Abstract
Purpose
To gain insight into factors involved in tumor progression and metastasis, we examined the role of non-coding RNAs (ncRNAs) in the biological characteristics of colorectal carcinoma (CRC), in paired samples of tumor together with normal mucosa from the same CRC patient. The tumor and healthy tissues samples were collected and stored under stringent conditions, thereby minimizing warm ischemic time.
Experimental Design
We focused particularly on distinctions among high stage tumors and tumors with known metastases, performing RNA-Seq analysis which quantifies transcript abundance and identifies novel transcripts.
Results
In comparing 35 CRCs, including 9 metastatic tumors (metastases to lymph nodes and lymphatic vessels (LN/LV)), to their matched healthy control mucosa, we found a distinct signature of MT-tRNAs and snoRNAs for metastatic and high stage CRC. We also found the following: 1) MT-TF (phenylalanine) and snord12B expression correlated with a substantial number of miRNAs and mRNAs in 14 CRCs examined; 2) a miRNA signature of oxidative stress, hypoxia and a shift to glycolytic metabolism in 14 CRCs, regardless of grade and stage, and 3) heterogeneous MT-tRNA/snoRNA fingerprints for 35 pairs.
Conclusions
These findings could potentially assist in more accurate and predictive staging of CRC including identification of those CRC likely to metastasize.
Keywords: MT-tRNAs, snoRNAs, microRNAs, mRNAs and tumor biomarkers
Introduction
CRC is the third most common cancer and also the third leading cause of cancer-related deaths in the United States (1). The prognosis in advanced cases is poor with more than one-third of the patients dying from progressive disease within 5 years (2). However, simultaneous evaluation of sRNA, miRNA and mRNA in matched pairs of CRC by RNA-Seq, a technology for high throughput sequencing of the whole transcriptome with some advantages over microarrays, has not been reported.
miRNAs are ncRNA molecules with 18-25 nucleotides (nts) in length and are known to repress mRNAs by inhibiting translation and stimulating mRNA degradation (3). Approximately 270 of such miRNAs have been shown to be dysregulated in CRC (3-5) by real-time PCR and microarray. sRNAs are approximately 60-300 nts in length which include transfer RNAs (tRNAs) and snoRNAs. MT-tRNAs, approximately 70-80 nts long, are structurally distinct from nuclear-encoded tRNAs by having one less loop than the nuclear canonical cloverleaf structure due to genome economization (6). Twenty-two MT-tRNAs and 2 ribosomal RNAs (rRNAs) are required for the synthesis of 13 out of the 84 essential protein subunits of NADH dehydrogenase, cytochrome reductase, cytochrome oxidase and ATP synthase which are essential for ATP production through oxidative phosphorylation in the mitochondria (7). Upregulated expression of such MT-tRNAs has been found in breast cancer (8). snoRNAs play a crucial role in ribosome biogenesis through methylation and pseudouridylation of rRNAs (9). A linkage between snoRNAs and carcinogenesis has been established for non-small-cell lung cancer (NSCLC) (10). mRNAs are RNA templates for protein synthesis and thus far, more than a dozen mRNAs have been identified as biomarkers for staging and prognosis of CRC (11).
Our study sought to investigate, using stringently collected and preserved tissue samples (12) and RNA-Seq analysis, whether expression of unique sRNA species correlated with and potentially contributed to the aggressive biological behavior of advanced stage colorectal cancers and cancers with known metastasis, as compared to lower grade tumors and healthy tissues.
Materials and Methods
Cohort
Thirty-five paired-tissues of pretreatment CRC were collected by Indivumed GmbH (Germany). Histologically, tumor content is 50-70% in tumors and 0% in normal tissues. Normal tissues were 5 cm away from tumors. Ischemia time to freeze was 6-11 minutes. The normal mucosa was collected in a distal part of the bowel close to the resection margins. Clinical and histopathological characteristics of the patients are summarized in table S1.
Small RNA and miRNA sequencing
All 35 pairs were sequenced at small RNA level and first 15 pairs were also sequenced at miRNA level. The RNA quality was assessed using the Agilent 2100 Bioanalyzer. Samples with a RNA Integrity Number (RIN) of 7 or higher were processed to generate libraries for small RNA sequencing following the Illumina®TruSeq™ Small RNA Sample Preparation protocols. In brief, 3’ and 5’ RNA adapters, specifically modified to target the 3’ hydroxyl group and 5’ phosphate group of most small RNA molecules, were ligated to the ends of sRNAs present in 1 μg of high quality total RNA. Reverse transcription was performed to generate the first-strand cDNAs followed by synthesis of the second-strand cDNAs. The small RNA libraries were loaded onto a 4% agarose gel after 11-cycle PCR amplification. DNA fragments of 145 to 160 bp containing miRNA inserts were excised from the gel and purified for micro RNA sequencing (miRNA-Seq), while those of 160 - 400 base pair (bp) containing longer inserts were similarly purified for small RNA (including MT-tRNAs and snoRNAs) sequencing (sRNA-Seq). The yield of small RNA library was quantified on Agilent 2100 Bioanalyzer. Twelve to twenty-four small RNA libraries were pooled, denatured and loaded onto one lane of a flow cell for cluster generation using the Illumina cBot. The flow cell was loaded onto Illumina HiSeq 2500 sequencer (Illumina, San Diego, CA, USA) and subjected to single-end, 50- and 100-cycle sequencing for miRNA and sRNA sequencing, respectively.
mRNA Sequencing
Fourteen pairs were further processed to generate mRNA-Seq libraries using Illumina TruSeq “stranded mRNA sample prep kit.” In this method, poly (A) tailed RNA was purified from 0.5 μg total RNA, fragmented and reverse-transcribed into cDNAs. Double strand cDNAs were adenylated at the 3’ ends and ligated to indexed sequencing adaptors, followed by limited-cycle (15) amplification. Paired-end sequencing was carried out on HiSeq 2500 sequencer (Illumina, San Diego, CA, USA) for 100×2 cycles.
Sequencing Data Analysis
The miRNA-Seq data analysis was performed on NIH Biowulf supercomputer. miRDeep2 was used to trim the adapter sequences, map the reads to the miRNA database, miRBase (version19), and quantify miRNA expression levels. Novel miRNAs were predicted using miRDeeps2 for aligning the reads against the reference human genome (hg19). For mRNA sequencing, Tophat V.2.0.11 and Cufflinks V.2.2.1 were used to align the reads in fastq files to the RefSeq UCSC human hg19 transcript reference genome annotation database and the quantification of relative abundance of each transcript was reported as Reads Per Kilo base per Million (RPKM). sRNA sequencing data was analyzed using the software CLC Genomics Workbench 5 for the mapping of trimmed reads to Ensemble GRCh37.75 noncoding RNA database and quantitation of the sRNA expression levels. Statistical analyses and gene clustering were carried out using R 2.15.3 (The R Project for Statistical Computing Program). The sRNA expression levels were normalized to the expression levels of all identified, known sRNAs. Paired Student t-test was conducted to identify sRNAs with differential expression between cancers and matched normal adjacent tissues (P < 0.05). Hierarchical clustering was conducted using Euclidean distance to cluster RNAs and samples. By standard detection criteria (13), 5,171 sRNAs, 1009 miRNAs and 13580 mRNAs with 1 RPKM in at least one sample were selected for analysis. Though paired t-test, 37 sRNAs, 204 miRNAs and 665 mRNAs were identified to have significant differential expression (p-value <0.05). The expression patterns of sRNAs, miRNAs and mRNAs are presented on clustering analysis, which were performed by the R package “gplots.” Principal component analysis (PCA) was conducted to visualize differences between samples and cluster samples on the gene expression profiles, with the majority of the data variation explained by PC1, PC2 and PC3. Pearson correlation analysis was done with EXCEL 2003 and correlation cutoff was 0.6 (r>0.6). Plots were made with Prism 6 software and EXCEL 2003.
