Abstract
Large conductance voltage and calcium-activated potassium (MaxiK) channels are activated by membrane depolarization and elevated cytosolic Ca2+. In the brain, they localize to neurons and astrocytes, where they play roles such as resetting the membrane potential during an action potential, neurotransmitter release, and neurovascular coupling. MaxiK channels are known to associate with several modulatory proteins and accessory subunits, and each of these interactions can have distinct physiological consequences. To uncover new players in MaxiK channel brain physiology, we applied a directed proteomic approach and obtained MaxiK channel pore-forming α subunit brain interactome using specific antibodies. Controls included immunoprecipitations with rabbit IgG and with anti-MaxiK antibodies in wild type and MaxiK channel knockout mice (Kcnma1−/−), respectively. We have found known and unreported interactive partners that localize to the plasma membrane, extracellular space, cytosol and intracellular organelles including mitochondria, nucleus, endoplasmic reticulum and Golgi apparatus. Localization of MaxiK channel to mitochondria was further confirmed using purified brain mitochondria colabeled with MitoTracker. Independent proof of MaxiK channel interaction with previously unidentified partners is given for GABA transporter 3 (GAT3) and heat shock protein 60 (HSP60). In HEK293T cells, both GAT3 and HSP60 coimmunoprecipitated and colocalized with MaxiK channel; colabeling was observed mainly at the cell periphery with GAT3 and intracellularly with HSP60 with protein proximity indices of ~0.6 and ~0.4, respectively. In rat primary hippocampal neurons, colocalization index was identical for GAT3 (~0.6) and slightly higher for HSP60 (~0.5) association with MaxiK channel. The results of this study provide a complete interactome of MaxiK channel the mouse brain, further establish the localization of MaxiK channel in the mouse brain mitochondria and demonstrate the interaction of MaxiK channel with GAT3 and HSP60 in neurons. The interaction of MaxiK channel with GAT3 opens the possibility of a role of MaxiK channel in GABA homeostasis and signaling.
Keywords: MaxiK interactome, BKCa, proteomics, mitochondria, neurons, mass spectrometry, GABA transporter, HSP60
Introduction
Large conductance calcium and voltage-activated potassium (MaxiK/BKCa/BK/KCa1.1/Slo) channels are known to play roles in diverse physiological functions such as neuroexcitability (Marrion and Tavalin, 1998, Raffaelli et al., 2004, Filosa et al., 2006), neurovascular coupling for the maintenance of vascular tone (Knot et al., 1998), and cell metabolism (Runden-Pran et al., 2002). Their minimal functional unit consists of four pore-forming α-subunits, each formed by 7 transmembrane domains (S0-S6), and a large C-terminus. At the plasma membrane, the pore forming region and the voltage sensor are located towards the extracellular N-terminus, while signaling sensors are mainly located in the cytoplasmic C-terminus including the Ca2+ sensor, phosphorylation sites, and binding motifs (Toro, 2007, Toro et al., 2014).
MaxiK channel is known to interact with proteins located at the plasma membrane as well as with proteins present in the cytosol, mitochondria, endoplasmic reticulum and nucleus. While some of these interactors regulate MaxiK channel traffic, others are involved in cell excitability, metabolism, development, transport and cell death implicating MaxiK channel in diverse cellular functions (Lu et al., 2006, Toro et al., 2014). For example, in neonatal astrocytes they associate with tubulin and their traffic from the microtubule network to the plasma membrane is modulated by Ca2+ and thromboxane A2 (Ou et al., 2009). In Purkinje cells, MaxiK channels cocluster with Ca2+ channels [Ca(v)2.1] supporting their role in the regulation of cellular excitability (Edgerton and Reinhart, 2003, Indriati et al., 2013).
Evidence for MaxiK channel localization in intracellular organelles keeps increasing [see recent reviews, (Singh et al., 2012b, Balderas et al., 2015)]. The first studies in mitochondria were performed in a glioma cell line (Siemen et al., 1999) and later in cardiomyocytes where MaxiK channel plays a cytoprotective role (Xu et al., 2002, Singh et al., 2012b, Singh et al., 2013). In the brain, its presence has been demonstrated in mitochondria of astrocytes, where it participates in the regulation of the permeability transition pore (Cheng et al., 2011), and in the nuclear envelope of hippocampal neurons, where it regulates gene expression by modulating nuclear calcium signaling (Li et al., 2014). This data indicates that localization in different subcellular compartments determines the multiple functions of the channel most likely due to its interactions with different subsets of proteins in each compartment. Even though splice variation is an important intrinsic mechanism for MaxiK channel differential localization (at least for endoplasmic reticulum and mitochondria) (Zarei et al., 2004, Ma et al., 2007, Singh et al., 2013), the cellular mechanisms determining the targeting of MaxiK channel to distinct organelles are not known. Thus, identification of binding partners of MaxiK channel can provide vital information not only about the intracellular transport, trafficking and targeting of MaxiK channel to intracellular organelles such as mitochondria and nucleus, but also on its diverse functions.
Few studies have been directed to identify the interactome of MaxiK channel and have focused to the cochlea (Kathiresan et al., 2009, Sokolowski et al., 2011) and brain cortical membranes (Gorini et al., 2010). However, the identification of MaxiK channel binding partners in the whole brain has not been addressed. In this study, we have carried out a MaxiK channel targeted proteomic analysis and discovered new partners of MaxiK channel in the brain. In addition, using alternative experimental approaches in HEK293T cells and primary hippocampal neurons, sodium and chloride dependent GABA transporter 3 (GAT3) and heat shock protein 60 (HSP60) were further established as their binding partners.
Material and Methods
Antibodies
Primary antibodies were: Anti Maxi-K pAb (APC021, lot AN12, Alomone labs), anti-MaxiK channel mAb (75-022, clone L6/60; UC Davis/NIH NeuroMab facility), anti-HSP60 (ab46798, Abcam), anti-GAT3 (480018, Invitrogen), Cox2 (A6407, Invitrogen) and anti-FLAG (F3165, Sigma). For Western blots, secondary antibodies were: IR Dye 800CW goat anti-mouse (926-32210, LI-COR), Alexa Fluor 680 goat anti-rabbit IR Dye (A21109, Invitrogen). For immunochemistry, secondary antibodies were: Alexa 488 (A-11008, Molecular Probes) and Atto 647N (50185, Sigma) goat anti-mouse IgGs, and Atto 647N goat anti-rabbit IgGs (15048, Active Motif).
Clones
Heat shock protein 60 kDa 1 (HSPD1 c-Myc-DDK-tagged, ORIGENE RC201281); Human solute carrier family 6 (SLC6A11 c-Myc-DDK-tagged, ORIGENE RC216392) and BKCa (N-terminal tagged with c-Myc epitope, pcDNA3) were used.
