Abstract
Although the critical role that our gastrointestinal microbes play in host physiology is now well established, we know little about the factors that influenced the evolution of primate gut microbiomes. To further understand current gut microbiome configurations and diet–microbe co-metabolic fingerprints in primates, from an evolutionary perspective, we characterized fecal bacterial communities and metabolomic profiles in 228 fecal samples of lowland and mountain gorillas (G. g. gorilla and G. b. beringei, respectively), our closest evolutionary relatives after chimpanzees. Our results demonstrate that the gut microbiomes and metabolomes of these two species exhibit significantly different patterns. This is supported by increased abundance of metabolites and bacterial taxa associated with fiber metabolism in mountain gorillas, and enrichment of markers associated with simple sugar, lipid and sterol turnover in the lowland species. However, longitudinal sampling shows that both species' microbiomes and metabolomes converge when hosts face similar dietary constraints, associated with low fruit availability in their habitats. By showing differences and convergence of diet–microbe co-metabolic fingerprints in two geographically isolated primate species, under specific dietary stimuli, we suggest that dietary constraints triggered during their adaptive radiation were potential factors behind the species-specific microbiome patterns observed in primates today.
Introduction
Studies of gut microbiome composition in primates have pointed to host phylogeny as a main driving force (Ochman et al., 2010; Yildirim et al., 2010). Nonetheless, evidence also suggests that specific microbiome arrangements may arise from environmental triggers, such as diet and geography (Amato et al., 2014a; Gomez et al., 2015). Thus, a reasonable approach to reconstruct the gut microbiomes of primates should consider both evolutionary (host phylogeny) and environmental perspectives (Sanders et al., 2014).
Members of the genus Gorilla, our closest evolutionary relatives after Pan (chimpanzees and bonobos), experienced particular ecological challenges over the course of evolution that resulted in their diversification into two species, around 1.75 million years ago (Doran and McNeilage, 1998; Scally et al., 2012). Important differences within each species' habitat in relation to geographical range, food availability and climate make them interesting models to test hypotheses of how primate gut microbiomes are shaped by host-phylogenetic and environmental factors.
Western lowland gorillas (G. g. gorilla) are the most numerous and widespread gorilla species, with more than 200 000 individuals distributed across west-central equatorial Africa (Doran-Sheehy and Boesch, 2004; Robbins, 2011). G. g. gorilla experience marked shifts in the availability of preferred resources yearlong (Rogers et al., 2004); they spend about 80% of their time consuming readily digestible fruit when seasonally available, but shift to a similar percent of time feeding on vegetation (terrestrial herbs and leaves) in drier periods of the year (Masi, 2007; Masi et al., 2009), while also incorporating fibrous fruit and bark (Remis, 1997b).
In contrast, mountain gorillas (G. b. beringei), whose numbers do not supersede 900 individuals in two populations—one in Uganda and the other spanning the Virunga Volcanoes in Uganda, Rwanda and the Democratic Republic of Congo—experience less seasonal fluctuation and year-round availability of terrestrial herbaceous vegetation in altitudes that range from 1450 to 3710 m, which makes their dietary choices less diverse (Stanford and Nkurunungi, 2003; Watts, 1984; Ganas et al., 2004). Although fruit availability is also modulated by seasonal changes, to some extent (Rothman et al., 2008; Rothman et al., 2011), ripe fruit is lower in abundance in the mountain gorilla environment compared with what's observed in the lowland gorilla habitat, which makes nutrient profiles of both species different (Rothman et al., 2014). Indeed, the annual diet of Bwindi mountain gorillas is about 15% fruit and 85% leaves, herbs and bark by wet weight mass (Rothman et al., 2007).
The ecological habitats in which gorillas evolved may have driven the main morphological and behavioral traits that currently characterize lowland and mountain gorillas (Doran and McNeilage, 1998; Robbins, 2011); differences in body size, dentition patterns, sociality and even locomotion are components of an adaptation centering on consumption of foods with high contents of fiber and difficult to digest foods such as leaves, bark and herbs, more relevant in the mountain specie niche (Doran and McNeilage, 1998). Fruit tends to have more easily digestible energy than vegetation, which is typically higher in less digestible structural carbohydrates such as hemicellulose, cellulose and lignin (Chapman et al., 2012). Thus, given the role of gut microbes in providing access to otherwise indigestible plant-based diets, it may be reasonable to expect that the gut microbiome evolved differently in the two gorilla species.
To understand the forces possibly involved in driving particular microbiome arrangements in different primate species, we performed comparative analyses on gut bacterial communities and host–diet–microbe co-metabolic markers (metabolomics) in western lowland and mountain gorillas. The analyses were carried out at different spatial and temporal scales: fecal samples were obtained from mountain gorillas in East Africa and from lowland gorillas in Central Africa. In addition, samples were collected across three different seasons in the lowland gorilla habitat: high fruit, low fruit, and a transition season between the two extremes. Based on these analyses, we suggest that the species-specific gut microbiome patterns seen in different primate hosts, including humans, were likely triggered when primates colonized diverse dietary niches during their adaptive radiation.
Materials and methods
Study site, subjects and sample collection
Western lowland gorilla (G.g.gorilla) fecal samples were collected near two research sites, Bai Hokou (2o50'N,16o28'E) and Mongambe (2o55'N,16o23'E), in the Dzanga sector of the Dzanga-Ndoki National Park, Dzanga-Sangha Protected Areas, Central African Republic. Data were collected in December of 2009 (N=40; Gomez et al., 2015), June and July of 2011 (N=85) and September 2012 (N=56). November and December usually correspond to periods of low fruit availability, whereas high fruit consumption takes place from May to July (Masi, 2007; Remis, 1997b; Masi et al., 2012; Rogers et al., 2004). Thus, September corresponds to a transition period between low and high fruit consumption. Samples were collected from individuals in several habituated and unhabituated groups either after defecation or from nest sites early in the morning, in the case of unhabituated individuals. Some known individuals from habituated groups were sampled more than once during the high fruit and transition seasons. Fecal samples from mountain gorillas (N=47) were collected from the night nests of gorilla groups in the Rushaga, Nkuringo and Buhoma areas of Bwindi Impenetrable National Park, southwestern Kigezi region of Uganda (0o53'–1o08'S, 29o35'–-29o50'E) in August 2013, which corresponded to a period of low fruit consumption (<15% of feeding scans contained fruit, Rothman, personal observation). These samples were collected from seven habituated groups and each sample represented a different individual. The fresh fecal samples (within 1–2 h of defecation) were collected from the center bolus of gorilla dung and placed in two vials, one containing RNAlater (Invitrogen, Life Technologies) for microbiome analyses and the other containing 95% ethanol, which was used for metabolomic profiling. Samples from the transition season in lowland gorillas could not be collected in 95% ethanol, and therefore, were not used for metabolomic profiling.
Microbial community analyses
DNA extraction, pyrosequencing of the V1–V3 16S rRNA region and processing of sequence reads using the online tool mothur and its standard 454 SOP (Schloss et al., 2011) were performed as described in Gomez et al. (2015). Sequence data were deposited in the MG-RAST server under project IDs # 6321 (Gomez et al., 2015; samples from the low fruit season) and # 13961 (samples from the high fruit and transition season and from mountain gorillas).