Verification of RNA-Seq fidelity and identification of colonic tissue house keeping genes (Information S1)
Results
1) Identification of MT-tRNAs and snoRNAs as biomarkers for tumor with LN/LV metastasis
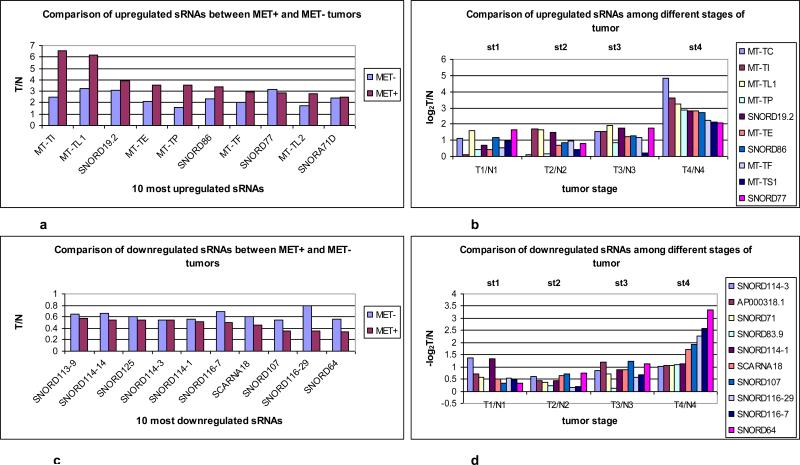
To evaluate the sRNA profile of tumors that had documented lymph node (LN) and lymphatic vessel (LV) metastases, the average ratios (Tumor/Normal: T/N) of sRNAs in tumors with metastases (MET+) (9 tumors: T8LN, T11LV, T14LN, T15LN, T20LV, T25LV, T26LN, T30LV and T35LN) (Table. S1) and tumors without documented metastases (MET−) (26 tumors) were compared. We found that 6 MtRNAs (MT-TI (isoleucine), MT-TL (leucine)1, MT-TE (glutamic acid), MT-TP (proline), MT-TF, MT-TL2 and 4 sRNAs (snord19.2, snord86, snord77 and snora71D) were among the top 10 upregulated genes with 2.5-6.5 fold increases in tumors with LN/LV metastasis compared to 1.5-3 fold increases in tumors without LN/LV metastases (Fig. 1a). Next, we evaluated whether such genes were also upregulated in advanced stage vs early stage tumors by comparing the average ratios (T/N) of sRNAs among tumors at stage 1 (5 tumors), stage 2 (6 tumors) and stage 3 (21 tumors) compared to stage 4 (3 tumors). We found that there was a 6-28 fold increased expression of 7 MT-tRNAs (MT-TC (cysteine), MT-TI, MT-TL1, MT-TP, MT-TE, MT-TF and MT-TS (serine)1) and 3 snoRNAs (snord19.2, snord86 and snord77) in stage 4 tumors compared to1-3 fold increases in all lower stage tumors (Fig. 1b). Overall, MT-tRNAs were found to be highly expressed in high grade tumors and in tumors with LN/LV metastases. Moreover, the upregulated expression of snord19.2, snord86, snord77 and snora71D in CRC has not been reported and their potential roles in malignant transformation/progression not known.
Fig. 1.
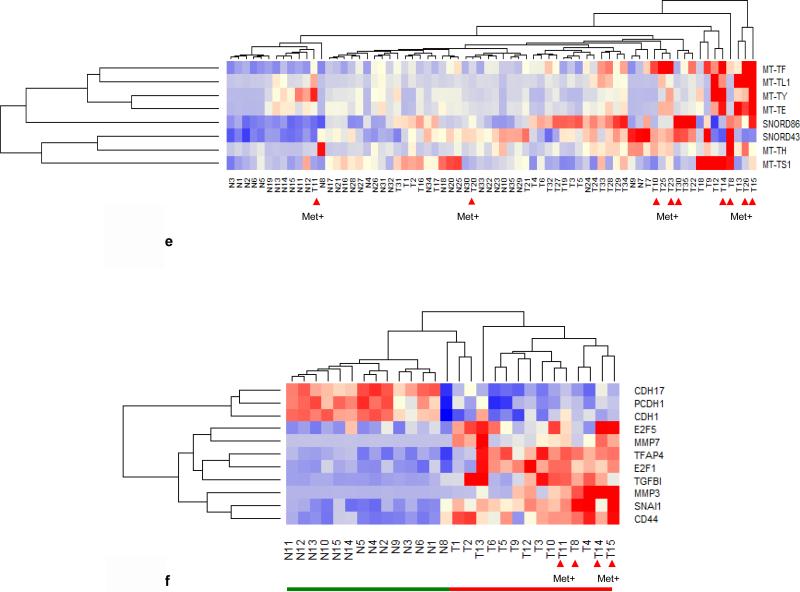
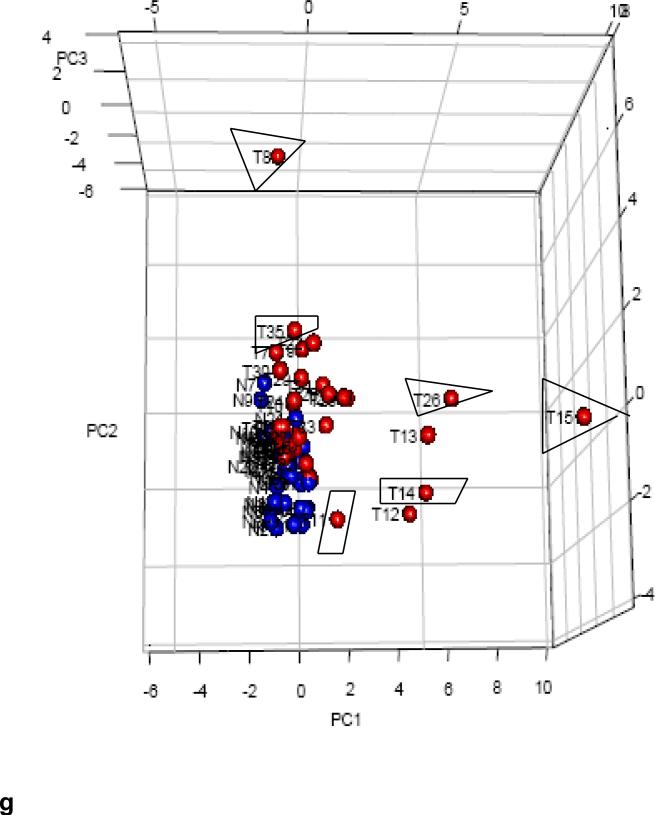
Identification of 10 most up/down regulated sRNAs between tumors with metastasis (Met+) and without metastasis (Met−). All 35 tumors were grouped into Met+ and Met−, T/N ratios were obtained, and log2 ratios of LN positive /negative ratio were sorted from high to low and plotted. (a) Six MT-tRNAs were among top 10 expressed sRNAs. The difference in the average ratio of the top 10 sRNAs expressed between Met+ and Met− tumors was highly significant, (P=0.00685 paired t-test). (b) Identification of most up regulated sRNAs by tumor stages. Tumors were grouped according to stage (T1, T2, T3 and T4) and the log2 ratios (T1/N1, T2/N2, T3/N3 and T4/N4) were obtained and plotted. Expression in stage 4 tumors differed significantly from all other stages: stage 1 (P<0.0001). There were no differences among stages 1, 2 and 3 (P>0.06) (ANOVA). (c) 10 snoRNAs were among the 10 most down regulated sRNAs in LN/LV positive tumors but not statistically significant (P=0.364) by student t- test. (d) Nine snoRNAs were among the 10 most downregulated sRNAs in stage 4 tumors. Stage 4 differed significantly from stage1/2/3 (P<0.0458) and stage1/2/3 had no significant difference among them (P>0.22) by ANOVA. (c) and (d) were plotted as –log2 ratio. (e)Hierarchical unsupervised clustering analysis of 6 MT-tRNAs/2 snoRNA expression RPKMs in 35 CRC pairs. Met+ tumors tended to cluster at higher expression side of the map. (f) Hierarchical unsupervised clustering analysis of mRNA expression of 11 markers associated with metastatic tumors by RPKMs in 14 CRC pairs. This 11-coding gene signature grouped T14/T15 together and separated all tumors from normal samples. Red arrows indicate Met+ tumor samples. Red and green bars delineate tumors from normal controls. Red in the heatmap indicates an expression level above the mean in all samples and blue indicates expression levels lower than the mean of all samples. (g) 3D PCA analysis of 6 MT-tRNAs/2 snoRNA (same sRNAs as in Fig. 1e) in 35 CRC pairs. Four Met+ tumors (T8LN, T14LN, T15LN, T26LN) (boxed) and 2 Met− tumors (T12, T13) were clearly separately from remaining tumors and normal controls.
