Abstract
Purpose
CSF3R mutations have been identified in the majority of chronic neutrophilic leukemia (CNL) and a smaller percentage of atypical chronic myeloid leukemia (aCML) cases. Although CSF3R point mutations (e.g. T618I) are emerging as key players in CNL/aCML, the significance of rarer CSF3R mutations is unknown. In this study we assess the importance of the CSF3R T640N mutation as a marker of CNL/aCML and potential therapeutic target.
Experimental Design
Sanger sequencing of leukemia samples was performed to identify CSF3R mutations in CNL and aCML. The oncogenicity of the CSF3R T640N mutation relative to the T618I mutation was assessed by cytokine independent growth assays and by mouse bone marrow transplant. Receptor dimerization and O-glycosylation of the mutants was assessed by western blot, and JAK inhibitor sensitivity was assessed by colony assay.
Results
Here we identify a CSF3R T640N mutation in two patients with CNL/aCML, one of whom was originally diagnosed with MDS and acquired the T640N mutation upon evolution of disease to aCML. The T640N mutation is oncogenic in cellular transformation assays and an in vivo mouse bone marrow transplantation model. It exhibits many similar phenotypic features to T618I, including ligand independence and altered patterns of O-glycosylation – despite the transmembrane location of T640 preventing access by GalNAc transferase enzymes. Cells transformed by the T640N mutation are sensitive to JAK kinase inhibition to a similar degree as cells transformed by CSF3R T618I.
Conclusions
Due to its similarities to CSF3R T618I, the T640N mutation likely has diagnostic and therapeutic relevance in CNL/aCML.
Keywords: CSF3R, chronic neutrophilic leukemia, atypical chronic myeloid leukemia, oncogenesis, kinase inhibitors
INTRODUCTION
Chronic neutrophilic leukemia (CNL) and atypical chronic myeloid leukemia (aCML) are two rare forms of leukemia characterized by high levels of neutrophils or neutrophil precursors (1). Diagnostic criteria for these malignancies include absence of BCR-ABL1, PDGFR or FGFR rearrangements and absence of underlying infections that would drive a non-neoplastic neutrophilia (1). As such, these malignancies have historically been difficult to diagnose. We have recently identified mutations in CSF3R as having high prevalence in CNL (~80%) and a lower frequency in aCML (2,3). Two classes of CSF3R mutations occur in these disorders, truncations of the cytoplasmic domain of the receptor, and membrane proximal threonine mutations (T618I (aka T595I) and T615A). These two classes of CSF3R mutations have different mechanisms of action. The CSF3R truncation mutations remove a portion of the cytoplasmic domain of the receptor, including endocytic and other negative regulatory motifs, resulting in increased cell surface expression (4–6). In contrast, the T618I and T615A mutations do not have increased expression, but instead exhibit constitutive dimerization (7) and ligand-independence (8,9). This increased dimerization is accompanied by a loss of O-glycosylation of the receptor resulting from mutation of threonine residues critical for addition of the O-glycosylation(10). The T618I mutation is by far the most common mutation in CNL/aCML (2).
The T618I mutation results in hyper-activation of the downstream Janus Kinase (JAK) pathway (2). Specimens obtained from a patient with the T618I mutation revealed sensitivity to JAK inhibitors, including ruxolitinib (Jakafi, Incyte), a JAK 1/2 inhibitor approved for treatment of myelofibrosis (2). In a mouse model of T618I driven leukemia, treatment of mice with ruxolitinib revealed a reduction in white blood cell counts and decreased splenomegaly (11). Additionally, a patient with CNL harboring the T618I mutation exhibited a marked decrease in leukemic white blood cells upon treatment with ruxolitinib (2), indicating that JAK inhibitor therapy for this patient population warrants further clinical investigation.
While the T618I mutation is the most prevalent CSF3R mutational event observed in this patient population, there are other rare mutations in CSF3R, and it will be important to understand the functional consequences of all CSF3R mutations for appropriate diagnostic and therapeutic decision-making. We recently identified the CSF3R T640N mutation in patients with CNL and aCML, which represents the first cases of this mutation in these disease entities. This mutation (also known as T617N, using a numbering scheme that does not include the signal peptide (12)) has been reported previously in a family with congenital neutrophilia and is found rarely in AML (7,13). The T640 residue is located within the transmembrane domain of CSF3R, thus precluding access of GalNAc transferase enzymes that regulate O-glycosylation to this portion of the receptor. The T640N mutation was shown to induce ligand-independent activation of CSF3R, and was predicted by molecular modeling to drive interaction of the transmembrane helices through increased hydrogen bonding (12,13). In this study we find that the T640N mutation is highly oncogenic, and has many features similar to the T618I mutation, including rapid transformation of Ba/F3 cells, ligand independence sensitivity to the JAK 1/2 inhibitor ruxolitinib, and lethality in a mouse bone marrow transplant model. Surprisingly, we also find alteration of O-glycosylation patterns similar to those observed with T618I indicating a more complex relationship between CSF3R dimerization and O-glycosylation than was previously perceived. Although rare, T640N is a mutation with oncogenic capacity that may have diagnostic and therapeutic relevance.
