Abstract
It has long been known that the interphase microtubule (MT) array is a key cellular scaffold that provides structural support and directs organelle trafficking in eukaryotic cells. Although in animal cells, a combination of centrosome nucleating properties and polymer dynamics at the distal microtubule ends is generally sufficient to establish a radial, polar array of MTs, little is known about how effector proteins (motors and crosslinkers) are coordinated to produce the diversity of interphase MT array morphologies found in nature. This diversity is particularly important in multinucleated environments where multiple MT arrays must coexist and function. We initiate here a study to address the higher ordered coordination of multiple, independent MT arrays in a common cytoplasm. Deletion of a MT crosslinker of the MAP65/Ase1/PRC1 family disrupts the spatial integrity of multiple arrays in Dictyostelium discoideum, reducing the distance between centrosomes and increasing the intermingling of MTs with opposite polarity. This result, coupled with previous dynein disruptions suggest a robust mechanism by which interphase MT arrays can utilize motors and crosslinkers to sense their position and minimize overlap in a common cytoplasm.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-2026-8) contains supplementary material, which is available to authorized users.
Keywords: Centrosome, Microtubules, MAP65/Ase1/PRC1, Dictyostelium, Interphase
Introduction
The centrosome is a landmark organelle found in nearly all eukaryotic animal cells [1]. Its primary function is to organize the assembly of polar MT arrays, which in turn, contribute to cell structure, division, and endomembrane distribution. During interphase, this aptly named “central body” is typically located near the cell center, a position largely due to a balance of forces acting on the MT array. It is well known that assembly/disassembly reactions at the MT distal ends create pushing and pulling forces that are sufficient to “self-center” the array, an action that can be recapitulated in synthetic environments and understood through modeling efforts [2, 3].
However, there are numerous examples of centrosome displacement or movements that indicate a higher ordered level to positioning in cells. Cortical pulling forces can act on astral MTs during division to displace the spindle and create asymmetric sized cells [4]; centrosomes can be drawn up near the cell periphery where they serve to nucleate axonemal structures [5]; centrosomes also orchestrate nuclear migration during cell growth by coupling distal forces to nuclei [6]. All of these activities involve molecular motors (e.g. dynein, kinesin) that push or pull against MTs and effect centrosome position (e.g. [7]). Yet despite a growing list of the molecular participants, including contributions from the actin-myosin machinery, our understanding is limited as to how motile MT events are locally controlled or coordinated.
In addition, there is an even larger scale of centrosome/MT organization that is understudied in animal cells. Many organisms exist or contain cells as syncytia, a common cytoplasm with multiple nuclei, centrosomes, and MT arrays. In many cases, syncytial nuclei appear evenly distributed throughout the cytoplasm, along with associated MT arrays. While one could attribute this distribution to simple properties of cytoplasm, we argue that this level of organization is also carefully orchestrated and important for cell survival. In some cases, MT arrays maintain their independence in a common cytoplasm, a feature that establishes cytoplasmic territories that may be useful in locally managing organelle traffic and polarity [8], as well as coordinate cell movement and growth.
In this paper we examine MT organization at this larger scale and reveal a new role for MT crosslinkers of the MAP65/Ase1/PRC1 family [9–12]. In the absence of Ase1A in D. discoideum, MT arrays no longer maintain spatial separation in a multinucleated environment. In addition to its well-known activity during mitosis, this MT crosslinker appears to play a role during interphase to exclude or minimize interdigitating MTs of opposite polarity. Our results emphasize that a multilevel coordination of motors and polymer dynamics is necessary to organize multiple MT arrays in a common cytosol.
Results
In multinucleated cells of the model organism, D. discoideum, each nucleus is tightly associated with a centrosome and corresponding MT array; these arrays generally remain distinct from each other and appear to maintain mutually exclusive territories during interphase (Fig. 1) [13–17]. MT plus ends minimally grow and shrink in D. discoideum, and thus conventional tip-based assembly dynamics do not significantly contribute to centrosome position [18]. Instead, microtubules undergo substantial lateral bending and sweeping motions, demonstrating there are robust pushing and pulling forces from motor machinery in the cytoplasm or at the cell cortex that influence centrosome position [17, 19–21]. Treatment of cells with 2.5 μg/ml nocodazole shrinks the distances between centrosomes, further indicating a MT-based component in their spacing (Fig. 2). To address how multiple arrays minimize their spatial overlap in a common cytoplasm, we focused attention on components that engage interdigitating MTs of opposite polarity and thus targeted the MAP65/Ase1/PRC1 protein family.
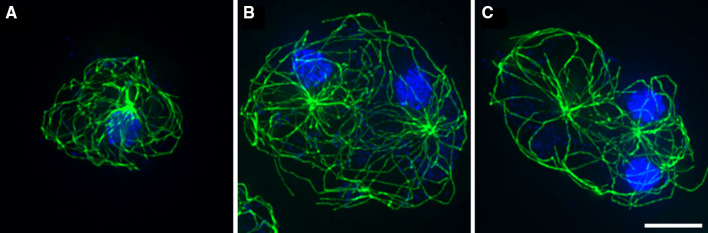
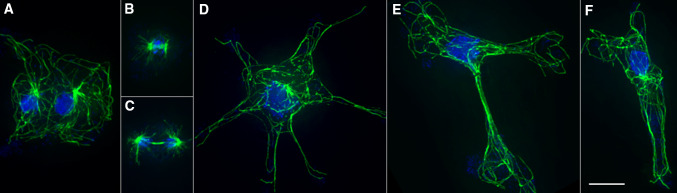
Fig. 1.
MT arrays maintain distinct territories in multinucleated D. discoideum cells. a Typical WT mononucleated cell, MTs are in green, nuclei in blue. b, c Binucleate examples of WT (b) and a kif9 − kinesin cell (c). In b and c, the MT arrays are largely separate from one another, with minimal overlap in the cytoplasm. Deletion of the kif9 kinesin detaches centrosomes from nuclei [54] and indicates here that centrosome position is independent of forces acting directly on nuclei. Bar 10 μm
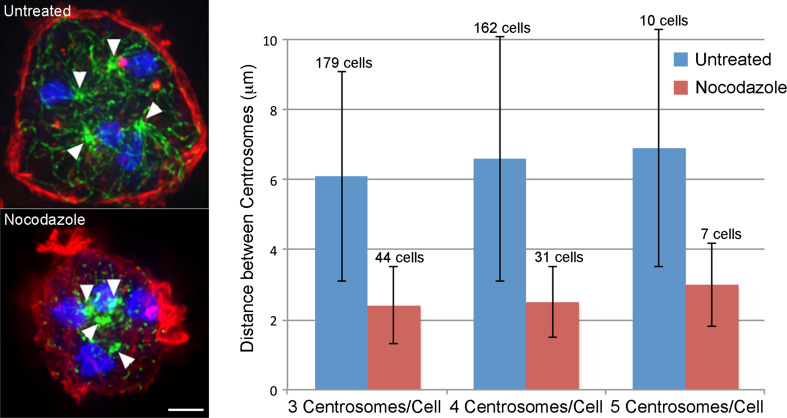
Fig. 2.