Thus to determine whether deregulated MT-tRNAs and snoRNAs can be used as a signature to distinguish some metastatic and high stage tumors, unsupervised clustering and PCA analysis of the selected 6 top upregulated MT-tRNAs (MT-TF, MT-TL1, MTTY (tyrosine), MT-TE, MT-TH (histidine), MT-TS1) and 2 top upregulated snoRNAs (snord43, snord86) were performed. The hierarchical clustering distinguished MET+ T8LN, T14LN, T15LN, T25LV, T26LN, T30LV and T35LN (Fig. 1e) while the PCA plot distinguished a subset of such MET+ tumors including T8LN, T14LN, T15LN and T26 LN (Fig. 1g). Among 9 MET+ tumors, 3 (T20LV T30LV and T35LN) had only upregulation of snord43 and snord86 but not MT-tRNAs. Furthermore, principal component analysis (PCA) of 22 MT-tRNAs also distinguished metastatic T11 LV, T14LN, T15LN and T26LN (Fig. S2a). Five MET− tumors (T9, T12, T13, T18, T22 and T23) that clustered with MET+ tumors (Fig. 1e, 1g, Fig. S2a) were moderately differentiated stage 2 or 3 tumors with high expression of MT-tRNAs (Fig. S3a). Thus, patients with tumors bearing such MT-tRNA profiles may be at higher risk for progression and metastasis.
We next evaluated sRNAs that were most prominently downregulated in MET+ as well as stage 4 tumors and found that the snord113-114-116 cluster, snord64, snord71, snord93, snord107, scarna18, CTD2651B20.6 (CTD: comparative toxicogenomics database) and AP000318.1 (AP: annotation process) were the 10 most downregulated sRNAs with 1.2-10 fold decreases (Fig. 1c, 1d) Interestingly, the snord113-114-116 cluster, snord116-7 and snord64 are deleted in PWS, a severe metabolic disorder causing excess adiposity which suggests a role for such dysregulation in energy storage in tumors (14). However, these snoRNAs have not previously been specifically linked to CRC malignant transformation or progression.
Since MET+ tumors clustered in multiple analyses of non-coding gene expression, we performed an unsupervised clustering analysis of 14 tumor pairs using 11 known metastatic mRNA biomarkers (15) including the following: liver-intestine cadherin 17 (CDH17), protocadherin 1 (PCDH1), transcription factor 5 (E2F5), matrix metalloproteases 3 and 7 (MMP3 and MMP7), transcription factor AP-4 (TFAP4), transcription factor 1 (E2F1), transforming growth factor, beta 1 (TGFB1), zinc finger transcriptional repressor 1 (SNAI1) and extracellular matrix receptor III (CD44). The resulting cluster analysis distinguished all MET+ tumors (T8LN, T14LN, T15LN and T11LV) in 14 tumors (Fig. 1f).
Taken together, the coding RNA data support ncRNAs signatures that distinguish MET+ CRC. Thus, such MT-tRNA/snoRNA signatures have the potential to more precisely identify these more aggressive and deadly tumors and suggest novel therapeutic targets.
2) Identification of sRNA as potential novel biomarkers for CRC
To evaluate the spectrum of sRNA biomarkers associated with tumors regardless of grade, stage and LN/LV metastases, as distinguished from normal tissue, the average ratios (T/N) of sRNAs of all 35 tumors and all 35 normal samples were compared. We again found a predominance of 9 MT-tRNAs (MT-TI, MT-TL1, MT-TL2, MT-Y, MTTC, MT-TE, MT-TF, MT-TH and MT-TD) among the top 20 upregulated genes with 1.4-5 fold increases (Fig. S2b, Table. S3a). Nineteen snoRNAs and AP000318.1 were among the 20 most downregulated genes in tumors with approximately 1.5-2.5 fold decreases relative to normal tissue (Fig. S2c). Overall, 41 sRNAs were found up or downregulated (Table. S2a, S2b, S2c, Fig. S3a, S3b, S3c). PCA analysis of these 32 differentially expressed snoRNAs distinguished 28 out of 35 tumors from normal controls (Fig. S2d). Among 41 differentially expressed sRNAs (9 MT-tRNAs, 32 snoRNAs), we found that the upregulation of snord12B expression in tumors bore the greatest statistically significant difference from normal samples (p value=3.830e-12) (Fig. S4a). Consistent with upregulation of snord12B, the host gene of this snoRNA, zinc finger nuclear transcription factor, x-box binding 1 anti-sense 1(ZNFX1-AS1) (16), a long non-coding RNA, was upregulated in all examined CRCs (Fig. S4b). There was an association strength (r=0.66) between the expression of snord12B and ZNFZ1-AS1 (Fig. S4c). Paradoxically, the expression of ZNFZ1-AS1 is down-regulated in mouse mammary tumors, and thus it may have distinct functions in different tissues or species. Although, snord12B was reported as upregulated in rectal cancer (17), the profound differences in snoRNA expression between CRC and normal mucosa have not hitherto been reported and suggest potentially important roles for these ncRNAs in malignant transformation, progression, or metastasis in CRC.