MATERIALS AND METHODS
Clinical Specimens
All clinical samples were obtained with informed consent by the patients. The study was approved by the institutional review boards at the Stanford University School of Medicine and Oregon Health and Science University (OHSU).
Sequencing
Sanger sequencing was performed on CSF3R as described previously (2). To determine the variant allele frequency of the CSF3R T640N and SETBP1 G870S mutations, we utilized the Ion Torrent AmpliSeq next-generation technology to quantify the frequency of these mutations relative to the wild-type amplicons. The library was prepared using a custom panel of 56 amplicon primer pools specific to the CDS of both CSF3R and SETBP1, respectively. The sample was sequenced on the Ion Torrent Personal Genome Machine (PGM) and the sequences were aligned and checked for known and novel variations usingthe Torrent Suite Software v.4.4.
Cloning
WT and T618I constructs in MSCF-IRES-GFP, p3XFLAG-CMV-14, pcDNA3.2/V5-DEST Gateway vector were described previously (10). The CSF3R T640N mutation was made using the Agilent Quikchange Site Directed Mutagenesis Kit using the following primers (CSF3R T640N-F gttccacagaggcagttgagcaacagcagga and CSF3R T640N-R tcctgctgttgctcaactgcctctgtggaac).
Cell Culture & Transfection
293T17 cells (ATCC) were maintained in DMEM (Invitrogen) supplemented with 10% FBS (Atlanta Biologicals), L-glutamine, penicillin/streptomycin (Invitrogen), and amphotericin B (HyClone). Ba/F3 cells were maintained in RPMI 1640 supplemented with 10% FBS, 15% WEHI-conditioned media (IL3 source), L-glutamine, penicillin/streptomycin, and amphotericin B. FuGENE 6 (Promega) was used for the transfection of 293T17 cells. Production of murine retrovirus was achieved by co-transfection of 293T17’s with CSF3R-MSCF-IRES-GFP constructs with pEcopac Helper and harvesting supernatants 48 hours post-transfection.
Cytokine-independent Growth Assays & Bone Marrow Transplantations
Ba/F3 cell cytokine-independence assays and mouse bone marrow colony assays were performed as described previously (2,10). Mouse bone marrow transplantations were performed as described previously (11). Briefly, BALB/c donor mice were treated with 5-fluorouracil and then whole bone marrow was extracted from mouse femurs after 5 days. Bone marrow was cultured overnight in medium containing IL-3, IL-6 and SCF and then spinoculated twice with WT CSF3R, CSF3R T618I or CSF3R T640N retrovirus (described above). Marrow was then transplanted into lethally irradiated BALB/c recipient mice (five per group), and blood counts including white blood cell counts and % granulocytes were monitored using a Vet ABC animal blood counter (Scil animal care company). All mouse work was performed with approval from the OHSU Institutional Animal Care and Use Committee.
Immunoblotting
Cells were lysed in cell lysis buffer (Cell Signaling Technologies) containing complete mini protease inhibitor tablets (Roche). To pellet cellular debris the lysates were spun at 12000 rpm, 4ºC for 10 minutes and subsequently mixed with 3X SDS sample buffer (75mM Tris (pH 6.8), 3% SDS, 15% glycerol, 8% β-mercaptoethanol, and 0.1% bromophenol blue). Samples were incubated at 95ºC for 5 minutes and run on Criterion 4–15% Tris-HCl gradient gels (Bio Rad). Gels were transferred to PVDF membranes and blocked in Tris-buffered saline with Tween (TBST) with 5% Bovine Serum Albumin (BSA). Blots were probed with the following primary antibodies: anti-GCSF receptor Rabbit antibody (Abcam, cat# ab126167), anti-Flag Mouse antibody (Sigma, cat# F3165-1MG), anti-V5 Mouse antibody (Invitrogen, cat# 46-0705), and anti-Actin Mouse antibody (CalBioChem, cat# CP01). HRP conjugate secondary antibodies against mouse IgG and rabbit IgG (Promega) were used followed by imaging of the blots on a Lummi imager (Roche Applied Science). Co-immunoprecipitation experiments to measure dimerization were performed asdescribed previously (10).
O-Glycosylation Labeling
CSF3R constructs (WT, T618I, T640N) were transfected into 293T17 cells in the presence of 50μM GalNAz (Thermo Scientific) and incubated for 48 hours (14). Cells were washed then incubated in 1ml PBS with 1% FBS and 30μM DBCO-sulfo-link-biotin conjugate (Click Chemistry Tools) for 1 hour. Cells were then lysed in cell lysis buffer (Cell Signaling Technologies) containing complete mini protease inhibitor tablets (Roche). Lysates were incubated overnight in the presence of streptavidin-agarose (Thermo Scientific) and the precipitated biotinylated proteins were subjected to immunoblot analysis for CSF3R (10).
Co-immunoprecipitations
Dimerization experiments were performed as described previously (10). Briefly, Flag and V5-tagged constructs were co-transfected into 293T17 cells. Flag-tagged constructs were immunoprecipated by incubating with Anti-Flag M2 Affinity Gel (Sigma-Aldrich) and then subjected to immunoblot analysis as described above using anti-Flag and anti-V5 antibodies. Immunoprecipitations were normalized to the starting input.