Brain lysate preparation and immunoprecipitation
Immunoprecipitations were carried out as illustrated in Fig 1 and previously described (Singh et al., 2009, Singh et al., 2013). Briefly, mouse brains from wild type and Kcnma1−/− mice were homogenized with mild lysis buffer (in mM, 50 Tris-HCl pH7.4, 150 NaCl, 5 EDTA, 10 Hepes, NP-40 0.1%, and Na-deoxycholate 0.25%). Lysates were centrifuged at 1,000 xg for 5 min at 4°C to remove unbroken cells and tissues. Supernatant was carefully aspirated to fresh tubes and incubated at 4°C on a rotatory shaker for 1 hr for further lysis. After incubation, samples were centrifuged at 12,000 xg for 10 min at 4°C and precleared with 25 μL protein A/G resin/mg protein (Pierce Biotechnology Inc.) for 1 hr at 4°C. Samples were centrifuged at 1000 xg for 5 mins at 4°C to remove protein A/G resin. Supernatants were collected, and protein concentration was estimated according to the manufacturer’s instructions (BIO-RAD). Anti-MaxiK antibodies (5 μg) or IgGs were incubated for 1 hr at 4°C with 50 μl protein A/G resin on a rotatory shaker to immobilize antibodies with beads. Unbound antibodies were removed by centrifugation at 1000 xg for 5 min at 4°C. Brain lysates (5 mg each) were added to 1.5 ml Eppendorf tubes containing immobilized anti-MaxiK and IgG antibodies on 30 μl of protein A/G resin, and incubated on a rotatory shaker for 1 hr at 4°C to allow binding of MaxiK channel to immobilized antibodies. Proteins bound to immobilized antibodies were washed three times with mild lysis buffer and centrifuged at 1000 xg for 5 min each at 4°C. 30 μL 3X Laemmli sample buffer was immediately added and samples were incubated at 37°C for 1 hr for elution. Eluted proteins were briefly centrifuged at 1000 xg and separated by SDS-PAGE using 4–20% precast gels at room temperature. Protein bands were stained with SYPRO-RUBY stain according to the manufacturer’s instructions (BIO-RAD).
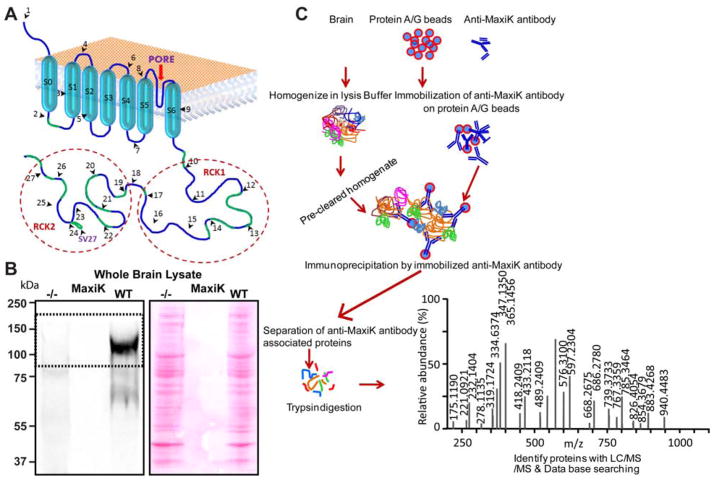
Figure 1. Schematic representation of the binary proteomic analysis of MaxiKα channel protein network in mouse brain mitochondria.
A. Cartoon representing structure of MaxiK channel α subunit. Transmembrane domains are labeled from S0–S6, exons are represented by black arrowheads and numbers, and pore is shown by a red arrow. Blue line represents amino acids and green line represents relative position of peptides identified by mass spectrometry. A peptide detected for spliced variant 27 (SV27) is marked with an extended loop before exon 24. B. Anti-MaxiK antibodies used for immunoprecipitations showed a specific band at ~125 kDa corresponding to MaxiK channel (dashed box) but no signal was detected in Kcnma1−/− mice brain. As control, Ponceau S-stained membrane is shown to indicate that equal amounts of whole brain lysate were loaded in both lanes. C. Brain was harvested and lysate was prepared from wild type and Kcnma1−/− mice. For IgG control experiments only wild type brain lysates were used. Brain lysates were precleared with protein A/G beads. In parallel anti-MaxiK and IgG antibodies were also immobilized on protein A/G beads. Precleared brain lysates were incubated with the immobilized anti-MaxiK antibodies or IgGs. Bound fraction was washed with the lysis buffer and immunoprecipitates were run on a SDS PAGE gel and stained with SYPRO Ruby. Gel bands (3 X 8 mm) from wild type, IgG control, and Kcnma1−/− lane were excised, trypsin digested and analyzed by mass spectrometry. Proteins present in IgG or knock out (Kcnma1−/−) lanes were discarded. A representative spectra shows the m/z scores for a MaxiK channel specific-peptide.
Mass spectrometry analysis of proteins
SYPRO-RUBY stained lanes for the wild type and Kcnma1−/−mice brain immunoprecipitates were excised using a sterile scalpel. Adjacent bands ranging from 10 kDa to 250 kDa were collected and digested with trypsin. Protein digestion and elutions were carried out as described earlier (Singh et al., 2009, Singh et al., 2013). LC/MS/MS was performed at the University of California Los Angeles W. M. Keck Proteomic Center using a Thermo LTQ-Orbitrap XL mass spectrometer equipped with an Eksigent Nano-LC-1D Plus liquid chromatography system and an Eksigent auto sampler as described (Singh et al., 2013). The LC/MS/MS spectra were searched against Swiss-Prot mouse database using MASCOT Daemon search engine (Matrix Science). The criteria used to accept a protein as authentic within a gel band was a minimum of two unique peptides ranked as number 1 with ion scores with a p<0.05. Unique proteins obtained for each gel band from wild type mice were compared with those obtained using Kcnma1−/− mice. All the peptides appearing in both animals were treated as non-specific associated proteins and not taken for further analysis. Unique proteins appearing only in the wild type mice lane were tabulated and categorized according to their cellular localization. In parallel experiments, proteins appearing in anti-MaxiK immunoprecipitates as well as IgG controls from wild type mice were considered as non-specific binding partners and were not further analyzed.
Cell culture and transfection
For immunolabeling, HEK293T cells were cultured on coverslips (0.17-mm thickness) coated with poly-L-lysine [0.1% (v/v)]. For immunoprecipitation, cells were cultured on 100-mm plates. Culture media was DMEM supplemented with 10% (v/v) FBS, 2 units/mL penicillin and streptomycin. Transient transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Primary neurons were derived from the hippocampi of rat pups via mechanical dissociation of papain (1 mg/ml for 20 minutes) digested tissue. Neurons were plated on poly-D-lysine coated coverslips (0.05 mg/ml) in MEM supplemented with 5% FBS, 0.1 mg/ml Transferrin, 2% B27, 0.5% Glucose, 24 mg/L insulin, 2 mM GlutamaxTM). 72 hours after plating cells, feeding medium was supplemented with 2 μM Cytosine β-D-Arabinofuranoside to avoid proliferation of astrocytes. Cells were maintained in a humidity-controlled 37°C CO2 [5% (v/v)] incubator.
Immunoprecipitation from transfected HEK293T cells
Cells were lysed 2 days after transfection with mild lysis buffer containing protease inhibitor tablets (Complete Protease Inhibitor Cocktail Tablets; Roche; one tablet/50 mL). Lysates were centrifuged at 13,000 xg for 10 min at 4°C and the supernatant was precleared with 10 μL protein A/G resin/mg protein for 1 h at 4°C on a rotatory shaker. The precleared lysates (1 mg protein) were incubated overnight at 4°C with 10 μL protein A/G resin preconjugated with 2 μg anti-MaxiK mAb antibody in a total volume of 500 μL. After incubation, samples were centrifuged at 5,000 xg for 2 min and washed for five times with mild lysis buffer. The immunoprecipitated proteins were eluted from the beads with 30 μL 3X Laemmli sample buffer at 37°C for 1 hr. Samples were centrifuged at 13,000 xg for 3 min at 4°C and separated on 4–20% SDS-PAGE along with whole cell lysates. Separated proteins were transferred to nitrocellulose membranes and immunoblotted with anti-MaxiK mAb (1 μg/mL) or anti FLAG (1 μg/mL) antibodies. Single- or double-labeled immunoblots were imaged with Odyssey Infrared Imaging system (LI-COR Biosciences).