3) Correlation of sRNAs with miRNAs and mRNAs in primary CRCs
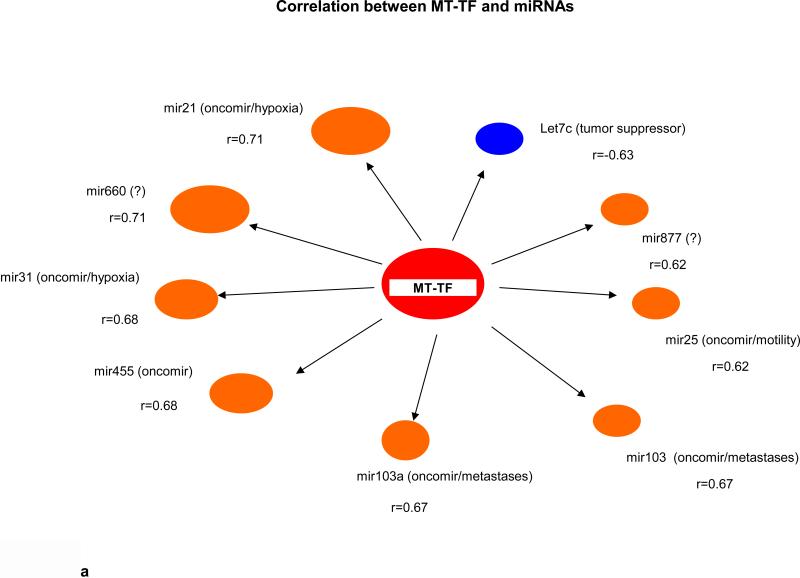
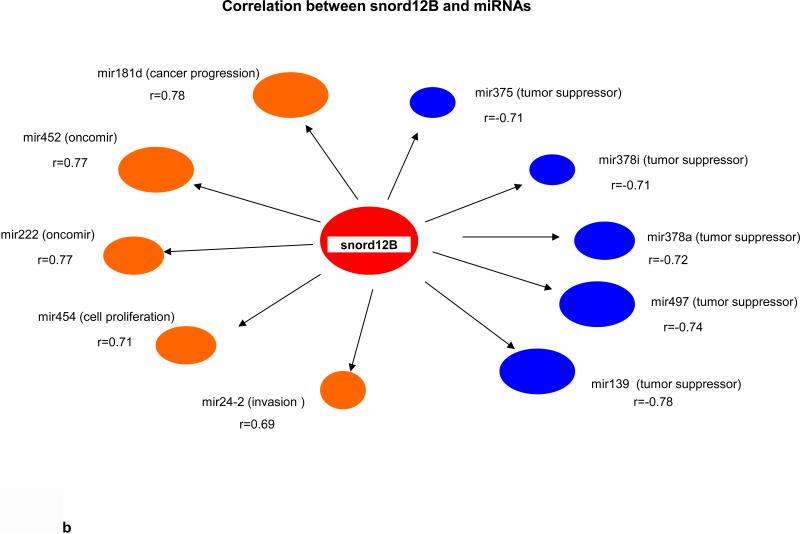
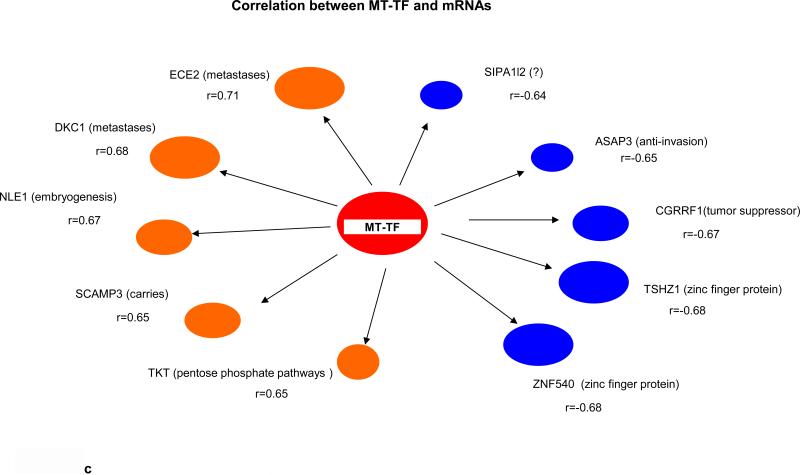
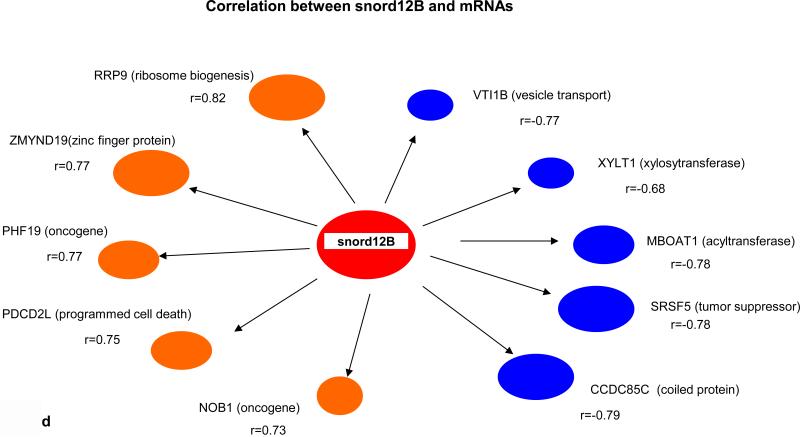
Key biological premises that underlie relationships of genes regarding “drivers’ and ‘passengers” consist of the following: highly co-expressed genes are more likely to be co-regulated; and those genes that display prominent connectivity patterns tend to play biologically influential or regulatory roles in disease-related processes (18-20). Measures of node centrality in biological networks may detect genes with critical functional roles. In gene co-expression networks, highly connected genes (i.e., candidate hubs) have been associated with key drivers of disease pathways and gene connectivity has been shown to be a measure of functional relevance (18-20). Thus to explore whether there were potential cancer “driver” sRNAs, we performed correlational analyses among 41 differentially expressed sRNAs (9 MT-tRNAs, 32 snoRNAs), 204 miRNAs, and 665 mRNAs in 14 tumor pairs. Interestingly, we found that expression of some sRNAs, such as MT-TF, snord12B and the snord114- cluster, correlated with expression of more than 100 to 450 miRNAs/mRNAs while expression of some sRNAs, such as MT-TY, snord19B and snord83.9 correlated with expression of less then 10 miRNAs/mRNAs (Table 1 and Information S2). Moreover, the upregulated expression of MT-TF and snord12B sRNAs positively correlated with upregulated hypoxia, metastatic and pentose phosphate pathway genes, such as mir21, mir181, ECE2, PHF19 and TKT, and negatively correlated with expression of tumor suppressor genes, such as let7c, mir139, CGRRF1 and SRSF5 (Fig. 2a, 2b, 2c, 2d). The downregulation of snord114-1 correlated with downregulation of tumor suppressors, such as let7c and AHNAK (Information. S2d). In addition, MT-TF, snord12B and snord114-1 were commonly deregulated in all 35 tumors (Table. S2a, S2b, S2c, Fig. S3a, S3b, S3c). Thus, our data suggest that deregulation of these 3 sRNAs may be causally linked to tumorigenesis, tumor proliferation or aggressiveness. The regulatory mechanisms controlling expression of these 3 sRNAs are not known.
Table. 1. The number of co-expressed miRNA/mRNA with sRNAs.
Pearson's correlation test was performed on RPKM value of each gene group cross 14 pairs. All genes showing correlation coefficient (r)> 0.6 with MT-tRNAs and snoRNAs (Information S2) were summarized in this table.
