1 prepared with 75% artificial seawater (22 g/l Instant Ocean, United Pet Group, Cincinnati, OH, USA), grown with shaking in A1 without agar (hereafter ‘A1') and cryopreserved at −80 °C with 10% glycerol.
DNA extraction, PCR and 16 S ribosomal RNA gene sequencing
All strains presumed to be actinomycetes based on morphology were grown in 7-ml A1 for 3–10 days while shaking at 160 rpm. DNA was extracted from 1 ml of the resulting culture according to the DNeasy protocol (Qiagen Inc., Valencia, CA, USA) with previously described changes (Gontang et al., 2007). For all other strains, colony PCR was performed by suspending a single colony in 5-μl dimethyl sulfoxide and using 1 μl as the PCR template. 16 S ribosomal RNA PCR primers are described in Supplementary Table S4. Each PCR consisted of a 25 μl mixture containing 10 × PCR Buffer (Applied Biosciences, Foster City, CA, USA), 2.5 mM MgCl2 (Applied Biosciences), 0.7% dimethyl sulphoxide, 10 mM dNTPs, 1.5 U AmpliTaq Gold DNA Polymerase (Applied Biosciences) and 10 μmol of each primer. PCR thermocycling conditions were as follows: 5 min of initial denaturation at 95 °C followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 1 min. Sequencing was performed by SeqXcel, Inc. (San Diego, CA, USA). Sequences were submitted to the National Center for Biotechnology Information Basic Local Alignment Search Tool (BLASTN) and identified based on the taxonomic assignment of the closest Basic Local Alignment Search Tool match.
Direct challenge assays
Salinispora cultures were inoculated from frozen stocks into 25 ml A1. Cultures were shaken at 160 rpm for 6 days (S. tropica) or 11 days (S. arenicola) after which 60 μl was transferred by pipet to square 150 × 150 mm petri plates containing 100 ml of A1 agar media. The cultures were spread with a sterile loop down the center of the plate to create a narrow (ca. 3–5 mm) lawn of growth and allowed to grow for 7 days (S. tropica) or 10 days (S. arenicola), based on the time required to reach confluence. These incubation periods correspond to the entry of liquid cultures into stationary phase (Supplementary Figure S1). Environmental isolates were then inoculated in triplicate from established plate colonies to within 1–2 mm of the Salinispora lawn (cross-streaking) with up to 50 perpendicular inoculations made per plate using sterile toothpicks (replicates inoculated onto different plates). Growth of the test strains was considered inhibited if a clearing zone ⩾5 mm was observed in the area adjacent to the Salinispora lawn in at least two of three replicate assays. The pH within the zones of inhibition was tested in comparison with a medium control using pH test strips (Micro Essential Laboratories Inc., New York, NY). Given that antibiotic production can be time-dependent (Bibb, 1996), a second series of assays was performed in which strains from both Salinispora spp. were allowed to grow on agar plates for 23 days prior to adding the bacterial strains. Only strains that showed no evidence of inhibition in the initial assays were tested at this second time point.
Interference vs exploitation competition assays
Follow-up assays were performed to distinguish between interference competition (the production of diffusible growth inhibitors) and exploitation competition (nutrient depletion) as the source of the growth inhibition detected in the direct challenge assays. Agar diffusion assays were performed in triplicate using two S. arenicola (CNY-679 and CNY-685) and two S. tropica (CNY-678 and CNY-681) strains. These strains were grown in the same manner used in the direct challenge assays, after which 1 cm2 agar blocks were cut from the area immediately adjacent to the culture and placed over a freshly inoculated lawn of an environmental isolate that previously tested positive for growth inhibition. The lawns were periodically checked (1–10 days) for zones of inhibition surrounding the agar block and scored as positive when a clear zone (⩾5 mm) was observed. The blocks were visually assessed throughout the tests to be free of Salinispora colonies, which are easily recognized based on morphology, so that any observed inhibition could be linked to the diffusion of compounds that were transferred with the agar blocks. All strains that were not inhibited in the agar diffusion assay were further analyzed to determine whether the cause of the activity observed in the direct challenge assay was due to iron depletion. In this case, the direct challenge assays were repeated using standard A1 and A1 supplemented with FeSO4 (10 μg ml−1 final concentration). If growth was inhibited on A1 but not on iron-replete A1, the inhibition was attributed to iron depletion.
Growth rates
Salinispora growth rates were determined by changes in liquid culture dry weight biomass over time. Salinispora cultures were inoculated from frozen bacterial stocks (1.8 ml) into 50 ml A1. Following 7 days of growth (25 °C, 160 rpm), 1-ml of this culture was inoculated into each of 24 glass tubes containing 10 ml A1 and shaken at 160 rpm (25 °C). On days 0, 3, 6, 9, 12, 15 and 18, triplicate tubes were filtered onto pre-weighed 47-mm glass fiber filters (Pall Corporation, Ann Arbor, MI, USA), dried overnight (32 °C) and weighed. Cell mass was calculated as mg dry weight per ml and growth curves generated by plotting the log of cell mass vs time. Growth rates were calculated as the change in biomass over time during the exponential phase of growth.
Chemical extractions and disc-diffusion assays
S. arenicola CNY-679 and S. tropica CNY-678 were grown on A1 agar plates for 10 and 23 days, respectively. Cell-free agar adjacent to Salinispora growth was removed and cut into small pieces using a sterile scalpel and extracted using methanol (500 ml, 160 rpm, 2 h). The volume of agar extracted was measured by solvent displacement. The extract was filtered (0.45 μm Whatman), dried in vacuo, dissolved in ca. 10 ml of water and extracted with an equal volume of ethyl acetate. The ethyl acetate layer was separated, filtered (0.45 μm Whatman), dried under N2, and weighed. A1 agar media control extracts were similarly prepared. Extracts were dissolved in methanol at 1 ×, 10 × and 100 × volumetrically equivalent concentrations with 1 × equal to the extract being dissolved in a volume of solvent equivalent to the volume of agar extracted. Extracts were tested for antibiotic activity against environmental isolates using standard disc-diffusion assays. For these assays, 15-μl of Salinispora extract, media extract or solvent controls (MeOH) were added to paper discs, allowed to dry and placed onto A1 agar plates, along with an antibiotic control disc (5 μg ciprofloxacin; BD, Sparks, MD, USA), onto which a bacterial strain had been inoculated as per the agar diffusion assays. Zones of inhibition were recorded as the diameter of clear halos surrounding the discs. Thirteen of the strains showing sensitivity to S. arenicola CNY-679 culture extracts were similarly tested for sensitivity to commercially available rifamycin SV (Sigma-Aldrich, St Louis, MI, USA) at concentrations of 0.01, 0.1, and 1 mg ml−1.