Intercentrosome distance in multinucleates is dependent on MT length. Left panel shows two tetranucleated WT cells ±2.5 μg/ml nocodazole treatment. MTs are in green, nuclei in blue, actin cortex is in red. Arrowheads mark centrosome positions. Right panel quantitates distance between all centrosome pairs in tri-, tetra- and penta-nucleated cells, ±nocodazole treatment. Note that the average distance decreases by at least half after nocodazole-induced MT depolymerization (p values for each comparison are <10−9). Bar 10 μm
The D. discoideum genome contains two isoforms of Ase1 (Fig. 3) [22], which we term A and B. The two sequences share a central core of approximately 219 residues that are 23 % identical and 47 % similar. This region corresponds to the spectrin-like microtubule binding domains representative of the gene family. As in other Ase1 isoforms, both polypeptides have extensive amino terminal regions predicted to form coiled-coil secondary structure. The two sequences are otherwise divergent in their similarity to reference human PRC1 and fungal Ase1 sequences of this family (Fig. 3). Ase1A most closely resembles the human and fungal isoforms, sharing moderate similarity (43–46 %) in the amino-terminal two-thirds of the coding sequence. Also similar to PRC1 [11], Ase1A contains two classical nuclear localization motifs, PIEKLKK beginning at residue320 and PNNKKKI at residue612. Ase1B is less related to PRC1 than is Ase1A, with sequence homology only in the carboxy-terminal one-third of the coding region, and absent any canonical nuclear localization motifs. As in PRC1 [23], Ase1B does contain two closely spaced consensus sequences for CDK phosphorylation (TPSK at residues721, 732); these motifs are not found in the Ase1A isoform. Ase1A and B were targeted for disruption by homologous recombination, via the strategy outlined in supplemental Fig. 1. Knockout strains of either gene are viable; cells accumulate in suspension cultures at rates similar to wild type (WT) growth (Sup. Fig. 1) and both produce viable spores when challenged to undergo a developmental cycle. However, disruptions of the two isoforms produce distinctly different phenotypes. Deletion of Ase1A impacts known mitotic functions and has a novel defect in organizing multiple MT arrays during interphase. Ase1B disruption only appears to affect MT bundling during interphase.
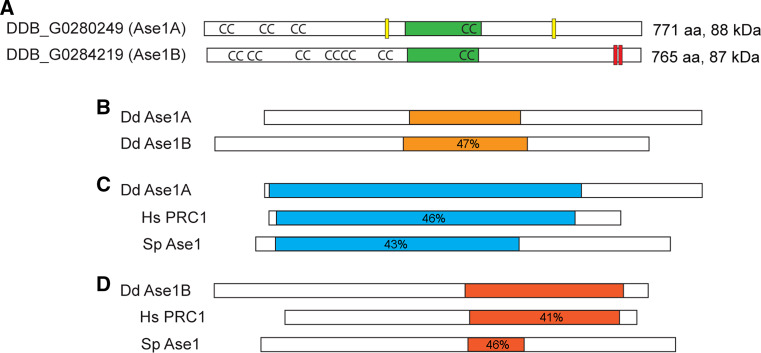
Fig. 3.
D. discoideum contains two isoforms of Ase1. a Domain organization based on sequence homologies, including position of the MT-binding domain (green), coiled-coil motifs (CC), nuclear localization signals (yellow), and CDK phosphorylation motifs (red). b–d Display sequence similarity between the two isoforms and Human PRC1 and S. pombe Ase1 proteins. The orange regions in b highlight the central core that is 23 % identical, 47 % similar between the two D. discoideum isoforms. c The relationship of isoform A with human PRC1 and S. pombe Ase1 proteins. The regions marked in blue are similar. d Same comparison but with the B isoform; similarity is shown in red. Percentage indicates degree of similarity to the D. discoideum isoform within the colored regions
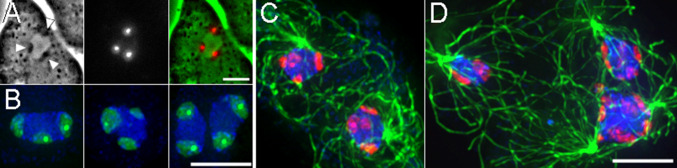
Previous work in other animal systems predicts that disruption of Ase1/PRC1 homologs will impact mitotic spindle assembly and cytokinesis [10–12]. In agreement with these studies, the central spindle that normally forms between separating spindle poles appears to either be substantially reduced or missing altogether in Ase1A− cells (Fig. 4). Quantitative analysis of spindle pole separation (Fig. 4) reveals two deviations from normal mitosis. In 74 % of the null cells tracked (28/38), centrosomes bolt apart just after their initial separation, at a burst rate close to 7 μm/min. GFP-labeled centromeres demonstrate similar timing (Sup. Fig. 2), indicating that chromosomes are connected to the daughter centrosomes early during division, and do not absolutely require a traditional metaphase stage for mitotic progression. In the remaining 26 % of Ase1A− cells (10/38), the duplicated poles stay adjacent to each other, largely failing to separate or complete karyokinesis over the observed time frame. We further noticed an increase in multinucleation of the Ase1A− cells. Under current growth conditions, only 12.4 % of WT cells had more than 1 nucleus (44/354). In the absence of Ase1A, 31.0 % of cells were multinucleated (113/364), suggesting defective cytokinetic processes. These results indicate that the Ase1A protein functions during mitosis in D. discoideum in a fashion similar to other organisms.
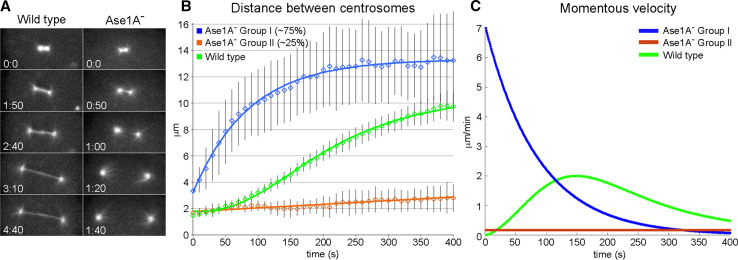
Fig. 4.