Isolation of brain mitochondria
Mouse brains were excised and minced into small pieces. Using a Potter-Elvejem homogenizer, brain samples were homogenized (7 rapid strokes) in an isolation buffer A (70 mM sucrose, 210 mM mannitol, 1 mM EDTA-Na2 and 50 mM Tris-HCl, pH 7.4). The homogenate was centrifuged at 1,300 xg for 3 min. The supernatant was carefully removed to a fresh tube and centrifuged at 12,000 xg for 10 min. The mitochondrial pellet was resuspended in 55 μL of isolation buffer A, and gently overlaid on 3 mL of 30% (v/v) Percoll solution in buffer B (250 mM sucrose, 10 mM Hepes, 1 mM EDTA-Na2, pH 7.4). Mitochondrial samples were centrifuged at 50,000 xg for 45 min at 4°C and three layers (M1, M2 and M3) of mitochondria were obtained as described and characterized earlier (Singh et al., 2012a). Purified M3 mitochondrial fraction was collected and centrifuged at 10,000 xg for 5 min. Mitochondrial pellet M3 was washed two times with 1 mL of isolation buffer A and resuspended in 100 μL of the same buffer A.
Immunolabeling
Rat primary hippocampal neurons were isolated according to the protocol described earlier (Chang et al., 2012). Cover glasses coated with poly-L-Lysine (0.1% (v/v) in PBS) were used for seeding isolated mitochondria, HEK293T cells or primary neurons. All mitochondrial samples were preloaded with 500 nM MitoTracker Red CMXRos (Singh et al., 2012a) for 60 min at 4°C, and kept in the dark at 4°C to prevent photo bleaching. All the samples were washed with ice cold PBS (2 mM KCl, 1.5 mM KH2PO4, 138 mM NaCl, 8.1 mM Na2HPO4) for 5 min each and fixed with 4% (w/v) paraformaldehyde in PBS for 10 min at room temperature. Samples were then permeabilized with 0.5% (v/v) Triton-X-100 in PBS for 10 min and blocked with 10% (v/v) normal goat serum containing 0.2% (v/v) Triton-X-100 for 15 min. Neuronal samples were incubated with primary antibodies (anti-MaxiK, 1 μg/mL mAb; anti-HSP60, 1 μg/mL; and anti-GAT3, 1 μg/mL), HEK293T cells with primary antibodies (anti-FLAG mAb 1 μg/mL; and anti-MaxiK, 1 μg/mL mAb) and mitochondrial samples with primary antibodies (anti-MaxiK 2 μg/ml pAb; and 0.2 μg/ml anti-Cox2) diluted in 0.2% Triton-X-100/PBS overnight at 4°C. After incubation, samples were washed with PBS for three times, and incubated with secondary antibodies conjugated with Atto647N (1μg/mL anti-mouse and anti-rabbit IgGs) or Alexa 488 (2 ug/mL anti-mouse IgG) in 1% (w/v) NGS, 0.2% (v/v) Triton-X-100 in PBS. Cover glasses were dried with a filter paper and mounted on to clean glass slides using ProLong Gold (Invitrogen). Confocal images were acquired with an Olympus confocal microscope at 56.65 nm/pixel.
Image Analysis
Confocal images of isolated mitochondria, HEK293T cells and primary neurons were median filtered (median intensities of 64 X 64-pixel and 32 X 32-pixel squares centered at the target pixel, respectively) and protein proximity indices were calculated as described earlier (Wu et al., 2010, Singh et al., 2012a).
Results
Immunoprecipitation of MaxiK channel from the whole brain
To identify the proteins that can form macromolecular complexes with MaxiK channel, we decided to use a proteomic approach on MaxiK channel immunoprecipitates obtained from mouse brain homogenates. First we tested the specificity of an anti-MaxiK polyclonal antibody (pAb) targeting the C-terminus of the pore-forming α subunit of the channel (Fig 1A). Anti-MaxiK pAb (Alomone) proved to be specific for its target as it detected a single band of the expected MaxiK channel α subunit mass at ~125 kDa in the wild type but not in the Kcnma1−/− mouse brain lysates (Fig 1B). Using this specific anti-MaxiK pAb, we have carried out immunoprecipitations from brain lysates of wild type (WT, n=3 brains) and Kcnma1−/− (n=3 brains) mice, the latter as one of our controls. The second control consisted in immunoprecipitations with IgG-instead of anti-MaxiK pAb- from WT brains (n=3 brains). As shown in the SYPRO Ruby stained gel of Fig 1C for WT and Kcnma1−/−, all immunoprecipitates were subjected to 1-D electrophoretic separation followed by systematic sectioning from top to bottom for in-gel trypsinization. Resulting peptides were subjected to LC/MS/MS for identification and further data mining. Peptides obtained were systematically analyzed with m/z ratio to verify amino acid sequences (Fig 1C). The anti-MaxiK antibody successfully immunoprecipitated MaxiK channel as LC/MS/MS analysis of bands ~ 100–125 kDa yielded a total of 14 unique MaxiK channel-specific peptides using WT brains (supplementary table 1) but not in the controls. We have also immunoprecipitated a peptide from a spliced variant exon 27 (SV27) (Fig 1A and supplementary table 1). These results give confidence to the identification of the coimmunoprecipitated proteins as being native partners of MaxiK channels.
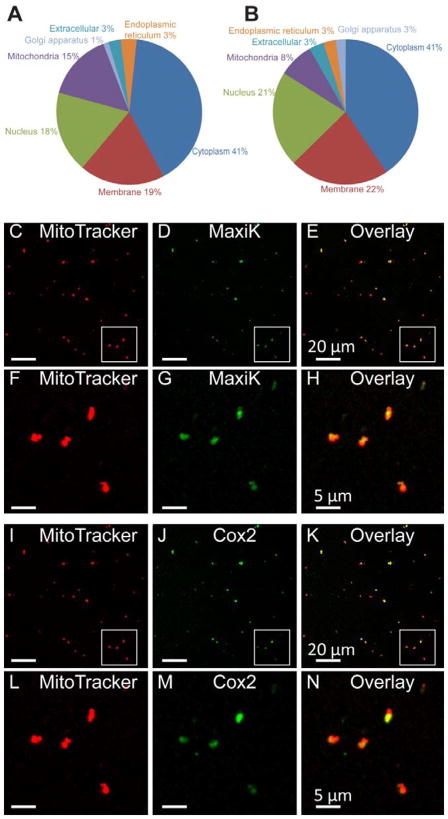
Proteomic analysis yielded 255 and 911 unique proteins as part of the MaxiK channel interactome after subtracting IgG and Kcnma1−/− results, respectively, to eliminate false positives. Proteins with scores of 20 and above, and as mentioned not appearing in the respective controls, were considered as MaxiK channel interacting proteins. The majority of MaxiK channel interactome belonged to proteins localized to the cytoplasm (39% and 41%), plasma membrane (19% and 24%), nucleus (18% and 19%), mitochondria (15% and 8%), extracellular region (3% and 3%), Golgi apparatus (1% and 2%), and endoplasmic reticulum (3% and 3%) with immunoprecipitation controls being IgGs for Fig 2A and Kcnma1−/− for Fig 2B. Several (2%) uncharacterized proteins, not attributed to any cellular compartment were also obtained after subtracting those found in Kcnma1−/− mouse brains. All the MaxiK channel interacting proteins are listed in supplementary table 2 (minus IgG results) and supplementary table 3 (minus Kcnma1−/− results). Overall the results support the idea that in addition to its presence at the plasma membrane where it interacts with cytosolic proteins (Toro et al., 2014), MaxiK channel may also be a significant resident of brain intracellular organelles like the nucleus and mitochondria.