Liquid chromatography-tandem mass spectrometry and MALDI-TOF imaging mass spectrometry
High-resolution liquid chromatography-tandem mass spectrometry was performed using an Agilent 6530 Accurate Mass Q-TOF coupled to an Agilent 1260 LC system (Santa Clara, CA, USA) as described in the Supporting Information. Imaging mass spectrometry was performed on agar plates prepared as per the direct challenge assays using S. arenicola CNY-679 and Kytococcus sp. CUA-766. The latter was chosen based on the consistently large zones of inhibition observed in the direct challenge assays with all four S. arenicola strains. The interaction zone, along with monocultures of both strains and a medium control, were processed for MALDI-based imaging mass spectrometry in positive mode using a Microflex Bruker Daltonics mass spectrometer as described in the Supporting Information.
Statistical analyses
A non-parametric PERMANOVA analysis was used to test for correlations between growth inhibition and the taxonomy of the strains used in the direct challenge assays (Anderson, 2001). This test was performed using the ‘adonis' function provided by the vegan package (Oksanen et al., 2014) and run in the statistical program R (R Core Team). Welch's two-sample t-tests (Welch, 1947) were performed in R to test for significant differences between growth rates (n=4 for each species), average zones of inhibition (n=4 for each species) and average percentage of strains inhibited (n=12 for S. arenicola, n=13 for S. tropica).
Genome sequencing and analysis
All genome sequences were generated as previously described (Ziemert et al., 2014) according to the guidelines of the Department of Energy Joint Genome Institute. Twenty-four genomes (Supplementary Table S3) were downloaded from the Joint Genome Institute website and submitted to antiSMASH for identification of genes associated with secondary metabolism (Medema et al., 2011). Gene clusters linked to rifamycin biosynthesis were submitted to NaPDoS (Ziemert et al., 2012) to confirm their identity based on a phylogenetic analysis of the associated ketosynthase domains. To assess the potential for siderophore biosynthesis, previously identified siderophore gene clusters (Supplementary Table S5) were extracted from the closed genomes of S. tropica CNB-440 and S. arenicola CNS-205 (Penn et al., 2009) using Geneious Pro 5.5.9 (created by Biomatters, available at http://www.geneious.com). These gene clusters were used as queries against a database created from 24 Salinispora genomes (12 S. arenicola and 12 S. tropica) using MultiGeneBlast 1.1.13 (Medema et al., 2013). Genome sequences were considered to contain a gene cluster if the sequence coverage and identity values were >50% to the query sequences.
Results
Strain isolation and identification
A total of 289 sediment samples were collected from 22 locations and processed for the selective cultivation of actinomycetes. These efforts yielded 22 S. arenicola and four S. tropica strains from seven locations ranging from the Dry Tortugas to the Yucatán peninsula. These results provide further support for the co-occurrence of the two Salinispora species (Jensen and Mafnas, 2006) and the higher relative abundance of S. arenicola (Mincer et al., 2005). Based on 16 S ribosomal RNA gene sequencing, the Salinispora strains all belong to previously identified sequence types (Freel et al., 2013). In addition, 127 taxonomically diverse marine bacteria were isolated from the same habitats that yielded the Salinispora strains (Supplementary Table S1). These bacteria were assigned to 23 families in four phyla (Supplementary Table S2) and used in a direct challenge assay designed to detect the ability of established Salinispora cultures to inhibit the growth of potential competitors. The genus Bacillus comprised the largest number of strains (32) followed by Streptomyces (phylum Actinobacteria; 12 strains), Erythrobacter (subphylum alphaproteobacteria; 11 strains) and the genera Pseudoalteromonas and Microbulbifer (both subphylum gammaproteobacteria; six strains). Forty-five strains had no matches at 100% identity to sequences in GenBank suggesting they have not previously been cultured (Supplementary Table S1).
Direct challenge assays
Four S. arenicola and four S. tropica strains were tested at two time points in a direct challenge assay designed to detect their ability to inhibit the growth of potential bacterial competitors (Figure 1). In total, growth inhibition was detected in 671 (38%) of the 1769 interactions tested. The vast majority of the 127 strains tested (119 or 93%) were sensitive to at least one Salinispora strain indicating the genus has the capacity to inhibit a broad spectrum of bacteria. The overall levels of inhibition were similar for the two species, with S. arenicola inhibiting on average slightly fewer strains (82) than S. tropica (87) when both time points were considered (Table 1). However, the potency of the inhibition, as measured by the average size of the zones of inhibition produced by each Salinispora strain (Supplementary Figure S1), was significantly greater for S. arenicola (P=0.008, Welch's two-sample t-test).
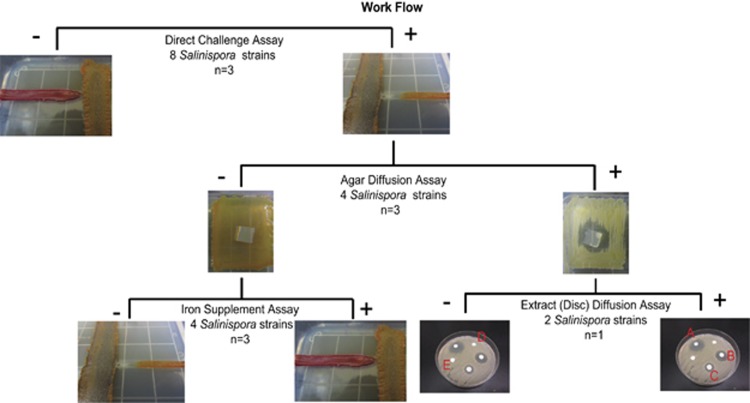
Figure 1.
Workflow. A direct challenge assay was used to detect the ability of established Salinispora cultures to inhibit the growth of co-occurring bacterial strains. All strains that were inhibited in the direct challenge assay (+) were tested further in an agar diffusion assay to determine whether the activity was due to a diffusible molecule. A positive result (+) was recorded when growth of the test strain was inhibited around the agar block but not around a medium control. Organic extracts generated from similar agar blocks were then tested against the sensitive strains in a disk-diffusion assay to determine whether the activity was organic soluble. Active organic extracts (a–c) were identified based on the detection of zones of inhibition around the discs. Strains that were inhibited in the direct challenge assay but not in the agar diffusion assay were tested further to determine whether the inhibition was due to iron depletion. Iron depletion was identified as the source of the inhibition when growth was restored on iron-supplemented media (+).