Ase1A− cells fail to form a proper midzone during mitosis. a The left column shows frames from a WT mitotic cell labeled with GFP-α-tubulin. Note the band of interdigitating MTs that connect the two spindle poles. The right column shows similar staged frames of an Ase1A− cell. Although there is a fluorescent MT bridge visible in the first two panels, this structure breaks early during the centrosome separation process. b Plots centrosome distance as a function of time during Ase1A− cell division. Blue and orange lines plot Ase1A− cell averages; green line traces the WT cell average [47]. c The momentous velocity of centrosome separation for the three classes of movement
In addition, the disruption of Ase1A produced a significant change in the interphase MT architecture of multinucleated cells (Fig. 5). Adjacent arrays visually appear to merge into each other, the radial character of MT patterns is diminished, and MTs appear tangled. Even in some mononucleated cells, the arrays appear less radial and more tangled. To quantitate centrosome convergence, we measured distances between centrosomes in WT and mutant binucleate cells. Under these conditions, the distance in WT cells averaged 6.7 ± 3.6 μm (n = 101); in Ase1A− cells, the distance decreased to 3.6 ± 1.3 μm (n = 98) (Fig. 5). This difference is statistically significant, with a p value <10−5. We further examined the dynamics of centrosomes over time (Fig. 6). Kymographs demonstrate oscillations in distance between centrosome pairs, but the WT organelles generally maintain a constant spacing. In contrast, there was greater positional variation and periodic convergence of centrosomes in Ase1A− cells. Finer analysis suggests there may be greater distance covered per transit in WT cells, but overall the frequency of movements were the same between WT and Ase1A− centrosome pairs (Sup. Fig. 3). These results indicate that in the absence of Ase1A, cells lose the ability to maintain spacing between multiple MT arrays.
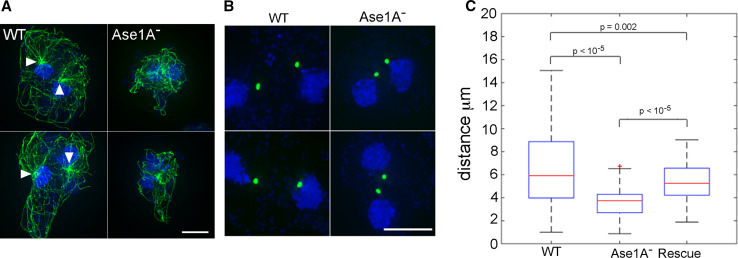
Fig. 5.
Deletion of Ase1A disrupts MT network integrity in multinucleates. a MT arrays in binucleate WT (left) and Ase1A− cells (right). While the two arrays are clearly distinguished in WT, centrosomes appear merged and MTs intertangled in Ase1A− cells. Bar 10 μm. b Centrosomes imaged with γ-tubulin-GFP (green), and c quantification of distance between binucleate centrosome pairs, displayed as box plots. The average distance between binucleate centrosomes in WT is 6.7 ± 3.6 μm; in the knockout, average distance is 3.6 ± 1.3 μm. In Ase1A− cells rescued with full-length Ase1A, the average distance is 5.4 ± 1.7 μm
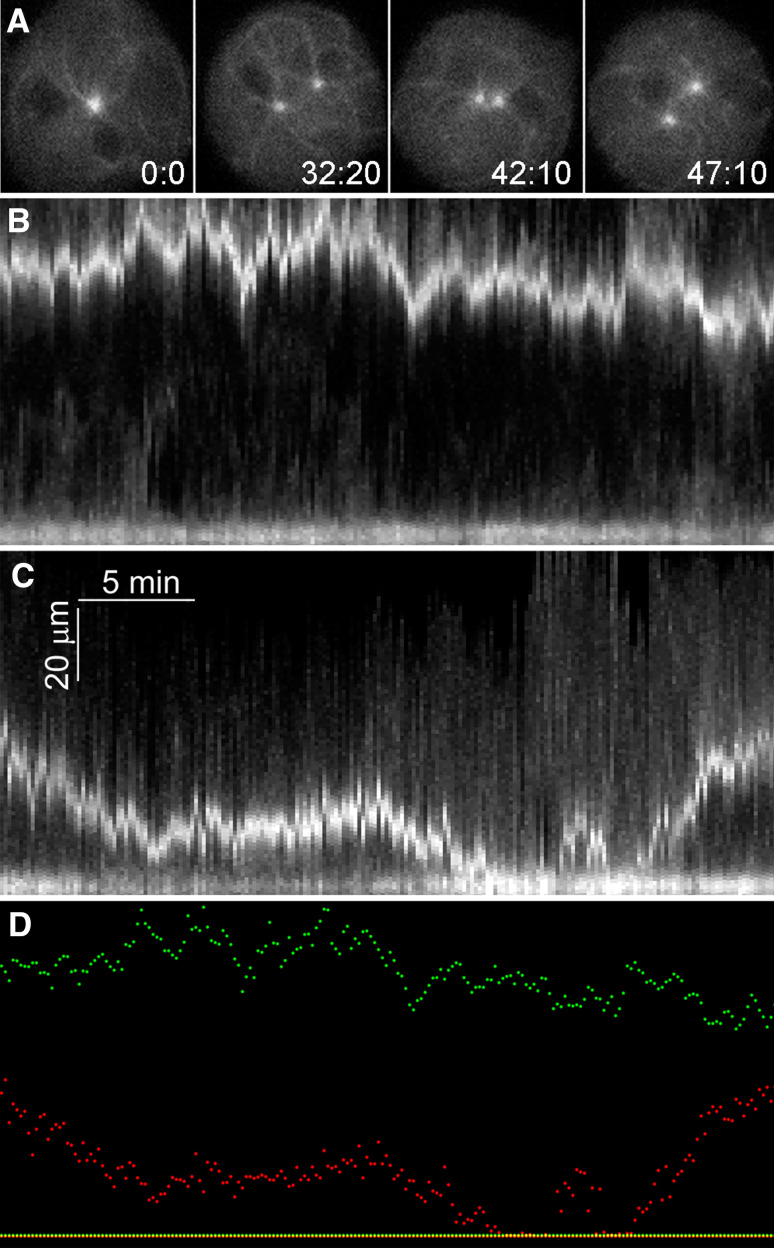
Fig. 6.