RESULTS
Identification of the CSF3R T640N mutation in patients with both CNL and aCML
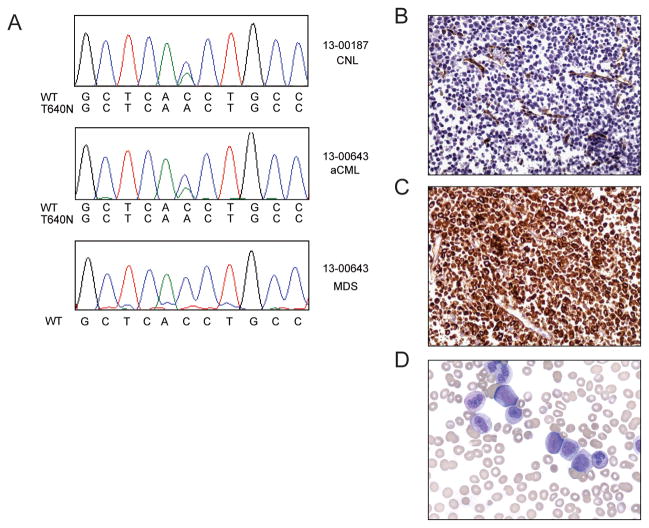
Sequencing of leukemic blasts from two patients with CNL and aCML revealed that that they carried the CSF3R T640N mutation (Figure 1A, B). These two patients were identified in a cohort of 38 CNL/aCML patients with CSF3R mutations that are known to be oncogenic, for an approximate frequency of T640N mutations at 5% of CSF3R mutation positive patients. Patient #13-000187 is a 52-year-old man who presented with abdominal pain, and an abdominal CT scan showed splenomegaly. His CBC revealed a WBC count of 36 × 109/L with 75% neutrophils and 2.5 × 109/L monocytes. Another CBC two weeks later showed a white count of 41 × 109/L, hemoglobin 13.6 g/dL, 78% neutrophils, and monocyte count 2.9 × 109/L. Serial CBC's showed a progressive increase in his WBC count to 102 × 109/L with 74 × 109/L neutrophils within a 3 month period. He also had significant night sweats, weight loss, and early satiety due to marked hepato/splenomegaly. A diagnostic bone marrow biopsy was consistent with CNL, without an increase in myeloblasts. The leukemic cells were BCR-ABL1 negative. Molecular analysis by Sanger sequencing revealed presence of the following mutations: CSF3R T640N, SETBP1 G870S (Figure 1A). The frequency of the CSF3R and SETBP1 mutations were further analyzed by Ion Torrent AmpliSeq sequencing and found to be present at 42.2 and 44.8% respectively. This is concurrent with the Illumina next-generation sequencing, which detected the CSF3R T640N mutation in 14 out of 35 reads (40%) and the SETBP1 mutation in 20 of 49 reads (41%) in this sample. Since material was not available for germline analysis of these samples, these mutations could not be definitively confirmed as germline or somatic, however a silent SNP in SETBP1 was present at 50.7%. Subsequent clinical testing, 7 months after the initial CSF3R and SETBP1 mutation analysis, also revealed the presence of a JAK2 V617F mutation at an allele fraction of 60%, although interestingly this mutation was not present at the time of initial CSF3R/SETBP1 analysis.
Figure 1. The CSF3R T640N mutation is found in patients with CML/aCML.
(A) The T640N mutation was identified in Patient 13-187 who was diagnosed with de-novo CNL. Electroferograms from Sanger sequencing are shown. Patient 13-643 initially had a diagnosis of MDS and was subsequently diagnosed with aCML. Electropherograms are shown from the time of diagnosis of aCML and from a retrospective sample from the time of the original MDS diagnosis. Electropherogram peaks correspond to the following nucleotides: A (green), T (red), C (blue), G (black). Histology from a patient with aCML showing a myeloid proliferation. Cervical node showing (B) myeloid hyperplasia with CD34 immunohistochemical (IHC) staining indicating a low proportion of myeloblasts as well as the presence of endothelial structures. (C) myeloid proliferation showing myeloperoxidase positive IHC staining. (D) Peripheral blood showing spectrum of immature myeloid cells and granulocytic dysplasia.