Table 1. Results from the direct challenge assay for all 8 Salinispora strains at each time point and for both time points combined.
| Species | Strain |
10-day time pt |
23-day time pt |
Combined time pts |
|||
|---|---|---|---|---|---|---|---|
| # Tested | # Inhibited (%) | # Tested | # Inhibited (%) | # Inhibited | % Inhibited | ||
| S. arenicola | CNY-679 | 127 | 59 (47) | 68 | 39 (31) | 98 | 77 |
| S. arenicola | CNY-680 | 127 | 83 (65) | 44 | 23 (18) | 106 | 84 |
| S. arenicola | CNY-685 | 127 | 58 (46) | 69 | 31 (24) | 89 | 79 |
| S. arenicola | CNY-694 | 127 | 21 (17) | 106 | 13 (10) | 34 | 27 |
| Avg.±SD | 55±26 (44±20) | 27±11 (21±9) | 82±33 | 65±26 | |||
| Species | Strain |
7-day time pt |
23-day time pt |
Combined time pts |
|||
|---|---|---|---|---|---|---|---|
| # Tested | # Inhibited (%) | # Tested | # Inhibited (%) | # Inhibited | % Inhibited | ||
| S. tropica | CNY-678 | 127 | 14 (11) | 113 | 59 (47) | 73 | 57 |
| S. tropica | CNY-681 | 127 | 20 (16) | 107 | 80 (63) | 100 | 79 |
| S. tropica | CNY-682 | 127 | 26 (20) | 101 | 71 (56) | 97 | 76 |
| S. tropica | CNY-684 | 127 | 21 (17) | 106 | 58 (46) | 79 | 62 |
| Avg.±SD | 20±5 (16±4) | 67±10 (53±8) | 87±13 | 69±10 | |||
Only strains that were not inhibited at the first time point were repeated at the second time point.
The patterns of inhibition varied both within and between Salinispora species (Figure 2). A PERMANOVA analysis showed no correlation between the taxonomy of the potential competitors and the likelihood they would be inhibited (P>0.05 for all taxonomic levels). The percentages of environmental isolates inhibited by all four Salinispora strains of either species were roughly the same (Supplementary Figure S2). However, in cases where bacteria were inhibited by at least one Salinispora strain, they were inhibited by all four strains in 42% of the cases for S. tropica relative to 28% for S. arenicola. This difference may largely be attributable to S. arenicola strain CNY-694, which grew poorly relative to the other strains (see below) and inhibited only 34 of the strains against which it was tested relative to an average of 98 for the other three S. arenicola strains (Table 1). An antiSMASH analysis of the secondary metabolite gene clusters in CNY-694 revealed a biosynthetic potential that is equivalent to other S. arenicola strains for which genome sequences were analyzed (Supplementary Table S3).
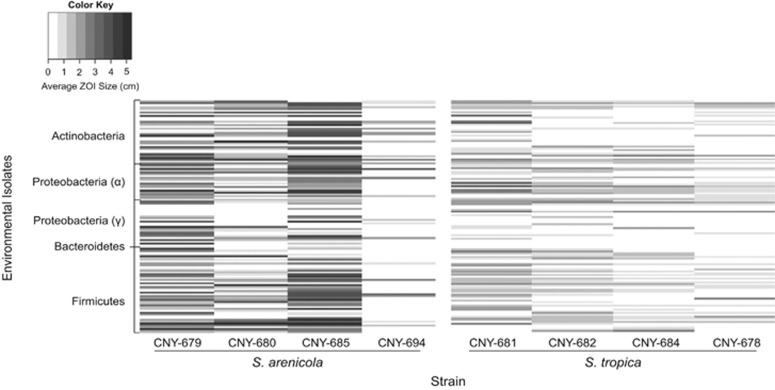
Figure 2.
Growth inhibition observed in the direct challenge assays over both time points. Each row corresponds to one bacterial strain tested against eight Salinispora strains. The bacteria are grouped by phyla, with the phylum Bacteriodetes represented by only one strain. The color intensity represents the average size (cm) of the zone of inhibition (ZOI) for triplicate assays.
Temporal variability
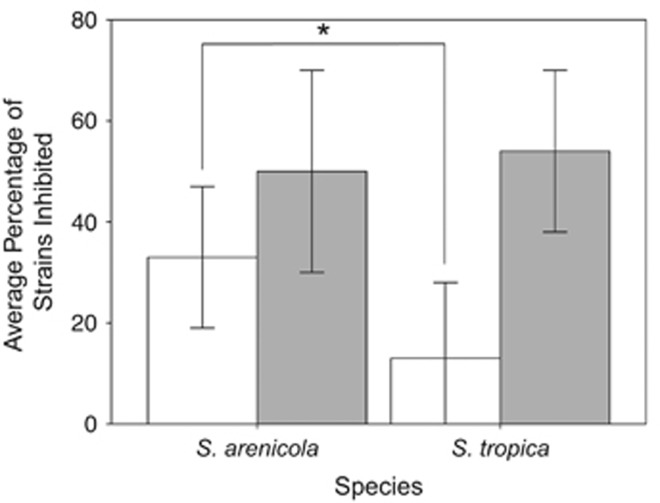
Although the total number of inhibitory interactions recorded for each Salinispora species was similar, there was a major difference in the temporal onset of the activities (Figure 3, Supplementary Figure S3). Most notably, S. arenicola exhibited on average 55 inhibitory interactions at the first time point relative to 20 for S. tropica (Table 1). The trend of greater activity at the first time point was also observed for S. arenicola strain CNY-694 despite the relatively low number of bacteria inhibited. To determine whether these patterns were consistent features of the two Salinispora species, eight additional S. arenicola and nine additional S. tropica strains were tested against a subset of 30 bacteria chosen to represent a range of the taxa encompassed by the original 127 strains (Supplementary Table S1). When the data from all assays were combined, S. arenicola remained significantly more active at the early time point (t-test; P=0.02), whereas there was no difference between the two species when the averages of the combined time points were compared (Figure 3) (t-test; P=0.49). Given that growth inhibition in direct challenge assays can be caused by factors such as nutrient depletion in addition to the production of inhibitory compounds, we next sought to distinguish between these two possibilities in an effort to better categorize the observed activities.
Figure 3.
Average percentage of strains inhibited by 12 S. arenicola and 13 S. tropica strains. White bars represent the results for the first time point (7 and 10 days for S. tropica and S. arenicola, respectively). Gray bars represent the combined percentage of strains inhibited at the first and second (23 days for both Salinispora species) time points. There was a significant difference between the average percentage of strains inhibited at the first time point by each species (*).
Interference vs exploitation competition
Growth inhibition due to the production of antibiotics or other inhibitory substances can be defined as interference competition, whereas inhibition owing to nutrient depletion can be considered exploitation competition (Hibbing et al., 2010). Agar diffusion assays were performed to determine whether the activities detected in the direct challenge assays could be linked to the presence of diffusible molecules (Figure 1) and thus indicative of antibiotic production. Agar taken at both time points from areas adjacent to two S. arenicola and two S. tropica strains was tested against all environmental isolates that showed sensitivity to these strains in the direct challenge assays. Together, these isolates represent 103 (95%) and 94 (87%) of the bacteria that displayed sensitivity to any of the S. arenicola or S. tropica strains, respectively. At the first time point, the inhibition generated by S. arenicola strains CNY-679 and CNY-685 was diffusible in 72% and 40% of the tests performed, respectively. In comparison, the inhibition generated by S. tropica strains CNY-678 and CNY-681 was diffusible in 9% and 14% of the assays, respectively (Figure 4). When the combined time points were considered, the inhibition generated by S. arenicola was diffusible in 51% of the assays compared with 20% for S. tropica. We next asked if the diffusible activities could be extracted with an organic solvent (Figure 1). At 10 × concentration, an extract generated from S. arenicola strain CNY-679 inhibited 32 of the 44 strains that were inhibited in the agar diffusion assay (Table 2). In contrast, an extract prepared from S. tropica strain CNY-678 did not exhibit any inhibition, even when tested at 100 × concentration.