Metabolomic analyses
Fecal samples from the low fruit season (Gomez et al., 2015; n=38), a subset form the high fruit season (n=39) and from mountain gorillas (n=46) were used for metabolomic profiling. All metabolomic analyses (metabolite extraction and derivatization) were performed as described in Gomez et al. (2015).
Statistical analyses
The Vegan package of R (Oksanen et al., 2012) was used to conduct multivariate community analyses: principal coordinate ordination, permutational multivariate analysis of variance, Mantel test and procrustes (used to determine the level of association between two data sets) and diversity analyses (Rarefied richness, Shannon diversity and multivariate dispersion). These tests were conducted on the relative abundance of each operational taxonomic unit (proportions), based on Bray–Curtis dissimilarity matrices (Clarke, 1993) and rarefying to the lowest number of pyrotag reads obtained for a given sample when necessary. Indicator species analysis (Dufrene and Legendre, 1997) was used to find discriminant taxa in each gorilla group, using the labdsv package of R (Roberts, 2012). The R packages psych (Revelle, 2014) and pgirmess (Giraudoux, 2014) were used to calculate Spearman correlations and Kruskal–Wallis tests adjusted for multiple comparisons. The ca package of R (Nenadic and Greenacre, 2007) was used to perform simple correspondence analysis on feeding behavior data. All remaining analysis and plots were completed using the basic stats package of R (R Core Team, 2014). GC/MS spectra data from metabolomic analyses were transformed as described in Gomez, et al. (2015). The metaboanalyst online tool (http://www.metaboanalyst.ca; Xia et al., 2009) was used to conduct partial-least squares discriminant analysis and to identify discriminant metabolites responsible for metabolome variation based on the variable influence on the projection (VIP) parameter. The VIP method relies on a weighted sum of squares of the principal least squares weight, which indicates the importance of each variable to the whole model. By calculating a VIP score for each variable, variables that increase the predicted ability of the model are retained (usually, with VIP values >1; Indahl et al., 2009). In this case, we kept variables with a VIP score >1.3 and showing significantly different profiles at P<0.05, according to non-parametric tests and false discovery rate adjustment (q-value). Network analyses and visualization were carried out with the open-source tool cytoscape (Shannon et al., 2003).
Feeding behavioral data
To confirm foraging seasonal differences in lowland gorillas, in light of the gut microbiome and metabolome data, the number of feeding bouts on particular food types (terrestrial herbaceous vegetation, fruits, leaves or bark) was recorded for all members of the habituated groups for 61, 57 and 55 days during low fruit, transition and high fruit seasons, respectively, before fecal sample collection. A feeding bout was defined as any manual manipulation of a given food type with the intention of foraging. Observation hours ranged from 9 to 12 h each day, depending on the time the groups were first found in the morning. Feeding data were expressed as the number of feeding bouts any habituated gorilla spent on a particular food type each day during each season and expressed in terms of percentages.
Results
The gut microbiome of Gorilla spp.
After sequence quality control, we obtained an average of 7426 (±9211) sequence reads per fecal sample. Gut microbiome composition analyses indicated that both lowland and mountain gorillas harbor significantly different gut bacterial communities, in terms of taxa sharing ≥97% 16 S rRNA sequence similarity (permutational multivariate analysis of variance, P<0.001, R2=0.17; Figure 1a). However, there was also a significant effect of season in which lowland gorilla fecal samples were collected (P<0.001, R2=0.05), with dry season (low fruit) samples, always forming a homogeneous separate group (Figure 1b).
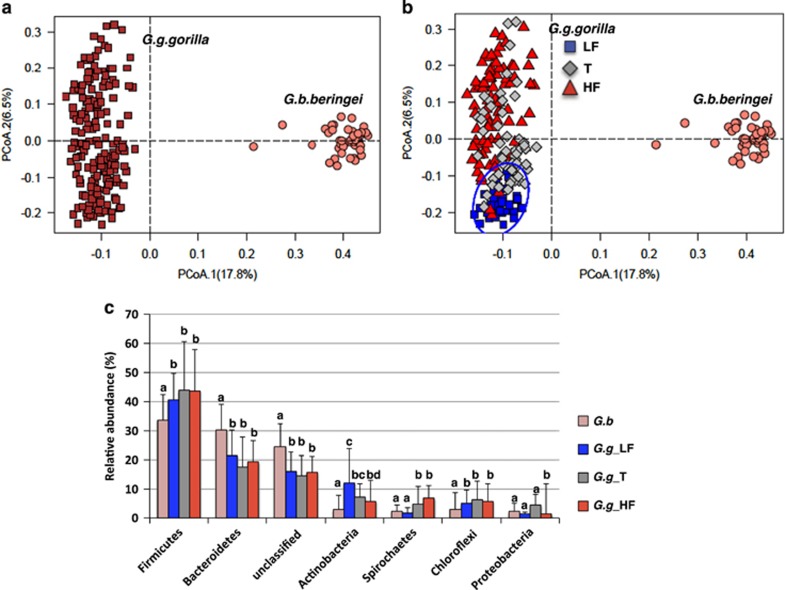
Figure 1.
Gut microbiome composition in lowland and mountain gorillas. (a) Principal coordinates analysis (PCoA) ordination (operational taxonomic units=97% 16S rRNA sequence similarity) based on a Bray–Curtis dissimilarity matrix showing significantly different microbiome composition between the two Gorilla spp., G. g. gorilla and G. b. beringei (permutational multivariate analysis of variance, P<0.001, R2=0.17). (b) PCoA ordination showing significantly different microbiome composition between the two Gorilla spp., and across seasons in lowland gorillas. The Blue circle highlights samples from lowland gorillas collected during the low fruit season and a 95% confidence interval. (c) Relative abundance of major phyla. Different letters (a, b, c) denote significant differences in the abundance of taxa between the microbiomes of both gorilla species in the low fruit (LF), transition (T) and high fruit (HF) seasons (P<0.05, Wilcoxon rank sum tests).
At phyla level, Bacteroidetes and unclassified bacteria were always more abundant in the gut microbiome of mountain gorillas, whereas western lowland gorillas were always more enriched for Firmicutes, Actinobacteria, Spirochetes and Chloroflexi, regardless of season (Wilcoxon rank sum test, adjusted for multiple corrections P<0.01; Figure 1c). Minor phyla such as Verrucomicrobia, Fibrobacteres and Planctomycetes were also more enriched in western lowland gorillas regardless of season (Supplementary Figure 1a).
When classifying operational taxonomic units at 97% rRNA sequence similarity, no differences in either microbiome diversity (Shannon's H') or rarefied richness were detected between lowland gorillas during the three seasons and mountain gorillas. However, at the genus level, lowland gorilla gut microbiomes were always more diverse (Shannon's H', P<0.001) and richer (P<0.001; Supplementary Figures 2c and d) than those of the mountain species, regardless of season (low fruit, transition and high fruit).
