Abstract
Protein-only RNase Ps (PRORPs) are a recently discovered class of RNA processing enzymes that catalyze maturation of the 5′ end of precursor tRNAs (pre-tRNAs) in Eukaryotes. PRORPs are found in the nucleus and/or organelles of most eukaryotic organisms. Arabidopsis thaliana is a representative organism that contains PRORP enzymes (PRORP1, PRORP2, PRORP3) in both its nucleus and organelles; PRORP2 and 3 localize to the nucleus, PRORP1 to the chloroplast and the mitochondria. Apart from their identification, almost nothing is known about the structure and function of PRORPs that act in the nucleus. Here we use a combination of biochemical assays and X-ray crystallography to characterize A. thaliana PRORP2. We solved the crystal structure of PRORP2 (3.2 Å) revealing an overall V-shaped protein and conserved metallonuclease active site structure. Our biochemical studies indicate that PRORP2 requires Mg2+ for catalysis and catalyzes the maturation of nuclear encoded substrates up to 10-fold faster than mitochondrial encoded precursor nad6 t-element under single turnover conditions. We also demonstrate that PRORP2 preferentially binds pre-tRNAs containing short 5′ leaders and 3′ trailers; however, leader and trailer length do not significantly alter the observed rate constants of PRORP2 in single turnover cleavage assays. Our data provide a biochemical and structural framework to begin understanding how nuclear localized PRORPs recognize and cleave their substrates.
Keywords: pre-tRNA, RNase P, RNA processing, metal dependence, t-element
Graphical abstract
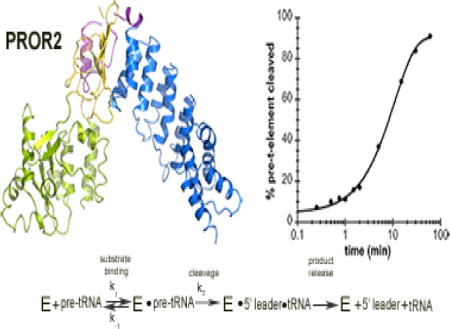
Introduction
RNase P catalyzes the removal of 5′ leader sequences from precursor tRNAs (pre-tRNAs) and is found in all three kingdoms of life. The canonical form of RNase P is a ribozyme associated with one to ten proteins depending on the organism1; 2. However, a second form of the enzyme, called protein only RNase P (PRORP), comprised solely of protein and lacking an RNA component has been discovered in Eukaryotes3; 4; 5. It is noteworthy that none of the known protein components of the “ribozyme-based” RNase P have any homology to PRORPs6. Beyond the bioinformatic identification of PRORPs in a wide variety of organisms3; 7; 8 little is known about how they recognize substrates. PRORPs can act either as single protein enzymes or as the catalytic subunit of multi-protein RNase Ps. Metazoans typically possess both RNA and protein based RNase Ps; retaining the traditional RNase P ribozyme in their nuclei, and using a PRORP transiently associated with MRPP1 and 2 (Mitochondrial RNase P Protein 1 and 2) in their mitochondria4. In contrast, land plants, exemplified by A. thaliana3; 9, and other unicellular Eukaryotic organisms, such as Trypanosoma bruceii5, appear to lack any RNA-based RNase P and instead utilize single protein versions of PRORP to catalyze the 5′ end cleavage of pre-tRNAs in both their nucleus and organelles. The single protein enzymes are related to the catalytic PRORP subunit of the multi-protein RNase Ps found in metazoan mitochondria and provide the simplest system for understanding how this new class of RNA processing enzymes function.