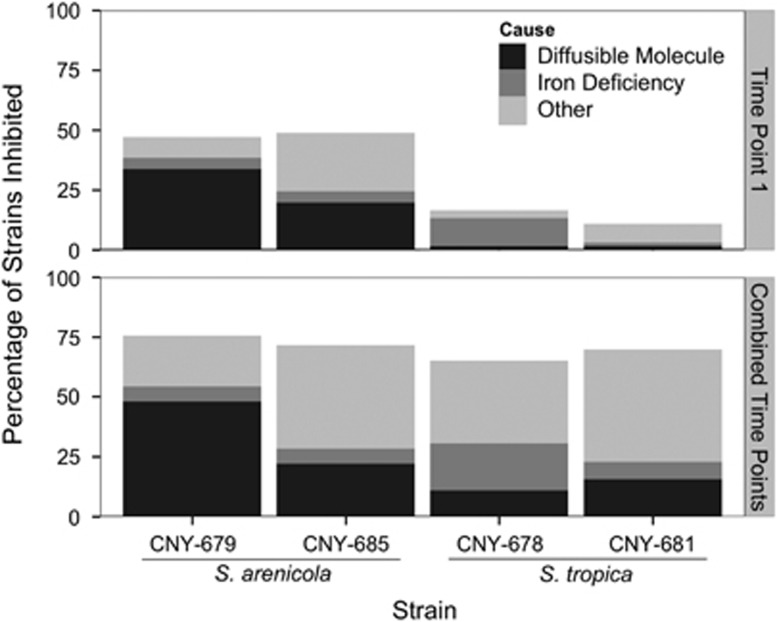
Figure 4.
Sources of growth inhibition. The top panel presents the results from the first time point (7 days for S. tropica, 10 days for S. arenicola), whereas the bottom panel presents cumulative results from the first and second time points (23 days for both species). Sensitive isolates were tested in an agar diffusion assay to determine whether activity could be linked to a diffusible molecule. If negative, further tests were performed to determine whether growth was restored when excess iron was added to the medium, in which case the activity was attributed to iron depletion. If both assays were negative, growth inhibition was attributed to ‘other' sources. Each Salinispora strain was tested against the environmental isolates they inhibited in the direct challenge assays.
Table 2. Number of strains inhibited by a diffusible molecule and organic extract.
| Inhibitory factor |
Strain extracted |
|
|---|---|---|
| S. arenicola CNY-679 | S. tropica CNY-678 | |
| Diffusible molecule | 44 (35) | 12 (9) |
| MeOH extract (⩾1 ×) | 11 | 0 |
| MeOH Extract (⩾10 ×) | 32 | 0 |
| MeOH Extract (100 ×) | 42 | 0 |
For the diffusible molecule test, percentage of total test panel is also included in parentheses. Cell-free areas adjacent to Salinispora cultures growing on agar media were extracted from S. arenicola and S. tropica strains and tested at three volumetric concentrations (1 ×, 10 × and 100 ×) against all environmental isolates that were inhibited in the agar diffusion assays. Extractions were performed at the time point that generated the most activity in the direct challenge assays (10 days for S. arenicola, 23 days for S. tropica).
In cases where the inhibitory activities could not be linked to a diffusible molecule, an iron supplementation assay was performed to determine whether iron depletion was the cause of the inhibition (Figure 1). On average over both time points, 9% of the inhibition generated by S. arenicola could be linked to iron depletion compared with 20% for S. tropica (Figure 4). The remaining inhibitory activities (on average, 40% and 60% for S. arenicola and S. tropica, respectively) were ascribed to ‘other' causes. There were no measurable differences in pH between the zones of inhibition and media controls (data not shown) suggesting that a change in pH was not the cause of the ‘other' inhibition.
Growth rates
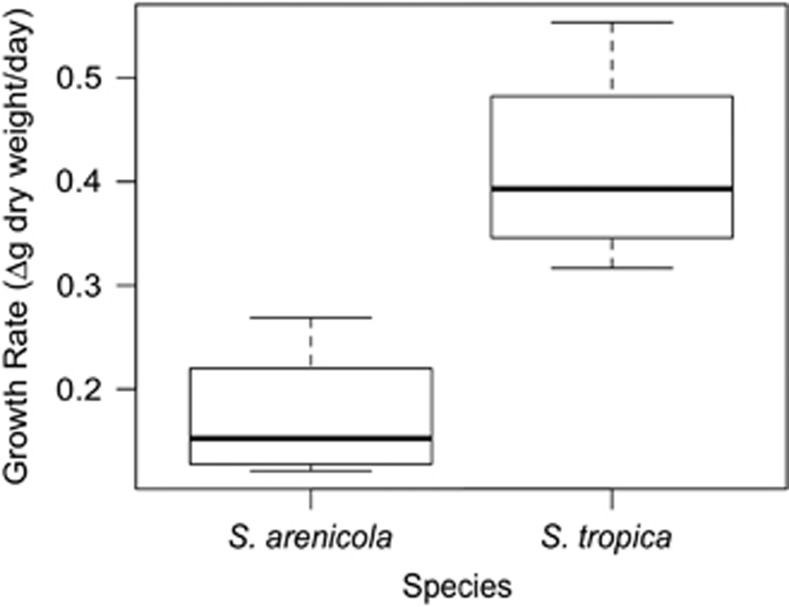
To explore the hypothesis that S. tropica preferentially employs exploitation competition as a competitive strategy, the growth rates of the two species were assessed. Growth curves generated for all eight strains revealed that S. tropica has faster growth rates as determined by changes in dry weight biomass over time, which were significantly different between the two species (Figure 5; t-test, P=0.02). In addition to faster growth rates, cellular biomass was greater on average in stationary phase S. tropica cultures (mean=4.13±SD 1.11 mg ml−1) than in stationary phase S. arenicola cultures (mean=2.98±SD 1.19 mg ml−1) (Supplementary Figure S4). S. arenicola CNY-694, which produced relatively few inhibitory interactions, also produced the least biomass per volume of any strain (Supplementary Figure S4).
Figure 5.
Salinispora growth rates. For each box, the dark horizontal bar represents the median value for the change in dry weight/day of the filtered cell biomass collected during exponential growth phase. The box edges represent the upper and lower quartiles of the data, and the whiskers represent the minimum and maximum values. The plot contains data from four strains of each species grown in triplicate.