Nonetheless, the abundance of some taxa in lowland gorillas during the low fruit season tended to resemble patterns seen in the mountain species. For instance, the abundance of Firmicutes was lower during the low fruit season in lowland gorillas, whereas Bacteroidetes tended to increase (Supplementary Figure 2a). Also, abundance of Spirochaetes was always higher in the high fruit season and lower during drier periods in the lowland species, resembling the values observed in mountain gorillas (Figure 1c). These observations suggested that temporal variation in the lowland gorilla niche, particularly during the low fruit season, caused convergence of microbiome patterns between both species.
Temporal variation and the gut microbiome of Gorilla spp
To take a closer look at patterns shared when lowland and mountain gorillas were constrained by low availability of fruit in their foraging, we looked at abundances of particular genera. The genus-based analyses confirmed that the gut microbiomes of lowland gorillas were more similar to those seen in the mountain species, when less ripe fruit was being consumed. Similarly, the incorporation of more ripe fruit in the lowland gorilla diet made both species' microbiomes more dissimilar (Figures 2a and b). Interestingly, the microbiomes of lowland gorillas during the transition period fell within an intermediate level of similarity to those of the mountain species. These inter-species relationships were also maintained when clustering operational taxonomic units at 97% 16S rRNA sequence similarity (Supplementary Figure 2b).
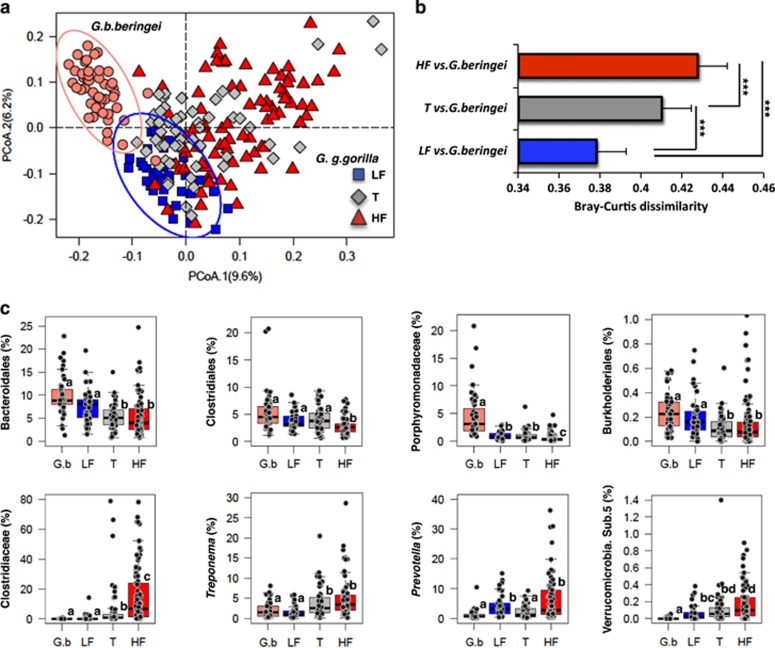
Figure 2.
Comparison of seasonal gut microbiome traits in lowland (G.g.gorilla) and mountain (G.b.beringei) gorillas. (a) Principal coordinates analysis (PCoA) ordination at genus level based on a Bray–Curtis dissimilarity matrix shows that low fruit (LF) lowland gorilla gut microbiomes are more similar to those of the mountain species. In contrast, lowland gorilla microbiomes in the high fruit (HF) season show the most dissimilarity to those of mountain gorillas. Microbiomes of lowland gorillas in the transition season (T) lie at an intermediate level of similarity between those of mountain gorillas and lowland gorillas in the HF season. Circles represent 95% confidence intervals for mountain and lowland gorilla samples collected during the LF season. Bar plots in b show mean Bray–Curtis dissimilarity between groups and asterisks denote significant differences (***P<0.001, Kruskal-Wallis tests adjusted for multiple comparisons). (c) Taxa that showed either decreasing or increasing abundance between mountain gorillas (G.b) and lowland gorilla microbiomes during LF, T and HF seasons. Different letters (a, b, c, d) denote significant differences according to Kruskal–Wallis tests adjusted for multiple comparisons. Actual abundances of each of these taxa can be seen in Supplementary Table 1. Boxplot showing differential abundance of Clostridiaceae represents the sum of the relative abundances obtained for unclassified Clostridiaceae, Anaerobacter, Clostridium sensu stricto and Sarcina.
We then proceeded to detect taxa causing both species microbiomes to converge or differ depending on how much ripe fruit lowland gorillas consumed. For instance, unclassified Bacteroidales, Clostridiales, Porphyromonadaceae and Rodocyclaceae were significantly more abundant in mountain gorillas. However, the abundances of these taxa in lowland gorillas increased during the low fruit season, resembling those levels seen in the mountain species, and significantly decreasing when lowland gorillas incorporated more ripe fruit in their diet (Figure 2c and Supplementary Table 1). An opposite trend was seen with Clostridiaceae (unclassified, Anaerobacter, Clostridium, Sarcina), Treponema, Prevotella and Verrucomicrobia subdivision 5. These taxa characterized the gut microbiomes of lowland gorillas when more ripe fruit was being consumed. Nonetheless, abundances of these taxa in lowland gorillas during the dry season were significantly lower and closer to the levels seen in the mountain species (Figure 2c and Supplementary Table 1). Bacterial taxa characterizing mountain gorillas and lowland gorillas across the three seasons sampled can be seen in Supplementary Table 1.
Another trait shared between both species when lowland gorillas were consuming less ripe fruit was low inter-individual gut microbiome variability. During the low fruit season, the microbiomes of lowland gorillas were less variable, similar to the levels observed in mountain gorillas (Supplementary Figure 2c). In contrast, when more ripe fruit was being consumed, inter-individual microbiome variability in lowland gorillas was significantly higher.
The gut metabolomes of Gorilla spp
A partial-least squares discriminant analysis model with 10-fold cross-validation (Figure 3a) indicated that the metabolomes of mountain and lowland gorillas could be significantly discriminated (permutation test, P<0.01, R2X=95.7%, Q2=93.3% for three components). Analysis of variables with the most significant influence in the partial-least squares discriminant analysis projection (VIP) revealed that the main discriminant trait driving metabolome differences between lowland and mountain gorillas was high abundance of lipids (long-chain fatty acids) as well as plant- and cholesterol-derived metabolites (stigmasterol, campesterol, sitosterol, coprostanol, cholestanol, cholesterol) in the lowland species (Figure 3b and Supplementary Table 2). However, these metabolites were low or absent in lowland gorillas during the low fruit season, closer to the patterns seen in mountain gorillas. Similar trends were observed for other metabolites such as deconjugated bile acids (ursodeoxycholic acid), organic phosphates and terpenoids (ursolic acid) and tyrosine metabolites (1H-indole-5-carboxilic acid).