Figure 2. Mass spectrometry analysis of brain MaxiK channel sub-proteome.
A. Pie chart showing distribution of MaxiK channel interactome in experiments using IgG as controls. B. Pie chart summarizing the distribution of MaxiK channel interactome using Kcnma1−/− as controls. MaxiK channel interactome pointed to proteins localized to mitochondria as MaxiK interactors. C–H. On probing with anti-MaxiK antibodies, MaxiK channel was found to be present in MitoTracker loaded brain mitochondria. I–N. As control, the same preparation of mitochondria loaded with MitoTracker was also probed with anti-cytochrome c oxidase IV subunit II (Cox 2) antibodies. Cox2 also localizes to the mitochondria (n= 3 independent experiments).
Recently, MaxiK channel has been visualized at the nuclear envelope of hippocampal neurons (Li et al., 2014). Although MaxiK channel activity has been recorded after reconstitution of brain mitochondria into lipid bilayers (Skalska et al., 2009) and in mitoplasts of astrocytes (Cheng et al., 2008) (for a recent review see Balderas et al., 2015) direct visualization in brain mitochondria has not been reported yet. Thus, we tested the localization of MaxiK channel in purified mitochondria (Singh et al., 2012a). Fig 2C,D show signals of purified brain mitochondria that have been loaded with MitoTracker and labeled with the specific anti-MaxiK antibody, respectively. The overlay of both images (Fig 2E) shows significant overlap demonstrating MaxiK channel presence in brain mitochondria. Colocalization between mitotracker and the mitochondrial protein cytochrome c oxidase IV subunit II (Cox 2) confirm the quality of our mitochondria preparation (Fig 2 I–K). For better detail, Figure 2F–H and L–N are corresponding magnifications of the regions (squares) in Fig 2C–E and 2I–K, respectively. The protein proximity index (PPI) of mitochondria and MaxiK channel was 0.55 ± 0.15 (n=3 independent experiments) indicating that approximately 55% of MaxiK channels colocalizes with MitoTracker labeled mitochondria.
MaxiK channel interacts with HSP60
Consistent with its localization in mitochondria, one of the partners of MaxiK channel found in our proteomic analysis is heat shock protein 60 (HSP60) which is known to play an important role in the folding of proteins in mitochondria but can also be present in the cytosol under stress conditions (Cheng et al., 1990). Therefore, we decided to obtain evidence independent of mass spectrometry and in a different expression system for the interaction of HSP60 with MaxiK channel.
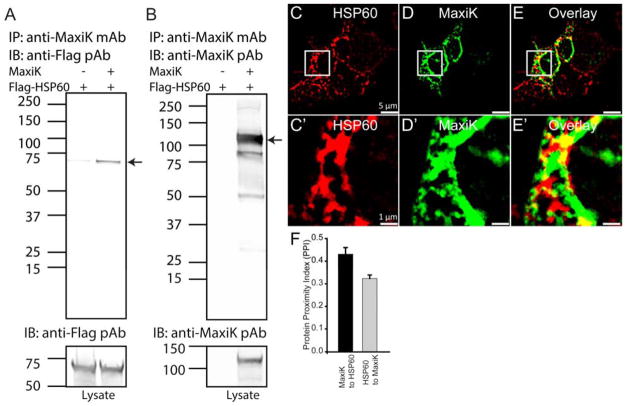
The independent assays we used were co-immunoprecipitation and immunolabeling. To this end, MaxiK α-subunit was co-transfected with N-terminus FLAG-tagged HSP60 (Flag-HSP60) in HEK293T cells. Using anti-MaxiK monoclonal antibodies (mAb) to immunoprecipitate MaxiK channel, we were able to pull down HSP60 which was recognized by anti-Flag pAb (Fig 3A) (n=4). As positive control, Fig 3B shows that MaxiK channel was indeed immunoprecipitated. In this case the protein was detected with anti-MaxiK pAb (Fig 3B). The bottom panels demonstrate appropriate expression of either MaxiK channel or HSP60 in the lysates.
Figure 3. Immunoprecipitation and colocalization of HSP60 with MaxiK channels.
Flag-HSP60 was transfected in HEK293T cells with or without MaxiK channel plasmid. A. Flag-HSP60 was transfected with or without MaxiK channel in HEK293T cells. Anti-flag antibody showed a signal for HSP60 in a lane cotransfected with MaxiK channel and HSP60. Lower panel shows presence of HSP60 in both lanes. B. MaxiK channel was immunoprecipitated with anti-MaxiK mAb and immunoblotted with anti-MaxiK pAb. MaxiK channel was seen migrating at the expected size in a lane containing cell lysates transfected with MaxiK channel. Lower panel shows presence of MaxiK channel. C–D. HEK293T cells transfected with Flag-HSP60 and labeled with anti-Flag antibodies. E. Overlay of C and D. C′–E′. Squared region from C, D and E. On close analysis MaxiK channel showed punctate association with HSP60. F. Bar graph representing PPI for MaxiK channel with HSP60 (black) and HSP60 with MaxiK channel (grey) in HEK293T cells.
Confocal microscopy and double immunolabeling of HSP60 and MaxiK channel are shown in Fig 3C–E. Fig 3C and C′ shows clear intracellular labeling of HSP60 with fluorophore-conjugated anti-Flag mAb; while Fig 3D and D′ displays the same cells but labeled with anti-MaxiK pAb. Note regions of strong coincidence in the overlaid image (Fig 3E and E′).
For closer inspection, squares in C–E were magnified in panels C′–E′. PPI analysis revealed score of MaxiK channel to HSP60 is 0.43±0.03 and HSP60 to MaxiK channel is 0.32±0.02 (n=5 independent experiments containing 16 individual cells). Together, the results demonstrate that HSP60 and MaxiK channels are indeed capable of associating with each other.
MaxiK channel interacts with GABA transporter 3
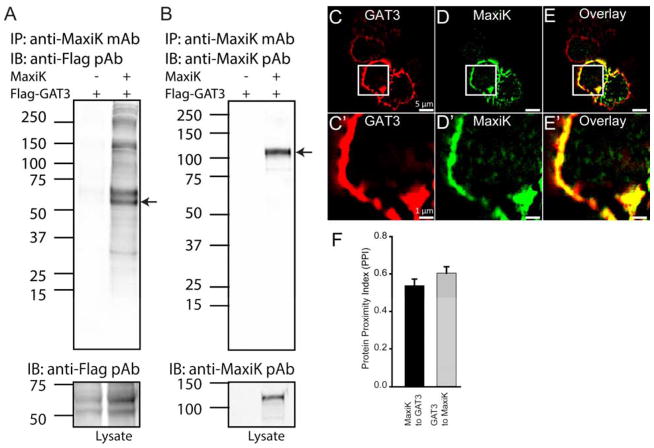
We next analyzed the list of previously unidentified MaxiK channel partners at the plasma membrane and found the GABA transporter 3 (GAT3) as a potentially important partner which is predominantly expressed in glial processes (Galvan et al., 2005). Interaction of MaxiK channel and GAT3 has not been reported earlier but both are known to be regulated by cytosolic calcium ions. Thus, we proceeded to demonstrate their association following the same strategy as for HSP60. To this end, N-terminus FLAG-tagged GAT3 and MaxiK channel were co transfected in HEK293T cells. When MaxiK channel was immunoprecipitated, it also pulled down GAT3 which was verified using an anti-Flag antibody (Fig 4A, lane 2). As negative control, no signal was observed if MaxiK channel was not transfected (Fig. 4A, lane 1). MaxiK channel immunoprecipitation with anti-MaxiK mAb was also verified by anti-MaxiK pAb (Fig 4B, lane 2). Lower panels show appropriate expression of both clones.