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| MT-tRNA | Correlated snoRNA # |
Correlated miRNA # |
Correlated mRNA # |
snoRNAs | Correlated miRNA # |
Correlated mRNA # |
snoRNAs | Correlated miRNA # |
Correlated mRNA # |
| MT-TF | 1 | 10 | 151 | SNORD12B | 27 | 451 | SNORD114-1 | 31 | 80 |
| MT-TC | 0 | 3 | 19 | SNORD12C | 16 | 184 | SNORD114-23 | 11 | 29 |
| MT-TL1 | 0 | 2 | 10 | SNORD12 | 18 | 90 | SNORD114-26 | 13 | 25 |
| MT-TS1 | 0 | 1 | 2 | SNORD78 | 13 | 60 | SNORD107 | 13 | 23 |
| MT-TI | 0 | 2 | 2 | SNORD77 | 12 | 53 | VTRNA1 | 5 | 10 |
| MT-TE | 1 | 5 | 1 | SNORD125 | 8 | 35 | SNORD114-14 | 3 | 8 |
| MT-TY | 0 | 5 | 0 | SNORA84 | 12 | 12 | SNORD114-25 | 4 | 7 |
| MT-TH | 0 | 2 | 0 | SNORD71 | 11 | 10 | SNORD64 | 12 | 3 |
| MT-TL2 | 0 | 7 | 0 | SNORD1A | 10 | 1 | SNORD116-29 | 0 | 3 |
| SNORD19B | 6 | 0 | SNORD113 | 13 | 1 | ||||
| SCARNA18 | 9 | 0 | SNORA71D | 5 | 1 | ||||
| SNORD54 | 5 | 0 | SNORD93 | 13 | 0 | ||||
| SNORD59A | 2 | 0 | SNORD83.9 | 8 | 0 | ||||
| CTD-2615B | 0 | 0 | AP000318.1 | 0 | 0 | ||||
| snord1A | 0 | 0 | SNORD116-7 | 0 | 0 | ||||
| snord86 | 0 | 0 | snord43 | 0 | 0 | ||||
Fig. 2.
(a) Nine upregulated and 1 downregulated miRNAs correlated with MT-TF. (b) Five top positive and negative correlated miRNAs with snord12B. Five top positive and negative correlated mRNAs for MT-TF (c) and snord12B (d). This figure was summarized from Information S2. Positive correlated genes are represented by brown oval dots and negative correlated genes are represented by blue oval dots. The correlation coefficient value for each gene pairs was showed and bigger dots had stronger correlation.
4) Evidence for Hypoxia and upregulation glucose metabolism in CRC
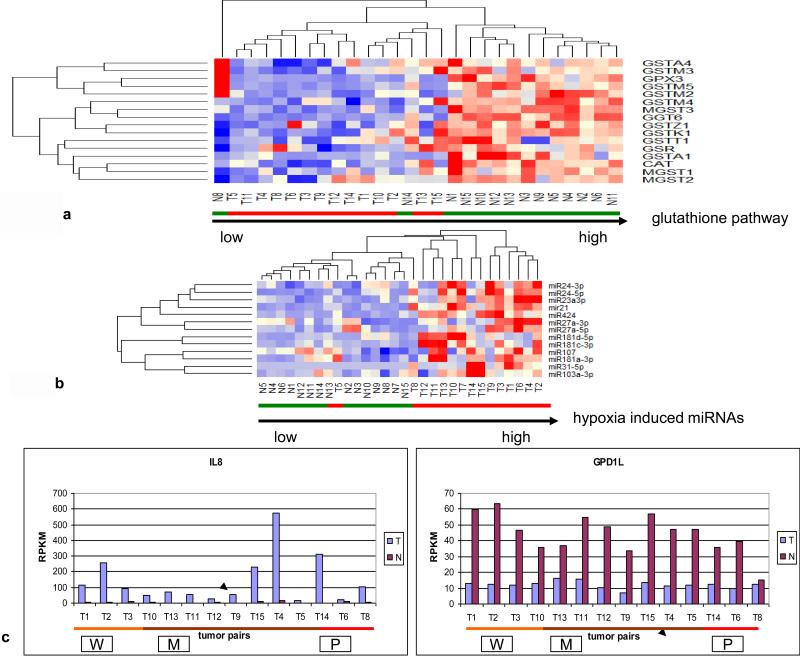
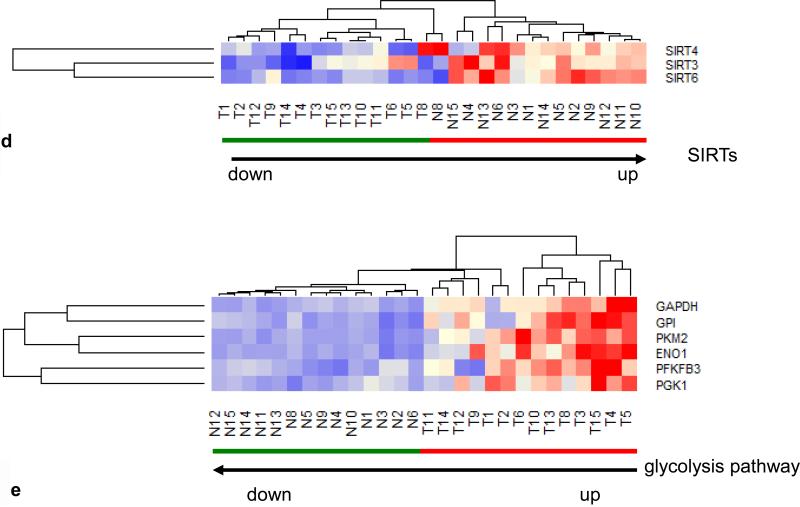
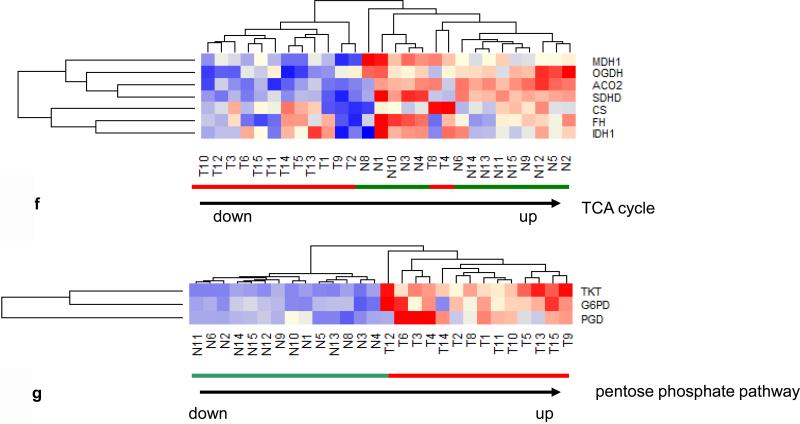
Since the prominent upregulation of MT-TF correlated with hypoxia biomarkers (mir21, mir31) (Fig. 2a), we examined whether enzymes to mitigate the oxidative effects of hypoxia were also affected. The glutathione anti-oxidative pathway consisting of 15 enzymes pertaining to glutathione S-transferase (GSTA), glutathione peroxidase (GPX), gamma-glutamyltransferase (GGT), glutathione reductase (GSR), microsomal/glutathione S-transferase (MGST) and catalase (CAT) in 14 CRCs were examined and downregulation of 9 out of 15 anti-oxidative enzymes (Fig. 3a) was found in all tumors as well as in 2 normal controls (N8, N14), which were associated with MET+ tumors. Furthermore, we also examined hypoxia induced miRNAs (21, 22, 23) and found 13 upregulated miRNAs pertaining to hypoxia in 14 out of 15 tumors (Fig. 3b), the sole exception being a poorly differentiated stage 3 tumor. Because IL-8 is known to be upregulated and glycerol-3-phosphate dehydrogenase 1-like (GPD1L) is known to be downregulated by hypoxia (24), we examined the mRNA expression profile of these 2 genes in the 14 tumor pairs to establish the validity of the hypoxia clustering analysis. Relative to healthy control tissue, IL-8 was upregulated 2-35 fold and GPD1L was downregulated 1.2-5.2 fold in all tumors examined (Fig 3c). Thus, these data are indicative of hypoxia in CRCs. Since hypoxia switches ATP production from oxidative phosphorylation to glycolysis in tumor cells (25, 26, 27), we analyzed the expression of 3 SIRTs, which are known to suppress glycolysis (28), and found all tumors had down regulation of all 3 SIRTs (Fig. 3d). Further, a clustering analysis of 6 genes directly pertaining to glycolytic enzymes including glyceraldehyde-3-phosphate dehydrogenase (GAPDH), glucose phosphate isomerase (GPI), pyruvate kinase2 (PKM2), enolase1 (ENO1), phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3) and phosphoglycerate kinase 1 (PGK1), clustered all 14 tumors together with upregulation of all 6 genes, except in one stage 1 and two stage 3 tumors (Fig. 3e), while a clustering analysis of 7 genes directly pertaining to TCA cycle enzymes, including malate dehydrogenase 1 (MDH1), oxoglutarate (alpha-ketoglutarate) dehydrogenase (OGDH), aconitase 2 (ACO2), succinate dehydrogenase (SDHD), citrate synthase (CS), fumarate hydratase (FH) and isocitrate dehydrogenase 1 (IDH1), grouped 12 out 14 tumors together (Fig. 3f). Among these 12 tumors, 6 high stage (3 or 4) tumors did not have downregulation of all 7 TCA cycle enzymes. One poorly differentiated stage 3 tumor (T4) and one poorly differentiated stage 4 tumor (T8) with LN metastases grouped with normal samples in this analysis. These two tumors showed the most upregulated mRNA expression of CS, a rate limiting enzyme in the TCA cycle, suggesting that while the glycolytic pathway is generally upregulated and the TCA pathway is generally downregulated in CRC, the most aggressive tumors may use both glycolysis and the TCA cycle for energy production. Alternatively, one pathway or the other may predominate in different cells within the heterogeneous tumor population or in