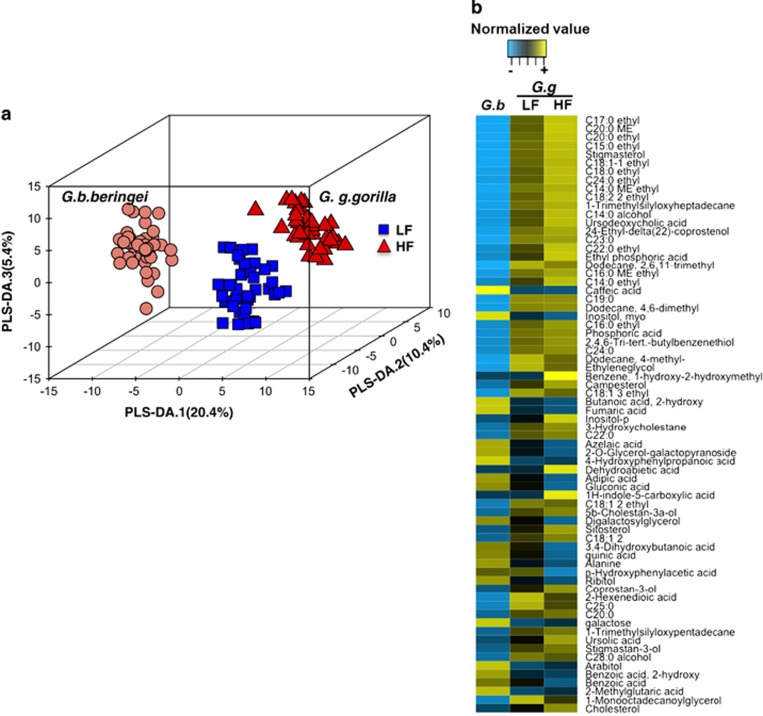
Figure 3.
Gut metabolomic profiles in lowland (G.g.gorilla) and mountain (G.b.beringei) gorillas. (a) Three-dimensional partial-least squares discriminant analysis (PLS-DA) score plot showing separation of the gut metabolomes of mountain and lowland gorillas in samples collected during low fruit (LF) and high fruit (HF) seasons (permutation test supported model variation P<0.001). The amount of the variation in the metabolite data set explained by the three component model was R2X=95.7%. Predictability of the model and statistical validity was Q2=93.3%. (b) Variables (metabolites) with influence on the PLS-DA projections (VIP) along component 1. The heat map shows the mean normalized abundance of metabolites within groups with VIP values >1.3 (false discovery rate: q<0.05, Kruskal–Wallis multiple comparisons) in mountain gorillas (G.b), LF and HF lowland gorilla samples. Highest VIP values are shown in decreasing order from C17:0 ethyl on top (VIP=2.15) to cholesterol (VIP=1.31) at the bottom. VIP values and normalized metabolite concentrations can be seen in Supplementary Table 2.
Metabolites characterizing the metabolomes of mountain gorillas were mainly classified into organic-aromatic acids (caffeic, azelaic, adipic, quinic and benzoic acids), branched-chain esters of butanoic propanoic and carboxylic acids, amino-acid metabolites (alanine and methyl glutaric acid) and sugar monomers (galactopyranosides, gluconic acid, digalactosyl glycerol, galactose, ribitol, arabitol and myo-inositol). Although these metabolites mainly characterized mountain gorillas, they were also abundant when lowland gorillas were consuming less fruit compared with when they were consuming high fruit diets (Figure 3b and Supplementary Table 2).
A Mantel test indicated a significant association between the microbiome and metabolomic data sets (r=0.54, P<0.001). Procrustes analysis was used to corroborate this association and suggested that metabolomic patterns could be predicted accurately from microbiome composition (m2 (sum of square deviations)=0.42, correlation in a symmetric rotation=0.75, P<0.001). Multiple correlation analysis between microbiome composition (at genus level) and metabolome profiles (Spearman correlation coefficient r>0.5 and r<0.68, false discovery rate: P<0.05), coupled with network visualization, indicated the presence of two sub-networks with biomarkers characterizing lowland and mountain gorillas, respectively (Figure 4).
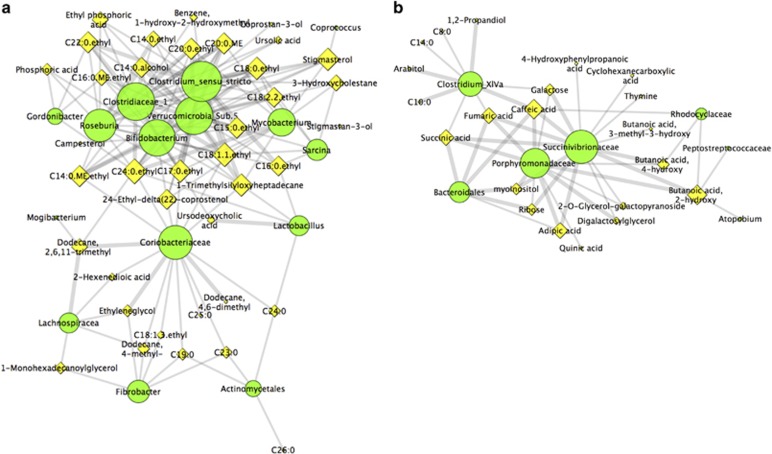
Figure 4.
Sub-network view of relationships between gut microbiome composition and gut metabolomic profiles in lowland (a) and mountain (b) gorillas. Green and yellow nodes represent bacteria and metabolites respectively. Node size represents the number of connections of a given taxa or metabolite within the network. Edges represent Spearman correlation coefficients (Rho>0.5). Edge thickness shows the strength of the correlation.
The sub-network representing the colonic ecosystem of lowland gorillas was characterized by showing a high number of associations between Clostridiales (Clostridiaceae, Roseburia, Clostridium, Sarcina, Lachnospiracea and Mogibacterium), long-chain fatty acids, cholesterol metabolites, sterols and ursodeoxycholic acid. Actinobacteria (particularly, Coriobacteriaceae and Gordonibacter), Lactobacillus, Bifidobacterium and Fibrobacter were also highly connected within this sub-network (Figure 4a). Furthermore, the lowland gorilla sub-network appears to be split in two components, one displaying biomarkers characterizing the high fruit season (more complex and connected) and another showing biomarkers highlighted in the low fruit season (less complex).
The sub-network representing the gut microbiome of mountain gorillas was less complex, with Succinivibrionaceae, Porphyromonadaceae and Bacteroidales showing a dominant role (higher number of connections). These taxa were highly connected to different organic, carboxylic and aromatic acids (fumaric, succinic, adipic, quinic butanoic, phenylpropanoic, caffeic) as well as to various sugars (galactose, ribose, galactopyranosides, inositol, arabitol). Clostridium XIVa and Rhodocyclaceae were also highlighted within this sub-network, with the former also forming connections with medium length fatty acids (C10, C14, C8; Figure 4b).