Figure 4. Immunoprecipitation and colocalization of GAT3 with MaxiK channels.
HEK293T cells transfected with FLAG-GAT3 and with or without MaxiK channel plasmid were lysed for immunoprecipitations. A. Flag-GAT3 was transfected with or without MaxiK channel in HEK293T cells. On probing with anti-Flag antibody, a signal corresponding to GAT3 molecular weight was detected only the lane co-transfected with MaxiK channels. Lower panel shows presence of GAT3 in both lanes. B. MaxiK channel was immunoprecipitated with anti-MaxiK mAb and immunoblotted with anti-MaxiK pAb. MaxiK channel was seen migrating at the expected size in a lane containing cell lysates transfected with MaxiK channel. Lower panel shows presence of MaxiK channel. C–D. Ectopically expressed MaxiK channel and GAT3 in HEK293T cells showed strong colocalization in the cell periphery. E. Overlay of C and D. C′–E′. Squared region from C, D and E. On close analysis MaxiK channel showed association with GAT3 at the periphery of the HEK293T cells. F. Bar graph representing PPI for MaxiK channel with GAT3 (black) and GAT3 with MaxiK channel (grey) in HEK293T cells.
To provide further evidence of the interaction of MaxiK channel with GAT3, MaxiK channel was co-transfected with N-terminus Flag-tagged GAT3 in HEK293 cells for immunolabeling of permeabilized cells. As shown in Fig 4C and D, both GAT3 and MaxiK channel are expressed at the cell periphery (likely plasma membrane) and the signals are highly coincidental (Fig. 4E, overlay). Magnification of the marked squares in Fig 4C–E highlights this property (Fig 4C′–E′). As expected from the visual inspection, image analysis (Fig 4F) yielded a high PPI score of MaxiK channel to GAT3 of 0.54±0.04 and of GAT3 to MaxiK channel of 0.60±0.03 (n=5 independent experiments containing 14 individual cells).
In summary, coimmunoprecipitation and colocalization analysis of MaxiK channel with GAT3 or HSP60 revealed that these proteins are located close to each other, which may facilitate a coordinated function.
Colocalization of MaxiK channels with GAT3 and HSP60 in primary hippocampal neurons
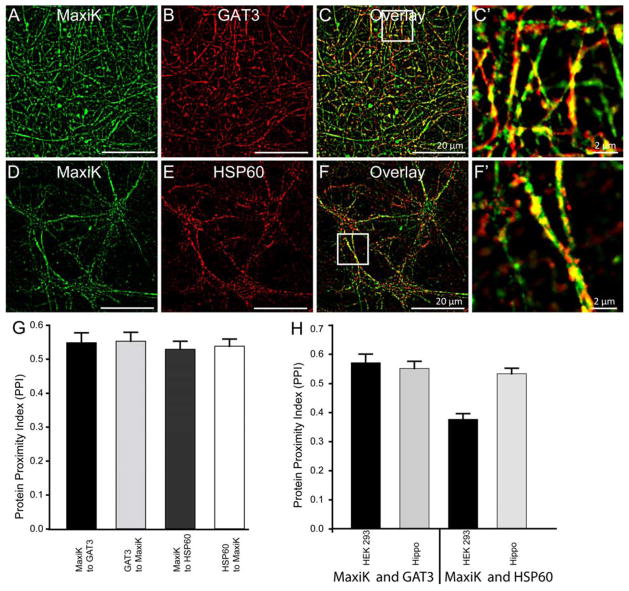
As heterologous expression may have a problem of overexpression perturbing natural interactions, we went ahead to determine whether GAT3 or HSP60 are found close enough in neuronal cells as to observe colocalization. To this end, we colabelled primary hippocampal neurons (7 days in vitro) with anti-MaxiK, anti-GAT3 and anti-HSP60 antibodies. Confocal images of hippocampal neurons showed a high level of MaxiK channel and GAT3 overlap (Fig 5A–C, C′) as well as a high degree of co-localization of MaxiK with HSP60 proteins in these cells (Fig 5D–F, F′). Image analysis revealed a PPI for MaxiK channel to GAT3, of 0.55±0.03 and for GAT3 to MaxiK channel of 0.55±0.03 (n=5 independent experiments containing 25 individual cells) (Fig 5G). Similarly, PPI of MaxiK channel to HSP60 is 0.53±0.02 and for HSP60 to MaxiK channel is 0.54±0.02 (n=5 independent experiments containing 21 individual cells) (Fig 5G). The values in hippocampal neurons for MaxiK channel and GAT3 are in close agreement with those obtained in cotransfected HEK293T cells (Fig 5H). The latter supports the idea that HEK293T cells results are indeed reflecting the normal ability of these protein partners to interact with MaxiK channel. However for MaxiK channel and HSP60, PPI in hippocampal neurons is 0.53±0.02 (n=5 independent experiments containing 21 individual cells) which is significantly higher (p<0.05) than PPI for MaxiK channel and HSP60 in HEK293 cells 0.38±0.02 (n=5 independent experiments containing 16 individual cells). This could be attributed to a preferential membrane localization of MaxiK channel in HEK293T cells.
Figure 5. MaxiK channel colocalizes with GAT3 and HSP60 in primary hippocampal neurons.
Primary hippocampal neurons were labeled with anti-MaxiK (A) and anti-GAT3 antibodies (B). C. Overlay image of A and B showed high degree of colocalization of MaxiK channel and GAT3 in neurons. C′ is enlarged squared region from C. Neurons were also colabelled with anti-MaxiK (D) and anti-HSP60 antibodies (E). As seen in HEK293T cells, HSP60 and MaxiK channel also colocalizes in primary neurons (F). F′ is an enlarged squared region from F. G. Bar graph representing colocalization index for MaxiK channel with GAT3 (black), GAT3 with MaxiK channel (light grey), MaxiK channel with HSP60 (dark grey) and HSP60 with MaxiK channel (white) in hippocampal neurons. H. Bar graph representing colocalization between MaxiK channel and GAT3 as well as MaxiK channel and HSP60 in HEK293 and hippocampal neurons, respectively.
Discussion
Since the initial discovery of MaxiK channels more than three decades ago, remarkable advancements have led to identification of key residues involved in functional units responsible for activation of channels and gating. At the same time, information on its complex molecular interactions that can affect its translocations, localization and subsequently its functional role is emerging. MaxiK channels are widely distributed in different tissues, intracellular compartments (both plasma membrane and intracellular organelles) and serve many diverse roles. In order to support its distribution and functional diversity, MaxiK channels are regulated by numerous mechanisms. Even though MaxiK channels are increasingly shown to be involved in vital physiological roles, there are very few attempts to generate a complete MaxiK interactome (Xia et al., 1998, Kathiresan et al., 2009, Gorini et al., 2010, Sokolowski et al., 2011, Peng et al., 2014). We have immunoprecipitated neuronal MaxiK channels from the mouse brain using two important controls involving IgG as a non-specific antibody binding control and Kcnma1−/− as a non-specific anti-MaxiK binding control. We have combined both approaches to minimize false positive and false negative discovery occurrences.
