Abstract
An octamer of histone proteins wraps about 200 base pairs of DNA into two super-helical turns to form nucleosomes found in chromatin. Although the static structure of the nucleosomal core particle has been solved, details of the dynamic interactions between histones and DNA remain elusive. We performed extensively long unconstrained, all-atom microsecond molecular dynamics simulations of nucleosomes including linker DNA segments and full-length histones in explicit solvent. For the first time we were able to identify and characterize the rearrangements in nucleosomes on a microsecond timescale including the coupling between the conformation of the histone tails and the DNA geometry. We found that certain histone tail conformations promoted DNA bulging near its entry/exit sites, resulting in the formation of twist-defects within the DNA. This led to a reorganization of histone-DNA interactions, suggestive of the formation of initial nucleosome sliding intermediates. We characterized the dynamics of the histone tails upon their condensation on the core and linker DNA and showed that tails may adopt conformationally constrained positions due to the insertion of “anchoring” lysines and arginines into the DNA minor grooves. Potentially, these phenomena affect the accessibility of post-translationally modified histone residues which serve as important sites for epigenetic marks (e.g. at H3K9, H3K27, H4K16), suggesting that interactions of the histone tails with the core and linker DNA modulate the processes of histone tail modifications and binding of the effector proteins. We discuss the implications of the observed results on the nucleosome function and compare our results to different experimental studies.
Keywords: Nucleosome dynamics, molecular dynamics simulations, epigenetics, chromatin, protein-DNA interactions
Graphical abstract
1. INTRODUCTION
Nucleosomes are elementary units of chromatin compaction in eukaryotic genomes spaced every 200 ± 40 bp along the DNA[1]. The main source of information about the atomistic structure of nucleosome comes from the X-ray studies of nucleosome core particles (NCP) (reviewed in [2]), which consistently yield highly similar structures of histones and DNA irrespective of histone sequence variants, mutations, post-translational modifications, DNA sequence, or presence of nucleosome binding proteins, peptides or chemical agents. For example, a superposition of different nucleosome structures using VAST+ algorithm [3] showed very small RMSD deviations of about 1 Å for the aligned parts of histone core regions. The consensus NCP structure is formed by wrapping ~145–147 bp of core DNA in ~1.7 left-handed superhelical turns around an octamer composed of four types of core histones (H3, H4, H2A, H2B) [4] and should be flanked by the linker DNA segments to yield a full nucleosome system. The NCP structure is characterized by the presence of C2 pseudo symmetry dyad axis, highly bent kinked DNA structure [5] and high positive charge of the histone octamer [6]. In addition, it includes a large number of water molecules permeating the nucleosome structure [7], as well as long histone tails protruding from the nucleosomal core and providing many sites for post-translational modifications (PTM).
On the other hand, the crucial role of nucleosomes in chromatin functioning (including delicate regulation of gene expression, DNA replication, repair and inheritance mediated by epigenetic mechanisms [8, 9]) relies on its dynamic nature and conformational transitions. Indeed, many biophysical (FRET, optical tweezers single-molecule experiments and others reviewed in [10]) and biochemical experiments (transcription factor binding assays [11], chemical crosslinking [12] and genome wide ChIP-exo analysis [13]) suggest that nucleosomes exhibit substantial conformational polymorphism both at local (DNA and histone tails conformations within nucleosome) and global (loss of histones, subnucleosomal particles, etc.) scales which in turn could be a result of equilibrium thermal fluctuations or could be actively induced by external forces within a cell.
Different functionally relevant modes of nucleosome dynamics have been suggested which include DNA breathing/unwrapping/opening, nucleosome splitting [14], as well as nucleosome sliding [15] and H2A–H2B dimer opening or loss [16]. At the same time, various conformational rearrangements of histone tails are required for intra- and inter-nucleosomal interactions [17], for multiple interactions with chromatin remodelers [18, 19], heterochromatin proteins [20], and with many effector proteins that contain histone modification-binding domains [21]. Nevertheless, the view of nucleosomes as dynamic entities [14] is still lacking insights at the atomistic level given the static view of the nucleosome provided by the crystallographic data.
In order to try to alleviate this apparent gap in our understanding of nucleosomal dynamics and function, a substantial amount of computational effort has been already invested, including all-atom molecular dynamics (MD) simulations [22] and coarse-grain simulations [23, 24]. However, due to considerably large size of the nucleosome by the atomistic simulation standards and extremely long timescales of its functional dynamics, previous studies had to resort to either coarse-grain modeling without atomistic details, or all-atom simulations without explicit solvent molecules, constraining the DNA motion [25]. Due to the computational limitations and the size of the system, previous MD simulations were performed on time scales not exceeding several hundreds of nanoseconds [26, 27]. Yet we know that changes in atomic interactions can have profound functional effects in nucleosomes on local and global scale. For example, post-translational modifications may alter intra- and inter-nucleosomal interactions or provide multiple binding sites for the so-called effector proteins [28], even a single acetylation of H4K16 can trigger chromatin fiber unfolding [29, 30].
In this paper our conceptual advance is to use the most robust all-atom explicit solvent approach in simulating the atomic details of unconstrained nucleosome dynamics and to extend this approach to a considerably longer microsecond timescale than previously reported. The microsecond timescale as well as nucleosome models with the realistic DNA linker segments and multiple comparative simulations enabled us to get insights into the functionally relevant rearrangements in nucleosome including the coupling between the conformations of histone tails and the DNA geometry. While many functionally relevant motions may occur on much longer time scales [10], several NMR studies recently suggested that histone tails showed sub-microsecond dynamics providing possibility to compare our computational results with the experimental data [31–33].
In this study we found that certain histone tail conformations promote the DNA bulging near its entry/exit sites, the formation of twist-defects within DNA and the rearrangement of histone-DNA interactions in the core, suggestive of the formation of initial nucleosome sliding intermediates. We characterized the dynamics of histone tails upon their condensation on the core and linker DNA at the atomistic level and showed that they could adopt conformationally constrained positions accompanied by the insertion of certain “anchoring” lysines and arginines into the DNA minor grooves. We reported that these processes affected the accessibility of post-translationally modified histone residues, which serve as important epigenetic marks (e.g. H3K9, H3K27, H4K16) suggesting that interactions with the core and especially linker DNA (for the H3 tail) may modulate the coupling between the accessibility of various sites on histone tails for binding of effector proteins.
RESULTS
Models and simulations
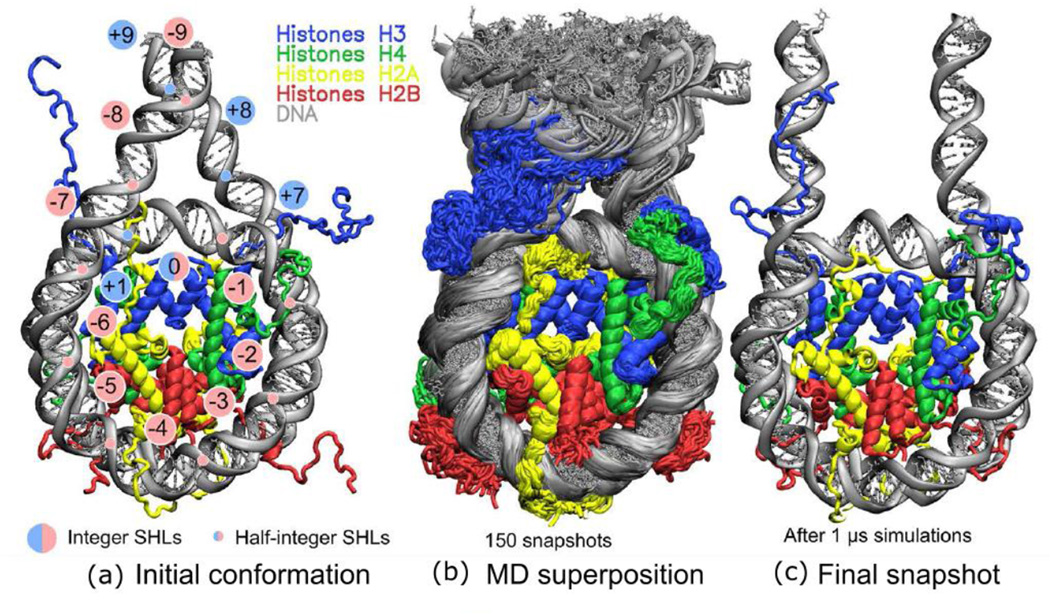
We used several simulation systems for our study, including a specially constructed full nucleosome system with 20 bp linker DNA strands flanking the core particle on every side (FN model) (Figure 1a), a minimalistic nucleosome core particle model with truncated histone tails (NCPm model) (Figure S1) and a set of supporting simulations with varying simulation conditions (see Table 1 for details). Throughout the manuscript, we mainly focus on the results obtained for the full nucleosome system for which extensive microsecond long simulations were performed. We admit that the nucleosome dynamics occurs on several timescales, and functionally relevant conformational transitions often exceed one microsecond timescale. Our simulations were limited to exploring the conformational ensembles around certain local quasi-equilibrium states on a microsecond timescale. Therefore while interpreting our simulation results we confirmed our findings by performing multiple supporting simulations. Moreover, to gain insights into the potential modes and stable patterns of histone-DNA interactions we focused on the detailed analysis of the local conformational ensembles sampled by the simulations, and used our observations to suggest generalized functionally relevant implications. In discussing results below we refer to key structural elements of histone sequences and nucleosome as outlined in Figures 2, S2–S3 (see also “Nucleosome structure description conventions” section below).
Figure 1.
Overview of structure and dynamics of the full nucleosome model (FN): (a) Initial structure of FN model, (b) superimposed conformations from the last frames (75%) of simulation trajectory, (c) the last snapshot after 1 µs of simulations, note the various conformations of H3 N-terminal tails on both sides.
Table 1.
Simulated systems and their details. Table S1 gives more details on the system composition.
| Model name |
Description |
Number of atoms |
Effective bulk salt concentration, mM |
Simulation box size, Å |
Simulated time, ns |
|---|---|---|---|---|---|
| FN | Full nucleosome together with linker DNA and full-length histone tails at physiological ionic strength |
366.8K | ~185 | 162×197×113 | 1000 |
| FN1M | Same as FN but in high (1M) salt concentration |
366.8K | ~1100 | 161×196×112 | 300 |
| FNbb | Same as FN, but in sufficiently bigger simulation box |
3.2M | ~160 | 322×358×274 | 50 |
| FNnt | Same as FN but with acetyl and N- methyl groups on N- and C-termini of histones, respectively |
366.8K | ~185 | 162×197×113 | 1000 |
| NCPm | Minimalistic NCP model: no linker DNA, truncated histone tails |
210.3K | ~200 | 145×141×101 | 1000 |
Figure 2.