Feeding behavior of habituated lowland gorilla groups during the sample collection period
To confirm that the gut microbiome and metabolome of the two Gorilla species converge under similar dietary constraints, we profiled feeding behavior in the lowland gorilla species during the high fruit, low fruit and transition seasons. A correspondence analysis based on the number of feeding bouts habituated gorillas were seen foraging on different food types, during each observation day showed that lowland gorilla diets during the low fruit season were mainly dominated by leaves (47.3% of days), terrestrial herbaceous vegetation (33.5%) and bark (7.8% Supplementary Figures 3a and b). All these values were significantly higher than those seen in the transition and high fruit seasons (Supplementary Figures 3c). In contrast, fruit was the dominant food characterizing the high fruit season (consumed 61% of days vs 47% and 11% seen in the transition and low fruit seasons, respectively). Percentage of days gorillas were seen foraging on specific food types can be seen in Supplementary Figures 3b and c with differences confirmed through Wilcoxon rank sum test adjusted for multiple comparisons (P<0.01).
Discussion
Our results present a novel perspective to understand the factors that gave rise to the gut microbiome patterns observed in primates today. As such, we present evidence that the phylogenetic signal that currently shapes the gut microbiome of different primate species might have originated when primates occupied diverse dietary niches during their adaptive radiation. This has been an important area of microbiome research since it was first stated that host phylogeny was the main factor explaining gut microbiome composition in different primate species (Ochman et al., 2010; Yildirim et al., 2010). Since then, further evidence has highlighted a more prominent role of diet and digestive modularity in defining gut microbiome composition in primates (Amato et al., 2014b; Gomez et al., 2015; Muegge et al., 2011; Ley et al., 2008; Moeller et al., 2013). However, for the first time, we present inter-species microbiome differences in two different members of the primate order in the context of seasonal dietary variability and microbe–metabolome interactions.
Diet as an important driver of gut microbiome composition in Gorilla spp.
These results provide insights into how dietary factors could have influenced the acquisition of particular gut microbiomes and diverse gut metabolic capabilities in two different, non-sympatric gorilla species. By combining functional, dietary and longitudinal data, we suggest that upon splitting from a common ancestor around 1.75 million years ago (Scally et al., 2012), western lowland gorillas evolved a gut microbiome with increased capacity to metabolize lipids, sterols and more digestible carbohydrate sources in a nutritionally diverse dietary niche. In contrast, it seems that mountain gorillas, constrained by low nutritional diversity, evolved distinct diet–microbe co-metabolic traits, with more active plant cell wall-processing roles.
This observation may be supported by the fact that several traits in both species' gut microbiomes and metabolomes converge when facing nutritional challenges influenced by low fruit availability and dietary diversity in their ecological niche. Lowland gorillas experience important dietary limitations when ripe fruit is not immediately available. That is, seasonal shifts in their habitat forces them to rely mainly on fibrous fruits, leaves and herbaceous vegetation during the low ripe fruit season (Masi et al., 2009). These fibrous foods are low in readily digestible energy, higher in hemicellulose, cellulose and lignin than ripe fruit (Remis, 1997a, 1997b, 1997c Remis et al., 2001; Rogers et al., 2004, 1990; Rothman et al., 2014), although in times of fruit scarcity fruits eaten are high in fiber and similar to vegetation (Remis et al., 2001). Thus, it is likely that bio-geographical and ecological factors across evolutionary timescales triggered both diversification of Gorilla spp., and the acquisition of distinct gut bacterial communities in mountain and lowland gorillas (Ley et al., 2008; Collins and Dubach, 2000; Gomez and Verdu, 2012; Sussman, 1991).
Indeed, all metabolites enriched in the mountain gorilla metabolome, and in lowland gorillas during the low ripe fruit season (that is, butanoic, propanoic, benzoic and fumaric acids as well as galactopyranosides) can be derived from microbial metabolism on plant cell walls (Pena et al., 2004; Garner et al., 2007). Moreover, we present evidence that the abundance of these metabolites and other components of the lignified portion of plant cell walls, such as caffeic, azelaic, quinic and adipic acid (Moco et al., 2012; Chung, 1997), co-vary along with the abundance of taxa related to unclassified Bacteroidales and Porphyromonadaceae, which could potentially be playing a fibrolytic role in the gut microbiome of mountain gorillas and the lowland species during the low ripe fruit season. Similarly, the shared abundance of taxa related to the Burkholderiales, a taxon with known aromatic compound degradation capabilities (Pérez-Pantoja et al., 2012), indicates potential metabolic activities on the lignified portion of cell walls by both species under high-fiber diets (Chung, 1997).
Convergence of microbiome traits between both species under low fruit consumption is also manifested in the reduced abundance of some taxa that were particularly enriched when lowland gorillas were consuming more fruit. Taxa related to the Clostridiaceae, Treponema, Prevotella and Verrucomicrobia follow this trend. The reasons for the enrichment of these taxa only during high fruit seasons in lowland gorillas is not immediately obvious, but it is likely that they relate to more digestible and diverse food substrates reaching the colon of lowland gorillas when consuming more ripe fruit.
It is also noteworthy that the gut microbiome of both mountain and lowland gorillas under low ripe fruit diets show significantly less inter-individual variability, compared with instances when lowland gorillas incorporated more fruit in their foraging. This observation may be related to the constraints imposed by high-fiber diets on feeding, and the fact that these limitations cause more cohesive foraging in Gorilla spp. (Remis, 1997a; Doran and McNeilage, 1998). In contrast, the fragmented distribution of fruit and the higher nutritional diversity of fruit substrates expand foraging options in gorilla groups, causing individuals within groups to consume more diverse diets when fruit is available (Masi et al., 2009; Rogers et al., 2004). This is an interesting example of how macroecological patterns may be also reflected at microecological scales. In addition, the microbial composition and functional machinery required to process fiber is limited to a few taxa (Flint et al., 2008; Flint and Bayer, 2008).
Despite the existence of shared patterns triggered by similar diets between Gorilla spp., clear differences remain, suggesting the potential acquisition of species-specific gut microbiome arrangements over evolutionary timescales. The gut microbiome and metabolome of mountain gorillas demonstrate different fiber-processing capabilities compared with those of the lowland species, even under the same dietary constraints. For instance, Succinivibrionaceae, the main discriminant taxon in mountain gorillas is almost absent in the lowland species. This taxon has previously been associated with succinate formation using fumarate as electron acceptor in the colonic ecosystem of wild wallabies (Macropus eugenii), known hindgut fermenters (Pope et al., 2011). This is consistent with our network analysis, in the sense that succinic acid (an intermediate in propionate synthesis) and fumaric acid (both discriminant metabolites in this species) were significantly correlated with abundances of this taxon. Thus, it is likely that propionigenic substrates and bacteria have a more important role in the colonic ecosystem of mountain gorillas than in the lowland species. In addition, the prevalence of Peptostreptococcaaeae and Rhodocyclace, only in mountain gorillas, may further confirm different fiber degradation machineries, on potentially different substrates.