different environmental niches. Furthermore, all 14 examined tumors also had evidence of upregulation of the pentose phosphate pathway as shown by upregulation of enzymes including transketolase (TKT), glucose 6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (PGD), (Fig. 3g). The pentose phosphate pathway utilizes glucose to generate ribonucleotides as well as NADPH and plays a pivotal role in anabolic processes and in combating oxidative stress in glycolytic cancer cells. Overall, these data suggest that there is increased utilization of glucose in CRCs via anaerobic glycolysis. Intriguingly, one normal sample (N8), paired to MET+ T8, was adjacent to tumors in various clustering analyses (Fig. 1f, 3a, 3d, 3f). Compared to other normal samples, this N8 had higher expression of oncogenes such as SNAI1 and CD44 as well as lower expression of anti-oxidative enzymes and tumor suppressors such as GST, GSR, CAT, SIRT3 and SIRT6 (Fig. 1f, 3a, 3d) at the mRNA level. These data suggest the potential presence of metastasis in this supposedly normal sample despite its designation as “normal” by histological diagnosis (Table. S1).
Fig. 3.
(a) Hierarchical unsupervised clustering analysis of the selected 16 anti-oxidative stress enzymes in 14 CRC pairs. All tumors and 2 normal controls (N8, N14) had downregulation of anti-oxidative stress pathway. (b) Hierarchical unsupervised clustering analysis of the selected 13 hypoxia induced upregulated miRNAs in 15 CRC pairs. Only one tumor (T5) was grouped with normal samples. (c) mRNA expression profiles of hypoxia upregulated IL8 (p=0.005) and hypoxia downregulated GPD1L (paired t-test: p=2.6-07E) in 14 CRC pairs. (d) Hierarchical unsupervised clustering analysis of 3 SIRT mRNAs in 14 CRC pairs. All tumors had lower mRNA levels of 3 SIRT and were separated from normal controls. (e) Hierarchical unsupervised clustering analysis of the 6 glycolytic enzyme mRNAs in 14 CRC pairs. All tumors had higher mRNA levels of glycolytic genes and were separated from normal controls. (f) Hierarchical unsupervised clustering analysis of the 7 TCA cycle mRNAs in 14 CRC pairs. Twelve of 14 tumors had lower mRNA levels of TCA cycle genes were separated from normal controls. (g) Hierarchical unsupervised clustering analysis of the 3 pentose phosphate pathway (PPP) mRNAs in 14 CRC pairs. All tumors had high mRNA levels of 3 PPP enzymes and were separated from normal controls. Red and green bars delineate tumors and normal controls, respectively. Red in the heatmap indicates an expression level above the mean in all samples and blue indicates expression levels lower than the mean of all samples. The clustering analysis was based genes’ RPKMs.
5) sRNA profiles in 35 CRCs
Finally, to compare and contrast expression profiles in tumors across the histologic spectrum, we established individual tumor sRNA fingerprints in paired matched samples and evaluated commonality and dissimilarity in sRNA expression. Since both the magnitude of the ratio (T/N) and of the subtractive difference (T-N) in gene expression may have biological significance, we established both relative ratio (T/N) and absolute difference (T-N) fingerprints for each tumor. We then selected the three most highly up and down regulated sRNAs based on values of (T/N) or (T-N) to build a sRNA fingerprint for each tumor. Relative ratio fingerprints identified the MT-tRNA family, CTD 2651B20.6 and snord19 cluster as the most frequently upregulated genes while the snord114 cluster, AP000318.1 and snord64 were identified as the most frequently down regulated genes (Table. 2a, Table. S3a), largely reflecting earlier analyses (Fig. S2b, S2c). As in the initial analysis of average fold increased or decreased expression of each sRNA in all MET+ tumors (Fig. 1a, 1c), the 3 most upregulated (8-20X) genes in 9 individual metastatic tumors were also either MT-tRNAs or the snord19 family while the 3 most downregulated (2-25X) genes again were in the snord113/114/116 cluster (Table. S3a). For absolute difference fingerprints, we found that in metastatic tumors, snord43, MT-tRNAs and the snord12 cluster were the most frequently upregulated genes while snord71, snord59A and the snord114 cluster were the most frequently down regulated genes among the top 3 up/downregulated genes (Table. 2b, Table. S3b). Met+ tumors(T8LN, T15LN, T25LV and T35LN) displayed the greatest level of absolute RPKM changes of snord43, snord12 cluster and snord113/114/116 cluster expression. The deregulation of these snoRNAs in CRC has thus far not been reported and their roles, if any, in tumorigenesis, proliferation, or metastasis are not known. Overall, each tumor had its own genomic signature, with some unique features and some common features. Based on the presence or absence of MT-tRNAs in the top 3 upregulated genes, 35 CRCs can be placed into two groups, regardless of their histological types and TNM classification (Table 2c).
Table. 2.
