Abstract
Beetroot (甜菜 tián cài) juice consumption is of current interest for improving aerobic performance by acting as a vasodilator and possibly through alterations in skeletal muscle metabolism and physiology. This work explored the effects of a commercially available beetroot supplement on metabolism, gene expression, and mitochondrial content in cultured myocytes. C2C12 myocytes were treated with various concentrations of the beetroot supplement for various durations. Glycolytic metabolism and oxidative metabolism were quantified via measurement of extracellular acidification and oxygen consumption, respectively. Metabolic gene expression was measured using quantitative reverse transcription–polymerase chain reaction, and mitochondrial content was assessed with flow cytometry and confocal microscopy. Cells treated with beetroot exhibited significantly increased oxidative metabolism, concurrently with elevated metabolic gene expression including peroxisome proliferator-activated receptor gamma coactivator-1 alpha, nuclear respiratory factor 1, mitochondrial transcription factor A, and glucose transporter 4, leading to increased mitochondrial biogenesis. Our data show that treatment with a beetroot supplement increases basal oxidative metabolism. Our observations are also among the first to demonstrate that beetroot extract is an inducer of metabolic gene expression and mitochondrial biogenesis. These observations support the need for further investigation into the therapeutic and pharmacological effects of nitrate-containing supplements for health and athletic benefits.
Keywords: dietary supplement, ergogenic, mitochondrial uncoupling protein 3 (UCP3), nitrate, mitochondrial efficiency, peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α)
Graphical abstract
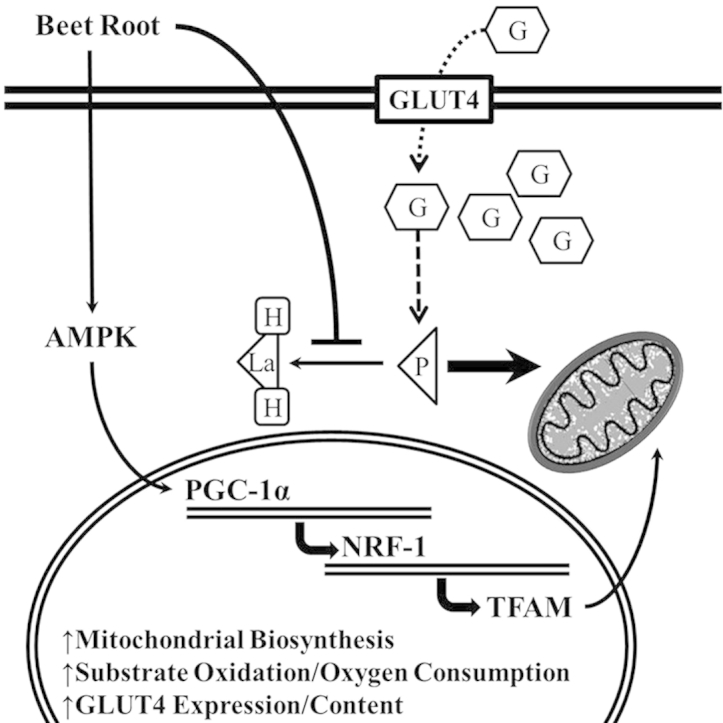
Effects of beet root supplement on myocyte metabolism. Treatment of myocytes with beetroot increases AMPK and PGC-1α expression, leading to heightened mitochondrial content and oxidative metabolism. Note. Dark bold arrow indicates increased substrate oxidation; light arrow indicates decreased substrate oxidation; short dashed line indicates glucose uptake; long dashed arrow indicates glucose metabolism. AMPK = 5′ adenosine monophosphate-activated protein kinase; G = glucose; GLUT4 = glucose transporter 4; La = lactate; NRF-1 = nuclear respiratory factor 1; P = pyruvate; PGC-1α = peroxisome proliferator-activated receptor γ coactivator 1α.
1. Introduction
Dietary sources of nitrate are found in a variety of foods including roots and green leafy vegetables, and more recently, some cured/processed meats. Dietary sources of nitrate have become of interest for ergogenic and metabolic purposes because of the ability of dietary nitrate to increase nitric oxide (NO) biosynthesis.1 Nitrate consumption results in heighted plasma nitrite, which has been previously recognized as an in vivo biomarker of NO production.2, 3 Consumption of nitrate-rich foods prior to endurance events including cycling, rowing, and running results in improved performance,4, 5, 6 resulting in increasing popularity of nitrate-containing foods.7, 8, 9, 10, 11, 12, 13 As a result, commercially available dietary products high in nitrate content have emerged as ergogenic aids purported to increase athletic performance.
