Abstract
Circulating tumor DNA (ctDNA) in the plasma or serum of cancer patients provides an opportunity for non-invasive sampling of tumor DNA. This ‘liquid biopsy’ allows for interrogations of DNA such as quantity, chromosomal alterations, sequence mutations and epigenetic changes, and can be used to guide and improve treatment throughout the course of the disease. This tremendous potential for real-time ‘tracking’ in a cancer patient has led to substantial research efforts in the ctDNA field. ctDNA can be distinguished from non-tumor DNA by the presence of tumor-specific mutations and copy number variations, and also by aberrant DNA methylation, with both DNA sequence and methylation changes corresponding to those found in the tumor. Aberrant methylation of specific promoter regions can be a very consistent feature of cancer, in contrast to mutations, which typically occur at a wide range of sites. This consistency makes ctDNA methylation amenable to the design of widely applicable clinical assays. In this review, we examine ctDNA methylation in the context of monitoring disease status, treatment response and determining the prognosis of cancer patients.
Keywords: cancer, prognosis, circulating DNA, plasma DNA, methylation
Methylated circulating DNA in cancer prognosis and monitoring
Genomics is anticipated to bring major improvements in the treatment of cancer patients. The capacity to quickly and relatively inexpensively sequence tumor DNA on a genome-wide scale allows the identification of potential molecular targets, assignment of cancer subtype and determination of patient prognosis, while also providing insights into cancer biology that can form the basis of further research. However, there are certain limitations to obtaining sequence information from solid cancers: due to the clonal evolution and heterogeneity of a tumor, a single biopsy may not represent the diversity of DNA changes present; metastasized disease may be difficult to identify and access; and repeated biopsies from a patient are not practicable.
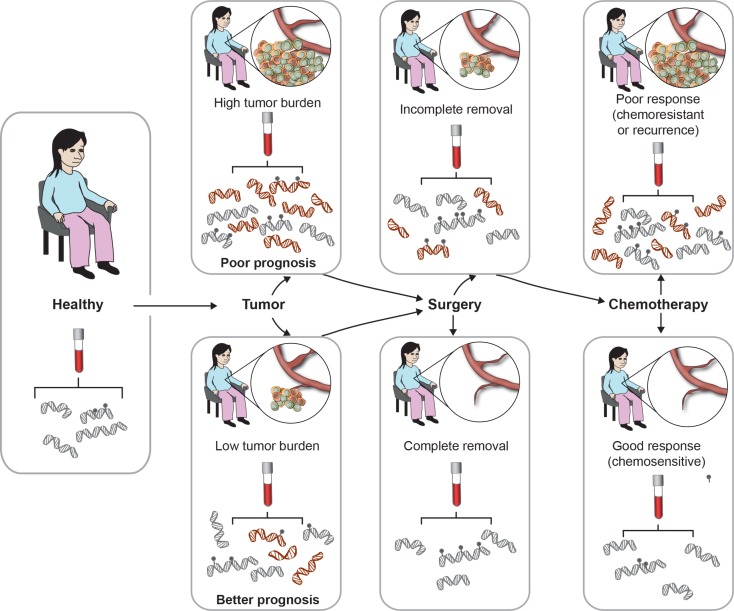
Circulating tumor DNA (ctDNA) containing the same molecular aberrations as the solid tumor is detectable in the bloodstream of many cancer patients (Ignatiadis & Dawson 2014), and sampling it via blood overcomes the problems related to tumor heterogeneity and accessibility. As illustrated in Fig. 1, sequential blood sampling is particularly appealing as it allows ongoing ‘real-time’ monitoring of patients following surgery and during treatment. ctDNA can be distinguished from circulating DNA from healthy cells by the presence of genomic aberrations that correspond to those found in the tumor, such as tumor-specific mutations or methylation. The feasibility of using tumor-specific mutations in ctDNA to monitor the response to therapy has been demonstrated in colorectal (Diehl et al. 2008), breast (Dawson et al. 2013, Murtaza et al. 2013), ovarian (Forshew et al. 2012, Murtaza et al. 2013) and lung (Murtaza et al. 2013) cancers; however, the highly individual nature of tumor DNA mutations (Vogelstein et al. 2013) makes this a very labor intensive approach.
Figure 1.
ctDNA can be used to trace tumor progression and patient response. All individuals carry circulating DNA in their blood. Upon tumor development, ctDNA carrying tumor-specific molecular alterations (such as DNA methylation) is released into the circulation, at levels relative to tumor burden. Following surgery, ctDNA levels reflect removal of the tumor. Throughout chemotherapy treatment, and upon completion, ctDNA can be used to monitor patient response and prognosis.
Methylation of CpG sites at selected DNA sequences provides a level of regulation over gene expression over that which is specified by DNA sequence alone. Genome methylation undergoes coordinated changes at defined stages of development and in response to environmental stimuli such as diet, chemical toxins and pollutants, and temperature stresses (reviewed in Feil & Fraga (2012)). For example, inadequate nutrition levels around early pregnancy decrease methylation of the insulin-like growth factor 2 gene (Heijmans et al. 2008), while exposure to benzene is associated with genome-wide hypomethylation and gene promoter hypermethylation in a pattern overlapping with acute myeloid leukemia, a cancer linked to this pollutant (Bollati et al. 2007). Molecular biology techniques applicable to detection and measurement of methylation have extensively reviewed (Laird 2003), including a recent review with a focus on detecting breast and ovarian cancer (Wittenberger et al. 2014).
Methylation changes are a common feature of different cancer types, and occur early in cancer development, typically repressing the expression of tumor suppressor genes (Baylin & Jones 2011). Aberrant DNA methylation may offer a more consistent and hence broadly applicable marker of tumor DNA in blood than mutations (Warton & Samimi 2015). For example, GSTP1 is methylated in >90% of prostate cancers (Meiers et al. 2007), STRATIFIN is methylated in 96% of breast cancers (Umbricht et al. 2001) and HOXA9 and EN1 are methylated in 95 and 80% of ovarian tumors respectively (Montavon et al. 2012). There is a very large amount of published information describing DNA methylation patterns in tumor tissue and their impact on patient prognosis (Heyn & Esteller 2012). When tumor DNA is shed into the bloodstream these patterns also become detectable in plasma and serum; these blood-based methylated ctDNA biomarkers are the focus of this review.
Cancer-specific ctDNA methylation can be used to quantitate tumor DNA, providing information about the level of tumor burden, as well as reveal the methylation patterns in the tumor. Currently, patients undergoing cancer therapy are routinely monitored by blood tests assaying protein-based biomarkers such as CA-125 for ovarian cancer and prostate-specific antigen (PSA) for prostate cancer (Ludwig & Weinstein 2005); hence DNA methylation-based biomarkers could be incorporated into patient care and management with only very minor changes to clinical practice. Here we consider recent applications of methylated ctDNA in determining cancer prognosis, and in disease monitoring following surgery or during chemotherapy treatment.
Methylated ctDNA as a marker of surgery outcome
ctDNA in the blood has a half-life of ∼2 h (Diehl et al. 2008), thus plasma or serum levels can provide a very rapid measure of changes in tumor status. The persistence of cancer DNA in blood following surgery to remove the tumor likely reflects residual tumor tissue in the body and has been linked to poor prognosis (Diehl et al. 2008; Fig. 1). One approach to determining whether tumor-derived DNA sequences are present is to assay circulating DNA for the presence of tumor-specific mutations. For example, a study by Diehl et al. (2008) found that of 18 subjects undergoing surgery for colorectal cancer, four had no detectable mutated DNA in plasma in the days following surgery and none of these four subjects experienced relapse. Conversely, of the remaining 12 who did have detectable ctDNA, all but one experienced relapse (Diehl et al. 2008).
The above study was based on mutations and used a highly personalized approach, first sequencing four selected genes (KRAS, APC, TP53 and PIK3CA) from each patient's FFPE tumor sample, and then designing probe-based PCR BEAMing assays for detection of the identified mutations in patient plasma (Diehl et al. 2008). While effective, this approach was found to be time consuming (Diehl et al. 2008), and difficult to apply to an extended patient population given the highly individual profile of cancer mutations (Vogelstein et al. 2013).