MaxiK channels are encoded by a nuclear gene Kcnma1 and after translation are transported to their target membranes. From synthesis to transportation, MaxiK channel is found in the cytoplasm and it is not surprising that most of its interacting partners are present in the cytoplasm. As expected, they included large 60S ribosomal proteins known to catalyze peptide bond formation and also contain the nascent polypeptide exit tunnel (Klinge et al., 2011). In addition, several cytoskeletal proteins such as dynamin 2 and 3, dynamin-like 120 KDa proteins, F-actin-capping proteins, actin, actin-like proteins, myosins, microtubule-associated proteins, and tubulins were found to be interacting with MaxiK channel. These cytoskeletal specific interactions could facilitate intracellular trafficking, regulation of channel activity and also maintain polarized expression of ion channels within specific membrane domains. One of the key challenges lying ahead is to establish the functional relationship between MaxiK channels and their association with cytoskeletal elements. Given that MaxiK channels also play relevant roles in the physiology of organelles such as nucleus and mitochondrion, elucidating their molecular interactors is a key element to enhance our understanding in the intricacies of their regulatory networks.
Recently, MaxiK channel in the nucleus was shown to be involved in the regulation of nuclear Ca2+-sensitive gene expression and in the promotion of dendritic arborization (Li et al., 2014). Interestingly, the majority of MaxiK channel organelle interactome was found to be localized to the nucleus. As shown in supplementary tables II and III, proteomic analysis of the brain showed that MaxiK channel can interact with several DNA polymerases, topoisomerases, repair proteins and RNA polymerases implying that MaxiK channel could be playing a direct role in replication and transcription. Another class of proteins which were present in MaxiK channel interactome was zinc finger proteins. Zinc finger proteins are amongst the most abundant proteins in mammalian genomes (Laity et al., 2001). Interaction of MaxiK channel with zinc finger proteins can implicate MaxiK channel in DNA recognition, RNA packaging, transcriptional activation, and lipid binding. Interaction with nucleoporin-like protein 2 indicates that MaxiK channel could also be involved in the regulation of export of mRNAs from the nucleus to the cytoplasm. All of these interactions need further experimental analysis.
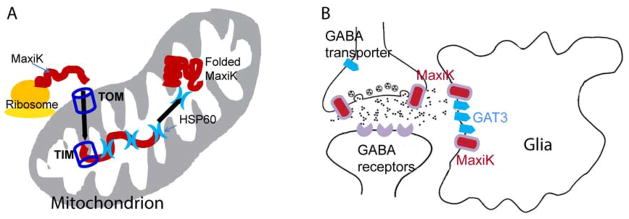
MaxiK channels have been shown to be present in the mitochondria and channel activity was recorded from the inner mitochondrial membrane (Xu et al., 2002) implying that the ~140 kDa protein encoded in the nucleus needs to unfold during shuttling to mitochondria and then refold to its native conformation. During the import to the mitochondrial matrix, classically proteins pass through the outer mitochondrial membrane (via the translocase of the outer membrane [TOM] system) and inner mitochondrial membrane (utilized the translocase of the inner membrane [TIM] system) (Neupert, 1997). In MaxiK channel interactome, we have immunoprecipitated chaperones (HSP 60, 70, 71, 90, 105) and TOM proteins 34 and 70 with MaxiK channel. Interestingly, HSP60 as well as HSP70 are known to participate in protein folding after import to the mitochondrion (Li and Srivastava, 2004). We have further evaluated the interaction of HSP60 with MaxiK channel and found that HSP60 does interact with MaxiK channel in HEK293T cells and colocalizes with the channel in primary neurons. Our current results support the hypothesis that the import of MaxiK channel to the mitochondria is facilitated via classical translocase pathways and that HSP60 and HSP70 may help MaxiK channel fold back to the functional form in mitochondria (Fig. 6A). However, specific roles of HSP60 and HSP70 in mitochondrial MaxiK channel import mechanisms via classical TOM and TIM pathway needs to be evaluated.
Figure 6. Potential roles of HSP60 and GAT3 with MaxiK channels.
A. MaxiK channel is present in mitochondria of cardiac and neuronal cells. However, its mechanism of translocation to mitochondria is not known. A hypothetical pathway is shown where TOM and TIM facilitate translocation of MaxiK channel to mitochondria. Since HSP60 immunoprecipitates with MaxiK channel, interacts with MaxiK channel in HEK293T and colocalizes with MaxiK channel in hippocampal primary neurons, it could be facilitating the refolding of MaxiK channel in the mitochondria. B. The interaction of MaxiK channel and GAT3 may indicate that MaxiK channel could be directly involved in GABAergic signaling. MaxiK channel and GAT3 are present in glia and their interaction could be playing a direct role in neuronal signaling.
MaxiK channels are ubiquitously expressed throughout the central nervous system but found preferentially in axons and presynaptic terminals (Knaus et al., 1995, Knaus et al., 1996, Faber and Sah, 2003). Depending on their localization and functional or physical association with other proteins in different neuronal cells, MaxiK channels are known to play complex roles in neuronal excitability as they can regulate high frequency firing, as well as the differential release of excitatory and non-excitatory neurotransmitters (Knaus et al., 1996, Faber and Sah, 2003, Lu et al., 2006, Kundu et al., 2009, Martire et al., 2010, Toro et al., 2014, Deng and Klyachko, 2015). In our studies, we have found several proteins involved in neuronal signaling that associate with MaxiK such as dopamine- and cAMP-regulated neuronal phosphoprotein (DARP-32), potassium voltage-gated channel subfamily KQT member 3, synaptosomal-associated protein 25, synaptic vesicle glycoprotein 2B, synaptotagmin-2, synapsin-2, neural cell adhesion molecule 1 and 2, and vesicular glutamate transporter 1 (supplementary table 2 and supplementary table 3). Further probing of some of these associations in specific regions of the brain can provide an insight into differential regulatory mechanisms of MaxiK channels.
MaxiK channels have also been shown to be present in murine cortical astrocytes, and associated with the microtubule network (Ou et al., 2009). Further, MaxiK channels have been implicated in the modulation of both excitatory and inhibitory synaptic transmission via suppression of glutamatergic synaptic depolarization and the consequent Cav channel activation in the retina (Grimes et al., 2009). IPSC experiments evoked by glutamate puff showed that Cav-mediated GABA release is diminished by MaxiK channel. It was proposed that L-type Cav channels, MaxiK channels, CP-AMPARs and CICR work in concert within single varicosities to provide an extended range of reciprocal feedback inhibition (Grimes et al., 2009). Our experiments place GAT3 and MaxiK channels together interacting with each other either directly or via secondary proteins. Because GAT3 is known to be preferentially expressed in glia (Galvan et al., 2005, Jin et al., 2011), together these results could indicate that association of MaxiK channels with GAT3, most likely in glia, may limit GABA release hence regulating the flow of excitatory synaptic transmission (Fig 6B). A direct functional test could provide information of this interaction.
The current effort to identify the MaxiK channel brain interactome has discovered potential binding partners of this channel protein that are known to form part of transcription, translation, protein sorting, translocation and localization protein networks. In this work, we have established beyond doubt the association of MaxiK channels with HSP60 and GAT3 opening new research avenues of MaxiK channel biology. Future work will determine the role of these protein interactions on modulation and transportation of MaxiK channels and our study provides an extensive database to identify signaling pathways involved in MaxiK channel-mediated cell signaling.