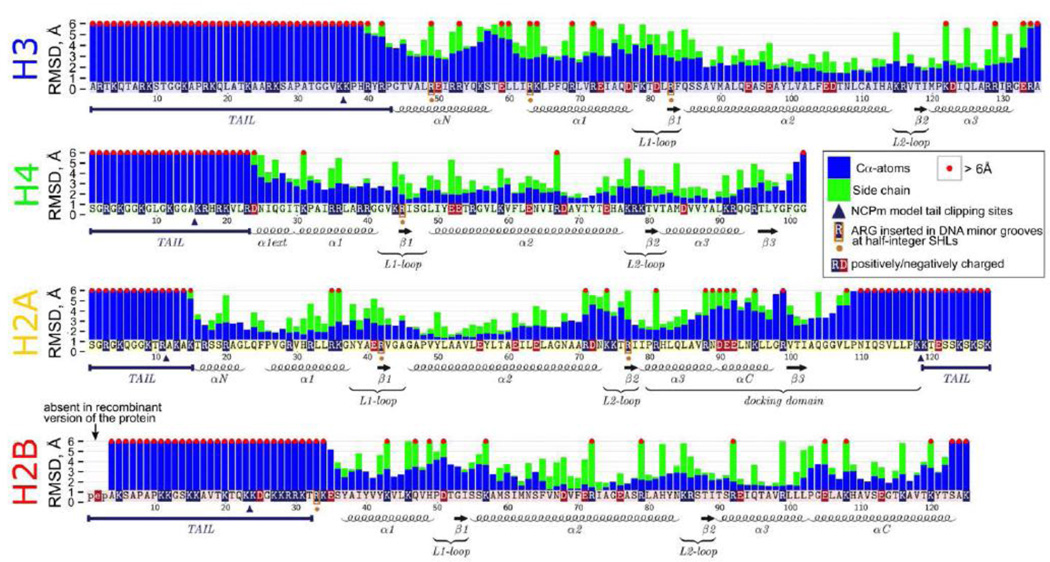
Maximum observed RMSD deviations of individual amino acids (Cα-atoms – blue bars, side chain atoms – green bars) during simulations with respect to their positions in the initial X-ray structure. Bars exceeding 6 Å are truncated at this value and marked by a red dot on top. The annotation of histone structural elements is given below the bar plots. For RMSF values relative to MD average structure see Figure S15.
Conformational stability and plasticity of histones and DNA
In agreement with the previous experimental studies, our simulations confirmed an overall stability of the nucleosome core with respect to the dissociation of nucleosome components or large-scale DNA unwrapping (see an overview of dynamics in Figure 1, S4 and Movie SM1). Overall, RMSD of the core region of histones remained on average within 1.5 Å from the crystallographic positions for all simulated models (Figure S5). This was true even for the minimalistic NCPm system with the truncated histone tails, suggesting that nucleosomes had sufficient stability margin with respect to histone tail clipping (Figure S4, Movie SM2). However, the detailed analysis of conformational dynamics, presented below, provided new insights into the conformational flexibility of nucleosomes and revealed conformational rearrangements within the histone core, histone tails, DNA core and linker regions, which happen at the microsecond timescale.
To provide a detailed picture of histone dynamics, we showed the maximum positional deviations encountered during dynamics by atoms of individual amino acid residues with respect to their positions in the initial model based on the X-ray structure (Figure 2). A time-expanded version (Figure S6, S7) allowed us to distinguish whether the observed deviations are caused by transient fluctuations or persistent rearrangements. As can be seen in Figure 2, relatively large conformational changes of more than 6 Å were observed for backbone atoms of all histone tails, even certain parts of histone folds exhibited high RMSD. Although, as suggested by the analysis of crystal structures, the H2A docking domain forms tight contacts with (H3-H4)2 tetramer [34], we demonstrated substantial conformational changes for the C-terminal part of this domain relative to the initial crystal structure (Figure S8). Conformational fluctuations of the backbone were frequently accompanied by the reorientations of the side chains. For example, the changes in αC-helix of the docking domain were accompanied by the transient formation of novel salt bridges between the H2A R88 and R99 flanking this helix and H3 E105, D106. The observed plasticity of the docking domain provides important insights about its function as a binding partner for chromatin proteins. In particular, recently it was shown that chaperone ANP32E can be responsible for deposition and eviction of H2A.Z histone variant into nucleosome. ANP32E may bind to the αC-helical region of the docking domain of H2A.Z [35], which in nucleosome is structurally very similar to the canonical H2A αC-helix. The histone-ANP32E binding is sterically incompatible with the full nucleosome structure and may trigger nucleosome disassembly and eviction when the ANP32E flexible region is inserted into the nucleosome core. The latter process is not very well understood, however, an increased conformational flexibility in this region, observed in our study, may provide important clues in this regard. Interestingly, Biswas et al. in earlier MD simulations of nucleosomes have shown potential coupling between the truncation of H3 histone tails and conformations of H2A R81 and R88 side chains [36]. Visual inspection of our simulations suggests that while H2A R88 side chain is highly flexible at microsecond timescale, H2A R81 side chain is mainly locked in its crystallographic orientation with occasional reorientations pointing away from the histone octamer, which may last for hundreds of nanoseconds.
In addition, significant backbone (more than 3 Å) and side chain displacements (more than 6 Å) were observed in the core regions of other histones as well, including H3 αN-helix, H3 α1-helix, L1-L2 binding site of H2A–H2B dimer proximal to the DNA entry/exit site, and H2B αC-helix) (Figures 2 and S8). As already mentioned, even if protein backbone did not show large changes from the crystallographic positions, significant changes of side chain conformers could still happen. In fact, all but one amino acid with the observed side chain reorientations of more than 6 Å belonged to charged residues. These reorientations often included the disruption and formation of new salt bridges within the histone core. Rearrangements in the NCPm model showed similar patterns (Figure S9).
Analysis of the conformational dynamics of core and linker DNA showed that DNA remained on average within 4 Å (5 Å for NCPm model) of its crystallographic positions as measured by RMSD of N1 and N9 atoms (Figure S5). In addition, DNA phosphate backbone was characterized by an oscillating flexibility (RMSF) profile (Figures S10–11), where parts of the DNA backbone in contact with histones manifested considerably lower fluctuations compared to those exposed to solution. On the other hand, the linker DNA segments in our simulations clearly showed much higher fluctuations than the core DNA regions (Figure 1b,c), concomitant with previous normal mode analyses [37]. The global DNA conformation was further studied in detail by the 2D projections of the polygons connecting the DNA base pair centers on the nucleosomal superhelical reference frame (see SI for definition, Figures 3 and S12). The positional fluctuations of the linker DNA segments spanned marginally overlapping cone-shaped regions in space with around ±45 degrees of angular span in both projections. No significant correlations in positional fluctuations of DNA linkers were detected. Average positions adopted by the linker DNA strands in MD were further apart from each other compared to their initial positions, which corresponded to the straight continuation of the core DNA termini (Figure 3a). A simulation of the same system, but in high 1 M salt solution (FN1M system in Figure S12) clearly showed that two DNA linkers came closer to each other and even interacted with their ends most of the time, suggesting that screening of electrostatic interactions significantly affected the DNA entry/exit angle. Interestingly, earlier experimental measurements of the linker end-to-end distances using FRET showed that the distance monotonically decreases with the increase in salt concentration [38].
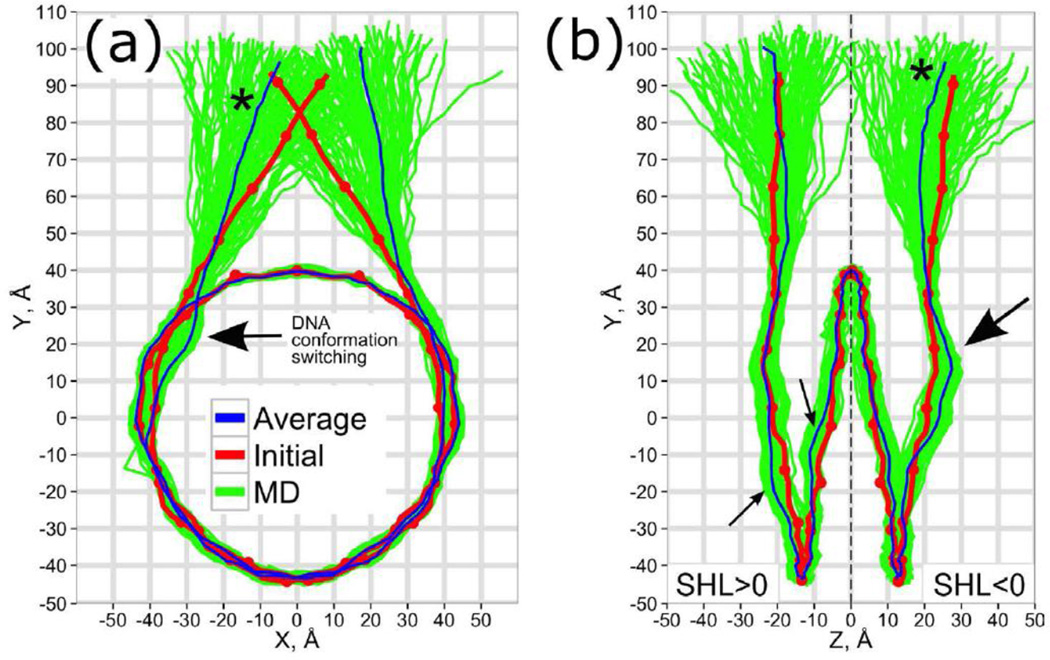
Figure 3.
The DNA conformational ensemble in nucleosome, (a) – front and (b) – side projections. The red dots mark the integer and half integer SHL values along the initial conformation, an asterisk marks the same linker DNA segment. Arrows indicate maximum deviations between initial and MD-average conformations.