The labor intensive process of developing circulating DNA assays to match tumor-specific mutations could be bypassed by using an existing library of validated assays representing the most common mutations (Diehl et al. 2008); however, another potential approach to quantitating post-surgery ctDNA is to measure tumor-specific methylation, which is less variable across tumors than mutation. A list of tumor-specific methylated sequences that have been shown to decrease in patient blood following surgery is presented in Table 1, and selected examples are discussed below.
Table 1.
Genes for which promoter methylation in serum or plasma has been shown to decrease following surgery
| Cancer type | Gene | Plasma or serum | Reference |
|---|---|---|---|
| Breast | RARB2 | Plasma | Liggett et al. (2011) |
| MSH2 | |||
| ESR1B | |||
| Gastric | RUNX3 | Serum | Sakakura et al. (2009) |
| Liver | CDKN2A | Plasma | Wong et al. (2003) |
| Esophageal | APC | Serum | Hoffmann et al. (2009) |
CDKN2A methylation is common in liver cancer and has been reported in 73% of hepatocellular carcinoma tumors (Wong et al. 1999). A study by Wong et al. (2003) found methylated CDKN2A in the pre-surgery plasma of 31% of liver cancer patients, with a median methylation index (methylated circulating CDKN2A/total circulating CDKN2A) of 35%. In contrast, the methylation index of eight samples collected postoperatively was 3.5%, reflecting the decrease in ctDNA following removal of the tumor (Wong et al. 2003).
APC methylation has been detected in the tumors of 44–68% of patients with esophageal cancer (Brock et al. 2003, Zare et al. 2009), and is indicative of poor patient outcome (Zare et al. 2009). The prognostic value of APC methylation in pre-operative and post-operative serum was examined in a study of 59 patients undergoing resection for esophageal cancer (Hoffmann et al. 2009). Consistent with previous reports, APC was methylated in 46% of pre-operative patient samples. Pre-operative methylated APC together with methylated DAPK predicted shorter overall survival, possibly by reflecting higher tumor burden at diagnosis. Detection of methylated APC in serum from blood collected 10 days following the operation was significantly associated with the presence of apparent residual tumor after surgery; however, impact on survival was not assessed (Hoffmann et al. 2009).
Liggett et al. (2011) used a 56 gene panel assay (MethDet-56) to identify methylated sequences in the plasma of breast cancer patients that decrease following surgery and tamoxifen treatment. Twenty patients with ER-positive breast cancer had plasma collected prior to and after surgery, and three genes (RARb2, MSH2 and ESR1B promoter) were found to be methylated in significantly more pre-surgery samples than post-surgery samples. RARb2 has also been previously identified as a potential biomarker for breast cancer detection (Hoque et al. 2006, Skvortsova et al. 2006), and a decrease in methylated RARB2 in plasma following surgery reflects the removal of the tumor.
RUNX3 has been reported to be a tumor suppressor gene in gastric cancer, with promoter hypermethylation contributing to tumorigenesis (Li et al. 2002). Sakakura et al. (2009) identified RUNX3 promoter methylation in 91% of gastric cancers and in 29% of patient serum samples. Pre-operative serum RUNX3 methylation was higher in late stage than in early stage cancers, and correlated with disease recurrence, most likely due to ctDNA tumor levels reflecting disease burden. The post-operative median methylation index for RUNX3 was 12-fold lower than the pre-operative median methylation index, and serum RUNX3 methylation was found to be a more sensitive indicator of cancer recurrence than CEA (Sakakura et al. 2009). High pre-operative serum methylated RUNX3 levels have also been shown to be indicative of subsequent recurrence in colorectal cancer (Nishio et al. 2010).
Studies in this area are in their early stages and it is yet to be determined how much information the detection of ctDNA adds to established prognostic factors such as tumor size, grade and lymph node status.
Methylated ctDNA as a marker of treatment response
For most metastatic malignancies, a minimum of three cycles of chemotherapy are currently required before treatment response can be assessed based on conventional imaging and biomarkers. This delay exposes many patients to unnecessary toxicity and delays access to other potentially effective therapies. Earlier detection of resistance to treatment is imperative to improving patient outcomes. ctDNA is a promising new approach for monitoring changes in tumor burden in response to therapy (Fig. 1). Several studies report the use of tumor-specific mutations to measure ctDNA dynamics at multiple time points during treatment. For example, Dawson et al. (2013) used mutations in tumor DNA identified via sequencing of FFPE samples to assay ctDNA and track patient response to chemotherapy during breast cancer treatment. Changes in the levels of ctDNA reflected disease status in 17 of 19 women studied, and in ten of these the increase in ctDNA could be detected 2–9 months before the identification of progressive disease by imaging.
As described above for determining surgery outcome, measuring tumor-specific methylation rather than mutation in blood can offer an alternative approach for tracking tumor response to therapy. A summary of gene promoter methylation that has been used to monitor response to treatment is presented in Table 2, and selected examples are discussed below.
Table 2.
Genes for which promoter methylation in serum or plasma has been shown to be associated with response to chemotherapy
| Cancer type | Gene | n | Notes | Reference |
|---|---|---|---|---|
| Breast | Ten gene panel (AKR1B1, ARHGEF7, COL6A2, GPX7, TM6SF1, TMEFF2, HOXB4, RASGRF2, RASSF1A, HIST1H3) | 29 patients (pre- and post-treatment sera) | Methylation decrease in patients having stable disease or therapeutic response | Fackler et al. (2014) |
| No methylation decrease in patients with progressive disease | ||||
| 13 patients (sera at three or more time points during therapy) | Ten out of 13 patients showed methylation levels reflective of tumor burden changes | |||
| BRCA1 | 30 patients (serum) | Significantly lower baseline methylation in responders to neoadjuvant therapy | Sharma et al. (2012) | |
| Significantly higher baseline methylation in non-respondents to neoadjuvant therapy | ||||
| STRATIFIN | 34 patients (serum) | Continuous decline in methylated STRATIFIN correspond to good prognosis | Zurita et al. (2010) | |
| Fluctuating STRATIFIN corresponded to poor prognosis | ||||
| RASSF1A | 148 patients (serum) | Loss of methylation during treatment linked to good survival | Fiegl et al. (2005) | |
| Gain of methylation during treatment linked to poor survival | ||||
| Prostate | GSTP1 | 35 patients (training set, plasma) | Baseline methylation a stronger predictor of overall survival than PSA change | Mahon et al. (2014) |
| 51 patients (validation set, plasma) | Undetectable methylation after one cycle of chemotherapy associated with PSA response | |||
| Lung | RARB2, RASSF1A | 43 patients (pre-treatment, post-neoadjuvant therapy, post-surgery) | Methylation index decreased during neoadjuvant chemotherapy and following surgery | Ponomaryova et al. (2013) |
| 26 patients (∼2 weeks, and 3, 6, 9 months post-surgery, plasma and cell-surface-bound circDNA) | Five patients relapsed during follow up period and all five showed raise in methylation of one or both genes | |||
| No patients without relapse showed increase in methylation in either gene | ||||
| CHFR | 308 patients (plasma) | Methylation was strongly predictive of response to EGFR tyrosine kinase inhibitor | Salazar et al. (2011) | |
| STRATI-FIN | 115 patients (serum) | Survival was significantly longer in the methylation positive group | Ramirez et al. (2005) | |
| SHOX2 | 36 patients (plasma) | Decrease in methylation 7–10 days after chemotherapy treatment identified responders | Schmidt et al. (2015) | |
| RASSF1A APC1 | 316 patients (plasma) | Methylation increase 24 h after chemotherapy associated with sensitivity | Wang et al. (2015) | |
| Melanoma | RASSF1A | 50 patients (serum) | Significantly less frequent methylation in pre-treatment of responders to biochemotherapy than non-responders | Mori et al. (2005) |
| Neuroblastoma | DCR2 | Five patients (serum) | During follow-up methylation was close to 0 in patients in remission, and raised in patients who experienced recurrence | Yagyu et al. (2008) |
GSTP1 is one of the most consistently methylated genes in prostate cancer, being methylated in >90% of tumors (Meiers et al. 2007). The presence of methylated GSTP1 DNA in plasma has been used to track the response of prostate cancer patients to chemotherapy (Mahon et al. 2014). In an exploratory cohort of 35 patients, receiving docetaxel or mitoxantrone treatment and followed for 2–38 (median 15) months, an increase in methylated GSTP1 in plasma after the first dose of chemotherapy was associated with subsequent PSA progression. This result was confirmed in a validation cohort of 51 patients, indicating the potential usefulness of plasma methylated GSTP1 as an early marker of resistance to treatment in prostate cancer. The level of methylated GSTP1 in plasma was a better predictor of overall survival than PSA (Mahon et al. 2014).