Patient #13-00643 was a 75 year-old male diagnosed with myelodysplastic syndrome (MDS), refractory anemia with multilineage dysplasia (RCMD) subtype. A CBC revealed a WBC count of 2.3 × 109/L, ANC 0.78 × 109/L, hemoglobin 13g/dL, hematocrit 37%, and platelet count of 111 × 109/L. The differential showed 27% neutrophils, 7% band forms, 49% lymphocytes, and 15% monocytes. Bone marrow biopsy was hypercellular with trilineage dysplasia and 1% myeloblasts with a normal karyotype. Three years later, the patient developed worsening dyspnea, splenomegaly and pancytopenia (hemoglobin 10.9 g/dL, platelets 67 × 109/L, WBC 2.6 × 109/L including 50% neutrophils, and a spectrum of myeloid immaturity, including 9% blasts). The bone marrow demonstrated trilineage dysplasia, and 12% myeloblasts, consistent with progression of the MDS to refractory anemia with excess blasts-2 (RAEB-2). In addition, cytogenetic analysis revealed a complex karyotype 46~57,XY,der(3)add(3)(p25)add(3)(q26)[cp4]/46,XY[18]. Twenty-two metaphase cells were analyzed, at least four of which demonstrated an incompletely characterized abnormal chromosome 3 [der(3)] with apparent rearrangement of both arms. These rearrangements involve addition of material of unknown origin. He was treated with 7 cycles of azacitidine (including pegfilgastrim immediately after each 7-day course). He was deemed ineligible for hematopoietic stem cell transplant due to symptomatic cryptogenic organizing pneumonia. With treatment he exhibited marked improvement in cytopenias, with a repeat marrow examination demonstrating a complete morphologic and cytogenetic remission.
Subsequently, the patient developed right cervical node enlargement. A biopsy demonstrated proliferation of a spectrum of myeloid cells that were negative for CD34 by immunohistochemical staining (Figure 1B), but positive for myeloperoxidase (Figure 1C). The findings were not consistent with a myeloid sarcoma, but were worrisome for extramedullary disease progression. At this time, a CBC showed a WBC count of 73 × 109/L, hemoglobin 10.2 g/dL, and platelet count 32 × 109/L. Over the next 3 months, the patient developed progressive leukocytosis, anemia and thrombocytopenia. The peripheral blood smear revealed a spectrum of immature myeloid cells with granulocytic dysplasia and increasing myeloblasts (Figure 1D). A repeat bone marrow biopsy showed myeloid hyperplasia and 10% myeloblasts. Mutation studies were negative for BCR-ABL1 and JAK2 V617F. Testing for mutations in other genes associated with myeloid neoplasms was negative (e.g. NPM1, CEBPA, KIT D816V, FLT3 ITD/ D835). Taken together, the diagnostic findings were consistent with atypical chronic myelogenous leukemia (aCML) arising after MDS. Given the recent identification of CSF3R mutations in CNL and aCML (2), sequencing of the peripheral blood was undertaken and revealed, a CSF3R T640N mutation (Figure 1A), which was not detectable by Sanger Sequencing in an archived bone marrow specimen taken at the time of RAEB-2 diagnosis (Figure 1A). This mutation was, however detectable by Illumina deep sequencing in 5.6% of the reads (1 mutant read out of 18 total reads) in this archival specimen, and was found in 50% of reads (20 mutant reads out of 40 total reads) from the specimen taken after evolution of disease to aCML.
The CSF3R T640N mutation is oncogenic and induces ligand independence
In order to determine the oncogenic capacity of the T640N mutation relative to T618I, we used a cytokine independent growth assay with the murine pro B-cell Ba/F3 cell line. These cells are normally dependent on the cytokine, IL3, for growth, but are able to grow in the absence of IL3 when a transforming oncogene, such as the CSF3R T618I mutation, is present (2). Similar to the CSF3R T618I mutation, CSF3R T640N exhibited transformative capacity of Ba/F3 cells to IL3-independent growth (Supplemental Figure 1A). The T640N mutation was reported previously to confer GCSF ligand independent growth in a murine bone marrow colony assay (12). To assess the relative capacity of CSF3R T640N versus T618I to promote ligand independent colony formation, we infected mouse bone marrow with a murine retrovirus harboring the CSF3R T618I, T640N, or WT constructs, and found that the T640N mutation led to GCSF-independent colony formation to a similar extent as T618I (Supplemental Figure 1B). The similar oncogenic potency of the T640N mutation to the T618I mutation in these models, indicates that it is likely an important driver of the CNL and aCML observed in these patients.
CSF3R T640N causes leukemia in a mouse bone marrow transplant model
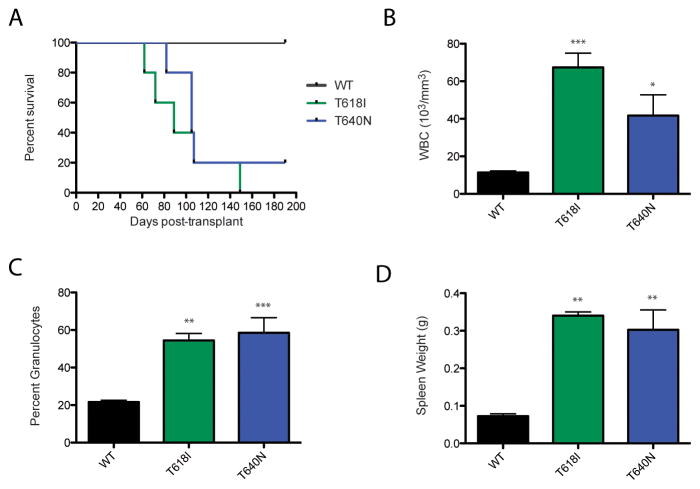
Since the T640N mutation appeared to have similar potency to CSF3R T618I in vitro (Supplemental Figure 1), we wanted to test whether it had similar potency to T618I in vivo. A mouse bone marrow transplant experiment revealed that the T640N mutation induces a lethal leukemia in vivo (Figure 2A), with similar latency to the CSF3R T618I mutation (slight differences in latency could be due to slight differences in transduction of the donor bone marrow as measured by GFP positivity of the cells: WT CSF3R 61.2%, CSF3R T618I 65.4% and CSF3R T640N 51.8%).