Supplementary Material
Highlights.
Identified the interactome of MaxiK channels in the mouse brain containing 1185 proteins using two independent approaches.
Established localization of MaxiK channels in the brain mitochondria.
Confirmed interaction of MaxiK channel with heat shock protein 60.
Confirmed interaction of MaxiK channel with GAT3.
Acknowledgments
We thank UCLA members Dr. Melissa Sondrej, Dr. Gregg Czerwieniec, and Dr. Arnab Chattopadhyay for their assistance and helpful discussion for mass spectrometry, and Dr. Felix E. Schweizer for providing hippocampal neurons for this study. This work was supported, in whole or in part, by National Institutes of Health Grants HL096740 and HL107418 (to E. S. and L. T.) and HL088640 (to E. S.). This work was also supported by American Heart Association National Scientist Development Award 11SDG7230059 (HS), Commonwealth Universal Research Enhancement (CURE) Program from the Pennsylvania Department of Health (HS), and American Heart Association Postdoctoral Fellowship 0825273F (to M. L.).
Abbreviations
- CICR
Calcium-induced calcium release
- Cox 2
cytochrome c oxidase IV subunit II
- CP-AMPARs
calcium-permeable α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors
- DMEM
Dulbecco’s Modified Eagle Medium
- EDTA
Ethylenediaminetetraacetic acid
- FBS
fetal bovine serum
- FLAG
DYKDDDDK
- GABA
γ-aminobutyric acid
- GAT3
GABA transporter 3
- HEK293T
human embryonic kidney 293 cells containing SV40 T-antigen
- Hepes
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HSP60
heat shock protein 60 kDa 1
- IgG
immunoglobulin G
- LC-MS/MS
Liquid chromatography-tandem mass spectrometry
- mAb
monoclonal antibody
- MaxiK (Kcnma1)
large conductance calcium and voltage activated potassium channel
- NGS
normal goat serum
- pAb
polyclonal antibody
- PBS
phosphate buffered saline
- PPI
protein proximity index
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TIM
translocase of the inner membrane
- TOM
translocase of the outer membrane
- Tris
Tris(hydroxymethyl)aminomethane
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Harpreet Singh, Email: Harpreet.singh@drexelmed.edu.
Min Li, Email: mli@mednet.ucla.edu.
Lyra Hall, Email: halll@kenyon.edu.
Scarlett Chen, Email: scarlett.chen2009@gmail.com.
Sowmya Sukur, Email: ss3773@drexel.edu.
Rong Lu, Email: ronglu@ucla.edu.
Anna Caputo, Email: annacaputo83@hotmail.it.
Andrea L. Meredith, Email: ameredith@som.umaryland.edu.
Enrico Stefani, Email: estefani@ucla.edu.
Ligia Toro, Email: ltoro@g.ucla.edu.
References
- Balderas E, Zhang J, Stefani E, Toro L. Mitochondrial BKCa channel. Frontiers in physiology. 2015;6:104. doi: 10.3389/fphys.2015.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JW, Arnold MM, Rozenbaum A, Caputo A, Schweizer FE, Huynh M, Mathern GW, Sarafian TA, Watson JB. Synaptoneurosome micromethod for fractionation of mouse and human brain, and primary neuronal cultures. Journal of neuroscience methods. 2012;211:289–295. doi: 10.1016/j.jneumeth.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Cheng MY, Hartl FU, Horwich AL. The mitochondrial chaperonin hsp60 is required for its own assembly. Nature. 1990;348:455–458. doi: 10.1038/348455a0. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Gu XQ, Bednarczyk P, Wiedemann FR, Haddad GG, Siemen D. Hypoxia increases activity of the BK-channel in the inner mitochondrial membrane and reduces activity of the permeability transition pore. Cell Physiol Biochem. 2008;22:127–136. doi: 10.1159/000149790. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Gulbins E, Siemen D. Activation of the permeability transition pore by Bax via inhibition of the mitochondrial BK channel. Cell Physiol Biochem. 2011;27:191–200. doi: 10.1159/000327944. [DOI] [PubMed] [Google Scholar]
- Deng PY, Klyachko VA. Genetic upregulation of BK channel activity normalizes multiple synaptic and circuit defects in a mouse model of fragile X syndrome. The Journal of physiology. 2015 doi: 10.1113/JP271031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgerton JR, Reinhart PH. Distinct contributions of small and large conductance Ca2+-activated K+ channels to rat Purkinje neuron function. The Journal of physiology. 2003;548:53–69. doi: 10.1113/jphysiol.2002.027854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber ES, Sah P. Calcium-activated potassium channels: multiple contributions to neuronal function. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. 2003;9:181–194. doi: 10.1177/1073858403009003011. [DOI] [PubMed] [Google Scholar]
- Filosa JA, Bonev AD, Straub SV, Meredith AL, Wilkerson MK, Aldrich RW, Nelson MT. Local potassium signaling couples neuronal activity to vasodilation in the brain. Nature neuroscience. 2006;9:1397–1403. doi: 10.1038/nn1779. [DOI] [PubMed] [Google Scholar]
- Galvan A, Villalba RM, West SM, Maidment NT, Ackerson LC, Smith Y, Wichmann T. GABAergic modulation of the activity of globus pallidus neurons in primates: in vivo analysis of the functions of GABA receptors and GABA transporters. Journal of neurophysiology. 2005;94:990–1000. doi: 10.1152/jn.00068.2005. [DOI] [PubMed] [Google Scholar]
- Gorini G, Ponomareva O, Shores KS, Person MD, Harris RA, Mayfield RD. Dynamin-1 co-associates with native mouse brain BKCa channels: proteomics analysis of synaptic protein complexes. FEBS letters. 2010;584:845–851. doi: 10.1016/j.febslet.2009.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes WN, Li W, Chavez AE, Diamond JS. BK channels modulate pre- and postsynaptic signaling at reciprocal synapses in retina. Nature neuroscience. 2009;12:585–592. doi: 10.1038/nn.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indriati DW, Kamasawa N, Matsui K, Meredith AL, Watanabe M, Shigemoto R. Quantitative localization of Cav2.1 (P/Q-type) voltage-dependent calcium channels in Purkinje cells: somatodendritic gradient and distinct somatic coclustering with calcium-activated potassium channels. J Neuroscience. 2013;33:3668–3678. doi: 10.1523/JNEUROSCI.2921-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin XT, Pare JF, Smith Y. Differential localization and function of GABA transporters, GAT-1 and GAT-3, in the rat globus pallidus. The European journal of neuroscience. 2011;33:1504–1518. doi: 10.1111/j.1460-9568.2011.07636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan T, Harvey M, Orchard S, Sakai Y, Sokolowski B. A protein interaction network for the large conductance Ca(2+)-activated K(+) channel in the mouse cochlea. Mol Cell Proteomics. 2009;8:1972–1987. doi: 10.1074/mcp.M800495-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- Knaus HG, Eberhart A, Koch RO, Munujos P, Schmalhofer WA, Warmke JW, Kaczorowski GJ, Garcia ML. Characterization of tissue-expressed alpha subunits of the high conductance Ca(2+)-activated K+ channel. JBiolChem. 1995;270:22434–22439. doi: 10.1074/jbc.270.38.22434. [DOI] [PubMed] [Google Scholar]