Centrosome position is dynamic in Ase1− cells. a Individual frames from live cell recording of a binucleate Ase1A− cell labeled with GFP-α-tubulin. Note the variable distance between the two centrosomes. b, c Kymograms of such movies where we fixed the position of one centrosome (lower tracing in each panel) to show distance variability between the two over time. b Displays a WT cell and c displays an Ase1A− cell. d Overlays the two tracings (WT in green, Ase1A− in red). Time in min:s
To examine the cellular distribution of Ase1A, we isolated a full-length coding sequence, developed an amino-terminus GFP tagged construct, and imaged its expression in wild type and in Ase1− cells (Fig. 7). Despite screening numerous transformants, we were only able to isolate clones where a fraction of cells were visibly expressing GFP at any one time (e.g. 99/252 cells, 39 %). On a population level, the full-length GFP-Ase1A polypeptide at least partially restores the centrosome distancing of WT binucleate cells (Fig. 5); individually, the construct expression does reestablish the spindle overlap region in dividing cells. Imaging during interphase reveals primarily an intranuclear distribution of Ase1A that appears restricted to the nucleolus (Fig. 7). Upon mitotic entry, the GFP-Ase1A protein redistributes throughout the nucleus and then concentrates onto the spindle region where MTs of opposite polarity interdigitate (Fig. 8). These results are consistent with previous studies in animal cells.
Fig. 7.
GFP-Ase1A localization in wild type (a, b) and Ase1A− cells (c, d). a Phase contrast, GFP-Ase1A fluorescence, and merged panels, revealing not just nuclear localization of the Ase1A protein, but restriction to the phase dense nucleolar structures (arrowheads in left frame). b Maximum intensity projections of four nuclei following deconvolution. GFP-Ase1A is in green; DNA is in blue. c, d Maximum intensity projections of a bi-, and a tri-nucleate cell that show MTs (in green), GFP-Ase1A (in red), and nuclear (blue) distributions. While nucleolar patches are readily apparent, cytoplasmic distribution of Ase1A is not seen in these cells. Bar 5 μm
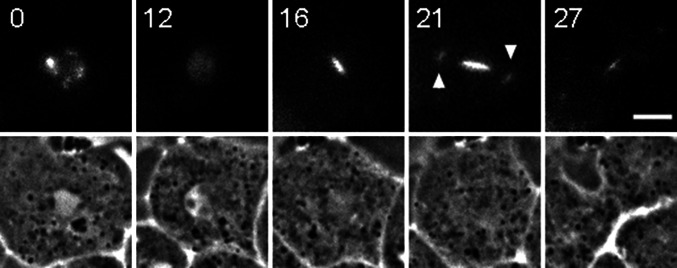
Fig. 8.
GFP-Ase1A labeling during mitosis in a wild type cell. Panel shows fluorescent Ase1A live cell images on top row, and corresponding phase contrast frames on bottom row. Time 0 captures nucleolar localization prior to mitosis; 12 min panel shows redistribution of this label as the cell enters division. Note the bright label of the spindle midzone in frames 16 and 21 min. Even the spindle poles show some staining (arrowheads). As the cell begins to cleave (frame 27), most of the spindle label is concentrated in the midbody. Time in min; bar 5 μm
Using conventional widefield imaging followed by deconvolution and maximum intensity projection (Fig. 7), we are as yet unable to detect Ase1A in the cytoplasm of interphase cells, particularly in the regions where MTs of opposite polarity would be expected to overlap. This result contrasts with similar imaging in yeast where the Ase1p is readily detectable in the interphase cytosol, bound to MTs [10, 24]. Immunoblot analysis does indicate a trace amount of GFP-Ase1A in the cytosolic fraction (Sup. Fig. 4), but we are as yet unable to determine its localization.
Disruption of the Ase1B isoform produces a phenotype that is markedly different than the Ase1A− (Fig. 9). Microtubule patterns in most fixed Ase1B− cells are indistinguishable from WT. These patterns include mono and multinucleated cells, as well as spindles in dividing cells. However, a minority of cells (~10 %) shows striking cell branching and protrusions that are rarely seen in WT. These hyperpolarized extensions contain MTs that extend out to the cell tips and often appear to be bundled. We interpret this result to indicate that the two Ase1 proteins participate in functionally independent activities.
Fig. 9.
Deletion of Ase1B enhances cell branching and polarization. a Binucleate Ase1B− cell with normal spacing between centrosomes. b, c Two mitotic cells demonstrating normal spindle assembly. d–f Examples of hyperpolarized cells. While cell elongation/polarization is typical in chemotactic cells migrating in cAMP gradients, this degree of branching and extension is not usually seen in vegetative growth conditions. MTs are in green, nuclei in blue. Bar 5 μm
Discussion
Our goal in this study is to understand how multiple MT arrays are organized in the common cytoplasm of syncytial animal cells. D. discoideum represents an attractive model to examine this process, for two reasons. First, the MT array is subject to very robust pushing and pulling forces that effect its position. Cytoplasmic dynein is an integral component of these forces, as disruptions to this motor result in the wide scale circulatory movement of the centrosome and entire MT network [20, 21, 25, 26]. Second, and despite these forces, multiple MT arrays are able to establish and maintain largely independent territories in a common cytoplasm. This effect is seen in WT cells that spontaneously fail cytokinesis, including those that accumulate large numbers of nuclei [13], and in mutants that impact cytokinesis machinery or centrosome replication (e.g. [14–17]). These observations raise the interesting question of how the cell is able to balance robust forces yet sense position to maintain radial, independent MT arrays.
Because of their known activities to engage MTs of opposite polarity, we focused attention on the MAP65/Ase1/PRC1 family of MT crosslinkers. The D. discoideum genome contains two isoforms of this family. Outside of the central MT-binding region, the two sequences are largely divergent and our results indicate that the polypeptides participate in distinct functional activities. Disruption of Ase1A, the isoform most closely related to the canonical PRC1/Ase1 proteins in animal cells, produces two defects, the expected disruption to the spindle midzone during division, and a novel disruption to the interphase MT network.
PRC1/Ase1 disruptions in human cells, yeasts, worm, and fly are well known to produce spindle midzone abnormalities [9–12, 27–29], and the Ase1A isoform in D. discoideum clearly functions in this capacity. Live cell imaging of Ase1A− cells demonstrates that despite improper spindle assembly, chromosomes distribute to the daughter centrosomes, and these structures separate to facilitate karyokinesis. These results highlight a longstanding contrast between force production mechanisms in anaphase B type movements, addressing to what extent the central spindle pushes poles apart vs. pole separation by astral MT mediated pulling mechanisms (e.g. [30–32]). Our results indicate that the astral MTs in D. discoideum play a major role in transducing a pulling force for pole separation during division. In the absence of a mature central spindle, the majority of daughter centrosomes separate at a significantly faster rate than in WT, suggesting that one function of the central spindle here is to moderate pole separation, perhaps to coordinate timing of the cytokinetic machinery. The minority of the cases where no pole separation is observed likely reflects either a failure to engage cytosolic force machinery or failure to generate vectorial forces that pull poles apart.