A paramount question of nucleosome dynamics is the amplitude, time scale and pathways of DNA breathing, unwrapping or opening from the histone octamer surface, which is thought to be crucial for DNA accessibility and transcription factor binding [11, 39]. Unfortunately, no complete picture is available to date, with different experimental setups, interpretation and terminology often supporting different views on this problem [10, 14]. In this study, we did not observe any large-scale unwrapping or opening of the core DNA neither in FN systems nor in NCPm systems. Such unwrapping, if it took place, would expose sufficient parts of DNA for transcription factor recognition. Our results are in line with experimental estimates of the timescales of unwrapping events, which lie in the sub-second range [11, 40]. However, if we consider more moderate fluctuations of the core DNA termini together with linker DNA (often referred to as “DNA breathing”), such processes may happen on shorter time scales[41]. For example, recent FRET measurements of distances between two dyes placed symmetrically on both segments of linker DNA 5 bp away from the DNA entry/exit sites suggested two different conformations with characteristic distances between two dyes of ~4.5 nm and ~6.5 nm (DNA breathing amplitude of around 2 nm), if a two state model was assumed [42]. In our simulations the distance between base pair centers of the corresponding positions of DNA linkers ranged from 4.5 nm to 6.8 nm (see Figure 3) (with mean of 5.6 nm and RMSF of 0.4 nm), showing that these distance spans are within the reach of rapid sub-microsecond fluctuations. In addition, we observed an asymmetry in average DNA conformations of two linkers as well as in their deviations from the initial crystal structure (indicated by arrows in Figure 3). Interestingly, if we assumed that the observed asymmetries corresponded to two distinct stable conformational states of DNA in a nucleosome, the transition between these states may already account for the combined DNA breathing motion of 0.5 nm at the distance of 5 bp from the entry/exit point, with no DNA unwrapping detected. However, we admit that the methodology for accurate comparison of various conformational transitions seen in simulations and results of FRET measurements has still to be developed and verified: as noted recently by Lenz et al. shape fluctuation of DNA in nucleosome can smear out FRET signals from intermediate conformational states of DNA in nucleosome [43].
We will demonstrate in the next sections that these DNA conformational asymmetries are triggered by the asymmetric condensation of histone tails. While we find that time needed for switching between different conformations of histone tails clearly exceeds microsecond timescale, we suggest that the DNA dynamics on microsecond timescale is dominated by interactions between histone tails and DNA, which, in turn, can affect DNA conformation on much longer scales.
Rapid condensation of histone tails on the core and linker DNA
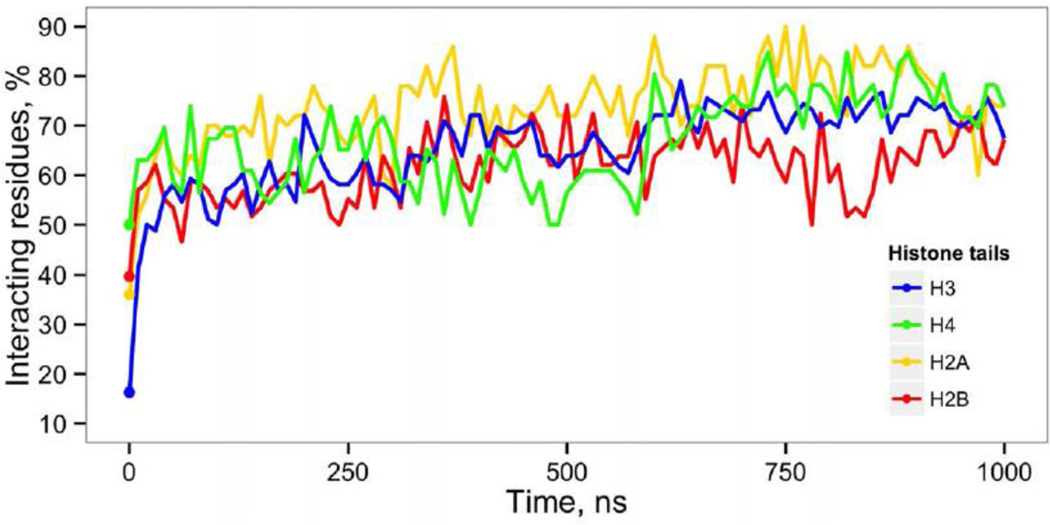
Positively charged histone tails have been extensively studied both experimentally and computationally, it is known that inter-nucleosomal interactions are mediated by tails and play important roles in chromatin compaction. Here we characterized the dynamics of interactions between histones and DNA at the atomic level and found that, on average, 60% of all contacts were contributed by the interactions of DNA with the histone tail regions. Histone tails that in the initial model were protruding into the bulk solvent (primarily H3 tails, Figure 1a), rapidly adsorbed onto the DNA within the first 20 ns of simulations. From 50 to 90% of all amino acids within these tail regions had direct or water mediated interactions with either core or linker DNA throughout the 1 µs simulation (Figure 4). We did not observe any substantial clustering of contacts towards the beginning or the end of the tails. Previous all-atom MD simulations of nucleosome core particles also pointed out a considerable association of histone tails with the core DNA at the 100 ns time scale [25, 36, 44]. However, certain concerns have been raised about the short simulation time scales as well as artifacts coming from periodic boundary conditions and lattice summation methods for electrostatic interactions. To address the latter concerns we performed 80 ns simulation in a sufficiently larger simulation solvent box (FNbb system) that should minimize the potential adverse effects of periodic boundary conditions. The FNbb simulation confirmed preferential association of histone tails with DNA and a rapid condensation of long H3 N-terminal tails (Figure S13). Thus we alleviate earlier raised concerns and show that histone tail condensation is not an artifact of specific techniques used in MD simulations.
Figure 4.
Condensation of histone tails on DNA. Proportion of histone tail residues that have direct or water mediated interactions with DNA is plotted as a function of simulation time. Initial values indicated by dots.
Several small angle X-ray scattering (SAXS) studies addressed the question of whether histone tails protruded from the nucleosome core away into solution or associated with the DNA at physiological ionic strength. A considerable increase in maximal diameters of nucleosomes was reported upon raising the monovalent salt concentration from 10 mM up to the physiological concentrations and higher [45]. This result was attributed to the process of extension of histone tails away from the nucleosome core. However, Yang et al. [46] pointed out that comparable increase in diameter may be explained by the nucleosome conformational changes including the DNA unwrapping. Our results support a model where histone tails are attached to the core or linker DNA most of the time on this timescale. If, for some reason, the tails become detached, characteristic time of their reattachment should be around several dozen of nanoseconds as evident from their fast condensation (Figure 4). Our theoretical estimates of gyration radii (shown together with the estimated X-ray scattering curves in Figure S14) suggest that both histone tails and linker DNA contribute to the increase in gyration radii.
To further elucidate the effect of salt concentration on tail behavior, we performed simulations at extremely high salt concentration of 1M (FN1M model). Although an octameric form of nucleosomes is known to be unstable at this salt concentration [47], we did not observe any nucleosome disassembly, suggesting that it happens on the time scale much longer than a microsecond. While the current force filed might somewhat overestimate sodium-phosphate interactions at high salt concentrations[48], it should not prevent the detachment of tails from DNA. Nevertheless it allows us to probe the effects of increased salt concentration on the dynamically responsive histone tails and linker DNA while the nucleosome core still remains in a compact state. Intriguingly, although it took tails (especially H3 tail) somewhat longer (70 ns compared to 20 ns at 150mM salt concentration) to establish contacts with the DNA from their starting conformation, histone tails still attached to DNA, despite considerable increase in electrostatic interactions screening induced by high salt concentration (Figure S13). This fact can be regarded as another evidence supporting the model of tight interactions between histone tails and DNA in nucleosomes, at least when the nucleosome core is in a compact assembled state.
There is some discussion in the literature concerning the extended or condensed states of histone tails in nucleosomes and in our opinion the basic physical principles support the latter view. As discussed in [49, 50] sufficiently long oligocations tend to almost completely associate with the highly charged DNA due to free energy gain upon the release of the condensed small monovalent ions.
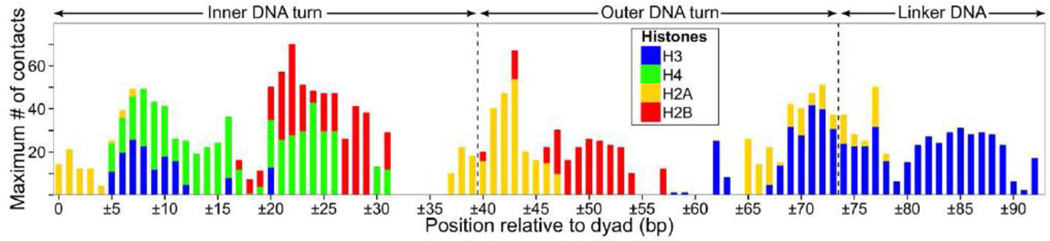
A combined ensemble of histone tail conformations sampled in different simulations showed a broad span of DNA sites accessible for interactions with the histone tails (Figure 5). For example, the 20 bp linker DNA segment was shown to be completely accessible to interactions with H3 N-terminal tails, and partially accessible to interactions with H2A C-terminal tail close to the DNA entry/exit point. In the FN and FNnt models one H3 N-terminal tail remained extended and stably bound to the nearest linker DNA segment (Figure 1c and 6a), while another tail formed an α-helix near the DNA entry/exit point (H3 21–28), or docked between the DNA gyres (Figure 6e,6f). This observed polymorphism of H3 tail conformations echoes alternative statements from different experimental studies. Binding of H3 tails with the linker DNA has been recently confirmed by a genome-scale ChIP-exo study [13]. On the other hand, NMR and hydrogen exchange studies concluded that in nucleosomal arrays H3 tails might form stable folded structures [31]. The possibility for H3 tail to condense onto the linker DNA suggests also its role in partial neutralization of linker DNA and competitive binding with other linker DNA interacting proteins (such as histone H1 or certain nucleosome remodeling complexes), which is important for understanding the chromatin functioning given the very high concentrations of nucleosomes in the nucleus (>200 mg/ml) [51].
Figure 5.
Accessibility of DNA for interactions with histone tails. Maximum number of atom-atom contacts between DNA and histone tails shown for all frames from different simulation systems (FN, FNnt and FNbb).