Similarly, although lowland gorillas also exploit important amounts of plant structural polysaccharides, some of the taxa involved in their fiber processing may be different. For instance, Fibrobacter, a known fiber degrader (Flint and Bayer, 2008), was almost absent in mountain gorillas and enriched in the lowland species across all seasons, particularly during low fruit periods. Nevertheless, it seems that fiber metabolism has a less prominent role in the diet–microbe co-metabolic landscape of lowland gorillas. The microbiome-metabolome profiles presented here suggest that lowland gorillas exhibit a colonic micro-ecosystem conducive to increased energy harvest. For instance, higher abundance of Firmicutes in relation to Bacteroidetes has been observed in gut microbiomes with increased capacity to process energy-dense diets (Turnbaugh et al., 2009). In addition, taxa related to Lachnospiracea, Blautia, Erysipelotrichaceae, Roseburia and Lactobacillus, prevalent in lowland gorillas across the three seasons sampled, are characterized as typically fermentative, rather than fibrolytic.
These observations may be related to higher availability of foods with more important contents of non-structural carbohydrates for lowland gorillas all year-round, compared with foods in the mountain gorilla niche (Remis et al., 2001; Rogers et al., 2004). Moreover, despite convergence of microbiome traits during seasons of low availability of ripe fruit, lowland gorillas in Central Africa include more species and plant parts in their foraging and have more dietary choices compared with mountain gorillas at Bwindi, even during seasons of fruit scarcity (Rothman et al., 2006; Rogers et al., 2004). This may be supported by the fact that lowland gorillas exhibit higher microbiome diversity at genus level, which may also reflect more diverse dietary substrates. Nonetheless, the extent to which the microbiome distinctions maintained between both species are due to differences in diets or host physiological (or phylogenetic) factors is still unclear.
Diet and gut microbes in Gorilla spp.: primate evolution in context
These results provide insight into the potential influence of ecological change in primate evolution. Climate change and its effect on the availability of dietary resources have been proposed to be a driving force behind hominin speciation, influencing bipedalism, cranial capacity, adaptability and cultural innovations (Behrensmeyer, 2006). Furthermore, recent reports on brain organization differences between lowland and mountain gorillas point to ecological distinctions as important driving factors (Barks et al., 2015). Here, we show that the acquisition of species-specific gut microbiome arrangements in primates may also follow a trend dependent on ecological change.
Thus, it is likely that the common ancestor of both mountain and lowland gorillas had a core set of gut microbes, which later changed to compensate for specific dietary demands as both species diverged in distinct ecological niches. Indeed, dietary constraints over evolutionary timescales are believed to be important factors behind primate diversification (Milton, 1993). Consequently, dietary differences during the adaptive radiation of primates could have caused simultaneous modifications in morphology, physiology and gut microbes. Gut microbiome differences could have been accentuated, as ecology and diet became permanent traits. Interestingly, however, gut microbiome composition is much more flexible than anatomy, and can vary on much shorter timescales in relation to diet and environment. This may explain why environmental signals such as those imposed by zoo diets can cause the microbiome of primates, including that of gorillas, to diverge from species-specific patterns (Ley et al., 2008), supporting the increased plasticity exhibited by mammalian gut microbiomes upon dietary change (David et al., 2014).
One important observation highlighted in these results, and that could provide information to understand the influence of gut microbes and diet in primate evolution, is the prevalence of lipid-derived metabolites and taxa potentially involved in lipid processing in lowland gorillas, predominantly during seasons of high fruit consumption. The abundance of these metabolites and taxa were decreased in low fruit seasons and almost absent in mountain gorillas. Although primate diets are typically very low in fat, fruits and seeds are important lipid sources (Rothman et al., 2012; Reiner et al., 2014). Indeed, taxa related to Coriobacteriaceae, Clostridium, Bifidobacterium and Lactobacillus, significantly enriched in lowland gorillas when ripe fruit was consumed, can also impact cholesterol and bile acid turnover in the colon (Gerard, 2010; Martin et al., 2007). This observation may explain the significant interactions observed between these taxa and abundances of coprostanol, cholestanol, hydrocholestane and ursodeoxycholic acid in the lowland gorilla metabolome.
Thus, these data may also provide hints to possible energy storage mechanisms in lowland gorillas, similar to those taking place in humans under high-caloric diets (Jones et al., 2008; Martin et al., 2007). Furthermore, fat deposition and storage are important evolutionary discriminant features between humans and non-human primates (Horrobin, 1999; Navarrete et al., 2011). Therefore, it seems that lowland gorillas maximize high-caloric diets during abundant ripe fruit seasons to later exploit fat depots when fruit is scarce (Knott, 1998). In humans, however, cultural and technological innovations could have made high-caloric intake and fat storage a permanent trait (Cordain et al., 2005). The way the metabolism of sterols and bile acids in colon impacts the nutritional and physiological status of lowland gorillas still needs to be determined. Nonetheless, these observations suggest that lipid and energy turnover is a key discriminant trait between the metabolic activities of both species' microbiomes and that high-caloric intake could have selected for specific microbe–metabolite profiles in some primate species (that is, humans).
Along these lines, these data motivate further research on the factors that gave rise to current microbiome patterns in humans. Here we present evidence pointing to an increasing gradient in the abundance of Prevotella and Treponema from mountain gorillas to lowland gorillas consuming more ripe fruit. High abundances of these taxa have also been reported in hunter-gatherers and non-western humans (Lin et al., 2013; Yatsunenko et al., 2012; Obregon-Tito et al., 2015). Prevalence of these taxa in humans seems to correlate with increased intake of starchy foods (Wu et al., 2011; Schnorr et al., 2014; David et al., 2014). Thus, these results show that the gut microbiome of lowland gorillas during high fruit consumption seasons shares important traits with that of traditional human groups, and that starchy foods and dietary shifts to more digestible carbohydrate sources may have had important impacts in the evolution of modern human microbiomes (Henry et al., 2011; McGrew, 2007).
Concluding remarks and future perspectives
In summary, we have shown distinctions and convergence of diet–microbe co-metabolic fingerprints in two geographically isolated primate species, under specific dietary stimuli. We specifically show that temporal dominance of fiber-rich foods or more digestible carbohydrate sources could have been important modulators of the species-specific microbiome arrangements seen in primates. Thus, it is likely that, from a common ancestor, the species-specific microbiome patterns observed in primates today were triggered when hosts occupied diverse dietary niches during their adaptive radiation. If this is the case, these data provide evidence on how seasonality and diet–microbiome interactions were important factors influencing gut microbiome composition and function in primates, with direct implications for unraveling the forces that impacted human evolution. Along these lines, we emphasize the need for detailed characterizations of changes in diet, host gene expression and diet–microbe metabolic interactions from ecological (temporal/spatial) frameworks. This approach has great promise to weigh the influence of extrinsic (environmental) and intrinsic (host-genetic) factors in shaping primate gut microbiomes, in light of human health and evolution.