The second and perhaps more interesting function for Ase1A supports the existence of machinery dedicated to maintaining spatially segregated interphase MT arrays. Cytoplasmic functions for MAP65/Ase1 isoforms are previously known in some organisms to stabilize MT bundles. For example, in Arabidopsis they play multiple roles in establishing MT bundle organization in the absence of dedicated MTOCs [33, 34]. In S. pombe, Ase1p crosslinks cytoplasmic MTs [10, 35] and provides MT rigidity for dynein-dependent horse tail-like oscillatory movements of nuclei during meiosis [24]. In Ashbya, Ase1 crosslinks opposite polarity MTs, providing a means to space nuclei apart [36].
In all these cases, the MAPs function to enhance/stabilize MT–MT interactions. The novel component to our study is that Ase1A appears to minimize opposite polarity MT–MT interactions. A similar scenario is presented in a recent cell free system constructed from Xenopus egg extracts [37]. In this case, mitotic MT asters adjacent to each other establish a midzone that blocks interpenetration of antiparallel MTs. The authors describe the recruitment of multiple midzone proteins to this aster interaction zone, including the kinesin Kif4A and an embryonic version of PRC1. Depletion of the Kif4A as well as another kinesin, Kif20AE resulted in widening or disorganization of the interaction zone suggesting that these motors play an active role in reducing antiparallel MT penetration, e.g. Fig. 2a in [37].
A related action may be at play in D. discoideum, but in an interphase environment with far fewer MT–MT engagements. One possible scenario is that Ase1A senses/identifies interphase MTs of opposite polarity and targets effector molecules to reduce this overlap. Two obvious effectors could be Kinesin-4 (Dd Kif8) and/or Kinesin-8 (Dd Kif10). Both of these motors have previously been implicated in opposing dynein-driven MT rotations in D. discoideum [38] and thus work synergistically with dynein to organize the MT array. Both motors are also known to participate in antiparallel MT–MT interactions [39–42]. Kinesin-8 has MT sliding and depolymerizing activities and has recently been modeled into a nuclear centering mechanism relevant to interphase [43]. Kinesin-4 is further known to interact directly with human PRC1 [39, 42, 44] where it also functions to regulate MT length during mitosis.
We have previously shown that depletion of the Dd Kinesin-4 isoform (Kif8), results in interphase MT disorganization similar to the Ase1A disruption (e.g. Sup Fig. 5) and reduces the whole-scale MT movements that occur in the absence of dynein function [38]. Kif4A disruption in the Xenopus egg extract system (Fig. 2a in [37]), results in a similar wavy, tangled MT appearance in the increased zone of interdigitation. Thus it is tempting to postulate that a combination of Ase1A and Kinesin-4 acts as part of a surveillance system to recognize and reduce MT interdigitation in multinucleated D. discoideum cells. Such a mechanism might include Ase1A marking overlapping MTs and recruiting Kinesin-4 or -8 to either to drive MTs apart or effect their depolymerization. In support of this idea, we can find examples where MTs from nearby centrosomes appear to snap together and then break apart (Fig. 10), providing at least proof in principle that MT trimming could occur. These actions, a surveillance mechanism that pushes or trims MTs, coupled with robust dynein driven cortical pulling represent two opposing forces that could orchestrate the arrangement of multiple MT networks in complex environments. How the two machineries coordinate their actions is certainly an interesting next step question in understanding how animal cells organize their cytoplasm.
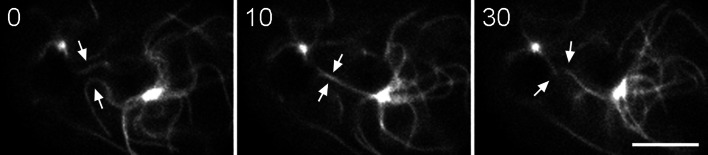
Fig. 10.
Antiparallel MT interactions. In this sequence, two GFP-MTs (arrows), one from each pole in a WT binucleate interphase cell appear to snap together in the middle panel, then fragment into shorter lengths in the right panel. Time in s; bar 5 μm
Materials and methods
Molecular genetics
Sequences of the Ase1 genes (DDB_G0280249, DDB_G0284219) were identified through comparative BLAST searches on NCBI (http://www.ncbi.nlm.nih.gov/) and obtained using the bioinformatic tools at dictyBase (http://dictybase.org/) [22]. Gene fragments used for disruption constructs and the full-length version of Ase1A were amplified from genomic D. discoideum AX-2 cell DNA using primer constructs outlined in Supplemental Fig. 1; specific primer details are available on request. All PCR products used for cloning were verified by sequencing. For gene knockouts, restriction enzyme sites were engineered into the ends of each primer (BamHI/XbaI and HindIII/BamHI) for cloning and to accommodate a blasticidin S resistance cassette (Bsr r) for isolating transformants.
For homologous recombination, we used the calcium phosphate method to transform AX-2 cells, as described [38, 45], and selected transformants in 10 μg/ml blasticidin S (MP Biomedicals). For Ase1A, 124 clones were screened by PCR using sets of upstream primers in the Bsr r cassette and downstream primers beyond the targeted insertion sites, each designed to amplify ~1 kb fragment if the insertion was properly targeted. Nine transformants showed evidence of marker insertion at the desired site. Four of the nine transformants were recloned; three showed the anticipated gene disruption via Southern blot and a lack of mRNA production by RT-PCR (Sup Fig. 1) using Qiagen RNeasy and One-Step RT-PCR kits according to the manufacturers instructions. For Ase1B, we screened 128 transformants, identifying four initial candidates. Three of these transformants were subcloned and similarly showed an absence of detectable mRNA by RT-PCR.
For cell growth measurements, triplicate room temperature shaking cultures were counted at 24 h intervals; Growth curves were calculated and displayed with Microsoft Excel.
Light microscopy
WT AX-2 and Ase1− cells were grown and processed for immunofluorescence following methods described in [38, 46, 47]. The α-tubulin antibody has been previously described [46], DNA was labeled with Hoechst 33342, and the GFP-γ-tubulin construct is described in [48]. The full-length Ase1A polypeptide was imaged by appending GFP in frame between the dynein heavy chain promoter [49] and the amino-terminus of the Ase1A coding region, on a plasmid containing a G418 selectable marker (e.g. [50]). Cells were transformed as above and selected using G418 and GFP expression criteria. 3-D image stacks were collected on a DeltaVision microscope workstation with an Olympus UPlan 100X 1.35 N.A. objective lens. For analysis and presentation, raw stacks were deconvolved with a lens-specific point-spread function using softWoRx 2.5 (Applied Precision). Maximum intensity projections were prepared in Fiji (http://fiji.sc/Fiji), figures were assembled in Adobe Photoshop. Live cell recordings were obtained for WT and Ase1A− cells that had been transformed with a GFP-α-tubulin expression plasmid [17], GFP-DdCenH3 [51], or GFP-Ase1A. For imaging, cells were washed in phosphate buffer (20 mM KCl, 2.5 mM Na2HPO4, 2.5 mM Na2HPO4, 0.24 MgCl2, pH 6.4) and placed under agarose in humidified Rose chambers [18]. Time-lapse images were collected at specified intervals in DIC and fluorescence on a Nikon TE2000 microscope, using a PlanApo 100X 1.4 N.A. objective lens and an Orca-R2 camera (Hamamatsu) controlled by IP Lab software (BD Biosciences).