NO is a potent cellular stimulus capable of increasing intracellular Ca2+ and cyclic guanosine monophosphate (cGMP) leading to activation of Ca2+/calmodulin-dependent protein kinase and 5′-adenosine monophosphate-activated protein kinase (AMPK). Dietary NO precursors including arginine and arginine-like metabolites increase NO, stimulating phosphorylation and activation of AMPK, which later acts to increase the expression and phosphorylation of peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α).14, 15 PGC-1α acts as a master metabolic regulator of energy metabolism and mitochondrial biogenesis by controlling the expression of nuclear respiratory factor (NRF) and its downstream target, mitochondrial transcription factor A (TFAM).16, 17, 18, 19 In adipocytes, arginine was shown to increase mitochondrial content under ambient conditions, and intensify cold-induced adipocyte browning.20
Despite the heightened interest in nitrate consumption, findings about the affects of beetroot (甜菜 tián cài; BR) supplementation have been inconsistent.10, 21, 22, 23 The primary aim of this work is to characterize the effects a commercially available BR supplement beverage on metabolism and related metabolic gene expression in skeletal muscle.
2. Materials and methods
2.1. Cell culture
Murine myocytes (C2C12) were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco's modified Eagle's medium containing 4500 mg/L glucose and supplemented with 10% heat-inactivated fetal bovine serum and 100 U/mL penicillin/streptomycin, in a humidified 5% CO2 atmosphere at 37 °C (standard conditions). Following overnight seeding, cells were treated with various concentrations of the nitrate-containing commercially available BEET IT Pro-Elite Shot from James White Drinks Ltd (Ipswich, UK) for 24 hours (ingredients are displayed in Table 1).
Table 1.
Food fact panel of BEET IT Pro-Elite Shot from James White Drinks Ltd (Ipswich, UK).
| Serving Size | 70ml/2.3fl oz |
| Servings per container | 1 |
| Energy/Calories | 306kj/72kcal |
| Protein | 2.5 g |
| Total Carbohydrates | 16 g |
| Sugars | 16 g |
| Total fat | <0.1 g |
| Fiber | <0.5 g |
| Sodium | <0.1 g |
| Dietary Nitrate | 0.4 g |
| Cholesterol | 0 g |
| Vitamin A | 0%* |
| Vitamin C | 7.20%* |
| Calcium | 1.20%* |
| Iron | 2.80%* |
* % daily values based on a 2000 calorie diet.
2.2. Determination of BEET-IT nitrate and nitrite content
BEET IT Pro-Elite's nitrate/nitrite content was quantified using the nitrate/nitrite quantification assay kit from Cayman Chemical (Ann Arbor, MI, USA). Nitrate and nitrite were quantified according to the manufacturer's protocol. Briefly, nitrate was quantified by diluting samples in an assay buffer, followed by 1 hour of incubation at room temperature with nitrate reductase and vitamin cofactors. Griess reagents were then added in equivocal volume to each well and incubated for 10 min at room temperature, and then sample absorbance was measured at 540 nm. The beverage nitrate concentrations were 5.312 ± 0.306μM, with undetectable nitrite. All treatments were standardized to determined nitrate concentrations of 20, 10, and 5μM, which correspond with 0.5%, 0.25%, and 0.125% BR, respectively, by volume (determined during preliminary cell viability and mitochondrial staining studies).
2.3. Metabolic assay
Cells were seeded overnight in 24-well culture plate from SeaHorse Bioscience (Billerica, MA, USA) at a density of 5 × 105 cells/well, and treated with either control medium or media containing BR in dilutions corresponding with 0.5%, 0.25%, and 0.125% BR by volume for 24 hours. Following treatment, culture media was removed and replaced with XF Assay Media from SeaHorse Bioscience containing 4500 mg/L glucose free of CO2 and briefly incubated at 37 °C. As per the manufacturer's protocol, SeaHorse injection ports were loaded with oligomycin, an inhibitor of ATP synthase that induces maximal glycolytic metabolism and reveals endogenous proton leak (mitochondrial uncoupling) at a final concentration of 1.0 μM. Oligomycin addition was followed by the addition of carbonyl cyanide p-[trifluoromethoxy]-phenyl-hydrazone, an uncoupler of electron transport that induces peak oxygen consumption (an indirect indicator of peak oxidative metabolism) at final concentration of 1.25μM. Rotenone was then added in 1.0μM final concentration to reveal nonmitochondrial respiration and end the metabolic reactions.24, 25 Extracellular acidification, an indirect measure of glycolytic capacity, and oxygen consumption, a measure of oxidative metabolism, was measured using the SeaHorse XF24 Extracellular Analyzer from SeaHorse Bioscience. SeaHorse XF24 Extracellular Analyzer was run using 8-minute cyclic protocol commands (mix for 3 minutes, let stand for 2 minutes, and measure for 3 minutes) in triplicate as previously described.26
2.4. Quantitative real-time polymerase chain reaction
Following a 24-hour treatment with either the control medium or media containing BR in dilutions corresponding with 0.5%, 0.25%, and 0.125% BR by volume for various durations up to 24 hours, total RNA was extracted using the RNeasy Kit from Qiagen (Valencia, CA, USA) and cDNA was synthesized using the Retroscript RT kit from Ambion (Austin, TX, USA) according to manufacturer's instructions. Polymerase chain reaction (PCR) primers were designed using the Primer Express software from Invitrogen (Carlsbad, CA, USA) and synthesized by Integrated DNA Technologies (Coralville, IA, USA). Amplifications of PGC-1α, NRF1, TFAM, glucose transporter 4 (GLUT4), and mitochondrial uncoupling protein 3 (UCP3) were normalized to the housekeeping gene, TATA binding protein (TBP). Table 2 summarizes the forward and reverse primers of each gene. Quantitative reverse transcription- polymerase chain reactions were performed in triplicate using the LightCycler 480 real-time PCR system from Roche Applied Science (Indianapolis, IN, USA). SYBR Green based PCR was performed in triplicate with final primer concentrations at 10μM in a total volume of 30 μL. The following cycling parameters were used: 95°C for 10 minutes followed by 45 cycles of 95°C for 15 seconds, and 60°C for 1 minute. Relative expression levels were determined using the ΔΔCp method and compared to the lowest expressing group as previously described.27
Table 2.