Acknowledgments
We thank the government of the Central African Republic, namely the Ministre de l'Education Nationale, de l'Alphabetisation, de l'Enseignement Superieur, et de la Recherche for providing research permits to conduct our work in the Central African Republic; World Wildlife Fund and the administration of DSPA for granting research approval and for assistance with obtaining permits; and the Primate Habituation Programme for providing logistical support in the field. We especially thank Dr Paul Garber (University of Illinois) and two anonymous reviewers for very important comments on this manuscript. Finally, we thank all of our field assistants and trackers for their help in the field. This work was supported by a Grant Agency of the Czech Republic (number 206/09/0927), NSF grant 0935347 and COLCIENCIAS (Colombian Agency for Science and Technology) and its doctoral fellowship program. This publication derives from the HPL-lab, Laboratory for Infectious Diseases Common to Humans and (non-Human) Primates, Czech Republic. This study was supported by the project ‘CEITEC- Central European Institute of Technology' (CZ.1.05/1.100/02.0068) from European Regional Development Fund and further co-financed from European Social Fund and state budget of the Czech Republic (project OPVK CZ.1.07/2.3.00/20.0300). The research in Bwindi was supported by Hunter College. We thank Hillary Musinguzi and the park rangers of Bwindi for assisting with sample collection, and Pontious Ezuma, Chemonges Amusa and Simplicious Gessa for logistical support. The Uganda Wildlife Authority and the Uganda National Council for Science and Technology gave JMR permission to conduct this research.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM et al. (2014. a). The gut microbiota appears to compensate for seasonal diet variation in the wild Black Howler Monkey (Alouatta pigra). Microb Ecol 69: 434–443. [DOI] [PubMed] [Google Scholar]
- Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM et al. (2014. b). The role of gut microbes in satisfying the nutritional demands of adult and juvenile wild, black howler monkeys (Alouatta pigra). Am J Phys Anthropol 155: 652–664. [DOI] [PubMed] [Google Scholar]
- Barks SK, Calhoun ME, Hopkins WD, Cranfield MR, Mudakikwa A, Stoinski TS et al. (2015). Brain organization of gorillas reflects species differences in ecology. Am J Phys Anthropol 156: 252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrensmeyer AK. (2006). Climate change and human evolution. Science(Washington) 311: 476–478. [DOI] [PubMed] [Google Scholar]
- Chapman C, Rothman J, Lambert J. (2012) Food as a selective force in primates. The evolution of primate societies. University Of Chicago Press: Chicago, pp 149–168. [Google Scholar]
- Chung K. (1997) Gastrointestinal Toxicology of Monogastrics. Mackie R, White B (eds).Gastrointestinal Microbiology. Heidelberg: Springer, 511–582. [Google Scholar]
- Clarke K. (1993). Nonparametric multivariate analyses of changes in community structure. Aust J Ecol 18: 117–143. [Google Scholar]
- Collins A, Dubach J. (2000). Biogeographic and ecological forces responsible for speciation in Ateles. Int J Primatol 21: 421–444. [Google Scholar]
- Cordain L, Eaton SB, Sebastian A, Mann N, Lindeberg S, Watkins BA et al. (2005). Origins and evolution of the Western diet: health implications for the 21st century. Am J Clin Nutr 81: 341–354. [DOI] [PubMed] [Google Scholar]
- Core Team R. (2014) R Core Team (2014). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: Austria. Available from: http://www.R-project.org/. [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran D, McNeilage A. (1998). Gorilla ecology and behavior. Evol Anthropol 6: 120–131. [Google Scholar]
- Doran-Sheehy D, Boesch C. (2004). Behavioral ecology of western gorillas: New insights from the field. Am J Primatol 64: 139–143. [DOI] [PubMed] [Google Scholar]
- Dufrene M, Legendre P. (1997). Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol Monogr 67: 345–366. [Google Scholar]
- Flint HJ, Bayer EA. (2008). Plant cell wall breakdown by anaerobic microorganisms from the mammalian digestive tract. Ann N Y Acad Sci 1125: 280–288. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. (2008). Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6: 121–131. [DOI] [PubMed] [Google Scholar]
- Ganas J, Robbins MM, Nkurunungi JB, Kaplin BA, McNeilage A. (2004). Dietary variability of mountain gorillas in Bwindi Impenetrable National Park, Uganda. Int J Primatol 25: 1043–1072. [Google Scholar]
- Garner CE, Smith S, de Lacy Costello B, White P, Spencer R, CSJ Probert et al. (2007). Volatile organic compounds from feces and their potential for diagnosis of gastrointestinal disease. FASEB J 21: 1675–1688. [DOI] [PubMed] [Google Scholar]
- Gerard P. (2010) Gastrointestinal Tract: Microbial Metabolism of Steroids In: Springer: Berlin, Heidelberg, pp 3133–3140. [Google Scholar]
- Giraudoux P. (2014), pgirmess: Data Analysis in Ecology http://CRAN.R-project.org/package=gplots R package version 1.5.9.
- Gomez A, Petrzelkova K, Yeoman CJ, Vlckova K, Mrázek J, Koppova I et al. (2015). Gut microbiome composition and metabolomic profiles of wild western lowland gorillas (Gorilla gorilla gorilla) reflect host ecology. Mol Ecol 24: 2551–2565. [DOI] [PubMed] [Google Scholar]
- Gomez JM, Verdu M. (2012). Mutualism with plants drives primate diversification. Syst Biol 61: 567–577. [DOI] [PubMed] [Google Scholar]
- Henry AG, Brooks AS, Piperno DR. (2011). Microfossils in calculus demonstrate consumption of plants and cooked foods in Neanderthal diets (Shanidar III, Iraq; Spy I and II, Belgium). Proc Natl Acad Sci USA 108: 486–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrobin DF. (1999). Lipid metabolism, human evolution and schizophrenia. Prostaglandins Leukot Essent Fatty Acids 60: 431–437. [DOI] [PubMed] [Google Scholar]
- Indahl UG, Liland KH, Næs T. (2009). Canonical partial least squares-a unified PLS approach to classification and regression problems. J Chemometrics 23: 495–504. [Google Scholar]
- Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR. (2008). Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA 105: 13580–13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott C. (1998). Changes in orangutan caloric intake, energy balance, and ketones in response to fluctuating fruit availability. Int J Primatol 19: 1061–1079. [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS et al. (2008). Evolution of mammals and their gut microbes. Science 320: 1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Bik EM, Costello EK, Dethlefsen L, Haque R, Relman DA et al. (2013). Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PloS one 8: e53838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FJ, Dumas M, Wang Y, Legido-Quigley C, IKS Yap, Tang H et al. (2007). A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Sys Biol 3: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masi S, Chauffour S, Bain O, Todd A, Guillot J, Krief S. (2012). Seasonal effects on great ape health: a case study of wild chimpanzees and Western gorillas. PLoS One 7: e49805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masi S, Cipolletta C, Robbins MM. (2009). Western lowland gorillas (Gorilla gorilla gorilla) change their activity patterns in response to frugivory. Am J Primatol 71: 91–100. [DOI] [PubMed] [Google Scholar]
- Masi S. (2007). , Seasonal influence on foraging [strategies], activity and energy budgets of Western Lowland gorillas (gorilla gorilla gorilla) in Bai-Hokou, Central African Republic. Ph.D. Thesis. Italy: University of Rome "La Sapienza"..