Figure 2. CSF3R T640N causes a lethal leukemia in a mouse model, similar to T618I.
A mouse bone marrow transplant was performed to determine whether the T640N mutation could cause leukemia in a mouse model. (A) While WT CSF3R does not lead to death of the animals, the mice transplanted with the T640N or T618I constructs succumb to disease. (B) The CSF3R T640N and T618I transplanted mice have higher white blood cell counts than the mice transplanted with WT CSF3R. Average white blood cell counts measured with an Animal Blood Counter (Scil) are shown for the last data point prior to death of the mice, or euthanization for moribund animals or at the completion of the experiment for the WT CSF3R transplanted mice. Statistical significance was assessed using a one way ANOVA followed by a Bonferroni’s Multiple Comparison Test. * p<.05, ** p<.01, *** p<.001, and shown for each mutant relative to WT CSF3R (C) The CSF3R T640N and T618I mice had a statistically significant higher percent granulocytes than their WT counterparts prior to death (T618I, T640N) or euthanization at the conclusion of the experiment (WT CSF3R). Each individual mouse is represented in the plot along with the mean and standard error. (D) The CSF3R T618I and T640N transplanted mice have significantly larger spleens than their WT CSF3R counterparts. Spleen weight is shown in grams.
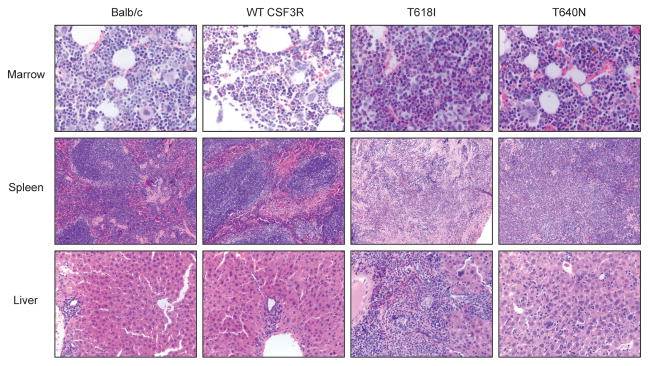
The T618I and T640N animals had statistically significant elevated white counts and an increased percentage of granulocytes relative to WT CSF3R transplanted mice at the time of death (Figure 2B, C). Both the T640N and T618I transplanted mice had statistically significant increased spleen weight relative to the WT CSF3R controls (Figure 2D). Histologic analysis of the T618I and T640N mutation revealed some hypercellularity of the bone marrow relative to the WT CSF3R and non-transplanted control mice (Figure 3). Furthermore, in the T640N mice there was some disorganization of the splenic architecture and infiltration of granulocytes into the liver, but again this was less pronounced than what was seen in with T618I.
Figure 3. Histological analysis of T640N mice reveals less dramatic hypercellularity of bone marrow that in the CSF3R T618I mice.
Femurs, spleen and livers were removed and fixed in 10% zinc formalin, section and stained with hematoxylin and eosin by the Histopathology Shared Resource at OHSU. Representative images are shown from the bone marrow (40X), spleen (10X) and liver (20X) of untransplanted Balb/c mice, WT CSF3R, T618I and T640N transplanted mice.
The CSF3R T640N mutation has an altered banding pattern and decreased O-glycosylation
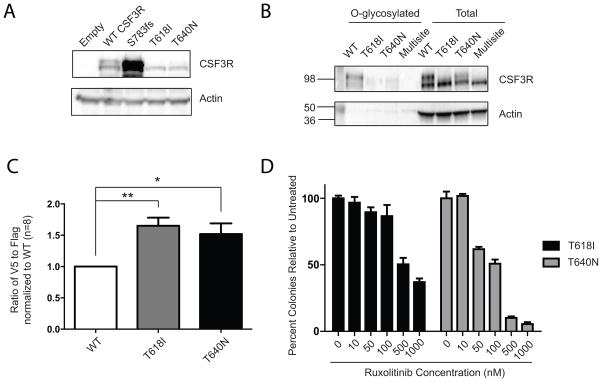
We next investigated similarities in the molecular mechanisms by which these two mutations activate CSF3R. Mutations in CSF3R studied thus far have exhibited two distinct mechanisms of action: either overexpression on the cell surface (truncation mutations) or loss of glycosylation, which results in an altered banding pattern by immunoblot (T618I/T615A). Interestingly, the CSF3R T640N mutation did not result in any changes in expression level relative to WT CSF3R, but did exhibit an alteration in banding pattern, consisting of mostly the lower molecular weight band (Figure 4A). To ascertain whether the alteration in banding pattern is due to altered posttranslational modification, we performed a direct O-glycosylation assay, in which an azide modified galactosamine (GalNAz) is fed to cells expressing WT or mutant versions of CSF3R. GalNAz then incorporates specifically into the site of O-glycosylation and the modified sugar can be labeled with biotin, purified, and detected by immunoblot. While expression of WT CSF3R results in substantial levels of GalNAz-labeled CSF3R (indicating O-glycosylation of the WT receptor), expression of T618I results in reduced levels, and expression of a multi-site mutant ablating seven threonine/serine residues in this membrane proximal region exhibits no detectable GalNAz incorporation. Interestingly, expression of the T640N mutation markedly reduced detectable O-glycosylation of CSF3R with GalNAz labeling, despite being outside of the region in which O-glycosylation of CSF3R is thought to occur (Figure 4B).