(a) The frequency of deregulated sRNAs in all 35 CRCs based on T/N ratio fingerprint. Gene ratios of each tumor pair (T/N) were calculated and sorted from high to low. Three top upregulated genes and 3 top downregulated genes were selected from each tumor to build tumor fingerprints. The actual ratios of each gene were shown in Table. S3a. (b) Frequency of deregulated sRNAs in 35 CRCs based on subtractive (T-N) RPKM fingerprint. The differences of each tumor pair (T-N) were calculated and sorted from high to low. Three top upregulated and downregulated genes were selected from each tumors and actual differential RPKM were shown in Table. S3b. Commonly deregulated genes presented in more than once in 35 CRCs while uniquely deregulated genes just presented only once in 35 CRCs. (c) Classification of CRCs based on the presence of MT-tRNAs in top 3 upregulated genes according to both ratio and subtractive fingerprints.
| Table. 2a Frequency of sRNAs/miRNA in 3 top up/downregulated genes in T/N ratio fingerprint | |||
|---|---|---|---|
| 13 top upregulated genes in 35 CRCs (T/N: 1.2 to 20X folds) | Total: 105/35 | 14 top downregulated genes in 35 CRCs (N/T: 1.4 to 50X folds) | Total :105/35 |
| MT-tRNA family | 45/35 | snord113-114-116 cluster | 45/35 |
| snord19 cluster | 16/35 | AP000318.1 | 16/15 |
| CTD2651 B20.6 | 13/35 | snord64 | 12/35 |
| snord77 | 8/35 | snord107 | 9/15 |
| snord12 cluster | 7/35 | VTRNA1 | 6/15 |
| snora71D | 6/35 | scarna18 | 4/15 |
| snord78 | 4/35 | snord49B | 3/15 |
| snord98 | 1/35 | snord123 | 3/15 |
| snord49A | 1/35 | mir3607 | 2/15 |
| snord15A | 1/35 | snord71 | 1/15 |
| RNY4 | 1/35 | snord125 | 1/15 |
| snord101 | 1/15 | snord93 | 1/15 |
| snord1A | 1/35 | snord65 | 1/35 |
| snord28 | 1/35 | ||
| Table. 2b Frequency of sRNAs in 3 top up/downregulated genes in T-N subtractive RPKM fingerprint | ||
|---|---|---|
| 5 upregulated sRNAs (T-N=0.5 to 15 × 106 RPKM) | T/N ratio profile | Frequency (total: 105/35) |
| snord12 cluster | 1.5-4 fold | 71/35 |
| snord43 | 1.5-2 folds | 16/35 |
| MT-tRNAs | 1.5-10 folds | 16/35 |
| snord19.2 | 3 fold | 1/35 |
| snord78 | 2 fold | 1/35 |
| 5 downregulated sRNAs (N-T=0.2 to 2 × 106 RPKM) | T/N ratio profile | Frequency (total: 105/35) |
|---|---|---|
| snord71 | 1.5-4 folds | 33/35 |
| snord59A | 1.5-2 folds | 33/35 |
| snord114 cluster | 1.5-4 folds | 20/35 |
| AP0000318.1 | 1.5-4 folds | 18/35 |
| snord107 | 1.5-4 folds | 1/35 |
| Table. 2c CRC subtypes based on both ratio and subtractive fingerprints | |
|---|---|
| Type I | Type II |
| snoRNAs only | MT-tRNAs + snoRNAs |
| T5, T16, T19, T20, T22,T27, T30, T31, T35 | T1, T2, T3, T7,T10, T13, T11, T12, T9, T15, T4, T14, T6, T8, T17, T18, T21, T23, T24, T25, T26, T28, T29, T32, T33, T34 |
Discussion
CRC is a disease with variegated genetic and epigenetic profiles (29). Thus advanced molecular profiling of CRC may improve staging and grading of tumors and thus predicting clinical course, but as well in defining specific tumor survival and proliferation pathways and thereby elucidate novel drug treatments. Currently there are a number of drugs that target such specific oncogenic pathways (30). In this study, we delineated sRNA for 35 CRCs. We found that all 35 CRCs had distinct combinations of common as well as uniquely deregulated genes. This new understanding of oncogenic mechanisms has begun to influence risk assessment, diagnostic categories, and therapeutic strategies, with increasing use of drugs and antibodies designed to counter the influence of specific molecular drivers.
Our study mainly focused on distinctions among tumors of high stage and tumors with metastases. In this study, we identified MT-tRNAs as the top upregulated sRNAs in LN/LV positive and high stage CRC, as well as a signature of sRNAs associated with such tumors and, in some cases, with their adjacent “normal” tissues which likely contain undetected metastasis. Furthermore, the distinct profiles of tumors with metastasis, as defined by both ncRNA and coding RNA signatures in CRC reveals some novel elements. Especially notable was the stunning downregulation of the snord113/114/116 clusters which are deleted in PWS, a genetically imprinted disease marked by profound obesity which may likewise enhance energy storage in cancer cells, but not described thus far in cancer. Whether these snoRNA clusters having tumor suppressor function need to be studied.
A multitude of data support the role of hypoxia in tumor physiology as hypoxia has been shown to upregulate glycolytic metabolism and MT-tRNAs (31) and downregulate enzymes involved in the TCA cycle. More generally, hypoxia has long been recognized to play a role in promoting the invasive and metastatic behavior of cancer cells and their ability to disseminate from established colonies and establish new colonies in distinct environments may depend on their ability to diversify metabolic pathways (21, 23). In this regard, the stringency of tumor harvest and preservation in our study is critical in assuring that these findings are not artifacts of prolonged ex-vivo time duration prior to storage (12). The superposition of upregulated MT-tRNAs, which may support generation of ATP through mitochondrial oxidative phosphorylation, on a profile of a shift to glycolytic metabolism, suggests that tumors with enhanced capacity for local and distant invasion are poised to use either oxidative phosphorylation or glycolytic metabolism, depending on the oxygenation circumstances in which they find themselves. Though downregulation of snoRNAs such as snord50A, h5sn2, snord43 and snord44, have been found in breast, prostate, brain and lung cancers (32, 33), the panel of downregulated snoRNAs, identified in this study, have not been reported previously and may be unique to colonic cancers. Interestingly, the host gene of the downregulated snord114 cluster (Fig. S5a) is the maternally expressed 3 RNA gene, MEG3 (34, 35). MEG3 inhibits cancer cell proliferation by both p53-dependent and p53 independent pathways (34, 35). Consistent with this finding, our sRNA/mRNA sequencing data revealed a strong expression correlation (downregulation of expression) between snord114-23/snord114-26 and MEG3 (rs>0.8) (Fig. S5b). Whether these imprinted snoRNAs have specific biological functions distinct from their host gene in malignant transformation needs to be further evaluated.
In conclusion, the 35 paired samples analyzed in this study serve as a training set that helps to identify sRNA signatures for metastatic and advanced CRC. To strengthen our findings, these putative sRNA markers will be evaluated and potentially validated by an independent study with larger numbers of samples. The hope is that such ncRNAs and coding RNAs will be validated as biomarkers and therapeutic targets which will ultimately benefit patients.
Supplementary Material
Statement of translational relevance.
The precise molecular profiling of colorectal cancer (CRC) should facilitate development of better staging systems as well as identify targets for development of novel treatments. Our study focused principally on non-coding RNAs in the characterization of tumors at high stage and with metastases. We identified mitochondrial transfer RNAs (MT-tRNAs) as the top upregulated small RNAs (sRNAs) in high stage CRC and in CRC with defined metastases. We also found that small nucleolar RNAs (snoRNAs) downregulated in Prader-Willi syndrome (PWS), a metabolic disorder characterized by obesity (energy storage), were commonly downregulated in CRC. These findings indicate that a hallmark of tumor aggressiveness/progression pertains to energy storage and endowment of mitochondria with enzymes critical for oxidative phosphorylation, despite a shift to glycolytic metabolism in such tumors.
Acknowledgments
The authors would like to thank Julia Berkson, Baolin Zhang and Ashutosh Rao for their critical review and comments on this manuscript.
Footnotes
Disclosure of Potential Conflicts of Interest
There are no current financial considerations to disclose.
Ethics Statement
Written informed consent was obtained from all the participants involved in the study via the procuring laboratory at Indivumed (Hamburg, Germany).