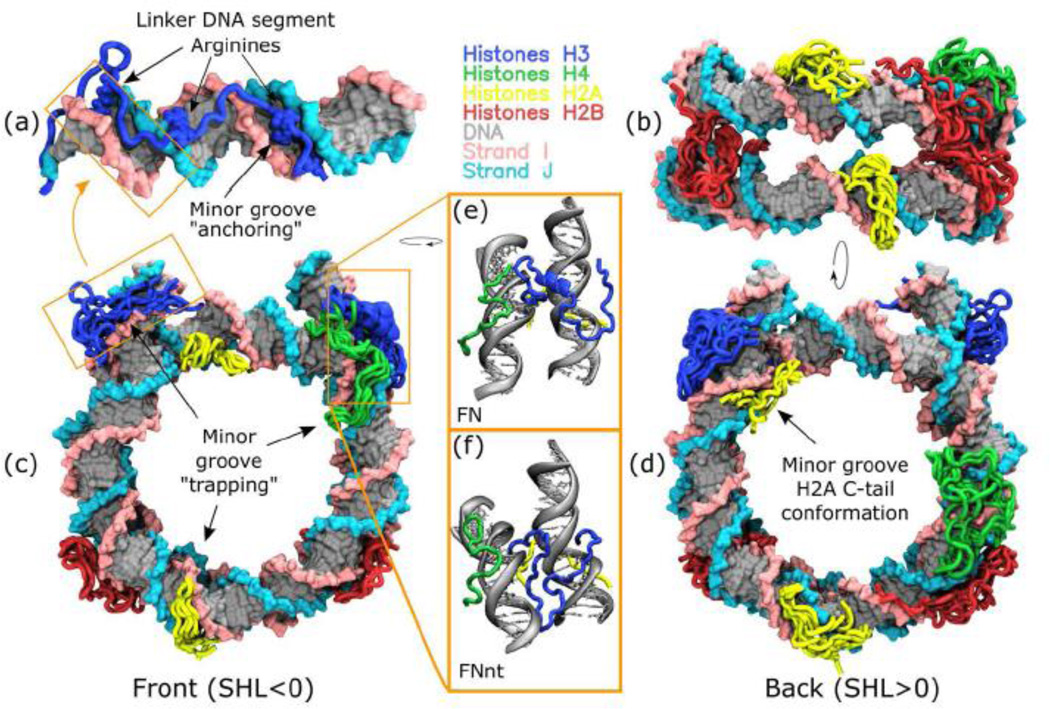
Figure 6.
Typical interaction patterns of histone tails with DNA observed in MD simulations. (a) A representative conformation of H3 N-terminal tail interacting with the linker DNA. (b) – bottom, (c) – front and (d) - back views of nucleosome with multiple superimposed histone tail conformation observed during 1 µs MD simulations (depicted every 100 ns with the first 250 ns frames discarded). (e) a zoom-in of a stable α-helix formed by residues 21–28 of one of H3 tails (chain A), (f) an alternative stable position of H3 tail “docked” between the DNA gyres from the supporting FNnt model simulation.
Histone tails are “trapped” in the DNA minor grooves and affect the accessibility of epigenetically modified sites
The condensation of histone tails onto DNA clearly altered the conformational dynamics of tails and DNA itself, in fact it resembled a 2D diffusion on the surface of DNA with the DNA minor grooves serving as kinetic traps. Despite extensive 1 µs simulations, we observed that the tails of two copies of the same histone often explored non-overlapping conformational space subsets, suggesting that switching between different conformational subspaces for such tails happened on a time scale longer than our simulation time (Figure 6). Such behavior corroborates cross-linking and NMR findings of asymmetrical conformations of tails (H2A N-terminal tail, for example) of two histone copies [12, 32]. Interestingly, in our microsecond simulations the tumbling rate of histone tails depended on their conformation and binding mode onto DNA (as illustrated by the RMSF profiles in Figure S15). For instance, the amplitude of fluctuations of the H3 tail on one side of nucleosome, which adsorbed on the linker DNA, was much higher (due to linker DNA flexibility) compared to the other H3 tail, which was compactly folded at the DNA entry/exit sites. The dynamical scales of histone tails in mononucleosomes and nucleosomal arrays were recently probed by a number of techniques including solution, solid state NMR and hydrogen exchange studies [31–33]. They reported rather controversial findings. In view of our results, we speculate that the dynamics of histone tails even in compacted nucleosomal arrays might be organized in a non-uniform fashion, with certain tails being stably folded, while others actively exploring the conformational space.
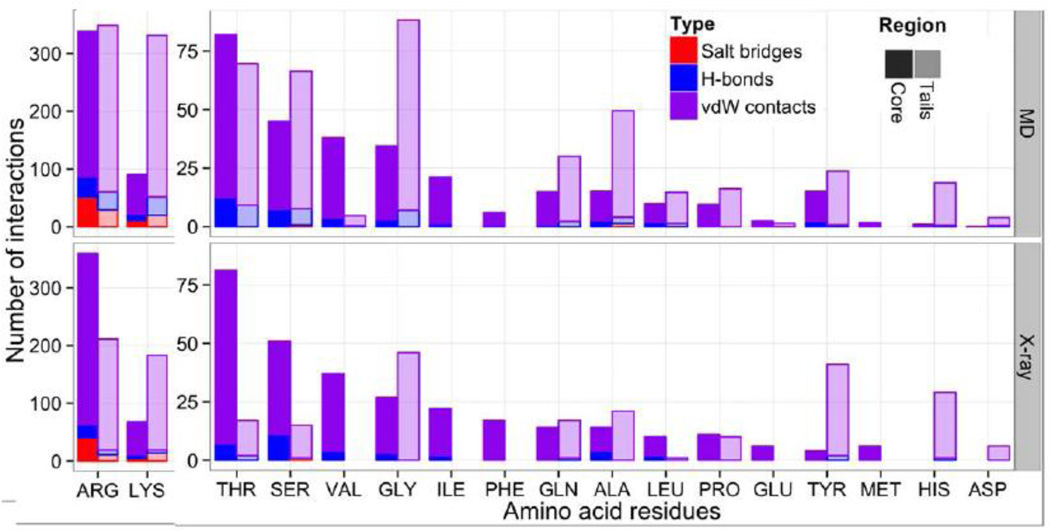
A detailed analysis of interactions between individual amino acid side chains and DNA bases (Figure S16–19) suggests that certain histone tail side chains might be deeply inserted into the DNA minor grooves, making contacts with the DNA bases and serving as anchors stabilizing the interactions and limiting the conformational dynamics. The largest number of contacts with the DNA bases was formed by arginines and lysines located within the histone tails. Namely, in the main (FN) model, the following residues showed high propensity to interact with the bases of DNA minor groove: R8 and R26 of histone H3; K16 and R17 of histone H4; R11, K13 and K126 of histone H2A, R29 and R30 of histone H2B. Interestingly, for H3 and H4 tails none of these interactions with the DNA bases were observed in the initial X-ray structure. While protein-DNA contacts within the histone core were dominated by the interactions with phosphates in FN model (82% - phosphates, 16% - sugars, 2% - bases), histone tails formed 17% of interactions with the DNA bases which represented a considerable increase compared to the NCP crystal structure with only 8% of contacts between DNA bases and histones (Figure S16).
It should be noted that many sites mentioned above potentially can undergo post-translational modifications including methylation of arginines and methylation and acetylation of lysines. For example, two of the most important epigenetically modified residues on histone H3 (H3K9 and H3K27) are located right next to the arginine residues (H3R8 and H3R26), which showed high tendency to be inserted into the DNA minor grooves (see Figure S17–18). Therefore, such arginine-lysine motifs within the histone tails may affect the accessibility of certain lysines to effector proteins and their ability to be post-translationally modified.
If we analyze those sites, which make direct contacts with the nucleotides in the DNA minor groove in our models, the most pronounced one is H4K16, which is docked to the DNA position SHL −1.5. It is a well-known acetylation site, which regulates the chromatin compaction and binding of ACF remodeler [52]. An NMR study of mononucleosomes suggested the flexibility for residues 1–15 of H4 tail, whereas residues 16–22 were found to be folded onto the nucleosome core and did not undergo sufficient tumbling on the relevant NMR time scale. Moreover, it was experimentally confirmed that acetylation mimic H4K16Q mutation, which, according to our study, would likely weaken the interaction of this residue with the DNA minor groove due to loss of side chain positive charge, induced structural disorder in this region [32]. Consistent with our observations, an experimental study showed that H4H18 and H4K16 may be cross-linked to the DNA positions SHL ±1.5 [53]. Our observation of H4K16 minor groove binding is also in agreement with earlier findings of Erler et al. in REMD simulations of nucleosomes [25]. More recently, Collepardo-Guevara et al. in a multiscale simulations showed that lysine acetylation increased secondary-structure content within the histone tail and decreased tail availability for crucial fiber compacting internucleosome interactions [54], consistent with earlier simulations of conformational ensemble of fragments of histone tails without DNA in solution [55, 56]. These results are also in line with the previous simulations of DNA interactions with H4-tail peptides [57, 58], which suggested that charged groups of arginines and lysines play major roles in the tail-mediated DNA-DNA attraction by forming bridges with phosphates and interacting with electronegative sites in the minor groove.
Our comparative analysis of histone-DNA interactions in different regions of nucleosome suggests that the majority of interactions in the histone core region are dominantly provided by arginine residues, while the interactions between histone tails and DNA are almost equally dominated by arginines and lysines residues (Figure 7). This is not the case for the starting conformation derived from the crystal structure of NCP, where protein-DNA interactions through lysine residues are largely underrepresented. Although it has to be noted that in the crystal structure of NCP additional histone-DNA contacts may be formed with the neighboring nucleosomes representing crystal packing interactions, which we do not include in our analysis. This later trend is consistent with the propensity of arginines to be inserted into the DNA minor grooves as was observed in crystal structures of many protein-DNA complexes [59]. As evident from Figure 7, glycine is also prevalent in H3, H2A and especially H4 N-terminal tails and its backbone carbonyl group may form non-specific contacts with phosphates of DNA backbone. It is important to note that, while direct contacts of proteins with the DNA bases (direct read-out) are generally thought to be almost absent in nucleosomes and do not contribute to the nucleosome positioning, this might be an oversimplified picture. For example, it was shown that histone tails may contribute to the sequence-dependent nucleosome positioning in view of repositioning of nucleosomes on 5S RNA gene upon tail cleavage [60]. However, this effect was not reproduced on high affinity sequences. Extensive contacts of histone tails with the DNA bases observed in our study may help to understand and interpret these experimental results.