Figure 4. Altered banding pattern and O-glycosylation of the CSF3R T640N mutation, increased dimerization and JAK kinase sensitivity.
(A) The CSF3R T640N has altered banding pattern relative to WT CSF3R, but is not overexpressed. The CSF3R T618I and a CSF3R truncation mutation (S783fs) are shown for comparison. (B) A direct glycosylation assay shows that the CSF3R T640N has reduced O-glycosylation when compared to WT CSF3R. 293T17 cells were transfected with WT CSF3R, T618I, T640N or a control multisite mutation that has 7 serines and threonines in the O-glycosylation cluster mutated to alanines and thus has no detectable O-glycosylation. They were labeled with GalNAz, which incorporates into O-glycosylation sites and then O-glycosylated proteins were pulled down using a biotinylated probe. The total protein is also shown. (C) Co-immunoprecipitation of a V5 and flag-tagged constructs reveals that both CSF3R T618I and T640N constructs have increased dimerization when compared to WT CSF3R. Each pair of V5 and Flag tagged constructs was transfected into 293T17 cells and then immunoprecipitated using anti-flag beads. The level of V5 and flag immunoprecipitation were quantified and then normalized to their respective inputs. The ratio of V5 (co-immunoprecipitated) to flag is expressed as a ratio relative to WT CSF3R. Statistical significance was assessed by a one way ANOVA followed by a Bonferroni’s Multiple Comparison Test. Eight replicates from three separate experiments are shown where *p<0.05 and **p<0.01. (D) The CSF3R T640N mutation is sensitive to JAK kinase inhibition. A mouse bone marrow colony assay shows that the CSF3R T618I and T640N constructs are both sensitive to the JAK kinase inhibitor ruxolitinib. Mouse bone marrow cells were infected with retrovirus expressing CSF3R T618I or CSF3R T640N constructs. Cells were plated in methylcellulose containing increasing doses of the JAK1/2 kinase inhibitor, ruxolitinib, at doses of 0, 10, 50, 100, 500, or 1000 nM.
To determine whether CSF3R T640N leads to increased receptor dimerization, we performed a co-immunoprecipitation study (Figure 4C). We made two tagged constructs for WT CSF3R and the T618I and T640N mutations. For each CSF3R variant, Flag and V5 tagged construct were co-expressed and then dimerization was quantified by immunoprecipitating the Flag-tagged construct and detecting the co-immunoprecipitated V5 partner. Both the T618I and T640N mutations had a statistically significant increase in dimerization relative to WT CSF3R. Although increased dimerization had been predicted in silico this is the first biochemical evidence of increased dimerization of the T640N mutated CSF3R.
The CSF3R T640N mutation confers sensitivity to JAK kinase inhibition
The activation of the JAK kinase pathway by the T640N mutation, as evidenced byupregulation of phospho-STAT3, led us to investigate whether this mutation confers sensitivity to the JAK inhibitor, ruxolitinib. We performed a mouse bone marrow colony assay, which revealed that colony formation driven by the CSF3R T640N mutation is similarly sensitive to colony formation driven by the T618I mutation (Figure 4D). To ensure that the effects of ruxolitinib were specific to CSF3R, we also tested the effects of ruxolitinib on an alternative oncogenic driver, BCR-ABL (Supplemental Figure 2). BCR-ABL causes chronic myeloid leukemia, a disease with many similar phenotypic features to atypical CML and CNL. BCR-ABL driven mouse bone marrow colony formation was not sensitive to inhibition with ruxolitinib, but was sensitive to the BCR-ABL kinase inhibitor, dasatinib. This indicates that our observed effects of ruxolitinib are not indicative of general inhibition of myeloid colony formation but are more specific to colonies driven by CSF3R mutations. These data indicate that JAK kinase inhibitors warrant further investigation for patients with the CSF3R T640N mutation.
DISCUSSION
The two main classes of CSF3R mutations, membrane proximal point mutations such as (T618I and T615A), and the truncation mutations in the cytoplasmic domain are clearly oncogenic. It is important to understand whether or not other mutational events in CSF3R are also oncogenic and have prognostic or therapeutic relevance. Certain CSF3R variants that have been reported at low frequencies in hematologic malignancies or found through sequencing, such as the P706C, P733T, E808K (2), and M696T (3,15), and R698C mutations, have not exhibited transformative capacity in the Ba/F3 cytokine independent model (Supplemental Figure 1A). It should be noted that these mutations could have functional consequences that are not apparent in this particular assay. In contrast, the T640N mutation is highly transformative in both cellular assays (Supplemental Figure 1) and in vivo (Figure 2). The T640N mutation resides within the transmembrane domain of CSF3R, and has been predicted to alter receptor dimerization (12). Mutations within the transmembrane region of other cytokine receptors have also been shown to confer ligand independent receptor dimerization, such as the W515L mutation of MPL, which is found in patients with myelofibrosis (16). Mutations have also been found in the IL7R alpha subunit in the juxtamembrane domain in patients with acute lymphoblastic leukemia (17,18). These mutations lead to the addition of an unpaired cysteine that results in the formation of a non-native disulfide bridge and covalent receptor dimerization (17,18).