Centrosomes were tracked manually with the standard plugins in FIJI and the resultant coordinates imported into MS Excel or MATLAB for calculations and statistical analysis. To ensure that kymograms (Fig. 6) accurately reflect changes in distances between the centrosomes, time-lapse series were rotationally and translationally aligned at individual time points to make one of the centrosomes stationary and restrict the movements of the second centrosome to only one axis [52]. Maximal intensity projections were then generated for each time point.
Biochemistry
Nuclei were isolated by detergent extraction, filtration through a 5 μm mesh polycarbonate filter (Whatman, GE Healthcare), and sedimentation through a sucrose cushion, as described in [53]. Aliquots of the supernatant (cytosol) and pellet (nuclei) resuspended in the same volume as original lysate were loaded on a 7.5 % polyacrylamide gel, electrophoretically separated, and transferred to nitrocellulose as described [46]. Blots were probed with a polyclonal antibody to GFP (Abcam) and imaged in a BioRad Gel Doc system using chemiluminescent chemistry (Amersham, GE Healthcare).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1. Targeted disruption of Ase1 isoforms by homologous recombination. The schematics in A diagram the strategy used to target insertion of a blasticidin S resistance cassette and disrupt gene transcription of Ase1A and Ase1B. Arrows show position of PCR primers used to screen clones for targeted inserts. Figure inserts show RT-PCR reactions in WT and KO strains. While ample message can be detected in WT cells, no gene products are seen in the Ase1A− or B− strains. Panel B shows growth curves of WT, Ase1A− and Ase1B− cells. (TIFF 2850 kb)
Supplemental Fig. 2. Centromeres rapidly segregate during division in Ase1A− cells. Sequential frames (20 s interval) of a cell labeled with the centromere marker, GFP-DdCenH3 [51]. Centromere clusters separate here at a rate comparable to the fast centrosome movements quantified in Fig. 4. Bar = 5 μm. (TIFF 88 kb)
Supplemental Fig. 3. Amplitude Spectrum of intercentrosome distance in WT vs. Ase1A− cells. Quantitation of movement between centrosome pairs, as presented in Fig. 6. Distance and frequency of 11 WT and 14 Ase1A− pairs were measured using a Fourier transform in a MATLAB environment. The trendlines are fitted to a cubic polynomial Eq. 1 px = 107 nm. (TIFF 2653 kb)
Supplemental Fig. 4. Immunoblot of soluble Ase1A. Left panel shows Coomassie-stained PAGE lanes of supernatant and pellet following cell lysis and nuclear sedimentation. Middle and right panels show immunoblots from the same samples probed with an antibody against GFP. The left blot is from cells expressing the ~ 116 kDa GFP-Ase1A fusion; the right blot is from cells expressing ~ 95 kDa GFP-histone H3 like variant (Sup Fig. 2, [51]). The arrowhead points out a trace amount of Ase1A that is present in the supernatant, while the bulk of the polypeptide copellets with intact nuclei. The H3v1 samples are provided to control for potential nuclear breakage; no H3v1 fusion product is detected in the soluble fraction. (TIFF 10777 kb)
Supplemental Fig. 5. MT disorganization in kinesin-4− cells. The left two panels show interphase binucleate cells where the Dd kinesin-4 family isoform (kif8) has been deleted [38]. Note a similar disorganization to the MT patterns as in Ase1A deletion. The two right panels show mitotic cells, demonstrating that a proper spindle forms in the absence of this kinesin. Bar = 10 μm (TIFF 495 kb)
Acknowledgments
We appreciate the use of Wadsworth Center’s Molecular Genetics Core for DNA sequencing. We thank Dr. Wolfgang Nellen for kindly providing the GFP-CenH3 expression plasmid, Dr. Manfred Schliwa for the GFP-γ-tubulin construct, Valentin Magidson for general light microscopy assistance, and Dr. Vitali Sikirzhytski for help with statistical analysis. We are grateful to the efforts at http://dictybase.org to archive and annotate Dictyostelium sequence information. This work was supported in part by the NSF (MCB-1051612 to M.P.K, DBI-1062963 for REU support of K.I), and the NIH (GM59363 to A.K).
Compliance with ethical standards
Ethical standards
The experiments presented in this manuscript submitted for publication were performed in compliance with the current laws of the United States.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Bornens M. The centrosome in cells and organisms. Science. 2012;335(6067):422–426. doi: 10.1126/science.1209037. [DOI] [PubMed] [Google Scholar]
- 2.Holy TE, Dogterom M, Yurke B, Leibler S. Assembly and positioning of microtubule asters in microfabricated chambers. Proc Natl Acad Sci (USA) 1997;94(12):6228–6231. doi: 10.1073/pnas.94.12.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malikov V, Cytrynbaum EN, Kashina A, Mogilner A, Rodionov V. Centering of a radial microtubule array by translocation along microtubules spontaneously nucleated in the cytoplasm. Nat Cell Biol. 2005;7(12):1213–1218. doi: 10.1038/ncb1332. [DOI] [PubMed] [Google Scholar]
- 4.McNally FJ. Mechanisms of spindle positioning. J Cell Biol. 2013;200(2):131–140. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawe HR, Farr H, Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J Cell Sci. 2007;120(1):7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- 6.Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152(6):1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, López M, Vale RD, Jülicher F, Reck-Peterson SL, Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148(3):502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mavrakis M, Rikhy R, Lippincott-Schwartz J. Cells within a cell: insights into cellular architecture and polarization from the organization of the early fly embryo. Commun Integr Biol. 2009;2(4):313–314. doi: 10.4161/cib.2.4.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan J, Jensen CG, Jensen LCW, Bush M, Lloyd CW. The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules. Proc Natl Acad Sci (USA) 1999;96(26):14931–14936. doi: 10.1073/pnas.96.26.14931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loiodice I, Staub J, Setty TG, Nguyen NP, Paoletti A, Tran PT. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol Bio Cell. 2005;16(4):1756–1768. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mollinari C, Kleman J-P, Jiang W, Schoehn G, Hunter T, Margolis RL. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol. 2002;157(7):1175–1186. doi: 10.1083/jcb.200111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuyler SC, Liu JY, Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. J Cell Biol. 2003;160(4):517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koonce MP, Khodjakov A. Dynamic microtubules in Dictyostelium . J Muscle Res Cell Motil. 2002;23(7–8):613–619. doi: 10.1023/A:1024446821701. [DOI] [PubMed] [Google Scholar]