Summary of qRT-PCR primers from Integrated DNA Technologies (Coralville, IA, USA).
| Primer Name | Forward Sequence | Reverse Sequence |
|---|---|---|
| GLUT4 | 5′-GGAGGGAGCCTTTGGTATTT-3′ | 5′-CAGGCGAGGACACTCATCTT-3′ |
| NRF1 | 5′-ACCCTCAGTCTCACGACTAT-3′ | 5′-GAACACTCCTCAGACCCTTAAC-3′ |
| PGC-1α | 5′-GACAATCCCGAAGACACTACAG-3′ | 5′-AGAGAGGAGAGAGAGAGAGAGA-3′ |
| TBP | 5′-GGGATTCAGGAAGACCACATA-3′ | 5′-CCTCACCAACTGTACCATCAG-3′ |
| TFAM | 5′-GAAGGGAATGGGAAAGGTAGAG-3′ | 5′-ACAGGACATGGAAAGCAGATTA-3′ |
GLUT4 = glucose transporter 4; NRF1 = nuclear respiratory factor 1; PGC-1α = peroxisome proliferator-activated receptor γ coactivator 1α; qRT-PCR = quantitative reverse transcription-polymerase chain reaction; TFAM = mitochondrial transcription factor A; TBP = TATA binding protein.
2.5. Flow cytometry
Cells were seeded in six-well plates at a density of 1.0 × 106 cells/well and treated with either control medium or media containing BR in dilutions corresponding with 0.5%, 0.25%, and 0.125% BR by volume for 24 hours. Following incubation, the medium was removed and the cells were resuspended in prewarmed media with 200nM Mitotracker Green from Life Technologies (Carlsbad, CA, USA) and incubated for 45 minutes in a humidified 5% CO2 atmosphere at 37°C. The cells were pelleted, the media with Mitotracker was removed, and the cells were suspended in prewarmed media. Group mean fluorescence was measured using Facscalibur filtering 488 nm.
2.6. Immunofluorescence and confocal microscopy
To investigate the protein expression of metabolic proteins PGC-1α, cytochrome C (Cyt C), GLUT4, and UCP3, cells were seeded in chamber slides obtained from BD Bioscience (Sparks, MD, USA) with 5000 cells/well and treated with the media containing 0.5% BR by volume for 24 hours. Cells were fixed using 3.7% formaldehyde in media, permeabilized with phosphate-buffered saline (PBS) with 0.1% Triton 100X from Sigma (St. Louis, MO, USA) for 10 minutes, and blocked for 1 hour with PBS with 0.1% Triton 100X and 3.0% bovine serum albumin (BSA) purchased from Sigma. Cells were stained with either an anti-PGC-1α, anti-Cyt C from Santa Cruz Biotechnologies (Santa Cruz, CA, USA), or an anti-GLUT4 or anti-UCP3 primary polyclonal-antibody from Abcam (Cambridge, MA, USA) at 1:200 dilution in PBS with 0.1% BSA overnight. The cells were rinsed with PBS with 0.1% Triton 100X and 3.0% BSA, and secondary AlexFluor 488 or AlexFluor 633 from Invitrogen was applied at 1:200 dilution for 2 hours. Slides were mounted with Prolong Gold with DAPI from Invitrogen, cured overnight, and imaged using the Axiovert 25 microscope with AxioCam MRc from Zeiss (Thornwood, NY, USA).