Interestingly, the T640N mutation that is predicted to enhance receptor dimerization through altered hydrogen bonding (12), also resulted in diminished O-glycosylation of the receptor. We were able to confirm biochemically that like CSF3R T618I, the T640N mutation also increases receptor dimerization (Figure 4). This was surprising since the T640 residue resides within the transmembrane domain, which should not be accessible by GalNAc transferase enzymes. Our finding suggests that there is an intimate link between dimerization and O-glycosylation. Unlike N-linked glycosylation, which happens co-translationally, O-linked glycosylation is added post-translationally in the secretory pathway (19). O-glycosylation is known to be highly sterically sensitive (19), raising the possibility that rapid dimerization of the transmembrane helices in the T640N variant of CSF3R in the biosynthetic pathway may prevent the GalNAc transferases from subsequently accessing the CSF3R glycosylation sites. Further structural and microscopy studies are needed to address this possibility.
CSF3R mutations are a hallmark of chronic neutrophilic leukemia, occurring in greater than 80% of patients (2,3). They also occur with much lower frequency in atypical CML, which has similar clinical presentation to CNL. These mutations are quite rare in other hematologic malignancies, with a frequency of approximately 0.5–1% in acute myeloid leukemia (2,7). The relative restriction of these mutations to CNL suggests that mutation of CSF3R may be a primary driver of the neutrophilic disease phenotype of CNL. A mouse model of the CSF3R T618I mutation revealed high white blood cell counts (primarily of the granulocytic lineage) and splenomegaly, which are both features of CNL/aCML (11). The case reported here of a patient with MDS whose disease evolved to aCML highlights the role of CSF3R mutations in driving this neutrophilic disease phenotype, since the T640N mutation was present at high frequency in the aCML sample, but was at a very low frequency in the prior sample taken during the MDS phase of disease, suggestive of a minor clone. It is interesting that the germline T640N mutation results in a relatively benign hereditary neutrophilia (12), but that this mutation also occurs in CNL/aCML which are much more aggressive hematological disorders, suggesting that other mutations may contribute to disease phenotype. In one of the patients in this study, the T640N mutation was concurrent with the SETBP1 G870S mutation, and a JAK2V617F mutation was detected in a later sample, but these abnormalities were not present in the other patient.
In conclusion, we show that the CSF3R T640N mutation has many features similar to the CSF3R T618I mutation, including ligand independence, and altered patterns of O-glycosylation. Although rare, given the oncogenic capacity and JAK kinase inhibitor sensitivity of the T640N mutation, it will be valuable to include this mutation for molecular testing for patients with CNL/aCML, especially since these patients may benefit from JAK inhibitor therapy.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Mutations in CSF3R are emerging as potential therapeutic targets in chronic neutrophilic leukemia (CNL) and atypical chronic myeloid leukemia (aCML). CSF3R mutations are highly prevalent in CNL (at approximately 80% frequency), and present in a minority of patients with aCML. In this study we found a mutational event in patients with CNL and aCML that had not been previously described in this patient population, CSF3R T640N (aka T617N). The T640N mutation exhibits many molecular features of the most common CSF3R mutation (T618I, aka T595I), including sensitivity to JAK kinase inhibitors. This mutation resides in exon 15, which is often not included in CSF3R sequencing assays. Although rare, the CSF3R T640N mutation may warrant inclusion in CSF3R mutation analysis due to its potential diagnostic and therapeutic relevance.
Acknowledgments
Financial Support: J.M. is supported by a Fellow Award from The Leukemia & Lymphoma Society and a Medical Research Foundation Early Clinical Investigator Award, and a K99/R00 award from NCI, CA190605-01. J.W.T. is supported by grants from the V Foundation for Cancer Research, The Leukemia & Lymphoma Society, the Gabrielle’s Angel Foundation for Cancer Research, and the National Cancer Institute (5R00CA151457-04; 1R01CA183974-01). JG receives support from the Charles and Ann Johnson Foundation.
Footnotes
Conflicts of Interest: JEM, SBL, JDM, JCP, PG, SB, SLS, MLA, CAE, MML, and EAS have no conflicts to disclose. JWT and JG receive research funding from Incyte; and JG sits on advisory boards and receives honoraria from Incyte.