- McGrew WC. (2007). Savanna chimpanzees dig for food. Proc Natl Acad Sci USA 104: 19167–19168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton K. (1993). Diet and primate evolution. Sci Am 269: 86–93. [DOI] [PubMed] [Google Scholar]
- Moco S, Martin FJ, Rezzi S. (2012). Metabolomics view on gut microbiome modulation by polyphenol-rich foods. J Proteome Res 11: 4781–4790. [DOI] [PubMed] [Google Scholar]
- Moeller A, Peeters M, Ndjango J, Li Y, Hahn B, Ochman H. (2013). Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome Res 23: 1715–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L et al. (2011). Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332: 970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete A, van Schaik CP, Isler K. (2011). Energetics and the evolution of human brain size. Nature 480: 91–93. [DOI] [PubMed] [Google Scholar]
- Nenadic O, Greenacre M. (2007). Correspondence analysis in R, with two- and three-dimensional graphics: The ca package. J Stat Software 20. [Google Scholar]
- Obregon-Tito A, Tito RY, Metcalf J, Sankaranarayanan K, Clemente JC, Ursell LK et al. (2015). Subsistence strategies in traditional societies distinguish gut microbiomes. Nat Commun 6: 6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, Hahn BH et al. (2010). Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol 8: e1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Guillaume Blanchet F, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2012), vegan: Community Ecology Package http://CRAN.R-project.org/package=vegan R package version 2.0-5.
- Pena MJ, Ryden P, Madson M, Smith AC, Carpita NC. (2004). The galactose residues of xyloglucan are essential to maintain mechanical strength of the primary cell walls in Arabidopsis during growth. Plant Physiol 134: 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pantoja D, Donoso R, Agulló L, Córdova M, Seeger M, Pieper DH et al. (2012). Genomic analysis of the potential for aromatic compounds biodegradation in Burkholderiales. Environ Microbiol 14: 1091–1117. [DOI] [PubMed] [Google Scholar]
- Pope PB, Smith W, Denman SE, Tringe SG, Barry K, Hugenholtz P et al. (2011). Isolation of Succinivibrionaceae Implicated in Low Methane Emissions from Tammar Wallabies. Science 333: 646–648. [DOI] [PubMed] [Google Scholar]
- Reiner WB, Petzinger C, Power ML, Hyeroba D, Rothman JM. (2014). Fatty acids in mountain gorilla diets: implications for primate nutrition and health. Am J Primatol 76: 281–288. [DOI] [PubMed] [Google Scholar]
- Remis MJ. (1997. a). Ranging and grouping patterns of a western lowland gorilla group at Bai Hokou, Central African Republic. Am J Primatol 43: 111–133. [DOI] [PubMed] [Google Scholar]
- Remis MJ. (1997. b). Western lowland gorillas (Gorilla gorilla gorilla) as seasonal frugivores: Use of variable resources. Am J Primatol 43: 87–109. [DOI] [PubMed] [Google Scholar]
- Remis M, Dierenfeld E, Mowry C, Carroll R. (2001). Nutritional aspects of western lowland gorilla (Gorilla gorilla gorilla) diet during seasons of fruit scarcity at Bai Hokou, Central African Republic. Int J Primatol 22: 807–836. [Google Scholar]
- Remis M. (1997. c). Western lowland gorillas (Gorilla gorilla gorilla) as seasonal frugivores: Use of variable resources. Am J Primatol 43: 87–109. [DOI] [PubMed] [Google Scholar]
- Revelle W. (2014). psych: Procedures for Psychological, Psychometric, and Personality Research. R package version 1.4.5.
- Robbins MM. (2011) Gorillas: diversity in ecology and behavior. In: Primates in Perspective. Oxford University Press: : New York, US, pp 326–339. [Google Scholar]
- Roberts DW. (2012). labsdv: Ordination and Multivariate Analysis for Ecology. Available from: http://ecology.msu.montana.edu/labdsv/R.
- Rogers ME, Abernethy K, Bermejo M, Cipolletta C, Doran D, McFarland K et al. (2004). Western gorilla diet: a synthesis from six sites. Am J Primatol 64: 173–192. [DOI] [PubMed] [Google Scholar]
- Rogers M, Maisels F, Williamson E, Fernandez M, Tutin C. (1990). Gorilla Diet in the Lope Reserve, Gabon - a Nutritional Analysis. Oecologia 84: 326–339. [DOI] [PubMed] [Google Scholar]
- Rothman JM, Nkurunungi JB, Shannon BF, Bryer MA. (2014). High altitude diets: implications for the feeding and nutritional ecology of mountain gorillas. High Altitude Primates. Springer: New York, NY, USA, pp 247–264. [Google Scholar]
- Rothman JM, Pell AN, Nkurunungi JB, Dierenfeld ES. (2006) Nutritional aspects of the diet of wild gorillas Primates of Western Uganda. Springer: New York, NY, USA, pp 153–169. [Google Scholar]
- Rothman JM, Raubenheimer D, Chapman CA. (2011). Nutritional geometry: gorillas prioritize non-protein energy while consuming surplus protein. Biol Lett 7: 847–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JM, Dierenfeld ES, Hintz HF, Pell AN. (2008). Nutritional quality of gorilla diets: consequences of age, sex, and season. Oecologia 155: 111–122. [DOI] [PubMed] [Google Scholar]
- Rothman JM, Plumptre AJ, Dierenfeld ES, Pell AN. (2007). Nutritional composition of the diet of the gorilla (Gorilla beringei): a comparison between two montane habitats. J Trop Ecol 23: 673–682. [Google Scholar]
- Rothman J, Chapman C, Soest P. (2012). Methods in Primate Nutritional Ecology: A Userâ•™s Guide. Int J Primatol 33: 542–566. [Google Scholar]
- Sanders JG, Powell S, Kronauer DJC, Vasconcelos HL, Frederickson ME, Pierce NE. (2014). Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol Ecol 23: 1268–1283. [DOI] [PubMed] [Google Scholar]
- Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J et al. (2012). Insights into hominid evolution from the gorilla genome sequence RID D-2982-2009. Nature 483: 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16 S rRNA-based studies. PLoS ONE 6: e27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G et al. (2014). Gut microbiome of the Hadza hunter-gatherers. Nat Commun 5: 3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. (2003). Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford CB, Nkurunungi JB. (2003). Behavioral ecology of sympatric chimpanzees and gorillas in Bwindi Impenetrable National Park. Int J Primatol 24: 901–918. [Google Scholar]
- Sussman RW. (1991). Primate origins and the evolution of angiosperms. Am J Primatol 23: 209–223. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE et al. (2009). A core gut microbiome in obese and lean twins. Nature 457: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DP. (1984). Composition and variability of mountain gorilla diets in the Central Virungas. Am J Primatol 7: 323–356. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y, Keilbaugh SA et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 333: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Psychogios N, Young N, Wishart DS. (2009). MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res 37: W652–W660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M et al. (2012). Human gut microbiome viewed across age and geography. Nature 486: 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim S, Yeoman CJ, Sipos M, Torralba M, Wilson BA, Goldberg TL et al. (2010). Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS One 5: e13963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.