Figure 7.
Quantification of protein-DNA interactions by type, histone region and participating amino acids. See Figure 2 for histone tails/core region definitions.
DNA bulging near the entry/exit points is modulated by the histone tail conformations
Now we turn our attention to the analysis of the coupling between conformation of histone tails and geometry of the core DNA. Several changes in the DNA conformation were evident (Figures 3ab, S20–23). The largest rearrangements included the bulging of the core DNA next to its entry/exit point around SHL ±6.5, which could only be observed on the time scales longer than 100 ns and was conditional on the various conformations of histone tails (Figure 6). Interactions in this DNA region (terminal 10 bp of the core DNA) were mostly formed with the histone tails and not with the histone core, suggesting that histone tails might have profound effects on the conformation of DNA leading to potential stabilization or destabilization of the whole nucleosome (Figure S16). Indeed, in FN model simulations the terminal core DNA region around SHL +6.5 formed many more contacts with the histone tails than its symmetric counterpart at SHL −6.5 (184 versus 64 contacts), which caused its stabilization and prevented its DNA bulging, otherwise observed at SHL −6.5. This stabilization apparently was caused by the extensive interactions with the H3 N-terminal tail that formed compact secondary structure near the DNA entry/exit site as well as by the insertion of H2A K126 into the DNA minor groove (Figure 6, S17). The latter observation provides a potential pathway for DNA stabilization by the H2A C-terminal tail. In fact, it was recently experimentally observed that partial deletion of the H2A C-terminal region resulted in unwrapping of the last 10 bp of the core DNA [61]. In agreement with this, fluctuations of the terminal parts of the core DNA were less constrained in our simulations for the NCPm system, which had truncated histone tails (Figure 3ab, S11, S12).
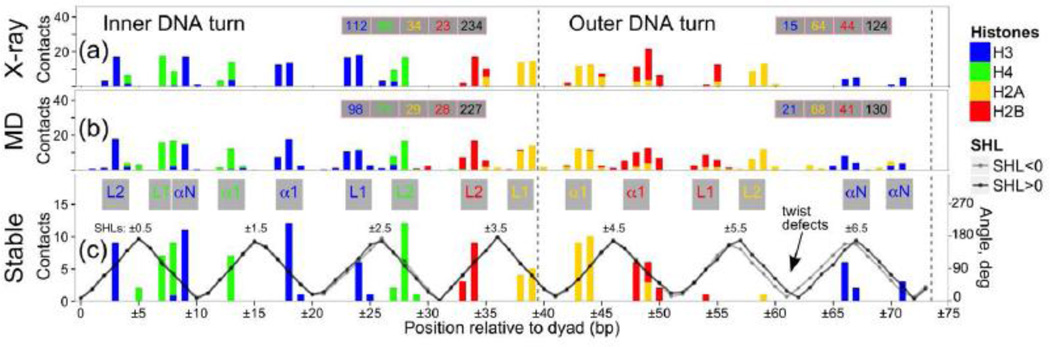
DNA binding sites in histone core show potential for rearrangement accompanied by the formation of twist-defects in DNA
In our previous sections we talked about the interactions between histone tails and DNA; here we study the histone-DNA interactions involving the histone core, and their coupling with the DNA geometry rearrangements. It is known from the analysis of NCP crystal structure that DNA has seven pairs of symmetrical binding sites, where the DNA minor groove faces the histone octamer [62]. At each binding site there is a key arginine inserted in the minor groove in majority of cases with the exception of canonical arginines R49 from both copies of H3 which are located too far away to be inserted into the minor groove around SHL ±6.5. Concomitantly, this location has a low number of contacts with the histone core (Figure 8a,b). Our analysis of the dynamics of histone-DNA interactions showed that this canonical H3R49 can actually be inserted into the corresponding minor groove due to the DNA bulging at SHL ±6.5 described previously. This distortion was apparently facilitated by the formation of additional and rearrangement of existing contacts between H3 αN-helix and DNA. To understand the implications of these distortions to an overall DNA geometry we plotted the effective rotation of DNA base pairs with respect to the superhelical axis location (Figures 8c, S11). We observed a shift in an overall DNA twisting by one base pair around SHL −6/−6.5, and in the case of NCPm model such shifts were observed in a region from SHL −5.5 up to the end of the core DNA (Figure S11). Similar shifts in the DNA twist geometry were previously named “twist-defects” [63]. Some experimental studies suggested that nucleosome exists in solution as a mixture of different twist-defect states and only some of them can be captured in crystal structures by varying the core DNA sequence, its length or by adding the intercalating agents [63]. In out study we were able for the first time to observe the formation of such defects in silico and describe the atomistic details behind the coupling between DNA geometry s and histone-DNA interactions.
Figure 8.
Protein-DNA interactions within the histone core region. (a) The contact profile for the X-ray structure averaged over two halves of the nucleosome. (b) The number of contacts averaged over all frames, and over symmetrical halves of nucleosome. A total number of contacts for respective DNA regions and histones is shown in pink-framed boxes for plots (a) and (b). (c) Number of stable protein-DNA contacts. At every DNA position the number of unique stable atom-atom contacts observed at either half of nucleosome is reported (i.e. symmetry related stable contacts observed in two halves of nucleosome were counted only once, asymmetric contacts contributed individually). The structural elements of the DNA binding sites are annotated on top of this plot, with different colors corresponding to different histone types. Black and grey curves show the rotational periodicity of DNA in the MD-average nucleosome structure.
To elucidate the patterns of interactions in different histone-DNA binding sites, we calculated a number of so-called stable contacts (see Methods). As can be seen in Figures 8a–c,S24–26 and Table 2, the number of contacts between the histone core and the inner DNA turn was almost twice as many as the number of contacts with the outer DNA turn. This is consistent with an easier unwrapping of the outer DNA turn in single molecule experiments [64, 65]. Moreover, Figure 8 shows a clear hierarchy between different canonical binding sites. Binding sites around SHL ±0.5 and SHL ±4.5 have the highest number of dynamically stable contacts in accordance with the high resolution mapping of protein-DNA interactions in nucleosome obtained by mechanical DNA unzipping. The latter experiments suggested that the strongest histone-DNA interactions occurred around the dyad and around the location of ±40 base pairs away from the dyad [64].
Table 2.
Number of stable contacts between histone core and DNA per binding site (please see Figure 8 captions for definitions).
| SHL | ±0.5 | ±1.5 | ±2.5 | ±3.5 | ±4.5 | ±5.5 | ±6.5 |
|---|---|---|---|---|---|---|---|
| Binding site type |
L1L2 αN | α1α1 | L1L2 | L1L2 | α1α1 | L1L2 | αN |
| Average number of contacts |
74 | 42.5 | 54 | 57.5 | 63.5 | 37.5 | 27.5 |
| Number of stable contacts |
38 | 20 | 22 | 21 | 33 | 2 | 12 |
The first histone-DNA binding site around SHL ±0.5 is formed by the L1/L2 loops of H3-H4 dimer, with an additional interaction coming from the N-terminus of the H3 αN-helix making this site very special in terms of its unique specific organization. Differences between histone-DNA binding patterns were observed in genome-wide nucleosome maps and a rogue signal with respect to A|T/G|C content of the nucleosomal DNA was recently observed nine base pairs from the nucleosome center by high throughput nucleosome positioning studies [66]. It was attributed to the insertion of H3R40 deep into the DNA minor groove possibly forming sequence-specific contacts with the bases [67], however, we did not observe such contacts on the microsecond time scale and H3R40 interacted with the DNA phosphates rather than bases (Figures S16–S17).
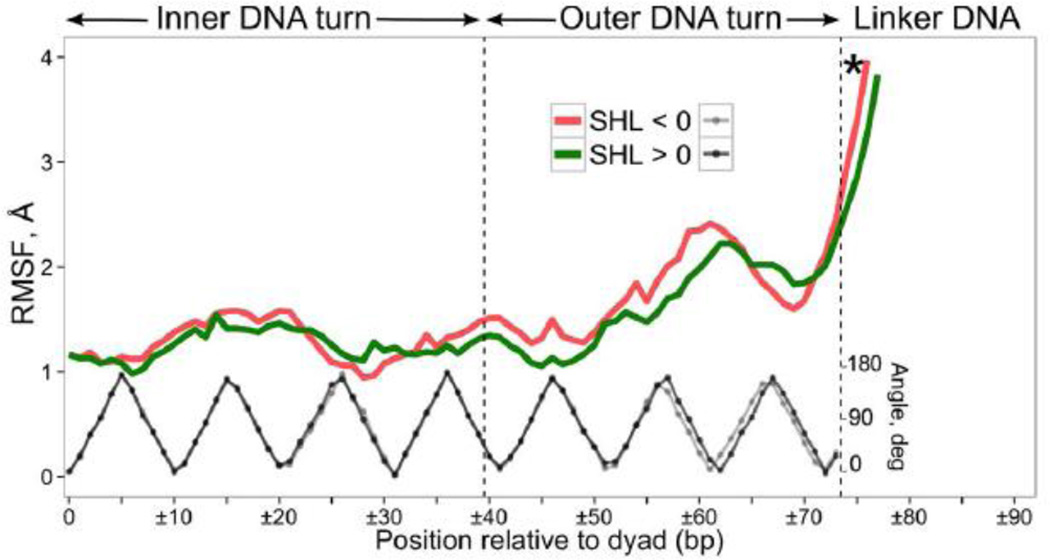
The second highly stable binding site around SHL ±4.5 is formed by the two α1-helices of H2A–H2B dimer whereas DNA binding sites with the least number of stable interactions are localized at SHL ±6.5, SHL ±5.5 and SHL ±1.5. The histone-DNA contact map is mimicked by the DNA RMSF fluctuation profile (Figures 9, S27) and DNA fluctuations show visible increase at SHL ±1.5. It suggests an increased potential to accommodate various DNA conformations at these sites upon binding of small molecules or proteins in light of recent experiments which showed that SHL ±1.5 was a hotspot for binding of heavy metal ions and intercalating antitumor agents [68]. The lack of stable interactions around SHL ±5.5/±6.5 also corresponds to an increase in the DNA fluctuations. The fluctuations at the very end of the core DNA are somewhat smaller, which is explained by their stabilization via the H3 N- and H2A C-terminal tails (see previous sections). The marginal stability and high flexibility of SHL ±5.5 and SHL ±6.5 binding sites might suggest that these sites present rather small energy barriers upon unwrapping of DNA from the nucleosome until SHL ±4.5 is reached. A biologically relevant process that relies on such gradual disruption of DNA histone contacts includes the transcription by RNA-polymerases which might occur without nucleosome eviction [69]. The nucleosomal barrier to transcription investigated by transcription pausing patterns showed that the first barrier is encountered by polymerase when the leading edge of polymerase enters the region approximately 40 bp away from the dyad [70], concomitant with the idea that the first two binding sites provide almost no barrier to transcription.