- Knaus HG, Schwarzer C, Koch RO, Eberhart A, Kaczorowski GJ, Glossmann H, Wunder F, Pongs O, Garcia ML, Sperk G. Distribution of high-conductance Ca(2+)-activated K+ channels in rat brain: targeting to axons and nerve terminals. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1996;16:955–963. doi: 10.1523/JNEUROSCI.16-03-00955.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knot HJ, Standen NB, Nelson MT. Ryanodine receptors regulate arterial diameter and wall [Ca2+] in cerebral arteries of rat via Ca2+-dependent K+ channels. The Journal of physiology. 1998;508(Pt 1):211–221. doi: 10.1111/j.1469-7793.1998.211br.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Kumar Y, Lu RW, Ou J, Sanchez-Pastor E, Li M, Stefani E, Toro L. BK Channels: Regulation of Expression and Physiological Impact. Department of Pharmacology, Yale University School of Medicine; New Haven, CT 06520, USA: Discovery Research, Knopp Neurosciences Inc; 2100 Wharton Street, Suite 615, Pittsburgh, PA 15203, USA: John Wiley & Sons, Inc; 2009. pp. 317–342. [Google Scholar]
- Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Current opinion in structural biology. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Li B, Jie W, Huang L, Wei P, Li S, Luo Z, Friedman AK, Meredith AL, Han MH, Zhu XH, Gao TM. Nuclear BK channels regulate gene expression via the control of nuclear calcium signaling. Nature neuroscience. 2014;17:1055–1063. doi: 10.1038/nn.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Srivastava P. Heat-shock proteins. In: Coligan John E, et al., editors. Current protocols in immunolog. 2004. Appendix 1:Appendix 1T. [DOI] [PubMed] [Google Scholar]
- Lu R, Alioua A, Kumar Y, Eghbali M, Stefani E, Toro L. MaxiK channel partners: physiological impact. The Journal of physiology. 2006;570:65–72. doi: 10.1113/jphysiol.2005.098913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Nakata T, Zhang G, Hoshi T, Li M, Shikano S. Differential trafficking of carboxyl isoforms of Ca2+-gated (Slo1) potassium channels. FEBS Lett. 2007;581:1000–1008. doi: 10.1016/j.febslet.2007.01.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrion NV, Tavalin SJ. Selective activation of Ca2+-activated K+ channels by co-localized Ca2+ channels in hippocampal neurons. Nature. 1998;395:900–905. doi: 10.1038/27674. [DOI] [PubMed] [Google Scholar]
- Martire M, Barrese V, D’Amico M, Iannotti FA, Pizzarelli R, Samengo I, Viggiano D, Ruth P, Cherubini E, Taglialatela M. Pre-synaptic BK channels selectively control glutamate versus GABA release from cortical and hippocampal nerve terminals. Journal of neurochemistry. 2010;115:411–422. doi: 10.1111/j.1471-4159.2010.06938.x. [DOI] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annual review of biochemistry. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Ou JW, Kumar Y, Alioua A, Sailer C, Stefani E, Toro L. Ca2+- and thromboxane-dependent distribution of MaxiK channels in cultured astrocytes: from microtubules to the plasma membrane. Glia. 2009;57:1280–1295. doi: 10.1002/glia.20847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Sakai Y, Kurgan L, Sokolowski B, Uversky V. Intrinsic disorder in the BK channel and its interactome. PloS one. 2014;9:e94331. doi: 10.1371/journal.pone.0094331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaelli G, Saviane C, Mohajerani MH, Pedarzani P, Cherubini E. BK potassium channels control transmitter release at CA3-CA3 synapses in the rat hippocampus. The Journal of physiology. 2004;557:147–157. doi: 10.1113/jphysiol.2004.062661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runden-Pran E, Haug FM, Storm JF, Ottersen OP. BK channel activity determines the extent of cell degeneration after oxygen and glucose deprivation: a study in organotypical hippocampal slice cultures. Neuroscience. 2002;112:277–288. doi: 10.1016/s0306-4522(02)00092-1. [DOI] [PubMed] [Google Scholar]
- Siemen D, Loupatatzis C, Borecky J, Gulbins E, Lang F. Ca2+-activated K channel of the BK-type in the inner mitochondrial membrane of a human glioma cell line. Biochem Biophys Res Commun. 1999;257:549–554. doi: 10.1006/bbrc.1999.0496. [DOI] [PubMed] [Google Scholar]
- Singh H, Lu R, Bopassa JC, Meredith AL, Stefani E, Toro L. mitoBKCa is encoded by the Kcnma1 gene, and a splicing sequence defines its mitochondrial location. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10836–10841. doi: 10.1073/pnas.1302028110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Lu R, Rodriguez PF, Wu Y, Bopassa JC, Stefani E, Toro L. Visualization and quantification of cardiac mitochondrial protein clusters with STED microscopy. Mitochondrion. 2012a;12:230–236. doi: 10.1016/j.mito.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Stefani E, Toro L. Intracellular BK(Ca) (iBK(Ca)) channels. The Journal of physiology. 2012b;590:5937–5947. doi: 10.1113/jphysiol.2011.215533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Warburton S, Vondriska TM, Khakh BS. Proteomics to identify proteins interacting with P2X2 ligand-gated cation channels. Journal of visualized experiments : JoVE. 2009 doi: 10.3791/1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalska J, Bednarczyk P, Piwonska M, Kulawiak B, Wilczynski G, Dolowy K, Kudin AP, Kunz WS, Szewczyk A. Calcium ions regulate K(+) uptake into brain mitochondria: the evidence for a novel potassium channel. International journal of molecular sciences. 2009;10:1104–1120. doi: 10.3390/ijms10031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski B, Orchard S, Harvey M, Sridhar S, Sakai Y. Conserved BK channel-protein interactions reveal signals relevant to cell death and survival. PloS one. 2011;6:e28532. doi: 10.1371/journal.pone.0028532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro L. BK Calcium Sensitive Potassium Channel. In: Enna SJ, David BB, editors. xPharm: The Comprehensive Pharmacology Reference. New York: Elsevier; 2007. pp. 1–18. [Google Scholar]
- Toro L, Li M, Zhang Z, Singh H, Wu Y, Stefani E. MaxiK channel and cell signalling. Pflugers Archiv : European journal of physiology. 2014;466:875–886. doi: 10.1007/s00424-013-1359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Eghbali M, Ou J, Lu R, Toro L, Stefani E. Quantitative determination of spatial protein-protein correlations in fluorescence confocal microscopy. Biophysical journal. 2010;98:493–504. doi: 10.1016/j.bpj.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Hirschberg B, Smolik S, Forte M, Adelman JP. dSLo interacting protein 1, a novel protein that interacts with large-conductance calcium-activated potassium channels. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1998;18:2360–2369. doi: 10.1523/JNEUROSCI.18-07-02360.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Liu Y, Wang S, McDonald T, Van Eyk JE, Sidor A, O’Rourke B. Cytoprotective role of Ca2+-activated K+ channels in the cardiac inner mitochondrial membrane. Science. 2002;298:1029–1033. doi: 10.1126/science.1074360. [DOI] [PubMed] [Google Scholar]
- Zarei MM, Eghbali M, Alioua A, Song M, Knaus HG, Stefani E, Toro L. An endoplasmic reticulum trafficking signal prevents surface expression of a voltage- and Ca2+-activated K+ channel splice variant. Proc Natl Acad Sci USA. 2004;101:10072–10077. doi: 10.1073/pnas.0302919101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.