2.7. Immunoblotting and protein expression
Cells were treated with and without media containing 0.5% BR by volume for 24 hours. Whole-cell lysates were prepared by harvesting the cells on ice in high salt lysis buffer (25mM Tris base, 8mM MgCl2, 1mM dithiothreitol, 15% glycerol, 0.1% Triton 100X) supplemented with protease inhibitor mix (Sigma), followed by incubation on ice for 60 minutes. The insoluble material was removed via centrifugation at 17,500 × g for 3 minutes, and protein concentrations were determined using Bradford assay (Protein Assay Dye Reagent Concentrate; Bio-Rad Laboratories, Hercules, CA, USA). Total protein (40 μg per sample) was size-separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and electrotransferred to nitrocellulose membranes. After blocking in TBST–5% nonfat milk powder for 1 hour, membranes were probed at 4°C for 24 h with an anti-PGC-1α or anti-AMPK primary polyclonal antibody from Santa Cruz Biotechnologies, and an anti-β-actin primary monoclonal antibody from Sigma in TBST–1% nonfat milk powder overnight. Bound antibodies were detected by horseradish peroxidase-conjugated secondary antibodies from Sigma and by chemiluminescence using the ECL Plus Western Blotting Detection kit from GE Healthcare Life Sciences (Little Chalfont, Buckinghamshire, UK). Signal intensities were obtained by densitometry using the ImageJ software (available from the NIH at http://rsbweb.nih.gov/ij/) by quantifying lane intensities followed by normalizing intensity with corresponding β-actin expression.
2.8. Cell viability
Cells were seeded in 96-well plates at a density of 5000 cells/well and grown overnight. Cells were treated and incubated with either control medium or media with BR ranging from 0.0025% to 25% by volume for 24 h hours. The medium and treatment were removed, and media containing 10% WST1 was added to each well and incubated. Fluorescence was measured 1 hour following WST1 addition using Wallac Victor3V 1420 Multilabel Counter from PerkinElmer (Waltham, MA, USA).
2.9. Statistical analyses
Metabolic measurements, flow cytometry, and cell viability data were analyzed using analysis of variance with Dunnett's pairwise comparisons. Gene expression was quantified by relative expression using the ΔΔCp method, and analyzed using analysis of variance with Dunnett's pairwise comparisons.27 Confocal microscopy images (performed using 3 randomly selected images), and Western blots were assessed for cellular fluorescence determined by image J quantification and analyzed using Student t tests. All data are presented as average ± standard deviation normalized to the control mean (control = 100) with *, **, and *** indicating p < 0.05, p < 0.01, and p < 0.001 statistical differences compared to control, respectively.
3. Results
3.1. BR enhances myocyte metabolism
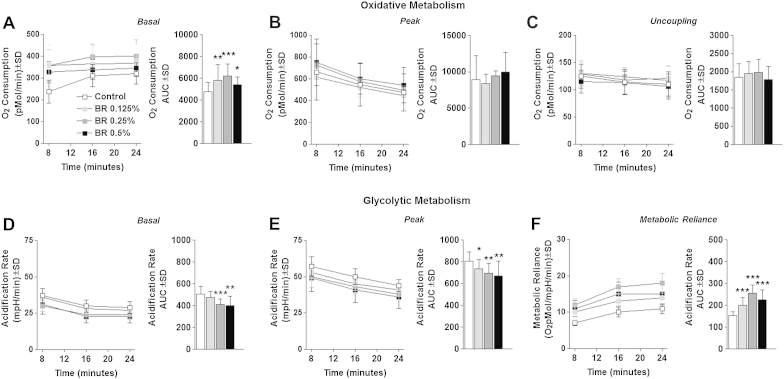
To investigate the effects of BR on myocyte metabolism, we first measured oxygen consumption rate (OCR) following treatment with and without BR in dilutions corresponding with 0.5%, 0.25%, and 0.125% BR by volume for 24 hours. Basal OCR was significantly elevated above control levels in cells treated with BR at all tested volumes, with 0.25% BR yielding the greatest increase in basal oxidative metabolism (Fig. 1A). Despite the significant elevations in basal oxygen consumption, BR-treated cells exhibited no change in chemically induced peak oxidative metabolism (Fig. 1B), or mitochondrial proton leak (uncoupling) (Fig. 1C). To further investigate the effects of BR supplementation on myocyte metabolism, we measured the extracellular acidification rate (ECAR), an indirect measure of glycolytic metabolism. Cells treated with 0.5% or 0.25% BR exhibited significantly reduced basal and peak glycolytic metabolism compared with control cells (Fig. 1D and E). To determine the effect of BR treatment on relative metabolic reliance, we compared the OCR/ECAR ratio of BR-treated cells to control cells. Cells treated with BR consistently displayed a significantly increased reliance on oxidative metabolism compared with control cells (Fig. 1F). Taken together, these data suggest that cells treated with BR and grown in high-glucose media develop a greater propensity to completely oxidize glucose, leading to increased oxygen consumption with concurrent reduction in lactate export (acidification).