Fackler et al. (2014) developed a biomarker panel of genes by identifying differentially methylated genes using genome-wide methylation arrays applied to breast tumor tissue, then narrowing their candidates through in silico validation against The Cancer Genome Atlas breast tumor methylation data, and finally by filtering against whole genome methylated sequences obtained from breast cancer and control sera. The final panel of ten genes (AKR1B1, COL6A2, GPX7, HIST1H3C, HOXB4, RASGRF2, TM6SF1, ARHGEF7, TMEFF2 and RASSF1) was chosen to consist of genes frequently methylated in ER-positive and ER-negative breast cancer, and rarely methylated in healthy control samples. Using the panel to track 29 breast cancer patients treated in clinical trials with docetaxel and imatinib or capecitabine, a decrease in serum methylation levels after 1–2 cycles of treatment was seen in patients having stable disease or partial response, but not in patients with progressive disease. Where sequential serum samples from patients undergoing treatment were available, the rise in methylated ctDNA was detectable prior to clinical disease progression being observed (Fackler et al. 2014).
Methylated serum RASSF1 is also an indicator of response to tamoxifen treatment in breast cancer (Fiegl et al. 2005). Serum from 148 breast cancer patients with resected localized disease was collected prior to adjuvant tamoxifen therapy, and again 1 year after commencement of therapy. Patients were followed up for 0.5–9.8 (median 4.0) years after collection of the second blood sample. No disease recurrences were seen in those women whose serum methylated RASSF1 was detectable at baseline but undetectable after 1 year of tamoxifen. Conversely, detectable serum methylated RASSF1 after 1 year of adjuvant tamoxifen treatment was an independent predictor of poor recurrence-free (RR 5.1, 95% CI 1.3–19.8) and overall survival (RR 6.9, 95% CI 1.9–25.9; Fiegl et al. 2005).
A similar relationship between methylated serum DNA and disease response was observed by Zurita et al. (2010), who used methylated serum STRATIFIN to track response to chemotherapy in 34 patients with metastatic breast cancer. The authors were able to classify patients into two groups: those that showed a continuous decline in methylated STRATIFIN in serum, and those whose levels fluctuated. The pattern of the measured changes was determined by response to chemotherapy treatment (Zurita et al. 2010). In a separate study, a high level of pre-treatment methylated serum STRATIFIN in serum has been reported to be associated with sensitivity to cisplatin-plus-gemcitabine treatment in non-small cell lung cancer patients (Ramirez et al. 2005); however, the study did not report on changes in methylation in response to treatment.
Sharma et al. (2012) analyzed the methylation of a panel of five genes (BRCA1, MGMT, GSTP1, STRATIFIN and MDR1) in six consecutive serum samples from breast cancer patients undergoing chemotherapy treatment. A correlation between reduction in total gene methylation and reduction in tumor volume in the respondents group was observed. At the single gene level, the frequency of BRCA1 methylation was significantly correlated with response to chemotherapy (Sharma et al. 2012).
RASSF1A and RARB2 methylation have been observed to be indicative of treatment response in lung cancer (Ponomaryova et al. 2013). Serum from 43 patients undergoing treatment for non-small cell lung cancer was collected prior to the initiation of neoadjuvant chemotherapy, then prior to surgery, and 10–15 days after surgery. Both RASSF1A and RARB2 showed a decrease in methylation index following chemotherapy, and then a further decrease following surgery. Of 26 patients who were tracked for 9 months after treatment, five relapsed and in all five the serum methylation of at least one gene returned to levels observed prior to starting treatment, while no patients without recurrence showed an increase in the methylation index (Ponomaryova et al. 2013).
Methylated SHOX2 was first tested as a plasma marker for lung cancer diagnosis and showed promising results with 60% sensitivity (95% CI: 53–67%) and 90% specificity (95% CI: 84–94%) (Kneip et al. 2011). It was then evaluated for efficacy in monitoring treatment effectiveness in patients receiving chemotherapy for lung cancer (Schmidt et al. 2015). Among 36 patients studied, there were 17 responders and all showed a decrease in plasma methylated SHOX2, with most displaying a drop at the first post-treatment blood draw 7–10 days after administration of chemotherapy. Only eight of the 19 non-responders showed a decrease in plasma methylated SHOX2 after therapy, and the difference was smaller than in the responders. Among patients with high baseline plasma methylated SHOX2, ROC curves of responders vs non-responders had an area under the curve exceeding 0.8 at the first post-treatment blood draw, which then increased to 0.939 at the fifth post-treatment blood draw (Schmidt et al. 2015). These data suggest that methylated plasma SHOX2 is able to identify patients who will benefit from chemotherapy early and with a high accuracy.
While the studies cited above link a decrease in methylated ctDNA to decreased tumor volume thus establishing chemosensitivity, Wang et al. (2015) took a different approach and instead used an increase in methylated ctDNA to evaluate the extent of tumor cell death induced by chemotherapy. ctDNA methylation of APC1 and RASSF1A in lung cancer patients was measured immediately prior to the administration of chemotherapy and again 24 h after, when DNA released from dying cells peaks (Jahr et al. 2001). An increase in circulating methylated RASSF1A or APC1 at the 24 h time point was shown to be associated with chemosensitivity and complete or partial response, while no change in the two markers following treatment was associated with stable or progressive disease (Wang et al. 2015). The different direction of methylated ctDNA biomarker change in chemosensitive patients described by the studies can be explained by an initial surge in the levels of ctDNA reflecting chemotherapy induced cell death, as observed by Wang et al, followed by a later decline in ctDNA reflecting tumor shrinkage in chemosensitive patients. In contrast to the study reporting increase after a 24 h interval (Wang et al. 2015), the lag between chemotherapy administration and methylated ctDNA measurement in studies reporting a decrease ranged from 1 week (Schmidt et al. 2015) to 1 year (Fiegl et al. 2005). These fluctuations underscore the importance of carefully characterizing ctDNA dynamics in response to chemotherapy as a part of bringing the biomarkers to the clinic for patient monitoring.
Gene methylation patterns in ctDNA linked to prognosis
Gene methylation patterns in tumor tissue can be indicative of tumor aggressiveness and likelihood of recurrence (Rodriguez-Paredes & Esteller 2011), and numerous studies correlate tissue methylation of both individual genes (Castro et al. 2010, Bradly et al. 2012, Li et al. 2014) and gene panels (Fischer et al. 2006, Haldrup et al. 2013, Garcia-Baquero et al. 2014) with patient survival. Methylation can facilitate tumor progression by silencing genes that directly regulate cell growth and metastatic potential, and it can also reflect tumor subtype, which is in turn linked to prognosis. Since tumors shed DNA into the blood, the methylation status of a tumor can be non-invasively assayed by analyzing ctDNA. In order to be informative, the DNA sequence must be unmethylated in hematopoietic cells, which contribute the bulk of circulating DNA (Lui et al. 2002).
Examples of prognostic methylated genes in serum or plasma include TIMP3 (Yu et al. 2014), XAF1 (Ling et al. 2013), ABPA2 (Han et al. 2014), SOX17 (Balgkouranidou et al. 2013, Fu et al. 2015) and RARb2 (Fujita et al. 2012, Mirza et al. 2012). A list of genes, separated by tumor type, for which methylation in plasma or serum has been shown to be prognostic in cancer is presented in Table 3. Selected genes with a known function in cancer-related processes such as cell proliferation, apoptosis and cell migration, that have also been shown to be prognostic when methylated in tissue, are discussed below.
Table 3.
Genes for which promoter methylation in serum or plasma has been shown to be prognostic in different cancer types
Methylation of the gene has a negative impact on prognosis.
No impact of gene methylation on prognosis was observed.
Methylation of the gene has a positive impact on prognosis.