- 14.Duleh SN, Collins JTB, Pope RK. Morphological and functional analysis of Rac1B in Dictyostelium discoideum . J Electron Microscopy. 2005;54(6):519–528. doi: 10.1093/jmicro/dfi070. [DOI] [PubMed] [Google Scholar]
- 15.Fukui Y, De Lozanne A, Spudich JA. Structure and function of the cytoskeleton of a Dictyostelium myosin-defective mutant. J Cell Biol. 1990;110(2):367–378. doi: 10.1083/jcb.110.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gräf R, Euteneuer U, Ho T-H, Rehberg M. Regulated expression of the centrosomal protein DdCP224 affects microtubule dynamics and reveals mechanisms for the control of supernumerary centrosome number. Mol Biol Cell. 2003;14(10):4067–4074. doi: 10.1091/mbc.E03-04-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neujahr R, Albrecht R, Kohler J, Matzner M, Schwartz JM, Westphal M, Gerisch G. Microtubule-mediated centrosome motility and the positioning of cleavage furrows in multinucleate myosin II-null cells. J Cell Sci. 1998;111(9):1227–1240. doi: 10.1242/jcs.111.9.1227. [DOI] [PubMed] [Google Scholar]
- 18.Brito DA, Strauss J, Magidson V, Tikhonenko I, Khodjakov A, Koonce MP. Pushing forces drive the comet-like motility of microtubule arrays in Dictyostelium . Mol Biol Cell. 2005;16(7):3334–3340. doi: 10.1091/mbc.E05-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hestermann A, Gräf R. The XMAP215-family protein DdCP224 is required for cortical interactions of microtubules. BMC Cell Biol. 2004;5:24. doi: 10.1186/1471-2121-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonce MP, Kohler J, Neujahr R, Schwartz JM, Tikhonenko I, Gerisch G. Dynein motor regulation stabilizes interphase microtubule arrays and determines centrosome position. EMBO J. 1999;18(23):6786–6792. doi: 10.1093/emboj/18.23.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rehberg M, Kleylein-Sohn J, Faix J, Ho TH, Schulz I, Gräf R. Dictyostelium LIS1 is a centrosomal protein required for microtubule/cell cortex interactions, nucleus/centrosome linkage, and actin dynamics. Mol Biol Cell. 2005;16(6):2759–2771. doi: 10.1091/mbc.E05-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basu S, Fey P, Pandit Y, Dodson R, Kibbe WA, Chisholm RL. DictyBase 2013: integrating multiple Dictyostelid species. Nucleic Acids Res. 2013;41(D1):D676–D683. doi: 10.1093/nar/gks1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang W, Jimenez G, Wells NJ, Hope TJ, Wahl GM, Hunter T, Fukunaga R. PRC1: a human mitotic spindle–associated CDK substrate protein required for cytokinesis. Mol Cell. 1998;2:877–885. doi: 10.1016/S1097-2765(00)80302-0. [DOI] [PubMed] [Google Scholar]
- 24.Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T. The roles of fission yeast Ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell. 2005;16(3):1378–1395. doi: 10.1091/mbc.E04-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koonce MP, Samsó M. Overexpression of cytoplasmic dynein’s globular head causes a collapse of the interphase microtubule network in Dictyostelium . Mol Biol Cell. 1996;7(6):935–948. doi: 10.1091/mbc.7.6.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma S, Triviños-Lagos L, Gräf R, Chisholm RL. Dynein intermediate chain mediated dynein–dynactin interaction is required for interphase microtubule organization and centrosome replication and separation in Dictyostelium . J Cell Biol. 1999;147(6):1261–1274. doi: 10.1083/jcb.147.6.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verbrugghe KJC, White JG. SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr Biol. 2004;14(19):1755–1760. doi: 10.1016/j.cub.2004.09.055. [DOI] [PubMed] [Google Scholar]
- 28.Vernì F, Somma MP, Gunsalus KC, Bonaccorsi S, Belloni G, Goldberg ML, Gatti M. Feo, the Drosophila homolog of PRC1, is required for central-spindle formation and cytokinesis. Curr Biol. 2004;14(17):1569–1575. doi: 10.1016/j.cub.2004.08.054. [DOI] [PubMed] [Google Scholar]
- 29.Lee K-Y, Esmaeili B, Zealley B, Mishima M. Direct interaction between centralspindlin and PRC1 reinforces mechanical resilience of the central spindle. Nat Commun. 2015;6:7290. doi: 10.1038/ncomms8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aist JR, Berns MW. Mechanics of chromosome separation during mitosis in Fusarium (Fungi imperfecti): new evidence from ultrastructural and laser microbeam experiments. J Cell Biol. 1981;91:446–458. doi: 10.1083/jcb.91.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pellman D, Bagget M, Tu YH, Fink GR, Tu H. Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae . J Cell Biol. 1995;130(6):1373–1385. doi: 10.1083/jcb.130.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waters JC, Cole RW, Rieder CL. The force-producing mechanism for centrosome separation during spindle formation in vertebrates is intrinsic to each aster. J Cell Biol. 1993;122:361–372. doi: 10.1083/jcb.122.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho C-MK, Hotta T, Guo F, Roberson RW, Lee Y-RJ, Liu B. Interaction of antiparallel microtubules in the phragmoplast is mediated by the microtubule-associated protein MAP65-3 in Arabidopsis . Plant Cell. 2011;23(8):2909–2923. doi: 10.1105/tpc.110.078204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucas JR, Courtney S, Hassfurder M, Dhingra S, Bryant A, Shaw SL. Microtubule-associated proteins MAP65-1 and MAP65-2 positively regulate axial cell growth in etiolated Arabidopsis hypocotyls. Plant Cell. 2011;23:1889–1903. doi: 10.1105/tpc.111.084970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janson ME, Loughlin R, Loïodice I, Fu C, Brunner D, Nédélec FJ, Tran PT. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 2007;128(2):357–368. doi: 10.1016/j.cell.2006.12.030. [DOI] [PubMed] [Google Scholar]