Fig. 1.
Cellular metabolism. (A) Basal oxygen consumption rate (OCR) of cells treated with 0.5%, 0.25%, or 0.125% beetroot (BR) by volume or control medium for 24 hours. (B) Peak OCR of cells treated as described in the main text. (C) Endogenous proton leak (uncoupling) of cells treated as described in the main text. (D) Basal extracellular acidification rate (ECAR) of cells treated as described in the main text. (E) Peak ECAR of cells treated as described in the main text. (F) Metabolic reliance expressed as the OCR/ECAR ratio of cells treated as described in the main text. Bar graphs are displayed as complete trial area under the curve (AUC). Note. Data are displayed as group mean ± standard deviation. *p < 0.05, **p < 0.01, ***p < 0.001 compared with control. †p < 0.05 between 0.5% and 0.125% BR by volume. ‡p < 0.05 between 0.5% and 0.25% BR by volume.
3.2. BR induces metabolic gene expression
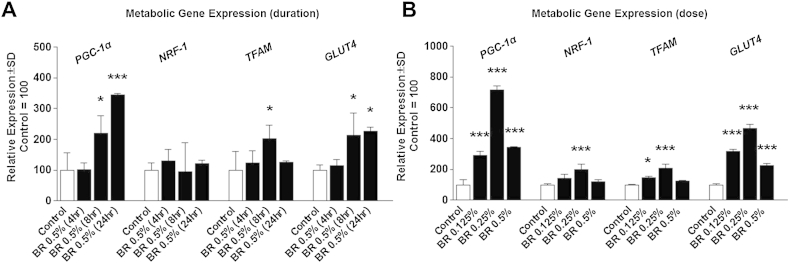
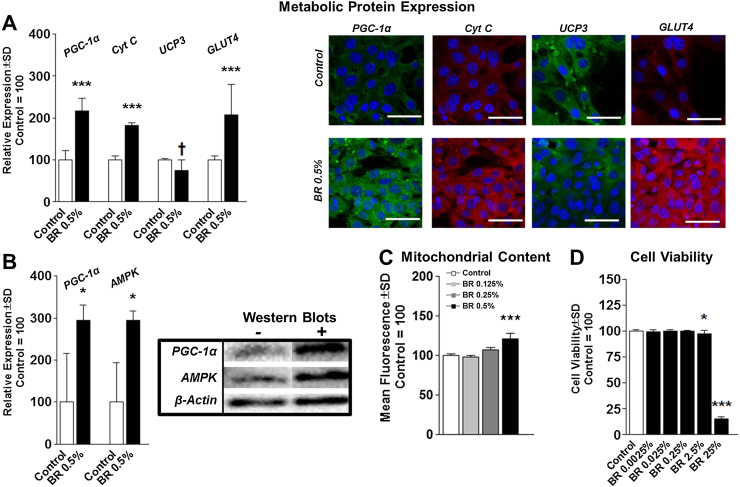
To investigate the effects of BR on metabolic gene expression, we treated cells with various percentages of BR for various durations and assessed the expression of PGC-1α, NRF1, TFAM, GLUT4, and UCP3, which were normalized to the housekeeping gene, TBP. Treatment of myocytes with 0.5% BR by volume induced PGC-1α and GLUT4 expression in a time-dependent manner compared with the corresponding control. TFAM expression increased 8 hours after treatment, prior to returning to basal levels at 24 hours after treatment (Fig. 2A). Interestingly, NRF1 expression was unaltered at all tested time points (Fig. 2A). Additionally, treatment with 0.5%, 0.25%, and 0.125% BR increased PGC-1α and GLUT4 expression following a 24-hour treatment (Fig. 2B). Furthermore, treatment with 0.25% and 0.125% BR increased TFAM expression, whereas only 0.25% BR increased NRF1 expression (Fig. 2B). Mitochondrial UCP3 expression was not altered by BR treatment at any of the tested doses or durations (data not shown). Similar to gene expression, PGC-1α protein expression was significantly elevated following BR treatment, which led to increased Cyt C content, an indirect indicator of increased mitochondrial content (Fig. 3A). Moreover, while UCP3 RNA expression was unchanged in BR-treated cells, UCP3 protein expression was significantly reduced in cells treated with BR (Fig. 3A). Also similar to RNA expression, GLUT4 expression was induced more than 2-fold greater than control cells (Fig. 3A). Additionally PGC-1α and AMPK protein expression were also significantly elevated in BR-treated cells (Fig. 3B). BR activation of metabolic gene expression also resulted in a dose-dependent increase in myocyte mitochondrial content (Fig. 3C). Following a 24-hour treatment, only treatment with 0.5% BR significantly increased mitochondrial content above control levels. In addition, cell viability was only significantly reduced in C2C12 myocytes following treatment with 2.5% BR and greater (Fig. 3D).