SOX17 plays a tumor suppressor role by regulating the WNT signaling pathway, and inhibition of SOX17 promotes tumorigenesis (Zhang et al. 2008, Jia et al. 2012). SOX17 methylation in tumor tissue is associated with poor prognosis in breast (Conway et al. 2014) and esophageal (Kuo et al. 2014) cancer. Two reports have examined the prognostic value of SOX17 methylation in blood. Fu et al. (2015) used plasma samples from 60 breast cancer patients to show that SOX17 methylation was associated with TNM stage, but was also an independent prognostic factor in multivariate analysis. Balgkouranidou et al. (2013) showed that in a cohort of 73 patients with gastric cancer, serum SOX17 methylation was correlated with tumor differentiation and overall survival.
RARb2, a retinoic acid receptor, has a complex role in regulating cell proliferation, and although it generally plays a role in tumor suppression (Yang et al. 2002), tumor promotion effects have also been described (Pappas et al. 2011). In tumors, methylation of RARb2 has been consistently shown to be associated with poor prognosis, and has been linked to poor outcome in colorectal cancer (Miladi-Abdennadher et al. 2010), breast cancer (Sharma et al. 2009, Tao et al. 2009) and lung cancer (Kim et al. 2015). The results obtained in tumor tissue are reflected in ctDNA, and methylated RARb2 in plasma or serum has been shown to be associated with worse patient outcomes in breast cancer (Fujita et al. 2012, Mirza et al. 2012), lung cancer (Ponomaryova et al. 2013) and mesothelioma (Fischer et al. 2006).
TIMP3 inhibits endothelial cell migration, thus limiting angiogenesis in tumors (Das et al. 2014) and methylation of the gene promoter is a known mechanism of carcinogenesis (Liu et al. 2011). In a study of 92 patients newly diagnosed with gastric cancer, detection of methylated TIMP3 promoter sequence in the serum was an independent predictor of poor disease-free survival (DFS; Yu et al. 2014).
XAF1 has a well-established role in limiting cancer progression through regulating apoptosis (Tu et al. 2010, Zou et al. 2012, Lee et al. 2014, Zhu et al. 2014) and inhibiting angiogenesis (Zhu et al. 2014). It is frequently methylated in various types of cancer, and tumor methylation is associated with poor prognosis (Chen et al. 2011). A study by Ling et al showed that XAF1 tumor methylation in gastric cancer is linked to decreased survival, with a median DFS of 23.4 months in patients with methylated XAF1, in contrast to a median DFS of 39.6 months in patients with unmethylated XAF1. Correspondingly, patients positive for serum XAF1 methylation had significantly lower DFS than patients negative for serum XAF1 methylation (Ling et al. 2013).
While promoter methylation silencing of tumor suppressor genes can directly confer a more aggressive tumor phenotype, broad methylation differences at hundreds of gene promoters can be reflective of different tumor subtypes, each with different prognosis. For example, gene methylation patterns distinguish three separate subgroups in triple-negative breast cancer, which have different clinical outcomes for patients (Stirzaker et al. 2015). It follows that the presence of a methylated sequence in blood could be indicative of tumor subtype, and hence prognosis. ER-positive breast cancer has a better prognosis than ER-negative breast cancer (Bishop et al. 1979), and circulating methylated ESR1 is an indicative of ER-negative breast cancer, thus corresponding with worse patient outcomes (Mirza et al. 2010, Martinez-Galan et al. 2014).
A particular cancer-specific methylated sequence need not be involved in processes directly linked to poor prognosis in order for its presence in blood to be informative. Detection and/or quantitation can simply be an indicative of the amount of ctDNA present in the circulation, which in turn reflects tumor burden. Circulating DNA typically occurs at low concentrations, often below 10 ng/ml (El Messaoudi et al. 2013, Warton et al. 2014). The proportion of circulating DNA which is derived from the tumor has been reported to vary from as high as 90% (Jahr et al. 2001) to <0.05% (Diehl et al. 2005). A high tumor burden and the presence of invasive tumor, which confers worse patient prognoses, also results in more tumor DNA in blood, and more easily detectable tumor-specific methylated sequences (Fig. 1). Hence, detection of target methylated sequences in serum or plasma can be indicative of aggressive phenotype and/or large volume of tumor, both of which correlate with poor prognosis. It is not always clear whether increased detection of particular circulating methylated genes in patients with poor outcomes reflects the impact of gene methylation on tumor biology, or simply increased ctDNA due to high tumor burden.
Challenges and future directions
There is overlap between the potential application of biomarkers for cancer diagnosis and cancer monitoring. Methylated ctDNA is being actively investigated as a substrate for cancer diagnostic blood tests (reviewed in Warton & Samimi (2015)), and a blood test for colorectal cancer based on SEPT9 methylation is currently under review by the FDA. Because a pre-requisite of diagnostic cancer tests being clinically useful is that the assay target is detectable in most people with the selected cancer, it follows that the diagnostic tests could be applicable to monitoring tumor DNA dynamics without the need for highly individualized assays being developed for each cancer patient.
Adapting diagnostic ctDNA methylation tests to cancer monitoring may be less challenging than developing the tests in the first place, as the problem of false positive results in healthy individuals is not applicable. These are analogous to elevated CA125 and PSA being effective for monitoring patients with ovarian and prostate cancer respectively, despite their limitations in population cancer screening due to the high false positive rate. Hence methylated ctDNA assays that are developed to meet the stringent criteria required for cancer screening may also be helpful for determining whether tumor DNA is still present in plasma after surgery or during treatment. One biomarker under development is methylated SHOX2, which has shown promise in blood-based diagnosis of lung cancer (Kneip et al. 2011), and has more recently been investigated as marker of early response to treatment in lung cancer patients (Schmidt et al. 2015). However, as far as we are aware from the published literature, a ctDNA methylation assay specifically developed for cancer screening has not yet entered clinical practice for monitoring cancer after diagnosis and treatment.
The collection of plasma or serum samples suitable for evaluating assays used in patient monitoring is more challenging than acquiring samples that can be used to evaluate diagnostic tests. Testing the relationship between the change in ctDNA and the response to surgery or therapy requires paired pre- and post-intervention blood samples, or even samples collected sequentially during the treatment, and these must be linked to information about treatment regimens and patient follow-up. This hurdle possibly accounts for the typically small numbers of patients monitored in the studies described in this review. Serum and plasma samples would ideally be acquired through prospective clinical trials, which collect detailed information about treatment regimens and patient outcomes as a matter of course, and in which the inclusion of biomarker sub-studies is rapidly becoming mandatory.
The National Cancer Institute (NCI) biomarker discovery and development pathway has attempted to standardize biomarker development and provide an efficient framework to move useful biomarkers into clinical practice. All large Phase III therapeutic studies should collect prospective samples that are rationally devised to test available biomarkers. While associated costs to cover sample collection, storage and testing are significant, appropriate biomarker studies are crucial for tailoring treatment and improving outcomes. In response to this need, the NCI has prioritized biomarker inclusion in large Phase II (≥100 patients) and III therapeutic trials by establishing the Biomarker, Imaging and Quality of Life Studies Funding Program (US 2014), which aims to provide funding support for associated biomarker sub-studies. However, funding is only provided for biomarkers that test a defined hypothesis, not for ancillary biomarkers that aim to generate hypotheses. As therapeutic trials take many years from conception, through accrual to full maturity, rational sample collection should also include samples for storage to accommodate possible novel uses in the future. This is difficult given the burgeoning technologies available and the fact that understanding often lags behind our ability to generate data. Methylated ctDNA has the advantage of being a relatively stable assay target that is compatible with sample freezing and requires relatively little in the way of specialized sample handling; however, to develop clinically valuable biomarkers, close collaboration is needed between clinicians devising therapeutic trials and scientists investigating biomarker development.
ctDNA is detectable in the blood of many cancer patients, with levels reflecting tumor load (Ignatiadis & Dawson 2014). Thus the abundance of tumor-specific methylated sequences in blood may provide a direct indication of the effect of drug treatment on the tumor. The advantages of rapidly identifying whether a tumor is responding to treatment are clear. Chemotherapy has significant associated toxicity and does not benefit every patient. Treating non-responders reduces the quality of life, incurs the medical costs and delays the initiation of other potentially effective therapies. Since elevated ctDNA levels can precede clinical establishment of progressive disease (Dawson et al. 2013), ctDNA may provide an early marker of disease resistance to allow prompt cessation of ineffective regimens, sparing chemotherapy-associated toxicities, and potentially providing an opportunity to try alternative treatments.