References
- 1.Constant S, Huang S, Wiszniewski L, Christophe M. Colon Cancer: Current Treatments and Preclinical Models for the Discovery and Development of New Therapies. Pharmacology, Toxicology and Pharmaceutical Science >> Drug Discovery >> “Drug Discovery”. 2013 Jan; [Google Scholar]
- 2.Zarate R, Boni V, Bandres E, Garcia-Foncillas J. MiRNAs and LincRNAs: Could they be considered as biomarkers in colorectal cancer? Int J Mol Sci. 2012;13(1):840–65. doi: 10.3390/ijms13010840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazeh H, Mizrahi I, Ilyayev N, Halle D, Brücher B, Bilchik A, et al. The Diagnostic and Prognostic Role of microRNA in Colorectal Cancer - a Comprehensive review. J Cancer. 2013;4(3):281–95. doi: 10.7150/jca.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oue N, Anami K, Schetter AJ, Moehler M, Okayama H, Khan MA, et al. High miR-21 expression from FFPE tissues is associated with poor survival and response to adjuvant chemotherapy in colon cancer. Int J Cancer. Apr 15. 2014;134(8):1926–34. doi: 10.1002/ijc.28522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25(4):469–83. doi: 10.1016/j.ccr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levinger L, Mörl M, Florentz C. Mitochondrial tRNA 3′ end metabolism and human disease. Nucleic Acids Res. 2004:5430–5441. doi: 10.1093/nar/gkh884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet. 2011;45:299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- 8.Pavon-Eternod M, 1, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37(21):7268–80. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu LH, Meng Q, Zhou H, Chen YQ. Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res. 2001;29(7):1623–1630. doi: 10.1093/nar/29.7.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao J, Yu L, Mei YP, Guarnera M, Shen J, Li R, et al. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer. 2010;9:198. doi: 10.1186/1476-4598-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galamb O, Sipos F, Solymosi N, Spisák S, Krenács T, Tóth K, et al. Diagnostic mRNA Expression Patterns of Inflamed, Benign, and Malignant Colorectal Biopsy Specimen and their Correlation with Peripheral Blood Results. Cancer Epidemiology Biomarkers $ Prevention. 2008;17:2835. doi: 10.1158/1055-9965.EPI-08-0231. [DOI] [PubMed] [Google Scholar]
- 12.Dang HT, Moore H, Player A, Wang YH, Kawasaki E, David K, et al. The NCI Biospecimen Research Network (BRN): The Influence of Warm Ischemic Time on Gene Expression Profiles for Colon Cancer National Cancer Institute, 2INDIVUMED GmbH, 3SAIC-Frederick, Inc. http://biospecimens.cancer.gov/meeting/brnsymposium/docs/2008pres/Poster7-Hien_Dang.pdf.
- 13.Jenjaroenpun P, Kremenska Y, Nair VM, Kremenskoy M, Joseph B, Kurochkin IV. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ. 2013 doi: 10.7717/peerj.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chenna R, Galiveti, Raabe Carsten A., Konthur Zoltán, Timofey S, et al. Differential regulation of non-protein coding RNAs from Prader-Willi Syndrome locus Scientific Reports Article number. :6445. doi: 10.1038/srep06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Human Tumor Metastasis PCR Array. http://www.sabiosciences.com/rt_pcr_product/HTML/PAHS-028A.html.
- 16.ME AA, French CJ, Smart CE, Smith MA, Clark MB, Ru K, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17(5):878–91. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaedcke J, Grade M, Camps J, Søkilde R, Kaczkowski B, Schetter AJ, et al. The rectal cancer microRNAome--microRNA expression in rectal cancer and matched normal mucosa. Clin Cancer Res. 2012;18(18):4919–30. doi: 10.1158/1078-0432.CCR-12-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azuaje FJ. Selecting biologically informative genes in co-expression networks with a centrality score. Biol Direct. 2014;9:12. doi: 10.1186/1745-6150-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penrod NM, Jason H, Moore JH. Influence networks based on coexpression improve drug target discovery for the development of novel cancer therapeutics. BMC Systems Biology. 2014;8:12. doi: 10.1186/1752-0509-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu Z, Liu J, Cao K, Zhang JF, Wang J. Centrality-based pathway enrichment: a systematic approach for finding significant pathways dominated by key genes. BMC Systems Biology. 2012;6:56. doi: 10.1186/1752-0509-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen G, Xiaobo, Jia YF, Piazza G, Xi YG. Hypoxia -regulated microRNAs in human cancer. Acta Pharmacologica Sinica. 2013;34:336–3411. doi: 10.1038/aps.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27(5):1859–67. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu CJ, Tsai MM, Hung PS, Kao SY, Liu TY, Wu KJ, et al. miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 2010;(4):1635–44. doi: 10.1158/0008-5472.CAN-09-2291. [DOI] [PubMed] [Google Scholar]
- 24.Rundqvist HJ. Curr Top Microbiol Immunol. 2010;345:121–39. doi: 10.1007/82_2010_77. [DOI] [PubMed] [Google Scholar]
- 25.Weljie AM, Jirik FR. Hypoxia-induced metabolic shifts in cancer cells: moving beyond the Warburg effect. Int J Biochem Cell Biol. 2011;(7):981–9. doi: 10.1016/j.biocel.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Bensinger SJ, Christofk HR. S New aspects of the Warburg effect in cancer cell biology. Semin Cell Dev Biol. 2012;(4):352–61. doi: 10.1016/j.semcdb.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Currie E, Schulze A, Zechner R, Walther TC, Farese RV., Jr Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18(2):153–61. doi: 10.1016/j.cmet.2013.05.017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL. Sirtuin-3 (SIRT3) and the Hallmarks of Cancer Genes Cancer. 2013;4(3-4):164–171. doi: 10.1177/1947601913486351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romano G. The Role of the Dysfunctional Akt-Related Pathway in Cancer: Establishment and Maintenance of a Malignant Cell Phenotype, Resistance to Therapy, and Future Strategies for Drug Development. Scientifica. 2013 doi: 10.1155/2013/317186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jian B, Wang D, Chen D, Voss J, Chaudry I, Raju R. Hypoxia-induced alteration of mitochondrial genes in cardiomyocytes: role of Bnip3 and Pdk1. Shock. 2010;(2):169–75. doi: 10.1097/SHK.0b013e3181cffe7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mannoor K, Liao J, Jiang F. Small nucleolar RNAs in cancer. Biochim Biophys Acta. 2012;1826(1):121–8. doi: 10.1016/j.bbcan.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 2012;12(2):84–8. doi: 10.1038/nrc3195. 19. [DOI] [PubMed] [Google Scholar]
- 34.Sun M, Xia R, Jin F, Xu T, Liu Z, De W, et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;(2):1065–73. doi: 10.1007/s13277-013-1142-z. [DOI] [PubMed] [Google Scholar]
- 35.Qin R, 1, Chen Z, Ding Y, Hao J, Hu J, Guo F. Long non-coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma. 2013;60(5):486–92. doi: 10.4149/neo_2013_063. [DOI] [PubMed] [Google Scholar]
- 36.Cummings M, Sarveswaran J, Homer-Vanniasinkam S, Burke D, Orsi NM. Glyceraldehyde-3-phosphate dehydrogenase is an inappropriate housekeeping gene for normalising gene expression in sepsis. Inflammation. 2014;7:1889–94. doi: 10.1007/s10753-014-9920-3. [DOI] [PubMed] [Google Scholar]
- 37.Bär M, Bär D, Lehmann B. Selection and Validation of Candidate Housekeeping Genes for Studies of Human Keratinocytes—Review and Recommendations. Journal of Investigative Dermatology. 2009;129:535–537. doi: 10.1038/jid.2008.428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.