Fig. 2.
Metabolic gene expression. (A) Expression of peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α), nuclear respiratory factor 1 (NRF-1), mitochondrial transcription factor A (TFAM), glucose transporter 4 (GLUT4), normalized to the housekeeping gene, TATA binding protein (TBP) following treatment of C2C12 myocytes with 0.5% beetroot (BR) by volume for 4, 8, or 24 hours. (B) Gene expression of C2C12 myocytes following treatment with 0.5%, 0.25%, or 0.125% BR by volume for 24 hours. Note. Data are presented as group mean ± standard deviation. *p < 0.05, **p < 0.01, ***p < 0.001 compared with control. †p < 0.05 between 0.5% and 0.125% BR by volume. ‡p < 0.05 between 0.5% and 0.25% BR by volume.
Fig. 3.
Protein expression and mitochondrial content. (A) Fluorescent protein expression of C2C12 cells treated with 0.5% BR by volume for 24 hours. (B) Quantification of PGC-1α and AMPK Western blots. (C) Mitochondrial content indicated by group mean log fluorescence measured by flow cytometry of C2C12 cells treated with0.5%, 0.25%, or 0.125% BR by volume for 24 hours. (D) Cell viability of C2C12 cells treated with BR beverage containing nitrate at various concentrations for 24 hours. AMPK = 5′ adenosine monophosphate-activated protein kinase; BR = beetroot; PGC-1α = proliferator-activated receptor gamma coactivator-1 alpha. Note. Data are presented as group mean ± standard deviation. *p < 0.05.
4. Discussion
Our observations are among the first to demonstrate that a BR supplement beverage can stimulate myocyte metabolism in a favorable fashion by increasing mitochondrial biogenesis and oxidative metabolism in a PGC-1α-specific fashion. Because our experiments were conducted under high glucose conditions, increased oxygen consumption with concurrent reductions in glycolytic acidification suggest that BR-treated myocytes undergo a more complete carbohydrate oxidation. Additionally, our data also demonstrate that BR can also promote GLUT4 expression, (and although speculative) nitrate-treated myocytes may have enhanced glucose transport and possibly insulin sensitivity in high glucose (diabetic) conditions (observations that should be verified experimentally in vivo). Our observations also demonstrate that BR containing nitrate increases myocyte mitochondrial efficiency in a dose-dependent manner, which is ultimately accompanied by an increase in total mitochondrial content; specifically increased basal mitochondrial metabolism without increased mitochondrial density following supplementation with 0.25% and 0.125% BR by volume. Similarly, previous observations by Larsen et al10 showed that inorganic dietary nitrate consumption in male human volunteers led to enhanced mitochondrial efficiency with suppressed UCP3 content in biopsied muscle. Despite reductions in UCP3 content, isolated mitochondria also displayed unaltered mitochondrial coupling indicated by oxygen consumption.10 Surprisingly, Larsen also reported no change in isolated mitochondrial oxygen consumption. Additionally, Larsen et al10 reported no change in PGC-1α or TFAM RNA content or mitochondrial content. Moreover and contrary to our findings, Larsen et al11 previously found that treatment of primary myotubes with sodium nitrate decreased oxygen consumption. Perhaps most interesting, sodium nitrate consumption reduced resting energy expenditure in healthy individuals.10 This is a meaningful finding because it suggests that despite some potentially favorable metabolic adaptations induced by nitrate-containing foods, dietary nitrate consumption (BR) may prove counterproductive to those with existing metabolic disease such as obesity.
Despite some similarities, Larsen et al's investigation used isolated mitochondria from primary myocytes, which may account for some of the discrepancies between the two findings. Moreover, Larsen and colleagues used inorganic nitrate consumed orally, likely explaining some of the other discrepancies between our findings. Previous in vitro experiments with dietary NO precursors demonstrate that NO increases AMPK activation, leading to heightened expression of PGC-1α, and potentially mitochondrial content.14, 15 On a related note, Mo et al28 previously demonstrated that treatment of primary rat aortic smooth muscle cells with 50μM nitrate mediates mitochondrial biogenesis via the activation of sirtuin 1 and PGC-1α by AMPK phosphorylation. Discrepancies between our observations and those made by Mo and colleagues may be a function of different experimental models or differences in nitrate type/source. One possible explanation for this difference may be related to nitrate bioactivation and subsequent bioavailability (considered biologically inert). Given that our experimental approach lacked commensal bacteria (microbes that express nitrate reductases catalyzing the conversation of the nitrate anion to nitrite), it is possible that an alternative mechanism exists that promotes nitrate-to-nitrite conversion of BR extract. Jansson et al29 reported that in vitro mammalian tissues (gastrointestinal, liver, kidney, heart, and lung) have nitrate reductase activities, notably higher in rodents. Using allopurinol (inhibitor of xanthine oxidoreductase), the group further documented reduced conversation of nitrate to nitrite, indicating a potential reduction mechanism in vitro.29 Another limitation of our experimental approach is that the BR used in our study was a blend of numerous other dietary food chemicals or properties (in addition to nitrate), any of which may have contributed to our findings. Despite this limitation, our observations document that BR reproducibly increases mitochondrial content and oxygen consumption in cultured myocytes.