Clinical trials of cancer treatments rely on overall survival time and relapse-free survival time to assess the efficacy of new drugs. While survival time and relapse-free survival are the gold standards in establishing treatment benefits, these data take a long time to accrue, prolonging the time it takes for new drugs to reach the clinic and adding considerably to trial costs. Surrogate endpoints, that is, clinical parameters related to disease progression that become apparent prior to relapse or death, have the potential to streamline drug development. However, surrogate endpoints have to date proved unreliable in evaluating cancer treatments. PSA, the most studied biomarker in prostate cancer, although useful for tracking individual response, does not correlate sufficiently well with overall survival times to effectively predict the outcome of treatment (Collette et al. 2005, Yap et al. 2012). Methylated GSTP1 in plasma was a better predictor of overall survival than PSA in prostate cancer patients (Mahon et al. 2014). If methylated ctDNA assays can improve on protein antigen-based biomarkers such as PSA and CA15.3, they may provide clinically useful surrogate biomarkers for prospective clinical trials to accelerate drug development.
Cancer monitoring by measuring tumor DNA dynamics in blood is a new and developing area, poised to advance rapidly both through the application of existing technology, and as a result of novel techniques that are constantly being pursued in the burgeoning field of molecular biology. The benefits can be anticipated to improve patient management, reduce unnecessary drug toxicity and accelerate data acquisition from clinical trials. Cancer-specific circulating DNA methylation offers a range of promising targets that can help to advance these aims.
Acknowledgements
The authors wish to thank Kate Patterson for preparation of Fig. 1.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this review.
Funding
The Ovarian Cancer Research Group acknowledges the support from the Gynaecological Oncology (GO) Fund of the Royal Hospital for Women Foundation, Sydney, Australia. G Samimi is a Cancer Institute NSW Career Development Fellow.
References
- Balgkouranidou I, Karayiannakis A, Matthaios D, Bolanaki H, Tripsianis G, Tentes AA, Lianidou E, Chatzaki E, Fiska A, Lambropoulou M, et al. Assessment of SOX17 DNA methylation in cell free DNA from patients with operable gastric cancer. Association with prognostic variables and survival. Clinical Chemistry and Laboratory Medicine. 2013;51:1505–1510. doi: 10.1515/cclm-2012-0320. [DOI] [PubMed] [Google Scholar]
- Balgkouranidou I, Chimonidou M, Milaki G, Tsarouxa EG, Kakolyris S, Welch DR, Georgoulias V, Lianidou ES. Breast cancer metastasis suppressor-1 promoter methylation in cell-free DNA provides prognostic information in non-small cell lung cancer. British Journal of Cancer. 2014;110:2054–2062. doi: 10.1038/bjc.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balgkouranidou I, Matthaios D, Karayiannakis A, Bolanaki H, Michailidis P, Xenidis N, Amarantidis K, Chelis L, Trypsianis G, Chatzaki E, et al. Prognostic role of APC and RASSF1A promoter methylation status in cell free circulating DNA of operable gastric cancer patients. Mutation Research. 2015;778:46–51. doi: 10.1016/j.mrfmmm.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Barault L, Amatu A, Bleeker FE, Moutinho C, Falcomata C, Fiano V, Cassingena A, Siravegna G, Milione M, Cassoni P, et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Annals of Oncology. 2015;26:1994–1999. doi: 10.1093/annonc/mdv272. [DOI] [PubMed] [Google Scholar]
- Bastian PJ, Palapattu GS, Lin X, Yegnasubramanian S, Mangold LA, Trock B, Eisenberger MA, Partin AW, Nelson WG. Preoperative serum DNA GSTP1 CpG island hypermethylation and the risk of early prostate-specific antigen recurrence following radical prostatectomy. Clinical Cancer Research. 2005;11:4037–4043. doi: 10.1158/1078-0432.CCR-04-2446. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome – biological and translational implications. Nature Review Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop HM, Blamey RW, Elston CW, Haybittle JL, Nicholson RI, Griffiths K. Relationship of oestrogen-receptor status to survival in breast cancer. Lancet. 1979;2:283–284. doi: 10.1016/S0140-6736(79)90295-2. [DOI] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Research. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- Bradly DP, Gattuso P, Pool M, Basu S, Liptay M, Bonomi P, Buckingham L. CDKN2A (p16) promoter hypermethylation influences the outcome in young lung cancer patients. Diagnostic Molecular Pathology. 2012;21:207–213. doi: 10.1097/PDM.0b013e31825554b2. [DOI] [PubMed] [Google Scholar]
- Brock MV, Gou M, Akiyama Y, Muller A, Wu TT, Montgomery E, Deasel M, Germonpre P, Rubinson L, Heitmiller RF, et al. Prognostic importance of promoter hypermethylation of multiple genes in esophageal adenocarcinoma. Clinical Cancer Research. 2003;9:2912–2919. [PubMed] [Google Scholar]
- Castro M, Grau L, Puerta P, Gimenez L, Venditti J, Quadrelli S, Sanchez-Carbayo M. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. Journal of Translational Medicine. 2010;8:86. doi: 10.1186/1479-5876-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YB, Shu J, Yang WT, Shi L, Guo XF, Wang FG, Qian YY. XAF1 as a prognostic biomarker and therapeutic target in squamous cell lung cancer. Chinese Medical Journal. 2011;124:3238–3243. [PubMed] [Google Scholar]
- Collette L, Burzykowski T, Carroll KJ, Newling D, Morris T, Schroder FH, European Organisation for Research and Treatment of Cancer, Limburgs Universitair Centrum, AstraZeneca Pharmaceuticals Is prostate-specific antigen a valid surrogate end point for survival in hormonally treated patients with metastatic prostate cancer? Joint research of the European Organisation for Research and Treatment of Cancer, the Limburgs Universitair Centrum, and AstraZeneca Pharmaceuticals. Journal of Clinical Oncology. 2005;23:6139–6148. doi: 10.1200/JCO.2005.08.156. [DOI] [PubMed] [Google Scholar]
- Conway K, Edmiston SN, May R, Kuan PF, Chu H, Bryant C, Tse CK, Swift-Scanlan T, Geradts J, Troester MA, et al. DNA methylation profiling in the Carolina Breast Cancer Study defines cancer subclasses differing in clinicopathologic characteristics and survival. Breast Cancer Research. 2014;16:450. doi: 10.1186/s13058-014-0450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AM, Seynhaeve AL, Rens JA, Vermeulen CE, Koning GA, Eggermont AM, Ten Hagen TL. Differential TIMP3 expression affects tumor progression and angiogenesis in melanomas through regulation of directionally persistent endothelial cell migration. Angiogenesis. 2014;17:163–177. doi: 10.1007/s10456-013-9385-2. [DOI] [PubMed] [Google Scholar]
- Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. New England Journal of Medicine. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Jr, Goodman SN, David KA, Juhl H, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. PNAS. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. Circulating mutant DNA to assess tumor dynamics. Nature Medicine. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating cell free DNA: preanalytical considerations. Clinica Chimica Acta. 2013;424:222–230. doi: 10.1016/j.cca.2013.05.022. [DOI] [PubMed] [Google Scholar]
- Ellinger J, El Kassem N, Heukamp LC, Matthews S, Cubukluoz F, Kahl P, Perabo FG, Muller SC, von Ruecker A, Bastian PJ. Hypermethylation of cell-free serum DNA indicates worse outcome in patients with bladder cancer. Journal of Urology. 2008;179:346–352. doi: 10.1016/j.juro.2007.08.091. [DOI] [PubMed] [Google Scholar]