Bioinformatic analyses
A prior bioinformatic analysis revealed that S. arenicola maintains a larger number of polyketide synthase and non-ribosomal peptide synthetase biosynthetic gene clusters than S. tropica (Ziemert et al., 2014). To more broadly assess the secondary metabolite potential of the two species, genome sequences from 12 strains of each species (including two S. tropica and three S. arenicola strains used in the direct challenge assays) (Supplementary Table S1) were analyzed for the presence of secondary metabolite gene clusters using antiSMASH, which can identify pathway types other than polyketide synthase and non-ribosomal peptide synthetase including those associated with lantibiotic and terpene biosynthesis (Medema et al., 2011). In addition, the genome sequences were investigated for gene clusters previously linked to siderophore biosynthesis in Salinispora spp. (Penn et al., 2009; Roberts et al., 2012). On average, 31.3 secondary metabolite gene clusters were identified in the S. arenicola strains in comparison with 23.1 for S. tropica (Supplementary Table S3). These results conform well with the 31 and 19 clusters detected in an earlier analysis of two closed S. arenicola and S. tropica genomes, respectively (Penn et al., 2009). The presence of the rifamycin (rif) gene cluster (Floss and Yu, 2005) was confirmed in all of the S. arenicola strains, whereas none of the pathways detected in S. tropica could be linked to the production of a known antibiotic. Although the total number of gene clusters is greater in S. arenicola, the trend is reversed when it comes to siderophore biosynthesis. In an earlier report, both S. arenicola CNS-205 and S. tropica CNB-440 were identified as possessing two siderophore-related gene clusters, one predicted to encode desferrioxamines (des) and a second (sid2) related to the gene cluster for yersiniabactin biosynthesis. However, two additional siderophore gene clusters (sid3 and sid4) were also identified in S. tropica CNB-440 (Penn et al., 2009). The production of desferrioxamines B and E was later confirmed for both S. tropica and S. arenicola and linked to the des locus (Roberts et al., 2012). A MultiGeneBlast analysis revealed that all 12 S. tropica strains for which genome sequences are available possess all four of the siderophore gene clusters, whereas the 12 S. arenicola genomes chosen for this study only possess the des and sid2 clusters (Supplementary Table S3).
Chemical analyses
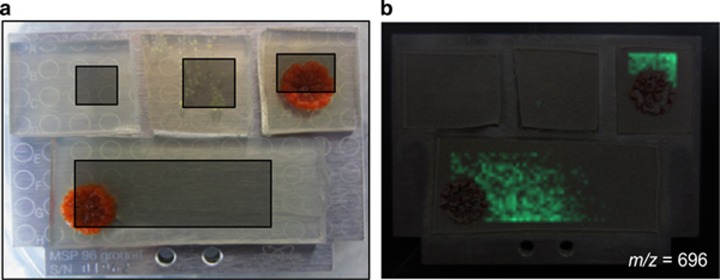
To determine whether any known antibiotics were produced by either Salinispora species, organic culture extracts of S. tropica CNY-678 and S. arenicola CNY-679 were examined by liquid chromatography-tandem mass spectrometry and the results compared with the AntiMarin database (Blunt et al., 2006) and an in-house database of previously identified Salinispora secondary metabolites. The S. arenicola extract contained the antibiotic rifamycin S (Supplementary Figure S5), which belongs to a class of compounds previously reported from this species (Kim et al., 2006; Jensen et al., 2007). In addition, a parent ion that matched rifamycin S was observed directly in a zone of inhibition between S. arenicola CNY-679 and Kytococcus sp. CUA-766 using MALDI-TOF imaging mass spectrometry (Figure 6). Thirteen strains showing sensitivity to S. arenicola culture extracts were tested for sensitivity to commercially available rifamycin SV (the quinone analog of rifamycin S) and found to be sensitive at concentrations between 10–100 μg ml−1 (data not shown), which is comparable to the MIC-50 values reported against a variety of enteropathogens (Farrell et al., 2011). These results provide evidence that the antibiotic rifamycin S is, at least in part, associated with the inhibitory activities generated by S. arenicola. Compounds in this class have been similarly linked to the activities observed between sponge-derived S. arenicola strains and Mycobacterium spp. (Izumi et al., 2010). No known antibiotics were detected in the S. tropica extract.
Figure 6.
MALDI-TOF imaging mass spectrometry of an S. arenicola CNY-679 interaction with Kytococcus sp. CUA-766. (a) MALDI plate setup with control samples in the top row, from left to right: media blank, Kytococcus sp. CUA-766 monoculture, S. arenicola CNY-679 monoculture. The sample in the bottom row contains the zone of inhibition between the two strains. The gray boxes define the areas chosen for imaging. (b) Spatial distribution of the m/z 696 ion, an exact match to rifamycin S (M+H), shown in green surrounding the S. arenicola colony and diffusing outwards toward the inhibited bacterial strain. This ion was observed in the S. arenicola monoculture but not in the medium blank or the Kytococcus sp. monoculture.
Discussion
Molecular surveys have revealed that bacteria exhibit extraordinary levels of phylogenetic diversity (Rappe and Giovannoni, 2003). Although the existence of this diversity is widely appreciated, establishing links between the clades observed in phylogenetic trees and the ecological and evolutionary processes that create and maintain them remains one of the great challenges in microbial ecology (Fuhrman, 2009; Cordero and Polz, 2014). It has been proposed that fine-scale phylogeny can be used to delineate bacteria into ecologically cohesive units or ‘ecotypes' (Cohan, 2002) and that in the few examples available, ecological populations represent ‘gene flow units' for which genome wide rates of homologous recombination are much greater within than between clusters (Cordero and Polz, 2014). A multilocus sequence analysis of Salinispora species supports the description of S. tropica and S. arenicola in accordance with what appear to be natural barriers to recombination (Freel et al., 2013). Although there are many processes that can account for these barriers (Cohan, 2002; Acinas et al., 2004; Thompson et al., 2005; Fraser et al., 2007; Hellweger et al., 2014), ecological differentiation has been proposed as the mechanism driving diversification between S. arenicola and the two sister species with which it co-occurs (Jensen et al., 2007; Freel et al., 2013). To date, the major adaptive traits that distinguish the three species relate to secondary metabolite production (Jensen et al., 2007), but experimental evidence linking these differences to divergent ecological strategies has been lacking. Here we provide evidence that S. tropica and S. arenicola employ fundamentally different competitive strategies that are mediated in part by secondary metabolites, with the former investing in growth or exploitation competition and the latter in interference competition via the production of antibiotics and at the expense of growth. The results create a link between the fine-scale phylogenetic relationships maintained by these two lineages and functional traits that establish them as distinct ecotypes.