5. Conclusion
Nitrate-containing supplements are rapidly becoming a topic of interest for many athletes, especially competitive endurance athletes. Our data support the hypothesis that BR (containing nitrate) consumption may promote a favorable metabolic phenotype in skeletal muscle cells, although the observations should be verified experimentally in vivo.
Conflicts of interest
The authors and contributors of this work declare no conflict of interest.
Acknowledgments
Funding for this study was provided by the University of New Mexico 2013–2014 College of Education Graduate Excellence Award, the University of New Mexico Summer 2012 Office of Graduate Studies Research, and the University of New Mexico Project and Travel Grant. We thank Dr Kristina Trujillo and the University of New Mexico Department of Biochemistry and Molecular Biology for their assistance in this work. We also thank the shared facilities available through the University of New Mexico Health Sciences Center: Flow cytometry data were generated in the Flow Cytometry Shared Resource Center supported by the University of New Mexico Health Sciences Center and the University of New Mexico Cancer Center. Images were generated in the and Cancer Center Fluorescence Microscopy Shared Resource, funded as detailed on: http://hsc.unm.edu/crtc/microscopy/acknowledgement.shtml.
Footnotes
Peer review under responsibility of The Center for Food and Biomolecules, National Taiwan University.
References
- 1.Hord N.G., Tang Y., Bryan N.S. Food sources of nitrates and nitrites: the physiologic context for potential health benefits. Am J Clin Nutr. 2009;90:1–10. doi: 10.3945/ajcn.2008.27131. [DOI] [PubMed] [Google Scholar]
- 2.Kleinbongard P., Dejam A., Lauer T. Plasma nitrite reflects constitutive nitric oxide synthase activity in mammals. Free Radic Biol Med. 2003;35:790–796. doi: 10.1016/s0891-5849(03)00406-4. [DOI] [PubMed] [Google Scholar]
- 3.Lauer T., Preik M., Rassaf T. Plasma nitrite rather than nitrate reflects regional endothelial nitric oxide synthase activity but lacks intrinsic vasodilator action. Proc Natl Acad Sci U S A. 2001;98:12814–12819. doi: 10.1073/pnas.221381098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond H., Morton L., Braakhuis A.J. Dietary nitrate supplementation improves rowing performance in well-trained rowers. Int J Sport Nutr Exerc Metab. 2012;22:251–256. doi: 10.1123/ijsnem.22.4.251. [DOI] [PubMed] [Google Scholar]
- 5.Cermak N.M., Gibala M.J., van Loon L.J.C. Nitrate supplementation's improvement of 10-km time-trial performance in trained cyclists. Int J Sport Nutr Exerc Metab. 2012;22:64–71. doi: 10.1123/ijsnem.22.1.64. [DOI] [PubMed] [Google Scholar]
- 6.Murphy M., Eliot K., Heuertz R.M., Weiss E. Whole beetroot consumption acutely improves running performance. J Acad Nutr Diet. 2012;112:548–552. doi: 10.1016/j.jand.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Bailey S.J., Winyard P., Vanhatalo A. Dietary nitrate supplementation reduces the O-2 cost of low-intensity exercise and enhances tolerance to high-intensity exercise in humans. J Appl Physiol. 2009;107:1144–1155. doi: 10.1152/japplphysiol.00722.2009. [DOI] [PubMed] [Google Scholar]
- 8.Lansley K.E., Winyard P.G., Bailey S.J. Acute dietary nitrate supplementation improves cycling time trial performance. Med Sci Sports Exerc. 2011;43:1125–1131. doi: 10.1249/MSS.0b013e31821597b4. [DOI] [PubMed] [Google Scholar]
- 9.Lansley K.E., Winyard P.G., Fulford J. Dietary nitrate supplementation reduces the O-2 cost of walking and running: a placebo-controlled study. J Appl Physiol. 2011;110:591–600. doi: 10.1152/japplphysiol.01070.2010. [DOI] [PubMed] [Google Scholar]
- 10.Larsen F.J., Schiffer T.A., Borniquel S. Dietary Inorganic nitrate improves mitochondrial efficiency in humans. Cell Metab. 2011;13:149–159. doi: 10.1016/j.cmet.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Larsen F.J., Schiffer T.A., Ekblom B. Dietary nitrate reduces resting metabolic rate: a randomized, crossover study in humans. Am J Clin Nutr. 2014;99:843–850. doi: 10.3945/ajcn.113.079491. [DOI] [PubMed] [Google Scholar]