- Fackler MJ, Lopez Bujanda Z, Umbricht C, Teo WW, Cho S, Zhang Z, Visvanathan K, Jeter S, Argani P, Wang C, et al. Novel methylated biomarkers and a robust assay to detect circulating tumor DNA in metastatic breast cancer. Cancer Research. 2014;74:2160–2170. doi: 10.1158/0008-5472.CAN-13-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nature Review Genetics. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Fiegl H, Millinger S, Mueller-Holzner E, Marth C, Ensinger C, Berger A, Klocker H, Goebel G, Widschwendter M. Circulating tumor-specific DNA: a marker for monitoring efficacy of adjuvant therapy in cancer patients. Cancer Research. 2005;65:1141–1145. doi: 10.1158/0008-5472.CAN-04-2438. [DOI] [PubMed] [Google Scholar]
- Fischer JR, Ohnmacht U, Rieger N, Zemaitis M, Stoffregen C, Kostrzewa M, Buchholz E, Manegold C, Lahm H. Promoter methylation of RASSF1A, RARβ and DAPK predict poor prognosis of patients with malignant mesothelioma. Lung Cancer. 2006;54:109–116. doi: 10.1016/j.lungcan.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, Dawson SJ, Piskorz AM, Jimenez-Linan M, Bentley D, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Science Translational Medicine. 2012;4:136ra168. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- Fu D, Ren C, Tan H, Wei J, Zhu Y, He C, Shao W, Zhang J. Sox17 promoter methylation in plasma DNA is associated with poor survival and can be used as a prognostic factor in breast cancer. Medicine. 2015;94:e637. doi: 10.1097/MD.0000000000000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Nakayama T, Yamamoto N, Kim SJ, Shimazu K, Shimomura A, Maruyama N, Morimoto K, Tamaki Y, Noguchi S. Methylated DNA and total DNA in serum detected by one-step methylation-specific PCR is predictive of poor prognosis for breast cancer patients. Oncology. 2012;83:273–282. doi: 10.1159/000342083. [DOI] [PubMed] [Google Scholar]
- Garcia-Baquero R, Puerta P, Beltran M, Alvarez-Mujica M, Alvarez-Ossorio JL, Sanchez-Carbayo M. Methylation of tumor suppressor genes in a novel panel predicts clinical outcome in paraffin-embedded bladder tumors. Tumour Biology. 2014;35:5777–5786. doi: 10.1007/s13277-014-1767-6. [DOI] [PubMed] [Google Scholar]
- Gobel G, Auer D, Gaugg I, Schneitter A, Lesche R, Muller-Holzner E, Marth C, Daxenbichler G. Prognostic significance of methylated RASSF1A and PITX2 genes in blood- and bone marrow plasma of breast cancer patients. Breast Cancer Research and Treatment. 2011;130:109–117. doi: 10.1007/s10549-010-1335-8. [DOI] [PubMed] [Google Scholar]
- Haldrup C, Mundbjerg K, Vestergaard EM, Lamy P, Wild P, Schulz WA, Arsov C, Visakorpi T, Borre M, Hoyer S, et al. DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer. Journal of Clinical Oncology. 2013;31:3250–3258. doi: 10.1200/JCO.2012.47.1847. [DOI] [PubMed] [Google Scholar]
- Han J, Lv P, Yu JL, Wu YC, Zhu X, Hong LL, Zhu WY, Yu QM, Wang XB, Li P, et al. Circulating methylated MINT2 promoter DNA is a potential poor prognostic factor in gastric cancer. Digestive Diseases and Sciences. 2014;59:1160–1168. doi: 10.1007/s10620-013-3007-0. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. PNAS. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nature Review Genetics. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- Hoffmann AC, Vallbohmer D, Prenzel K, Metzger R, Heitmann M, Neiss S, Ling F, Holscher AH, Schneider PM, Brabender J. Methylated DAPK and APC promoter DNA detection in peripheral blood is significantly associated with apparent residual tumor and outcome. Journal of Cancer Research and Clinical Oncology. 2009;135:1231–1237. doi: 10.1007/s00432-009-0564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque MO, Feng Q, Toure P, Dem A, Critchlow CW, Hawes SE, Wood T, Jeronimo C, Rosenbaum E, Stern J, et al. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. Journal of Clinical Oncology. 2006;24:4262–4269. doi: 10.1200/JCO.2005.01.3516. [DOI] [PubMed] [Google Scholar]
- Horning AM, Awe JA, Wang CM, Liu J, Lai Z, Wang VJ, Jadhav RR, Louie AD, Lin CL, Kroczak T, et al. DNA methylation screening of primary prostate tumors identifies SRD5A2 and CYP11A1 as candidate markers for assessing risk of biochemical recurrence. Prostate. 2015;75:1790–1801. doi: 10.1002/pros.23052. doi:1710.1002/pros.23052 [DOI] [PubMed] [Google Scholar]
- Hoshimoto S, Kuo CT, Chong KK, Takeshima TL, Takei Y, Li MW, Huang SK, Sim MS, Morton DL, Hoon DS. AIM1 and LINE-1 epigenetic aberrations in tumor and serum relate to melanoma progression and disease outcome. Journal of Investigatory Dermatology. 2012;132:1689–1697. doi: 10.1038/jid.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatiadis M, Dawson SJ. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Annals of Oncology. 2014;25:2304–2313. doi: 10.1093/annonc/mdu480. [DOI] [PubMed] [Google Scholar]
- Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Research. 2001;61:1659–1665. [PubMed] [Google Scholar]
- Jia Y, Yang Y, Zhan Q, Brock MV, Zheng X, Yu Y, Herman JG, Guo M. Inhibition of SOX17 by microRNA 141 and methylation activates the WNT signaling pathway in esophageal cancer. Journal of Molecular Diagnostics. 2012;14:577–585. doi: 10.1016/j.jmoldx.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Kim Y, Jin D, Lee BB, Cho EY, Han J, Shim YM, Kim DH. RARβ2 hypermethylation is associated with poor recurrence-free survival in never-smokers with adenocarcinoma of the lung. Clinical Epigenetics. 2015;7:32. doi: 10.1186/s13148-015-0066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneip C, Schmidt B, Seegebarth A, Weickmann S, Fleischhacker M, Liebenberg V, Field JK, Dietrich D. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. Journal of Thoracic Oncology. 2011;6:1632–1638. doi: 10.1097/JTO.0b013e318220ef9a. [DOI] [PubMed] [Google Scholar]
- Koyanagi K, Mori T, O'Day SJ, Martinez SR, Wang HJ, Hoon DS. Association of circulating tumor cells with serum tumor-related methylated DNA in peripheral blood of melanoma patients. Cancer Research. 2006;66:6111–6117. doi: 10.1158/0008-5472.CAN-05-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo IY, Chang JM, Jiang SS, Chen CH, Chang IS, Sheu BS, Lu PJ, Chang WL, Lai WW, Wang YC. Prognostic CpG methylation biomarkers identified by methylation array in esophageal squamous cell carcinoma patients. International Journal of Medical Sciences. 2014;11:779–787. doi: 10.7150/ijms.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW. The power and the promise of DNA methylation markers. Nature Review Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- Lecomte T, Berger A, Zinzindohoue F, Micard S, Landi B, Blons H, Beaune P, Cugnenc PH, Laurent-Puig P. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. International Journal of Cancer. 2002;100:542–548. doi: 10.1002/ijc.10526. [DOI] [PubMed] [Google Scholar]
- Lee MG, Han J, Jeong SI, Her NG, Lee JH, Ha TK, Kang MJ, Ryu BK, Chi SG. XAF1 directs apoptotic switch of p53 signaling through activation of HIPK2 and ZNF313. PNAS. 2014;111:15532–15537. doi: 10.1073/pnas.1411746111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/S0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Li FF, Yang Y, Wang XL, Hong YY, Wang NF, Chen ZD. Promoter methylation of DAPK gene may contribute to the pathogenesis of nonsmall cell lung cancer: a meta-analysis. Tumour Biology. 2014;35:6011–6020. doi: 10.1007/s13277-014-1796-1. [DOI] [PubMed] [Google Scholar]