Figure 9.
Characterization of DNA dynamics. RMSF of base pair centers for two symmetric nucleosome halves (red and green curves); black and grey curves show the rotational periodicity of the DNA superhelix. The data points exceeding RMSF of 4 Å are not shown. See Figure 3 captions for the asterisk definition.
DISCUSSION
Here we presented a molecular dynamics study of a full nucleosome model with the full-length histone tails and linker DNA regions. We used the robust all-atom explicit solvent approach in simulating the atomic details of free nucleosome dynamics and extended this approach to the microsecond timescale. The extensively long microsecond timescale, atomic-level models and multiple comparative simulations enabled us to get insights into the functionally relevant rearrangements in nucleosome conformations including the coupling between histone tails and DNA.
The mechanistic insights obtained in our study suggest that the conformational rearrangements within the core DNA depend on the patterns of DNA interactions with the histone core regions as well as histone tails. In particular, we observe that the H3 N-terminal and H2A C-terminal tails make many contacts with the core DNA and may stabilize the geometry of its terminal region, suppressing the formation of the localized twist-defect states within DNA. On the other hand, the conformation of the tails that protrude away from the nucleosome favor the formation of the twist-defect states, that can be important intermediates in nucleosome sliding and remodeling [15]. We suggest a mechanism by which the observed DNA over-twisting within the terminal DNA region can represent an initial twist-defect state that can be stabilized by the subsequent rearrangements of DNA interactions within the core region of histones. Subsequent nucleosome sliding may occur via the propagation of this defect further down the nucleosome core. It is interesting to note that several in vitro mononucleosome crosslinking studies have previously shown that histone tails occupy different preferential conformations in intact NCPs, nucleosomes with linkers and nucleosomes with H1 histone systems [71, 72], suggesting that histone tails are highly responsive to the presence/absence of DNA linkers, H1 histone and changes in the nucleosomal environment.
In accordance with the cross-linking studies [72, 73] we found that histone tails readily adsorbed onto the linker (mostly H3 tails) and core DNA (mostly H4, H2B and H2A tails) and the majority of histone-DNA contacts occurred with the histone tails. A possibility of the H3 N-terminal tails to interact with the linker DNA was reported earlier [13, 72]. In fact, the efficiency of cross-linking in NCP was found to be 3–4 times lower for H2A, H2B and H4 and 10–12 times lower for H3 compared to the full nucleosomes with the DNA linkers [73]. Moreover, our detailed study of histone tail behavior showed that their microsecond scale dynamics is characterized by the limited diffusion on the DNA surface and kinetic trapping of tail regions in the DNA minor grooves of core and linker DNA regions. The observed stable attachment of histone tails to the core and linker DNA suggests profound implications for the interactions of nucleosomes with other chromatin proteins. Indeed, when these proteins bind to histone tails or DNA, first they have to outcompete the DNA-histone tail interactions and displace the respective partner. For example, Pilotto et al. recently experimentally demonstrated an elegant practical illustration of this concept using LSD1-CoREST complex as an example, which acts as H3K4 demethylase [74]. It was suggested that this complex binds to the nucleosome via its CoREST subunit, which displaces the H3 tail from the DNA. This displacement is critically needed to render the methylated H3K4 available for the interactions with its LSD1 subunit. It can be hypothesized that such mechanisms may be utilized to orchestrate a cross-talk between multiple binding sites on histone tails including the PTM sites and their respective binding partners [28, 75].
The atomistic representation of the histone tail binding to DNA revealed by our study further showed that this process was facilitated by the insertion of the anchoring arginine and lysine side chains into the DNA minor grooves. This in turn affected those residues that serve as epigenetically modified sites in histone tails or impacted residues located right next to them (e.g., H3K9, H3K27, H4K16 and H3R8, H3R26, etc.). The presence of multiple binding sites on one histone tail and side chain occlusion by the minor grooves may give rise to the cooperativity effects. Namely, binding of one partner to its respective DNA or histone tail site may trigger a histone tail displacement from the DNA and thus facilitate the binding of another partner. Similar effects may be triggered by post-translational modifications such as arginine methylation and especially lysine acetylation that would disfavor the insertion of these residues into the minor groove and thus may facilitate the conformational flexibility and binding availability of the histone tails. The proposed effects suggest further design of experimental studies that would elucidate the complex dynamical interplay between histone post-translational modifications and histone tail accessibility for effector protein binding.
MATERIALS AND METHODS
Initial model construction
As a starting point we used high resolution (1.9 Å) X-ray crystal structure of nucleosome core particle formed by recombinant variants of X. laevis canonical core histones and modified human α-satellite DNA (PDB ID 1kx5 [7]). To create a full nucleosome model with the linker DNA segments, a straight 20 bp long B-DNA duplex (AGTC)5 was constructed using the NAB software [76]. The used linker DNA sequence is balanced in the number of flexible and rigid base pair steps [77]. It was attached to the core DNA at both ends of NCP. One of the H3 histone tails was slightly rotated to avoid steric clashes with the linker DNA (ψ angle of Lys36 was set to −35 degrees) (Figure 1a). The minimalistic model of NCP (NCPm) was obtained from the same crystal structure by clipping histone tails at the sites specified in Figure 2 by triangles, additional details are provided in Supplementary Information (SI). All models were explicitly solvated in a rectangular box with a minimum distance between the solute and the box boundaries of 20 Å, except for the FNbb (Table 1) system for which 100 Å threshold was used. Sodium ions were added to the system for neutralization, and then additional sodium and chloride ions were added at a concentration of 150 mM with respect to the volume of water, except for the FN1M system where 1 M concentration was used. The exact corresponding bulk ion concentration was estimated directly from the simulation data (see “Ion concentration reevaluation” section in SI). Crystallographic water molecules were retained in the system, while all crystallographic ions were removed. Protonation states of amino acids were assigned based on their solution pK values at neutral pH, histidine residues were considered neutral and protonated on ε-nitrogens. The FNnt model was identical to FN model but had neutrally charged protein termini to study robustness of MD simulations with respect to small perturbations. The structures of initial models are presented in Figure 1a and S1. The choice of NaCl as the salt for our simulations was influenced by the common use of it in nucleosome in vitro experiments as well as nucleosome reconstitution protocols [78].
Simulation protocols and force field choice
The CHARMM36 force field was used for DNA and protein [79, 80], TIP3P parameters for water molecules and adjusted ion parameters from Luo and Roux [81]. A choice of the force field is always a delicate issue due to certain known and unknown limitations in the force field accuracy balanced by the continuous efforts on force field improvement. In the case of nucleosome the problem is complicated by the need to combine accurate protein and DNA representations as well as to provide the realistic modeling of the interactions with the solvent. Below we briefly discuss several recent state-of-the-art studies about the behavior of different force filed in nucleosome simulations.
The accuracy of the protein force field seems to be especially essential to model the behavior of intrinsically disordered histone tails. Systematic studies of biomolecular force fields [82] suggested that certain force fields (e.g. CHARMM27, AMBER ff03) overstabilize the helical conformation of peptides and therefore this problem was addressed in the CHARMM36 protein force field revisions [79]. In addition, several recent studies questioned the applicability of protein force fields to specifically model the histone tails [25, 54, 55]. For example, Erler et al. showed that different versions of AMBER force fields may result in different dynamical behavior of histone tails [25]. Collepardo-Guevara et al. compared the results of simulations using state-of-the-art force fields, including CHARMM36, with the NMR data on histone tail dynamics and concluded that all force fields yield near identical results [54].
The DNA force field is essential to correctly reproduce the DNA conformational transitions in nucleosome simulations while the parameterization of the nucleic acids force fields proved to be more complicated than for protein force fields. Currently, both CHARMM and AMBER force fields are known to reproduce the stable dynamics of DNA at microsecond time scale and beyond [83]. However, accurate reproduction of the experimental results on the sequence dependent DNA deformability using the available force fields still remains an unsolved problem [84] (see additional discussion on force field choice in SI).
The simulation systems were prepared with the VMD program [85] and MD simulations were performed with the NAMD 2.9 package [86]. Langevin dynamics with 2 fs integration step, damping parameter of 0.5 ps−1 and T=310K were used as means to perform constant temperature simulations. Pressure coupling was implemented via Langevin piston method and set to 1 atm. Simulations were performed with the rigid covalent bonds and Van der Waals interactions were gradually switched off over the distance between 10 and 12 Å. Electrostatic calculations employed PME method with grid spacing of 1 Å, cubic interpolation, 12 Å real space cutoff and direct space tolerance of 10−6. Periodic boundary conditions were used. To remove the nucleosome diffusion, slight constraints of 0.003 kcal/mol/A2 were applied to C-α atoms of H3 histone folds (residue numbers 64-78, 86-114, 121-131). To avoid base pair fraying at DNA ends in NCPm simulation a restricting artificial wall potential was used to keep the distance between the centers of mass of bases in the terminal base pairs within 120% of the initial.
All systems were subjected to energy minimization and initial equilibration protocol as described in Supporting information. The production simulations were then performed up to the simulation time of 1 µs. The trajectory frames were saved every 100 ps. We run simulations in parallel on high performance computer clusters / supercomputers using effective parallelization available in NAMD. The simulation speed varied depending on the simulated system, number of CPUs and machine architecture. As a reference, the FN model system was simulated in parallel on 384 CPU cores for 120 days progressing at a pace of ~8 ns/day.