The two Salinispora species exhibited similar overall levels of growth inhibition against a diverse panel of co-occurring marine bacteria. However, temporal differences in the onset of inhibition suggested there could be fundamental differences in the mechanisms by which these activities were generated. Subsequent tests made it possible to distinguish between inhibition due to the production of allelopathic molecules and that caused by other factors, including nutrient depletion. The observation that S. arenicola generated more than twice the number of inhibitory activities linked to the production of diffusible substances relative to S. tropica provided the initial line of evidence that these two species differentially invest in interference and exploitation competition, respectively. Additional evidence comes from the identification of the antibiotic rifamycin S in S. arenicola culture extracts and directly in the zones of inhibition, whereas no antibiotic activities were detected in the S. tropica culture extracts. S. arenicola also averaged 31 gene clusters related to secondary metabolism relative to 23 for S. tropica, indicating greater genetic investment in this functional trait. The distinction between interference and exploitation competition was further supported by the significantly faster growth rates recorded for S. tropica. One notable result is the large number of activities that could not be linked to diffusible molecules or iron limitation (Figure 4). These activities require further study and may be linked to the depletion of nutrients other than iron-including carbon and nitrogen sources. The prevalence of non-diffusible growth inhibition suggests that exploitation competition may be widely overlooked as the source of growth inhibition in direct challenge assays. This is not to imply, however, that S. tropica does not also engage in interference competition via the production of inhibitory compounds; they were simply not detected in the assays employed.
Although S. arenicola maintains a larger number of gene clusters devoted to secondary metabolism, this trend is reversed when it comes to siderophore biosynthesis. Siderophores are an important mechanism by which bacteria acquire growth-essential iron (Neilands, 1995) and their production has been linked to antagonism in Vibrio (Pybus et al., 1994) and Pseudomonas spp. (Simões et al., 2008). Although siderophores are secreted secondary metabolites, it can be argued they have a functional role in exploitation competition as opposed to more traditional allelopathic agents such as antibiotics, which function in interference competition. S. tropica genome sequences maintain two gene clusters predicted to encode siderophore biosynthesis (sid3 and sid4) that are not found in S. arenicola (Supplementary Table S3 and S5), suggesting an additional investment in iron uptake by this species. Although the inactivation of key genes in the S. tropica sid3 and sid4 clusters did not affect growth in iron-limited media (Roberts et al., 2012), these pathways may have a role in the acquisition of other limiting metals (Bellenger et al., 2008) or be regulated by factors other than iron limitation, and thus enhanced siderophore production could help support higher growth rates in this species. The conservation of sid3 and sid4 among all 12 S. tropica strains for which genome sequences were available suggests that there are strong selective pressures to maintain the functions provided by the products of these pathways. The observation that secondary metabolites can have a role in exploitation competition by facilitating nutrient acquisition as well as interference competition via antibiotic production emphasizes the importance of distinguishing between these two competitive mechanisms when addressing the ecological functions of secondary metabolites.
Considerable intra-specific variation was observed in the inhibitory activities generated by both species. This supports a growing body of evidence in which inhibitory activities are strain-specific (Grossart et al., 2004; Rypien et al., 2010; Long et al., 2013) or occur at low frequencies within a population (Vetsigian et al., 2011). It also supports the concept that secondary metabolite gene cluster evolution is a dynamic process (Medema et al., 2014) with high levels of plasticity within a single species (Ziemert et al., 2014). One rationalization for this plasticity is that the targets of the antibiotics produced by any one strain may matter less than the ability of the population as a whole to minimize the total number of competitors. Intra-specific variability also provides a rational approach to ensure that at least some individuals will remain competitive as new challengers are encountered either when spores are dispersed or when new resources become available at an existing site. It also provides an effective strategy to avoid resistance, as in the application of combinatorial drug therapy, and the subsequent need to enter into a co-evolutionary arms race (Kinkel et al., 2013). Regardless of the ecological benefits, this variability can likely be linked to recent horizontal gene transfer events and the concept that bacteria frequently ‘sample' gene clusters from the local gene pool (Ziemert et al., 2014), with conservation observed for only those clusters whose products provide sufficient selective advantage (Jensen et al., 2007).
The bacteria used in the direct challenge assays were all heterotrophic and originated from similar, if not the same, sediment samples as the Salinispora isolates, thus making them potential competitors. As with any culture-based study, these strains represent only a small fraction of the total bacterial community and do not include classes such as the Deltaproteobacteria and the Planctomycetacia, which can represent major components of sediment communities (Schauer et al., 2010). Nonetheless, these strains encompass a wide range of taxonomic diversity, including common marine families such as the Rhodobacteraceae and Pseudoalteromonadaceae (Acinas et al., 2004; Gilbert et al., 2012), and share similar growth requirements with Salinispora spp., further supporting their ecological relevance. Although colony growth on agar plates is not the natural state of sediment bacteria, the encroachment of an established colony by competing bacteria represents a scenario that may be important in dictating defensive strategies in marine sediments. Structured habitats such as sediments are prime locations for interference competition as has been suggested for colicinogenic bacteria grown in soft agar matrices (Chao and Levin, 1981). The mycelium growth form exhibited by Salinispora spp. may facilitate the accumulation of antibiotics at concentrations that achieve functional levels in the surrounding microenvironment. However, translating the results obtained here to what occurs in nature will require additional studies aimed toward gaining a spatial context for competitive interactions and more accurate estimates for in situ compound concentrations.
Salinispora strains largely occur as dormant spores and represent relatively rare members of the sediment microbial community (Mincer et al., 2005). Thus, it is possible for actively growing, localized populations to originate from individual spores. This may help explain the apparent lack of ‘social cheaters' benefiting from the production of antibiotics by con-specifics, as has been reported in marine Vibrionaceae (Cordero et al., 2012). The consistent production of antibiotics in the rifamycin class by S. arenicola (Jensen et al., 2007) suggests there are strong selective pressures to maintain the functions of specific compounds and that non-producers or potential cheaters are rapidly selected against. The large diversity of Salinispora secondary metabolites that do not possess antibiotic activity (Fenical and Jensen, 2006) suggests that many of these compounds provide ecological functions other than allelopathy.
Methodological limitations and a poor understanding of the scale at which microbial interactions occur have hampered ongoing efforts to resolve genetic diversity in the context of ecological interactions. Advances in microscopy and mass spectrometry have enhanced our ability to observe physical interactions within limited microbial consortia (Orphan et al., 2001; Malfatti et al., 2010; Valm et al., 2011) but the detailed visualization of interactions in complex environments like sediments remains out of reach. The imaging mass spectrometry technique applied here provided clear evidence that rifamycins were present in the zones of inhibition between Salinispora strains and potential bacterial challengers. The ability to resolve specific compounds in a more natural setting provides expanded opportunities to decipher their roles in mediating interactions among microbes, including those that occur at sub-lethal concentrations (Jauri et al., 2013).
The resource availability hypothesis states that natural selection favors slow growth rates and high levels of defense in low resource habitats (Coley et al., 1985). Although the genus Salinispora belongs to a larger group of actinomycetes that are widely recognized for slow growth and secondary metabolite production, it is interesting that, within this broader ecological strategy, lineages can be identified that preferentially invest in growth or defense. Although preliminary evidence indicates that S. tropica is the less abundant of the two species, higher growth rates and more effective nutrient acquisition suggest it may experience ephemeral blooms in response to nutrient pulses, a concept that can be tested in future studies. Such fine-scale differentiation also provides evidence for co-evolutionary strategies within a microdiverse cluster. Synergistic evolution has been suggested to boost the ecological success of co-occurring species, particularly those that employ secondary metabolites as a form of chemical defense (Cordero and Polz, 2014). Although the extensive arsenal of secondary metabolites available to these two species suggest their ecological functions are more complex than simply distinguishing between two competitive strategies, the results provide the first experimental evidence for ecological divergence in the genus Salinispora.