- Liggett TE, Melnikov AA, Marks JR, Levenson VV. Methylation patterns in cell-free plasma DNA reflect removal of the primary tumor and drug treatment of breast cancer patients. International Journal of Cancer. 2011;128:492–499. doi: 10.1002/ijc.25363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling ZQ, Zhao Q, Zhou SL, Mao WM. MSH2 promoter hypermethylation in circulating tumor DNA is a valuable predictor of disease-free survival for patients with esophageal squamous cell carcinoma. European Journal of Surgical Oncology. 2012;38:326–332. doi: 10.1016/j.ejso.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Ling ZQ, Lv P, Lu XX, Yu JL, Han J, Ying LS, Zhu X, Zhu WY, Fang XH, Wang S, et al. Circulating Methylated XAF1 DNA Indicates Poor Prognosis for Gastric Cancer. PLoS ONE. 2013;8:e67195. doi: 10.1371/journal.pone.0067195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WB, Cui ZH, Ao L, Zhou ZY, Zhou YH, Yuan XY, Xiang YL, Liu JY, Cao J. Aberrant methylation accounts for cell adhesion-related gene silencing during 3-methylcholanthrene and diethylnitrosamine induced multistep rat lung carcinogenesis associated with overexpression of DNA methyltransferases 1 and 3a. Toxicology and Applied Pharmacology. 2011;251:70–78. doi: 10.1016/j.taap.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nature Review Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- Lui YY, Chik KW, Chiu RW, Ho CY, Lam CW, Lo YM. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clinical Chemistry. 2002;48:421–427. [PubMed] [Google Scholar]
- Mahon KL, Qu W, Devaney J, Paul C, Castillo L, Wykes RJ, Chatfield MD, Boyer MJ, Stockler MR, Marx G, et al. Methylated Glutathione S-transferase 1 (mGSTP1) is a potential plasma free DNA epigenetic marker of prognosis and response to chemotherapy in castrate-resistant prostate cancer. British Journal of Cancer. 2014;111:1802–1809. doi: 10.1038/bjc.2014.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Galan J, Torres-Torres B, Nunez MI, Lopez-Penalver J, Del Moral R, Ruiz De Almodovar JM, Menjon S, Concha A, Chamorro C, Rios S, et al. ESR1 gene promoter region methylation in free circulating DNA and its correlation with estrogen receptor protein expression in tumor tissue in breast cancer patients. BMC Cancer. 2014;14:59. doi: 10.1186/1471-2407-14-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiers I, Shanks JH, Bostwick DG. Glutathione S-transferase pi (GSTP1) hypermethylation in prostate cancer: review 2007. Pathology. 2007;39:299–304. doi: 10.1080/00313020701329906. [DOI] [PubMed] [Google Scholar]
- Miladi-Abdennadher I, Abdelmaksoud-Damak R, Ayadi L, Khabir A, Frikha F, Kallel L, Amouri A, Frikha M, Sellami-Boudawara T, Gargouri A, et al. Hypermethylation of RARβ2 correlates with high COX-2 expression and poor prognosis in patients with colorectal carcinoma. Tumour Biology. 2010;31:503–511. doi: 10.1007/s13277-010-0063-3. [DOI] [PubMed] [Google Scholar]
- Mirza S, Sharma G, Parshad R, Srivastava A, Gupta SD, Ralhan R. Clinical significance of Stratifin, ERα and PR promoter methylation in tumor and serum DNA in Indian breast cancer patients. Clinical Biochemistry. 2010;43:380–386. doi: 10.1016/j.clinbiochem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Mirza S, Sharma G, Parshad R, Srivastava A, Gupta SD, Ralhan R. Clinical significance of promoter hypermethylation of ERβ and RARβ2 in tumor and serum DNA in Indian breast cancer patients. Annals of Surgical Oncology. 2012;19:3107–3115. doi: 10.1245/s10434-012-2323-5. [DOI] [PubMed] [Google Scholar]
- Misawa A, Tanaka S, Yagyu S, Tsuchiya K, Iehara T, Sugimoto T, Hosoi H. RASSF1A hypermethylation in pretreatment serum DNA of neuroblastoma patients: a prognostic marker. British Journal of Cancer. 2009;100:399–404. doi: 10.1038/sj.bjc.6604887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon C, Gloss BS, Warton K, Barton CA, Statham AL, Scurry JP, Tabor B, Nguyen TV, Qu W, Samimi G, et al. Prognostic and diagnostic significance of DNA methylation patterns in high grade serous ovarian cancer. Gynecologic Oncology. 2012;124:582–588. doi: 10.1016/j.ygyno.2011.11.026. [DOI] [PubMed] [Google Scholar]
- Mori T, O'Day SJ, Umetani N, Martinez SR, Kitago M, Koyanagi K, Kuo C, Takeshima TL, Milford R, Wang HJ, et al. Predictive utility of circulating methylated DNA in serum of melanoma patients receiving biochemotherapy. Journal of Clinical Oncology. 2005;23:9351–9358. doi: 10.1200/JCO.2005.02.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Martinez SR, O'Day SJ, Morton DL, Umetani N, Kitago M, Tanemura A, Nguyen SL, Tran AN, Wang HJ, et al. Estrogen receptor-alpha methylation predicts melanoma progression. Cancer Research. 2006;66:6692–6698. doi: 10.1158/0008-5472.CAN-06-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- Nishio M, Sakakura C, Nagata T, Komiyama S, Miyashita A, Hamada T, Kuryu Y, Ikoma H, Kubota T, Kimura A, et al. RUNX3 promoter methylation in colorectal cancer: its relationship with microsatellite instability and its suitability as a novel serum tumor marker. Anticancer Research. 2010;30:2673–2682. [PubMed] [Google Scholar]
- Okegawa T, Nutahara K, Higashihara E. Association of circulating tumor cells with tumor-related methylated DNA in patients with hormone-refractory prostate cancer. International Journal of Urology. 2010;17:466–475. doi: 10.1111/j.1442-2042.2010.02502.x. [DOI] [PubMed] [Google Scholar]
- Pappas JJ, Toulouse A, Basik M, Levesque L, Bradley WE. Knockdown of RARB2 identifies a dual role in cancer. Genes, Chromosomes & Cancer. 2011;50:700–714. doi: 10.1002/gcc.20892. [DOI] [PubMed] [Google Scholar]
- Philipp AB, Stieber P, Nagel D, Neumann J, Spelsberg F, Jung A, Lamerz R, Herbst A, Kolligs FT. Prognostic role of methylated free circulating DNA in colorectal cancer. International Journal of Cancer. 2012;131:2308–2319. doi: 10.1002/ijc.27505. [DOI] [PubMed] [Google Scholar]
- Ponomaryova AA, Rykova EY, Cherdyntseva NV, Skvortsova TE, Dobrodeev AY, Zav'yalov AA, Bryzgalov LO, Tuzikov SA, Vlassov VV, Laktionov PP. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer. 2013;81:397–403. doi: 10.1016/j.lungcan.2013.05.016. [DOI] [PubMed] [Google Scholar]
- Ramirez JL, Rosell R, Taron M, Sanchez-Ronco M, Alberola V, de Las Penas R, Sanchez JM, Moran T, Camps C, Massuti B, et al. 14-3-3sigma methylation in pretreatment serum circulating DNA of cisplatin-plus-gemcitabine-treated advanced non-small-cell lung cancer patients predicts survival: The Spanish Lung Cancer Group. Journal of Clinical Oncology. 2005;23:9105–9112. doi: 10.1200/JCO.2005.02.2905. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nature Medicine. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Sakakura C, Hamada T, Miyagawa K, Nishio M, Miyashita A, Nagata H, Ida H, Yazumi S, Otsuji E, Chiba T, et al. Quantitative analysis of tumor-derived methylated RUNX3 sequences in the serum of gastric cancer patients. Anticancer Research. 2009;29:2619–2625. [PubMed] [Google Scholar]