Trajectory analysis
Trajectory analysis and visualization were performed using a set of in-house developed scripting libraries written in TCL, Python and R which utilized the capabilities of VMD [85] and 3DNA [87] for general and DNA-specific structure analyses. For nucleosome structural analyses individual trajectory frames were superimposed onto the initial crystal structure to minimize the root mean square deviation (RMSD) values between the positions of C-α atoms of histone folds helices. The analysis of the DNA conformation was performed with respect to the nucleosomal superhelical reference frame determined by its dyad axis and superhelical axis (defined in SI). Rotational periodicity of DNA in nucleosome was determined by calculating the angle between the base pair vector (connecting N1 and N9 atoms of juxtaposed bases in the base pair) and the nucleosomal superhelical axis. Maxima and minima of rotational periodicity values corresponded to the integer and half-integer SHL values.
Detailed analyses of histone-DNA interactions were performed for every trajectory frame (first 250 ns frames were disregarded as an initial conformational equilibration period) by analyzing the positions of corresponding atoms. Contacts (SC) between two atoms of histone and DNA were defined as those between non-hydrogen atoms at a distance less than 3.9 Å. Contacts were further classified as following: salt bridges (SB) involved two charged non-hydrogen atoms at a distance less than 3.9 Å; hydrogen bonds (HB) were defined as bonds between donor (D) and acceptor (A) atoms with a hydrogen in-between (D-H… A), where the distance between D and A was less than 3.5 Å and D-H…A angle was larger than 150 degrees; van der Waals contacts (vdW) were defined as contacts between the atoms that were neither hydrogen bonded nor formed a salt bridge; and water mediated interactions (WM) were defined between non-hydrogen atoms of DNA or histones that formed hydrogen bond(s) with the same water molecule. We also introduced the notion of stable contacts between DNA and protein to describe contacts that persisted during the MD simulations. Namely, they were defined as individual pairs of atomic contacts between DNA and protein molecules that were present in more than 80% of trajectory frames after the initial period of conformational equilibration.
Nucleosome structure description conventions
Positions of DNA base pairs were numbered relative to the central base pair (referred to as dyad), its location was assumed to be zero. Rotational orientation of DNA double helix is semi-quantitatively described by the superhelical location (SHL) parameter (Figure 1a), which we extend to include not only core DNA (SHL from 0 to ±7), but also the linker DNA (SHL up to ±9) segments. The original 147 DNA base pairs of the NCP are referred to as “core DNA”, while regions where core DNA connects to the linker DNA are referred to as DNA entry/exit sites. We distinguish two parts in every histone: the tail region(s) (as labeled in Figure 2) and the rest, termed the histone core region(s). The key elements of core regions are histone fold helices α1, α2 and α3 [88] and L1, L2 loops as shown in Figure 2 (Figure S2 also shows the letter codes of the individual histone chains, Figure S3 shows locations of common PTM sites).
Supplementary Material
Highlights.
Mono-nucleosome dynamics is the key to decipher the chromatin function and epigenetic regulation.
All-atom dynamics of full nucleosome with DNA linkers is probed on the microsecond timescale.
Coupling between DNA geometry and reorganization of the histone-DNA interactions is observed;
DNA bulging and twist-defect formation are detected near the entry/exit site, suggestive of nucleosome sliding mechanisms
Accessibility of important epigenetically modified sites is governed by the patterns of histone tails adsorption on both core and linker DNA.
Acknowledgments
This research was supported by the Intramural Research Programs of the National Library of Medicine and the National Cancer Institute, National Institutes of Health; and Russian Science Foundation [grant No. 14-24-00031] (development of nucleosome visualization algorithms). Funding for open access charge: Intramural Research Program, NIH. AS was supported by the US-Russia Collaboration in the Biomedical Sciences NIH visiting fellows program. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Md. (http://biowulf.nih.gov). The reported study was supported in part by the Supercomputing Center of Lomonosov Moscow State University and with computational resources of Hexagon Cray XE6m-200 supercomputer, operated by the University of Bergen and the Norwegian metacenter for High Performance Computing (NOTUR).
Abbreviations used
- SI
Supporting Information
- RMSD
root-mean-square deviation
- RMSF
root-mean-square fluctuation
- SHL
superhelical location
- NCP
nucleosome core particle
- PTM
post-translational modification
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.McGhee JD, Felsenfeld G. Nucleosome structure. Annual review of biochemistry. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- 2.Dechassa ML, Luger K. Nucleosomes as Control Elements for Accessing the Genome. Genome Organization and Function in the Cell Nucleus: Wiley-VCH Verlag GmbH & Co. KGaA. 2011:55–87. [Google Scholar]
- 3.Madej T, Lanczycki CJ, Zhang D, Thiessen PA, Geer RC, Marchler-Bauer A, et al. MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic acids research. 2014;42:D297–D303. doi: 10.1093/nar/gkt1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marino-Ramirez L, Levine KM, Morales M, Zhang S, Moreland RT, Baxevanis AD, et al. The Histone Database: an integrated resource for histones and histone fold-containing proteins. Database : the journal of biological databases and curation. 2011;2011 doi: 10.1093/database/bar048. bar048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tolstorukov MY, Colasanti AV, McCandlish DM, Olson WK, Zhurkin VB. A novel roll-and-slide mechanism of DNA folding in chromatin: implications for nucleosome positioning. Journal of molecular biology. 2007;371:725–738. doi: 10.1016/j.jmb.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenley AT, Adams DA, Onufriev AV. Charge state of the globular histone core controls stability of the nucleosome. Biophysical journal. 2010;99:1577–1585. doi: 10.1016/j.bpj.2010.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. Journal of molecular biology. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 8.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nature reviews Genetics. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 9.Petty E, Pillus L. Balancing chromatin remodeling and histone modifications in transcription. Trends in genetics : TIG. 2013;29:621–629. doi: 10.1016/j.tig.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choy JS, Lee TH. Structural dynamics of nucleosomes at single-molecule resolution. Trends in biochemical sciences. 2012;37:425–435. doi: 10.1016/j.tibs.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nature structural & molecular biology. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 12.Lee KM, Hayes JJ. The N-terminal tail of histone H2A binds to two distinct sites within the nucleosome core. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:8959–8964. doi: 10.1073/pnas.94.17.8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee HS, Bataille AR, Zhang L, Pugh BF. Subnucleosomal structures and nucleosome asymmetry across a genome. Cell. 2014;159:1377–1388. doi: 10.1016/j.cell.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zlatanova J, Bishop TC, Victor JM, Jackson V, van Holde K. The nucleosome family: dynamic and growing. Structure. 2009;17:160–171. doi: 10.1016/j.str.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 15.Mueller-Planitz F, Klinker H, Becker PB. Nucleosome sliding mechanisms: new twists in a looped history. Nature structural & molecular biology. 2013;20:1026–1032. doi: 10.1038/nsmb.2648. [DOI] [PubMed] [Google Scholar]
- 16.Shaytan AK, Landsman D, Panchenko AR. Nucleosome adaptability conferred by sequence and structural variations in histone H2A–H2B dimers. Current opinion in structural biology. 2015;32C:48–57. doi: 10.1016/j.sbi.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pepenella S, Murphy KJ, Hayes JJ. Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma. 2014;123:3–13. doi: 10.1007/s00412-013-0435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang WL, Deindl S, Harada BT, Zhuang X. Histone H4 tail mediates allosteric regulation of nucleosome remodelling by linker DNA. Nature. 2014 doi: 10.1038/nature13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Racki LR, Naber N, Pate E, Leonard JD, Cooke R, Narlikar GJ. The histone H4 tail regulates the conformation of the ATP-binding pocket in the SNF2h chromatin remodeling enzyme. Journal of molecular biology. 2014;426:2034–2044. doi: 10.1016/j.jmb.2014.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F, Li G, Altaf M, Lu C, Currie MA, Johnson A, et al. Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8495–8500. doi: 10.1073/pnas.1300126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Current opinion in genetics & development. 2012;22:148–155. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biswas M, Langowski J, Bishop TC. Atomistic simulations of nucleosomes. Wiley Interdisciplinary Reviews: Computational Molecular Science. 2013;3:378–392. [Google Scholar]
- 23.Arya G, Schlick T. Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collepardo-Guevara R, Schlick T. Chromatin fiber polymorphism triggered by variations of DNA linker lengths. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:8061–8066. doi: 10.1073/pnas.1315872111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erler J, Zhang R, Petridis L, Cheng X, Smith JC, Langowski J. The role of histone tails in the nucleosome: a computational study. Biophysical journal. 2014;107:2902–2913. doi: 10.1016/j.bpj.2014.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Materese CK, Savelyev A, Papoian GA. Counterion atmosphere and hydration patterns near a nucleosome core particle. Journal of the American Chemical Society. 2009;131:15005–15013. doi: 10.1021/ja905376q. [DOI] [PubMed] [Google Scholar]
- 27.Ruscio JZ, Onufriev A. A computational study of nucleosomal DNA flexibility. Biophysical journal. 2006;91:4121–4132. doi: 10.1529/biophysj.106.082099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nature reviews Molecular cell biology. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. Journal of molecular biology. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 30.Norouzi D, Zhurkin VB. Topological polymorphism of the two-start chromatin fiber. Biophysical journal. 2015;108:2591–2600. doi: 10.1016/j.bpj.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato H, Gruschus J, Ghirlando R, Tjandra N, Bai Y. Characterization of the N-terminal tail domain of histone H3 in condensed nucleosome arrays by hydrogen exchange and NMR. Journal of the American Chemical Society. 2009;131:15104–15105. doi: 10.1021/ja9070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou BR, Feng H, Ghirlando R, Kato H, Gruschus J, Bai Y. Histone H4 K16Q mutation, an acetylation mimic, causes structural disorder of its N-terminal basic patch in the nucleosome. Journal of molecular biology. 2012;421:30–37. doi: 10.1016/j.jmb.2012.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao M, Nadaud PS, Bernier MW, North JA, Hammel PC, Poirier MG, et al. Histone H3 and H4 N-terminal tails in nucleosome arrays at cellular concentrations probed by magic angle spinning NMR spectroscopy. Journal of the American Chemical Society. 2013;135:15278–15281. doi: 10.1021/ja407526s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nature structural biology. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 35.Obri A, Ouararhni K, Papin C, Diebold ML, Padmanabhan K, Marek M, et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature. 2014;505:648–653. doi: 10.1038/nature12922. [DOI] [PubMed] [Google Scholar]