Acknowledgments
This research was supported by the National Science Foundation (OCE-1235142). We acknowledge J Pawlik for facilitating the field collections and the captain and crew of the R/V Walton Smith (U Miami).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL et al. (2004). Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430: 551–554. [DOI] [PubMed] [Google Scholar]
- Ahmed L, Jensen PR, Freel KC, Brown R, Jones AL, Kim BY et al. (2013). Salinispora pacifica sp. nov., an actinomycete from marine sediments. Antonie Van Leeuwenhoek 103: 1069–1078. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46. [Google Scholar]
- Bellenger JP, Wichard T, Kustka AB, Kraepiel AML. (2008). Uptake of molybdenum and vanadium by a nitrogen-fixing soil bacterium using siderophores. Nat Geosci 1: 243–246. [Google Scholar]
- Berdy J. (2005). Bioactive microbial metabolites. A personal view. J Antibiot 58: 1–26. [DOI] [PubMed] [Google Scholar]
- Bibb M. (1996). The regulation of antibiotic production in Streptomyces coelicolor A3 (2). Microbiology 142: 1335–1344. [DOI] [PubMed] [Google Scholar]
- Blunt J, Munro M, Laatsch H. (2006) AntiMarin Database. University of Canterbury: Christchurch, New Zealand. [Google Scholar]
- Case TJ, Gilpin ME. (1974). Interference competition and niche theory. Proc Natl Acad Sci USA 71: 3073–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L, Levin BR. (1981). Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc Natl Acad Sci USA 78: 6324–6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan F. (2002). What are bacterial species? Annu Rev Microbiol 56: 457–487. [DOI] [PubMed] [Google Scholar]
- Coley PD, Bryant JP, Chapin FS III. (1985). Resource availability and plant antiherbivore defense. Science 230: 895–899. [DOI] [PubMed] [Google Scholar]
- Cordero OX, Wildschutte H, Kirkup B, Proehl S, Ngo L, Hussain F et al. (2012). Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science 37: 1228–1231. [DOI] [PubMed] [Google Scholar]
- Cordero OX, Polz MF. (2014). Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol 12: 263–273. [DOI] [PubMed] [Google Scholar]
- Czárán TL, Hoekstra RF, Pagie L. (2002). Chemical warfare between microbes promotes biodiversity. Proc Natl Acad Sci USA 99: 786–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell DJ, Putnam SD, Biedenbach DJ, Moro L, Bozzella R, Celasco G et al. (2011). In vitro activity and single-step mutational analysis of rifamycin SV tested against enteropathogens associated with traveler's diarrhea and Clostridium difficile. Antimicrob Agents Chemother 55: 992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenical W, Jensen PR. (2006). Developing a new resource for drug discovery: marine actinomycete bacteria. Nat Chem Biol 2: 666–673. [DOI] [PubMed] [Google Scholar]
- Ferris MJ, Kuhl M, Wieland A, Ward DM. (2003). Cyanobacterial ecotypes in different optical microenvironments of a 68C hot spring mat community revealed by 16 S-23 S rRNA internal transcribed spacer region variation. Appl Environ Microbiol 69: 2893–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamholz A, Noor E, Bar-Even A, Liebermeister W, Milo R. (2013). Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc Natl Acad Sci 110: 10039–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss H, Yu T. (2005). Rifamycin-mode of action, resistance, and biosynthesis. Chem Rev 105: 621–632. [DOI] [PubMed] [Google Scholar]
- Fraser C, Hanage WP, Spratt BG. (2007). Recombination and the nature of bacterial speciation. Science 315: 476–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. (2009). The bacterial species challenge: making sense of genetic and ecological diversity. Science 323: 741–746. [DOI] [PubMed] [Google Scholar]
- Freel KC, Nam SJ, Fenical W, Jensen PR. (2011). Evolution of secondary metabolite genes in three closely related marine actinomycete species. Appl Environ Microbiol 77: 7261–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freel KC, Edlund A, Jensen PR. (2012). Microdiversity and evidence for high dispersal rates in the marine actinomycete 'Salinispora pacifica'. Environ Microbiol 14: 480–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freel KC, Millan-Aguinaga N, Jensen PR. (2013). Multilocus sequence typing reveals evidence of homologous recombination linked to antibiotic resistance in the genus Salinispora. Appl Environ Microbiol 79: 5997–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA. (2009). Microbial community structure and its functional implications. Nature 459: 193–199. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B et al. (2012). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontang EA, Fenical W, Jensen PR. (2007). Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl Environ Microbiol 73: 3272–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossart HP, Schlingloff A, Bernhard M, Simon M, Brinkhoff T. (2004). Antagonistic activity of bacteria isolated from organic aggregates of the German Wadden Sea. FEMS Microbiol Ecol 47: 387–396. [DOI] [PubMed] [Google Scholar]
- Hellweger FL, van Sebille E, Fredrick ND. (2014). Biogeographic patterns in ocean microbes emerge in a neutral agent-based model. Science 345: 1346–1349. [DOI] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB. (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA et al. (2007). Microbial population structures in the deep marine biosphere. Science 318: 97–100. [DOI] [PubMed] [Google Scholar]
- Hugoni M, Taib N, Debroas D, Domaizon I, Jouan Dufournel I, Bronner G et al. (2013). Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc Natl Acad Sci USA 110: 6004–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. (2008). Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320: 1081–1085. [DOI] [PubMed] [Google Scholar]
- Hutchinson GE. (1961). The paradox of the plankton. Am Nat 95: 137–145. [Google Scholar]
- Izumi H, Gauthier MEA, Degnan BM, Ng YK, Hewavitharana AK, Shaw PN et al. (2010). Diversity of Mycobacterium species from marine sponges and their sensitivity to antagonism by sponge-derived rifamycin-synthesizing actinobacterium in the genus Salinispora. FEMS Microbiol Lett 313: 33–40. [DOI] [PubMed] [Google Scholar]
- Jarvis BB. (1995). Secondary metabolites and their role in evolution. An Acad Bras Cienc 67(Suppl 3): 329–345. [PubMed] [Google Scholar]
- Jauri PV, Bakker MG, Salomon CE, Kinkel LL. (2013). Subinhibitory antibiotic concentrations mediate nutrient use and competition among soil Streptomyces. PLoS One 8: e81064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W. (2005). Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ Microbiol 7: 1039–1048. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Mafnas C. (2006). Biogeography of the marine actinomycete Salinispora. Environ Microbiol 8: 1881–1888. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Williams PG, Oh DC, Zeigler L, Fenical W. (2007). Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol 73: 1146–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PR. (2010). Linking species concepts to natural product discovery in the post-genomic era. J Ind Microbiol Biotechnol 37: 219–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EM, Chisholm SW. (2006). Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311: 1737–1740. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hewavitharana AK, Shaw PN, Fuerst JA. (2006). Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Appl Environ Microbiol 72: 2118–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkel LL, Schlatter DC, Xiao K, Baines AD. (2013). Sympatric inhibition and niche differentiation suggest alternative coevolutionary trajectories among streptomycetes. ISME J 8: 249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppel A, Perry EB, Sikorski J, Krizanc D, Warner A, Ward DM et al. (2008). Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc Natl Acad Sci USA 105: 2504–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchman E, Klausmeier CA, Schofield OM, Falkowski PG. (2007). The role of functional traits and trade-offs in structuring phytoplankton communities: scaling from cellular to ecosystem level. Ecol Lett 10: 1170–1181. [DOI] [PubMed] [Google Scholar]