- 12.Larsen F.J., Weitzberg E., Lundberg J.O., Ekblom B. Effects of dietary nitrate on oxygen cost during exercise. Acta Physiol. 2007;191:59–66. doi: 10.1111/j.1748-1716.2007.01713.x. [DOI] [PubMed] [Google Scholar]
- 13.Larsen F.J., Weitzberg E., Lundberg J.O., Ekblom B. Dietary nitrate reduces maximal oxygen consumption while maintaining work performance in maximal exercise. Free Radic Biol Med. 2010;48:342–347. doi: 10.1016/j.freeradbiomed.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Fu W.J.J., Haynes T.E., Kohli R. Dietary l-arginine supplementation reduces fat mass in Zucker diabetic fatty rats. J Nutr. 2005;135:714–721. doi: 10.1093/jn/135.4.714. [DOI] [PubMed] [Google Scholar]
- 15.Lira V.A., Brown D.L., Lira A.K., Kavazis A.N., Soltow Q.A., Zeanah E.H. Nitric oxide and AMPK cooperatively regulate PGC-1 alpha in skeletal muscle cells. J Physiol Lond. 2010;588:3551–3566. doi: 10.1113/jphysiol.2010.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bostroem P., Wu J., Jedrychowski M.P. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–U472. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knutti D., Kaul A., Kralli A. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol Cell Biol. 2000;20:2411–2422. doi: 10.1128/mcb.20.7.2411-2422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H., Kanatous S.B., Thurmond F.A. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- 19.Wu Z.D., Puigserver P., Andersson U. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 20.Petrovic V., Korac A., Buzadzic B. Nitric oxide regulates mitochondrial re-modelling in interscapular brown adipose tissue: ultrastructural and morphometric–stereologic studies. J Microsc. 2008;232:542–548. doi: 10.1111/j.1365-2818.2008.02132.x. [DOI] [PubMed] [Google Scholar]
- 21.Bescos R., Ferrer-Roca V., Galilea P.A. Sodium nitrate supplementation does not enhance performance of endurance athletes. Med Sci Sports Exerc. 2012;44:2400–2409. doi: 10.1249/MSS.0b013e3182687e5c. [DOI] [PubMed] [Google Scholar]
- 22.Peacock O., Tjonna A.E., James P. Dietary nitrate does not enhance running performance in elite cross-country skiers. Med Sci Sports Exerc. 2012;44:2213–2219. doi: 10.1249/MSS.0b013e3182640f48. [DOI] [PubMed] [Google Scholar]
- 23.Wilkerson D.P., Hayward G.M., Bailey S.J., Vanhatalo A., Blackwell J.R., Jones A.M. Influence of acute dietary nitrate supplementation on 50 mile time trial performance in well-trained cyclists. Eur J Appl Physiol. 2012;112:4127–4134. doi: 10.1007/s00421-012-2397-6. [DOI] [PubMed] [Google Scholar]
- 24.Giulivi C., Ross-Inta C., Horton A.A., Luckhart S. Metabolic pathways in Anopheles stephensi mitochondria. Biochem J. 2008:415. doi: 10.1042/BJ20080973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wikstrom J.D., Sereda S.B., Stiles L. A hovel high-throughput assay for islet respiration reveals uncoupling of rodent and human islets. Plos One. 2012;7 doi: 10.1371/journal.pone.0033023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaughan R.A., Garcia-Smith R., Gannon N.P., Bisoffi M., Trujillo K.A., Conn C.A. Leucine treatment enhances oxidative capacity through complete carbohydrate oxidation and increased mitochondrial density in skeletal muscle cells. Amino Acids. 2013;45(4):901–911. doi: 10.1007/s00726-013-1538-5. [DOI] [PubMed] [Google Scholar]
- 27.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mo L., Wang Y., Geary L. Nitrite activates AMP kinase to stimulate mitochondrial biogenesis independent of soluble guanylate cyclase. Free Radic Biol Med. 2012;53:1440–1450. doi: 10.1016/j.freeradbiomed.2012.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jansson E.A., Huang L., Malkey R. A mammalian functional nitrate reductase that regulates nitrite and nitric oxide homeostasis. Nat Chem Biol. 2008;4:411–417. doi: 10.1038/nchembio.92. [DOI] [PubMed] [Google Scholar]