- 36.Biswas M, Voltz K, Smith JC, Langowski J. Role of histone tails in structural stability of the nucleosome. PLoS computational biology. 2011;7:e1002279. doi: 10.1371/journal.pcbi.1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pachov GV, Gabdoulline RR, Wade RC. On the structure and dynamics of the complex of the nucleosome and the linker histone. Nucleic acids research. 2011;39:5255–5263. doi: 10.1093/nar/gkr101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toth K, Brun N, Langowski J. Chromatin compaction at the mononucleosome level. Biochemistry. 2006;45:1591–1598. doi: 10.1021/bi052110u. [DOI] [PubMed] [Google Scholar]
- 39.Mirny LA. Nucleosome-mediated cooperativity between transcription factors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:22534–22539. doi: 10.1073/pnas.0913805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomschik M, van Holde K, Zlatanova J. Nucleosome dynamics as studied by single-pair fluorescence resonance energy transfer: a reevaluation. Journal of fluorescence. 2009;19:53–62. doi: 10.1007/s10895-008-0379-1. [DOI] [PubMed] [Google Scholar]
- 41.Gansen A, Valeri A, Hauger F, Felekyan S, Kalinin S, Toth K, et al. Nucleosome disassembly intermediates characterized by single-molecule FRET. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15308–15313. doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nurse NP, Jimenez-Useche I, Smith IT, Yuan C. Clipping of flexible tails of histones H3 and H4 affects the structure and dynamics of the nucleosome. Biophysical journal. 2013;104:1081–1088. doi: 10.1016/j.bpj.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenz L, Hoenderdos M, Prinsen P, Schiessel H. The influence of DNA shape fluctuations on fluorescence resonance energy transfer efficiency measurements in nucleosomes. Journal of physics Condensed matter : an Institute of Physics journal. 2015;27:064104. doi: 10.1088/0953-8984/27/6/064104. [DOI] [PubMed] [Google Scholar]
- 44.Roccatano D, Barthel A, Zacharias M. Structural flexibility of the nucleosome core particle at atomic resolution studied by molecular dynamics simulation. Biopolymers. 2007;85:407–421. doi: 10.1002/bip.20690. [DOI] [PubMed] [Google Scholar]
- 45.Mangenot S, Leforestier A, Vachette P, Durand D, Livolant F. Salt-induced conformation and interaction changes of nucleosome core particles. Biophysical journal. 2002;82:345–356. doi: 10.1016/S0006-3495(02)75399-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang C, van der Woerd MJ, Muthurajan UM, Hansen JC, Luger K. Biophysical analysis and small-angle X-ray scattering-derived structures of MeCP2-nucleosome complexes. Nucleic acids research. 2011;39:4122–4135. doi: 10.1093/nar/gkr005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilhelm FX, Wilhelm ML, Erard M, Duane MP. Reconstitution of chromatin: assembly of the nucleosome. Nucleic acids research. 1978;5:505–521. doi: 10.1093/nar/5.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoo J, Aksimentiev A. Improved Parametrization of Li+, Na+, K+, and Mg2+ Ions for All-Atom Molecular Dynamics Simulations of Nucleic Acid Systems. The journal of physical chemistry letters. 2012;3:45–50. [Google Scholar]
- 49.Iwaki T, Saito T, Yoshikawa K. How are small ions involved in the compaction of DNA molecules? Colloids and Surfaces B: Biointerfaces. 2007;56:126–133. doi: 10.1016/j.colsurfb.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 50.Korolev N, Vorontsova OV, Nordenskiold L. Physicochemical analysis of electrostatic foundation for DNA-protein interactions in chromatin transformations. Progress in biophysics and molecular biology. 2007;95:23–49. doi: 10.1016/j.pbiomolbio.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Dehghani H, Dellaire G, Bazett-Jones DP. Organization of chromatin in the interphase mammalian cell. Micron. 2005;36:95–108. doi: 10.1016/j.micron.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 52.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 53.Weng L, Zhou C, Greenberg MM. Probing Interactions between Lysine Residues in Histone Tails and Nucleosomal DNA via Product and Kinetic Analysis. ACS chemical biology. 2014 doi: 10.1021/cb500737y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collepardo-Guevara R, Portella G, Vendruscolo M, Frenkel D, Schlick T, Orozco M. Chromatin Unfolding by Epigenetic Modifications Explained by Dramatic Impairment of Internucleosome Interactions: A Multiscale Computational Study. Journal of the American Chemical Society. 2015;137:10205–10215. doi: 10.1021/jacs.5b04086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Potoyan DA, Papoian GA. Energy landscape analyses of disordered histone tails reveal special organization of their conformational dynamics. Journal of the American Chemical Society. 2011;133:7405–7415. doi: 10.1021/ja1111964. [DOI] [PubMed] [Google Scholar]
- 56.Potoyan DA, Papoian GA. Regulation of the H4 tail binding and folding landscapes via Lys-16 acetylation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17857–17862. doi: 10.1073/pnas.1201805109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korolev N, Nordenskiold L. H4 histone tail mediated DNA-DNA interaction and effects on DNA structure, flexibility, and counterion binding: a molecular dynamics study. Biopolymers. 2007;86:409–423. doi: 10.1002/bip.20749. [DOI] [PubMed] [Google Scholar]
- 58.Korolev N, Yu H, Lyubartsev AP, Nordenskiold L. Molecular dynamics simulations demonstrate the regulation of DNA-DNA attraction by H4 histone tail acetylations and mutations. Biopolymers. 2014;101:1051–1064. doi: 10.1002/bip.22499. [DOI] [PubMed] [Google Scholar]
- 59.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Z, Zheng C, Hayes JJ. The core histone tail domains contribute to sequence-dependent nucleosome positioning. The Journal of biological chemistry. 2007;282:7930–7938. doi: 10.1074/jbc.M610584200. [DOI] [PubMed] [Google Scholar]
- 61.Shukla MS, Syed SH, Goutte-Gattat D, Richard JL, Montel F, Hamiche A, et al. The docking domain of histone H2A is required for H1 binding and RSC-mediated nucleosome remodeling. Nucleic acids research. 2011;39:2559–2570. doi: 10.1093/nar/gkq1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 63.Edayathumangalam RS, Weyermann P, Dervan PB, Gottesfeld JM, Luger K. Nucleosomes in solution exist as a mixture of twist-defect states. Journal of molecular biology. 2005;345:103–114. doi: 10.1016/j.jmb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 64.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nature structural & molecular biology. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brogaard K, Xi L, Wang JP, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davey CA. Does the nucleosome break its own rules? Current opinion in structural biology. 2013;23:311–313. doi: 10.1016/j.sbi.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 68.Tan S, Davey CA. Nucleosome structural studies. Current opinion in structural biology. 2011;21:128–136. doi: 10.1016/j.sbi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Studitsky VM, Kassavetis GA, Geiduschek EP, Felsenfeld G. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–1963. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- 70.Jin J, Bai L, Johnson DS, Fulbright RM, Kireeva ML, Kashlev M, et al. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nature structural & molecular biology. 2010;17:745–752. doi: 10.1038/nsmb.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee KM, Hayes JJ. Linker DNA and H1-dependent reorganization of histone-DNA interactions within the nucleosome. Biochemistry. 1998;37:8622–8628. doi: 10.1021/bi980499y. [DOI] [PubMed] [Google Scholar]
- 72.Angelov D, Vitolo JM, Mutskov V, Dimitrov S, Hayes JJ. Preferential interaction of the core histone tail domains with linker DNA. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6599–6604. doi: 10.1073/pnas.121171498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stefanovsky V, Dimitrov SI, Russanova VR, Angelov D, Pashev IG. Laser-induced crosslinking of histones to DNA in chromatin and core particles: implications in studying histone-DNA interactions. Nucleic acids research. 1989;17:10069–10081. doi: 10.1093/nar/17.23.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pilotto S, Speranzini V, Tortorici M, Durand D, Fish A, Valente S, et al. Interplay among nucleosomal DNA, histone tails, and corepressor CoREST underlies LSD1-mediated H3 demethylation. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2752–2757. doi: 10.1073/pnas.1419468112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nishi H, Demir E, Panchenko AR. Crosstalk between signaling pathways provided by single and multiple protein phosphorylation sites. Journal of molecular biology. 2015;427:511–520. doi: 10.1016/j.jmb.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Macke Thomas J, Case David A. Modeling Unusual Nucleic Acid Structures. Molecular Modeling of Nucleic Acids: American Chemical Society. 1997:379–393. [Google Scholar]
- 77.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods in enzymology. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 79.Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. Journal of chemical theory and computation. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hart K, Foloppe N, Baker CM, Denning EJ, Nilsson L, Mackerell AD., Jr Optimization of the CHARMM additive force field for DNA: Improved treatment of the BI/BII conformational equilibrium. Journal of chemical theory and computation. 2012;8:348–362. doi: 10.1021/ct200723y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luo Y, Roux B. Simulation of Osmotic Pressure in Concentrated Aqueous Salt Solutions. The journal of physical chemistry letters. 2010;1:183–189. [Google Scholar]
- 82.Lindorff-Larsen K, Maragakis P, Piana S, Eastwood MP, Dror RO, Shaw DE. Systematic validation of protein force fields against experimental data. PloS one. 2012;7:e32131. doi: 10.1371/journal.pone.0032131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Galindo-Murillo R, Roe DR, Cheatham TE., 3rd Convergence and reproducibility in molecular dynamics simulations of the DNA duplex d(GCACGAACGAACGAACGC) Biochimica et biophysica acta. 2015;1850:1041–1058. doi: 10.1016/j.bbagen.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perez A, Luque FJ, Orozco M. Frontiers in molecular dynamics simulations of DNA. Accounts of chemical research. 2012;45:196–205. doi: 10.1021/ar2001217. [DOI] [PubMed] [Google Scholar]
- 85.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 86.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. Journal of computational chemistry. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nature protocols. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baxevanis AD, Arents G, Moudrianakis EN, Landsman D. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic acids research. 1995;23:2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shaytan AK, Armeev GA, Goncearenco A, Zhurkin VB, Landsman D, Panchenko AR. Trajectories of microsecond molecular dynamics simulations of nucleosomes and nucleosome core particles. Data in Brief . doi: 10.1016/j.dib.2016.04.073. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.