- Little AE, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. (2008). Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol 62: 375–401. [DOI] [PubMed] [Google Scholar]
- Long RA, Azam F. (2001). Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol 67: 4975–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RA, Eveillard D, Franco SL, Reeves E, Pinckney JL. (2013). Antagonistic interactions between heterotrophic bacteria as a potential regulator of community structure of hypersaline microbial mats. FEMS Microbiol Ecol 83: 74–81. [DOI] [PubMed] [Google Scholar]
- Maldonado LA, Fenical W, Jensen PR, Kauffman CA, Mincer TJ, Ward AC et al. (2005). Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol 55: 1759–1766. [DOI] [PubMed] [Google Scholar]
- Malfatti F, Samo TJ, Azam F. (2010). High-resolution imaging of pelagic bacteria by atomic force microscopy and implications for carbon cycling. ISME J 4: 427–439. [DOI] [PubMed] [Google Scholar]
- Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA et al. (2011). antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucl Acids Res 39: 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Takano E, Breitling R. (2013). Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol Biol Evol 30: 1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Cimermancic P, Sali A, Takano E, Fischbach MA. (2014). A systematic computational analysis of biosynthetic gene cluster evolution: Lessons for engineering biosynthesis. PLoS Comp Biol 10: e1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mincer TJ, Jensen PR, Kauffman CA, Fenical W. (2002). Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl Environ Microbiol 68: 5005–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mincer TJ, Fenical W, Jensen PR. (2005). Culture-dependent and culture-independent diversity within the obligate marine actinomycete genus Salinispora. Appl Environ Microbiol 71: 7019–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilands JB. (1995). Siderophores - structure and function of microbial iron transport compounds. J Biol Chem 270: 26723–26726. [DOI] [PubMed] [Google Scholar]
- Nicholson AJ. (1954). An outline of the dynamics of animal populations. Aust J Zool 2: 9–65. [Google Scholar]
- Oakley BB, Carbonero F, van der Gast CJ, Hawkins RJ, Purdy KJ. (2010). Evolutionary divergence and biogeography of sympatric niche-differentiated bacterial populations. ISME J 4: 488–497. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2014). Vegan: Community Ecology Package. R package version 2.2-0. http://CRAN.R-project.org/package=vegan.
- Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. (2001). Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293: 484–487. [DOI] [PubMed] [Google Scholar]
- Ottesen EA, Young CR, Eppley JM, Ryan JP, Chavez FP, Scholin CA et al. (2013). Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci USA 110: E488–E497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP et al. (2009). Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J 3: 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Gutiérrez RA, López-Ramírez V, Islas Á, Alcaraz LD, Hernández-González I, Olivera BC et al. (2013). Antagonism influences assembly of a Bacillus guild in a local community and is depicted as a food-chain network. ISME J 7: 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus V, Loutit MW, Lamont IL, Tagg JR. (1994). Growth-inhibition of the salmon pathogen Vibrio ordalii by a siderophore produced by Vibrio anguillarum strain Vl4355. J Fish Dis 17: 311–324. [Google Scholar]
- Rao D, Webb JS, Kjelleberg S. (2005). Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl Environ Microbiol 71: 1729–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappe MS, Giovannoni SJ. (2003). The uncultured microbial majority. Annu Rev Microbiol 57: 369–394. [DOI] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Roberts AA, Schultz AW, Kersten RD, Dorrestein PC, Moore BS. (2012). Iron acquisition in the marine actinomycete genus Salinispora is controlled by the desferrioxamine family of siderophores. FEMS Microbiol Lett 335: 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rypien KL, Ward JR, Azam F. (2010). Antagonistic interactions among coral-associated bacteria. Environ Microbiol 12: 28–39. [DOI] [PubMed] [Google Scholar]
- Schauer R, Bienhold C, Ramette A, Harder J. (2010). Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J 4: 159–170. [DOI] [PubMed] [Google Scholar]
- Shank EA, Klepac-Ceraj V, Collado-Torres L, Powers GE, Losick R, Kolter R. (2011). Interspecies interactions that result in Bacillus subtilis forming biofilms are mediated mainly by members of its own genus. Proc Natl Acad Sci USA 108: E1236–E1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski J, Nevo E. (2005). Adaptation and incipient sympatric speciation of Bacillus simplex under microclimatic contrast at "Evolution Canyons" I and II, Israel. Proc Natl Acad Sci USA 102: 15924–15929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões M, Simões LC, Pereira MO, Vieira MJ. (2008). Antagonism between Bacillus cereus and Pseudomonas fluorescens in planktonic systems and in biofilms. Biofouling 24: 339–349. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored "rare biosphere". Proc Natl Acad Sci 103: 12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svanbäck R, Bolnick DI. (2007). Intraspecific competition drives increased resource use diversity within a natural population. Proc Biol Sci 274: 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J et al. (2005). Genotypic diversity within a natural coastal bacterioplankton population. Science 307: 1311–1313. [DOI] [PubMed] [Google Scholar]
- Valm AM, Mark Welch JL, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R et al. (2011). Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci USA 108: 4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetsigian K, Jajoo R, Kishony R. (2011). Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol 9: e1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD et al. (2012). Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA 109: E1743–E1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch BL. (1947). The generalization of student's' problem when several different population variances are involved. Biometrika 34: 28–35. [DOI] [PubMed] [Google Scholar]
- Weller DM, Raaijmakers JM, Gardener BBM, Thomashow LS. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens 1. Annu Rev Phytopathol 40: 309–348. [DOI] [PubMed] [Google Scholar]
- Williams DH, Stone MJ, Hauck PR, Rahman SK. (1989). Why are secondary metabolites (natural products) biosynthesized? J Nat Prod 52: 1189–1208. [DOI] [PubMed] [Google Scholar]
- Yawata Y, Cordero OX, Menolascina F, Hehemann J-H, Polz MF, Stocker R. (2014). Competition-dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci 111: 5622–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR. (2012). The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One 7: e34064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Lechner A, Wietz M, Millan-Aguinaga N, Chavarria KL, Jensen PR. (2014). Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc Natl Acad Sci 111: E1130–E1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.