- 36.Anderson CA, Eser U, Korndorf T, Borsuk ME, Skotheim JM, Gladfelter AS. Nuclear repulsion enables division autonomy in a single cytoplasm. Curr Biol. 2013;23(20):1999–2010. doi: 10.1016/j.cub.2013.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen PA, Groen AC, Loose M, Ishihara K, Wühr M, Field CM, Mitchison TJ. Spatial organization of cytokinesis signaling reconstituted in a cell-free system. Science. 2014;346(6206):244–247. doi: 10.1126/science.1256773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nag DK, Tikhonenko I, Soga I, Koonce MP. Disruption of four kinesin genes in Dictyostelium . BMC Cell Biol. 2008;9:21. doi: 10.1186/1471-2121-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bieling P, Telley IA, Surrey T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142(3):420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 40.Hu C-K, Coughlin M, Field CM, Mitchison TJ. Kif4 regulates midzone length during cytokinesis. Curr Biol. 2011;21:815–824. doi: 10.1016/j.cub.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su X, Arellano-Santoyo H, Portran D, Gaillard J, Vantard M, Thery M, Pellman D. Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat Cell Biol. 2013;15(8):948–957. doi: 10.1038/ncb2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian R, Ti S-C, Tan L, Darst SA, Kapoor TM. Marking and measuring single microtubules by PRC1 and kinesin-4. Cell. 2013;154(2):377–390. doi: 10.1016/j.cell.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glunčić M, Maghelli N, Krull A, Krstić V, Ramunno-Johnson D, Pavin N, Tolić IM. Kinesin-8 motors improve nuclear centering by promoting microtubule catastrophe. Phy Rev Lett. 2015;114(7):078103. doi: 10.1103/PhysRevLett.114.078103. [DOI] [PubMed] [Google Scholar]
- 44.Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23(16):3237–3248. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egelhoff TT, Titus MA, Manstein DJ, Ruppel KM, Spudich JA. Molecular genetic tools for study of the cytoskeleton in Dictyostelium. In: Vallee RB, editor. Methods in enzymology. Amsterdam: Academic Press; 1991. pp. 319–334. [DOI] [PubMed] [Google Scholar]
- 46.Koonce MP, McIntosh JR. Identification and immunolocalization of cytoplasmic dynein in Dictyostelium . Cell Motil Cytoskel. 1990;15(1):51–62. doi: 10.1002/cm.970150108. [DOI] [PubMed] [Google Scholar]
- 47.Tikhonenko I, Nag DK, Martin N, Koonce MP. Kinesin-5 is not essential for mitotic spindle elongation in Dictyostelium . Cell Motil Cytoskel. 2008;65(11):853–862. doi: 10.1002/cm.20307. [DOI] [PubMed] [Google Scholar]
- 48.Ueda M, Gräf R, MacWilliams HK, Schliwa M, Euteneuer U. Centrosome positioning and directionality of cell movements. Proc Natl Acad Sci USA. 1997;94(18):9674–9678. doi: 10.1073/pnas.94.18.9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koonce MP, Grissom PM, Lyon M, Pope T, McIntosh JR. Molecular characterization of a cytoplasmic dynein from Dictyostelium . J Eukaryot Microbiol. 1994;41(6):645–651. doi: 10.1111/j.1550-7408.1994.tb01528.x. [DOI] [PubMed] [Google Scholar]
- 50.Tikhonenko I, Magidson V, Gräf R, Khodjakov A, Koonce MP. A kinesin-mediated mechanism that couples centrosomes to nuclei. Cell Mol Life Sci. 2013;70:1285–1296. doi: 10.1007/s00018-012-1205-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dubin M, Fuchs J, Gräf R, Schubert I, Nellen W. Dynamics of a novel centromeric histone variant CenH3 reveals the evolutionary ancestral timing of centromere biogenesis. Nucleic Acid Res. 2010;38(21):7526–7537. doi: 10.1093/nar/gkq664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sikirzhytski V, Magidson V, Steinman JB, He J, Le Berre M, Tikhonenko I, Ault JG, McEwen BF, Chen JK, Sui H, Piel M, Kapoor TM, Khodjakov A. Direct kinetochore–spindle pole connections are not required for chromosome segregation. J Cell Biol. 2014;206(2):231–243. doi: 10.1083/jcb.201401090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulz I, Reinders Y, Sickmann A, Gräf R. An improved method for Dictyostelium centrosome isolation. Methods Mol Biol. 2006;346:479–489. doi: 10.1385/1-59745-144-4:479. [DOI] [PubMed] [Google Scholar]
- 54.Tikhonenko I, Nag DK, Robinson DN, Koonce MP. Microtubule-nucleus interactions in Dictyostelium discoideum mediated by central motor kinesins. Eukaryot Cell. 2009;8(5):723–731. doi: 10.1128/EC.00018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Targeted disruption of Ase1 isoforms by homologous recombination. The schematics in A diagram the strategy used to target insertion of a blasticidin S resistance cassette and disrupt gene transcription of Ase1A and Ase1B. Arrows show position of PCR primers used to screen clones for targeted inserts. Figure inserts show RT-PCR reactions in WT and KO strains. While ample message can be detected in WT cells, no gene products are seen in the Ase1A− or B− strains. Panel B shows growth curves of WT, Ase1A− and Ase1B− cells. (TIFF 2850 kb)
Supplemental Fig. 2. Centromeres rapidly segregate during division in Ase1A− cells. Sequential frames (20 s interval) of a cell labeled with the centromere marker, GFP-DdCenH3 [51]. Centromere clusters separate here at a rate comparable to the fast centrosome movements quantified in Fig. 4. Bar = 5 μm. (TIFF 88 kb)
Supplemental Fig. 3. Amplitude Spectrum of intercentrosome distance in WT vs. Ase1A− cells. Quantitation of movement between centrosome pairs, as presented in Fig. 6. Distance and frequency of 11 WT and 14 Ase1A− pairs were measured using a Fourier transform in a MATLAB environment. The trendlines are fitted to a cubic polynomial Eq. 1 px = 107 nm. (TIFF 2653 kb)
Supplemental Fig. 4. Immunoblot of soluble Ase1A. Left panel shows Coomassie-stained PAGE lanes of supernatant and pellet following cell lysis and nuclear sedimentation. Middle and right panels show immunoblots from the same samples probed with an antibody against GFP. The left blot is from cells expressing the ~ 116 kDa GFP-Ase1A fusion; the right blot is from cells expressing ~ 95 kDa GFP-histone H3 like variant (Sup Fig. 2, [51]). The arrowhead points out a trace amount of Ase1A that is present in the supernatant, while the bulk of the polypeptide copellets with intact nuclei. The H3v1 samples are provided to control for potential nuclear breakage; no H3v1 fusion product is detected in the soluble fraction. (TIFF 10777 kb)
Supplemental Fig. 5. MT disorganization in kinesin-4− cells. The left two panels show interphase binucleate cells where the Dd kinesin-4 family isoform (kif8) has been deleted [38]. Note a similar disorganization to the MT patterns as in Ase1A deletion. The two right panels show mitotic cells, demonstrating that a proper spindle forms in the absence of this kinesin. Bar = 10 μm (TIFF 495 kb)