References
- 1.Gotlib J, Maxson JE, George TI, Tyner JW. The new genetics of chronic neutrophilic leukemia and atypical CML: implications for diagnosis and treatment. Blood. 2013;122:1707–11. doi: 10.1182/blood-2013-05-500959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maxson JE, Gotlib J, Pollyea DA, Fleischman AG, Agarwal A, Eide CA, et al. Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. The New England journal of medicine. 2013;368:1781–90. doi: 10.1056/NEJMoa1214514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pardanani A, Lasho TL, Laborde RR, Elliott M, Hanson CA, Knudson RA, et al. CSF3R T618I is a highly prevalent and specific mutation in chronic neutrophilic leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2013;27:1870–3. doi: 10.1038/leu.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward AC, van Aesch YM, Schelen AM, Touw IP. Defective internalization and sustained activation of truncated granulocyte colony-stimulating factor receptor found in severe congenital neutropenia/acute myeloid leukemia. Blood. 1999;93:447–58. [PubMed] [Google Scholar]
- 5.van de Geijn GJ, Gits J, Aarts LH, Heijmans-Antonissen C, Touw IP. G-CSF receptor truncations found in SCN/AML relieve SOCS3-controlled inhibition of STAT5 but leave suppression of STAT3 intact. Blood. 2004;104:667–74. doi: 10.1182/blood-2003-08-2913. [DOI] [PubMed] [Google Scholar]
- 6.Dong F, Qiu Y, Yi T, Touw IP, Larner AC. The carboxyl terminus of the granulocyte colony-stimulating factor receptor, truncated in patients with severe congenital neutropenia/acute myeloid leukemia, is required for SH2-containing phosphatase-1 suppression of Stat activation. J Immunol. 2001;167:6447–52. doi: 10.4049/jimmunol.167.11.6447. [DOI] [PubMed] [Google Scholar]
- 7.Beekman R, Valkhof M, van Strien P, Valk PJ, Touw IP. Prevalence of a new auto-activating colony stimulating factor 3 receptor mutation (CSF3R-T595I) in acute myeloid leukemia and severe congenital neutropenia. Haematologica. 2013;98:e62–3. doi: 10.3324/haematol.2013.085050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beekman R, Valkhof MG, Sanders MA, van Strien PM, Haanstra JR, Broeders L, et al. Sequential gain of mutations in severe congenital neutropenia progressing to acute myeloid leukemia. Blood. 2012;119:5071–7. doi: 10.1182/blood-2012-01-406116. [DOI] [PubMed] [Google Scholar]
- 9.Mehta HM, Glaubach T, Long A, Lu H, Przychodzen B, Makishima H, et al. Granulocyte colony-stimulating factor receptor T595I (T618I) mutation confers ligand independence and enhanced signaling. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2013 doi: 10.1038/leu.2013.164. [DOI] [PubMed] [Google Scholar]
- 10.Maxson JE, Luty SB, MacManiman JD, Abel ML, Druker BJ, Tyner JW. Ligand independence of the T618I mutation in the colony-stimulating factor 3 receptor (CSF3R) protein results from loss of O-linked glycosylation and increased receptor dimerization. The Journal of biological chemistry. 2014;289:5820–7. doi: 10.1074/jbc.M113.508440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischman AG, Maxson JE, Luty SB, Agarwal A, Royer LR, Abel ML, et al. The CSF3R T618I mutation causes a lethal neutrophilic neoplasia in mice that is responsive to therapeutic JAK inhibition. Blood. 2013;122:3628–31. doi: 10.1182/blood-2013-06-509976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plo I, Zhang Y, Le Couedic JP, Nakatake M, Boulet JM, Itaya M, et al. An activating mutation in the CSF3R gene induces a hereditary chronic neutrophilia. The Journal of experimental medicine. 2009;206:1701–7. doi: 10.1084/jem.20090693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes LV, Gale RE, Pizzey A, Pouwels K, Nathwani A, Linch DC. An activating mutation in the transmembrane domain of the granulocyte colony-stimulating factor receptor in patients with acute myeloid leukemia. Oncogene. 2002;21:5981–9. doi: 10.1038/sj.onc.1205767. [DOI] [PubMed] [Google Scholar]
- 14.Hang HC, Yu C, Kato DL, Bertozzi CR. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14846–51. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosmider O, Itzykson R, Chesnais V, Lasho T, Laborde R, Knudson R, et al. Mutation of the colony-stimulating factor-3 receptor gene is a rare event with poor prognosis in chronic myelomonocytic leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2013;27:1946–9. doi: 10.1038/leu.2013.182. [DOI] [PubMed] [Google Scholar]
- 16.Pikman Y, Lee BH, Mercher T, McDowell E, Ebert BL, Gozo M, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS medicine. 2006;3:e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shochat C, Tal N, Bandapalli OR, Palmi C, Ganmore I, te Kronnie G, et al. Gain-of-function mutations in interleukin-7 receptor-alpha (IL7R) in childhood acute lymphoblastic leukemias. The Journal of experimental medicine. 2011;208:901–8. doi: 10.1084/jem.20110580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zenatti PP, Ribeiro D, Li W, Zuurbier L, Silva MC, Paganin M, et al. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nature genetics. 2011;43:932–9. doi: 10.1038/ng.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van den Steen P, Rudd PM, Dwek RA, Opdenakker G. Concepts and principles of O-linked glycosylation. Critical reviews in biochemistry and molecular biology. 1998;33:151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.