- Salazar F, Molina MA, Sanchez-Ronco M, Moran T, Ramirez JL, Sanchez JM, Stahel R, Garrido P, Cobo M, Isla D, et al. First-line therapy and methylation status of CHFR in serum influence outcome to chemotherapy versus EGFR tyrosine kinase inhibitors as second-line therapy in stage IV non-small-cell lung cancer patients. Lung Cancer. 2011;72:84–91. doi: 10.1016/j.lungcan.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Schmidt B, Beyer J, Dietrich D, Bork I, Liebenberg V, Fleischhacker M. Quantification of cell-free mSHOX2 Plasma DNA for therapy monitoring in advanced stage non-small cell (NSCLC) and small-cell lung cancer (SCLC) patients. PLoS ONE. 2015;10:e0118195. doi: 10.1371/journal.pone.0118195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma G, Mirza S, Yang YH, Parshad R, Hazrah P, Datta Gupta S, Ralhan R. Prognostic relevance of promoter hypermethylation of multiple genes in breast cancer patients. Cell. 2009;31:487–500. doi: 10.3233/CLO-2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma G, Mirza S, Parshad R, Srivastava A, Gupta SD, Pandya P, Ralhan R. Clinical significance of promoter hypermethylation of DNA repair genes in tumor and serum DNA in invasive ductal breast carcinoma patients. Life Sciences. 2010;87:83–91. doi: 10.1016/j.lfs.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Sharma G, Mirza S, Parshad R, Gupta SD, Ralhan R. DNA methylation of circulating DNA: a marker for monitoring efficacy of neoadjuvant chemotherapy in breast cancer patients. Tumour Biology. 2012;33:1837–1843. doi: 10.1007/s13277-012-0443-y. [DOI] [PubMed] [Google Scholar]
- Skvortsova TE, Rykova EY, Tamkovich SN, Bryzgunova OE, Starikov AV, Kuznetsova NP, Vlassov VV, Laktionov PP. Cell-free and cell-bound circulating DNA in breast tumours: DNA quantification and analysis of tumour-related gene methylation. British Journal of Cancer. 2006;94:1492–1495. doi: 10.1038/sj.bjc.6603117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirzaker C, Zotenko E, Song JZ, Qu W, Nair SS, Locke WJ, Stone A, Armstong NJ, Robinson MD, Dobrovic A, et al. Methylome sequencing in triple-negative breast cancer reveals distinct methylation clusters with prognostic value. Nature Communications. 2015;6:5899. doi: 10.1038/ncomms6899. [DOI] [PubMed] [Google Scholar]
- Tao MH, Shields PG, Nie J, Millen A, Ambrosone CB, Edge SB, Krishnan SS, Marian C, Xie B, Winston J, et al. DNA hypermethylation and clinicopathological features in breast cancer: the Western New York Exposures and Breast Cancer (WEB) Study. Breast Cancer Research and Treatment. 2009;114:559–568. doi: 10.1007/s10549-008-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu SP, Sun YW, Cui JT, Zou B, Lin MC, Gu Q, Jiang SH, Kung HF, Korneluk RG, Wong BC. Tumor suppressor XIAP-Associated factor 1 (XAF1) cooperates with tumor necrosis factor-related apoptosis-inducing ligand to suppress colon cancer growth and trigger tumor regression. Cancer. 2010;116:1252–1263. doi: 10.1002/cncr.24814. [DOI] [PubMed] [Google Scholar]
- Umbricht CB, Evron E, Gabrielson E, Ferguson A, Marks J, Sukumar S. Hypermethylation of 14-3-3 sigma (stratifin) is an early event in breast cancer. Oncogene. 2001;20:3348–3353. doi: 10.1038/sj.onc.1204438. [DOI] [PubMed] [Google Scholar]
- (US) NCI. Biomarker, Imaging and Quality of Life Studies Funding Program. US Department of Health and Human Services, National Institutes of Health; Bethesda, MD, USA: 2014. [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang B, Chen D, Xia W, Zhang J, Wang F, Xu J, Zhang Y, Zhang M, Zhang L, et al. Real-time monitoring efficiency and toxicity of chemotherapy in patients with advanced lung cancer. Clinical Epigenetics. 2015;7:119. doi: 10.1186/s13148-015-0150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warton K, Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Frontiers in Molecular Biosciences. 2015;2:13. doi: 10.3389/fmolb.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warton K, Lin V, Navin T, Armstrong NJ, Kaplan W, Ying K, Gloss B, Mangs H, Nair SS, Hacker NF, et al. Methylation-capture and Next-Generation Sequencing of free circulating DNA from human plasma. BMC Genomics. 2014;15:476. doi: 10.1186/1471-2164-15-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberger T, Sleigh S, Reisel D, Zikan M, Wahl B, Alunni-Fabbroni M, Jones A, Evans I, Koch J, Paprotka T, et al. DNA methylation markers for early detection of women's cancer: promise and challenges. Epigenomics. 2014;6:311–327. doi: 10.2217/epi.14.20. [DOI] [PubMed] [Google Scholar]
- Wong IH, Lo YM, Zhang J, Liew CT, Ng MH, Wong N, Lai PB, Lau WY, Hjelm NM, Johnson PJ. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Research. 1999;59:71–73. [PubMed] [Google Scholar]
- Wong IH, Zhang J, Lai PB, Lau WY, Lo YM. Quantitative analysis of tumor-derived methylated p16INK4a sequences in plasma, serum, and blood cells of hepatocellular carcinoma patients. Clinical Cancer Research. 2003;9:1047–1052. [PubMed] [Google Scholar]
- Wong TS, Kwong DL, Sham JS, Wei WI, Kwong YL, Yuen AP. Quantitative plasma hypermethylated DNA markers of undifferentiated nasopharyngeal carcinoma. Clinical Cancer Research. 2004;10:2401–2406. doi: 10.1158/1078-0432.ccr-03-0139. [DOI] [PubMed] [Google Scholar]
- Yagyu S, Gotoh T, Iehara T, Miyachi M, Katsumi Y, Tsubai-Shimizu S, Kikuchi K, Tamura S, Tsuchiya K, Imamura T, et al. Circulating methylated-DCR2 gene in serum as an indicator of prognosis and therapeutic efficacy in patients with MYCN nonamplified neuroblastoma. Clinical Cancer Research. 2008;14:7011–7019. doi: 10.1158/1078-0432.CCR-08-1249. [DOI] [PubMed] [Google Scholar]
- Yang Q, Sakurai T, Kakudo K. Retinoid, retinoic acid receptor β and breast cancer. Breast Cancer Research and Treatment. 2002;76:167–173. doi: 10.1023/A:1020576606004. [DOI] [PubMed] [Google Scholar]
- Yap TA, Swanton C, de Bono JS. Personalization of prostate cancer prevention and therapy: are clinically qualified biomarkers in the horizon? EPMA Journal. 2012;3:3. doi: 10.1007/s13167-011-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JL, Lv P, Han J, Zhu X, Hong LL, Zhu WY, Wang XB, Wu YC, Li P, Ling ZQ. Methylated TIMP-3 DNA in body fluids is an independent prognostic factor for gastric cancer. Archives in Pathology and Laboratory Medicine. 2014;138:1466–1473. doi: 10.5858/arpa.2013-0285-OA. [DOI] [PubMed] [Google Scholar]
- Zare M, Jazii FR, Alivand MR, Nasseri NK, Malekzadeh R, Yazdanbod M. Qualitative analysis of Adenomatous Polyposis Coli promoter: hypermethylation, engagement and effects on survival of patients with esophageal cancer in a high risk region of the world, a potential molecular marker. BMC Cancer. 2009;9:24. doi: 10.1186/1471-2407-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Glockner SC, Guo M, Machida EO, Wang DH, Easwaran H, Van Neste L, Herman JG, Schuebel KE, Watkins DN, et al. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Research. 2008;68:2764–2772. doi: 10.1158/0008-5472.CAN-07-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LM, Shi DM, Dai Q, Cheng XJ, Yao WY, Sun PH, Ding Y, Qiao MM, Wu YL, Jiang SH, et al. Tumor suppressor XAF1 induces apoptosis, inhibits angiogenesis and inhibits tumor growth in hepatocellular carcinoma. Oncotarget. 2014;5:5403–5415. doi: 10.18632/oncotarget.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou B, Chim CS, Pang R, Zeng H, Dai Y, Zhang R, Lam CS, Tan VP, Hung IF, Lan HY, et al. XIAP-associated factor 1 (XAF1), a novel target of p53, enhances p53-mediated apoptosis via post-translational modification. Molecular Carcinogenesis. 2012;51:422–432. doi: 10.1002/mc.20807. [DOI] [PubMed] [Google Scholar]
- Zurita M, Lara PC, del Moral R, Torres B, Linares-Fernandez JL, Arrabal SR, Martinez-Galan J, Oliver FJ, Ruiz de Almodovar JM. Hypermethylated 14-3-3-sigma and ESR1 gene promoters in serum as candidate biomarkers for the diagnosis and treatment efficacy of breast cancer metastasis. BMC Cancer. 2010;10:217. doi: 10.1186/1471-2407-10-217. [DOI] [PMC free article] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a