A. thaliana contains three differentially localized PRORPs (PRORP1, PRORP2, PRORP3) that share a high level of protein sequence identity (48–80%). PRORP1 is found in the mitochondria and the chloroplast, and PRORP2 and PRORP3 are present in the nucleus9. Thus far, PRORP1 is the best-characterized PRORP from any organism and the only PRORP for which there is a crystal structure where all of its domains are intact8; 10; 11. A recent study evaluating the in vitro pH profile and metal dependence of PRORP1 cleavage activity proposed that, like the “ribozyme-based” RNase P, PRORP1 uses a metal-bound water nucleophile to cleave pre-tRNA substrates12. The precise molecular recognition strategies used by PRORPs remain largely unknown. The crystal structure of PRORP1 revealed three distinct domains forming an overall V-shaped molecule (pentatricopeptide repeat (PPR) domain, central domain and metallonuclease domain)8. The PPR domain, consisting of five PPR motifs, is one of the putative binding sites for pre-tRNA. Canonical PPR motifs are ~35 amino acid long sequences that form two antiparallel α-helices joined by a short loop known to be involved in RNA binding13. Truncation of the four PRORP1 N-terminal PPR motifs reduces the ability of the enzyme to bind and cleave pre-tRNAs8. The Nedd4-BP1, YacP nuclease (NYN) domain (metallonuclease domain) houses the active site that catalyzes 5′ pre-tRNA cleavage. The functional importance of this domain has been validated in mutagenesis studies where mutations of conserved aspartates to alanines resulted in catalytically inactive PRORP18; 14. The central domain holds the PPR and the metallonuclease domain together in such a way that the two functionally relevant domains face towards each other. The central domain also harbors a structurally important zinc-binding site. Several studies have shown that recombinant PRORP1 is able to bind and cleave pre-tRNAs derived from A. thaliana and E. coli3; 8; 9; 15. Meanwhile, little is known about the nuclear localized PRORPs found in A. thaliana or other organisms. Recently, it was shown that A. thaliana PRORP2 and PRORP3 can catalyze the maturation of the 5′ ends of two nuclear pre-tRNA substrates in vivo, confirming their predicted roles as RNase Ps in the nucleus9. However, there are no structural and limited mechanistic data available for nuclear localized PRORPs.
While the investigations of PRORP1 provide an important first step towards understanding how PRORPs work8; 12; 15; 16 the study of a single enzyme cannot reveal how PRORPs generally function; comparison of the structure and function of multiple proteins is necessary to understand any given class of enzymes. Here, we present the first x-ray structure and biochemical characterization of a nuclear localized PRORP, A. thaliana PRORP2. Our crystal structure of PRORP2 (3.2 Å) provides the only structural information available on any nuclear localized PRORP and reveals an overall structure that, while similar to the mitochondria and chloroplast localized PRORP1, exhibits a more open conformation. Thermodynamic binding studies provide insight into the differences between proteinaceous and ribozyme RNase Ps in recognition of the 5′ and 3′ end of pre-tRNAs. Furthermore, our work demonstrates that PRORP2 exhibits a modest preference against catalyzing maturation of the mitochondrial encoded nad6 t-element, suggesting that there are subtle but important distinctions in how differentially localized PRORPs recognize and process a sub-set of substrates. The data presented here provide a basis for understanding how nuclear localized PRORPs function, and allow us to begin understanding how PRORPs act as a new class of RNA processing proteins to bind and cleave their substrates.
Results
PRORP2 uses Mg2+ as a catalytic co-factor
Given that the only mechanistic information available on PRORPs comes from studies of PRORP112; 15 little is known about how the nuclear localized PRORPs recognize and mature their substrates. Here, we begin by assessing how PRORP2 catalyzes pre-tRNA cleavage using single turnover kinetic assays. In these assays, we measure the rate constants (kobs) for single turnover reactions with limiting substrate (nuclear pre-tRNAGly, < 100 nM) and saturating enzyme concentrations (5 μM) (Figure 1A, 1B). These conditions were selected because the kobs values are independent of PRORP2 concentration (Figure 1B). The kobs values report on steps in the mechanism that occur after substrate binding and before, and including, pre-tRNA cleavage (Scheme 1).
Figure 1. Metal dependence of PRORP2.
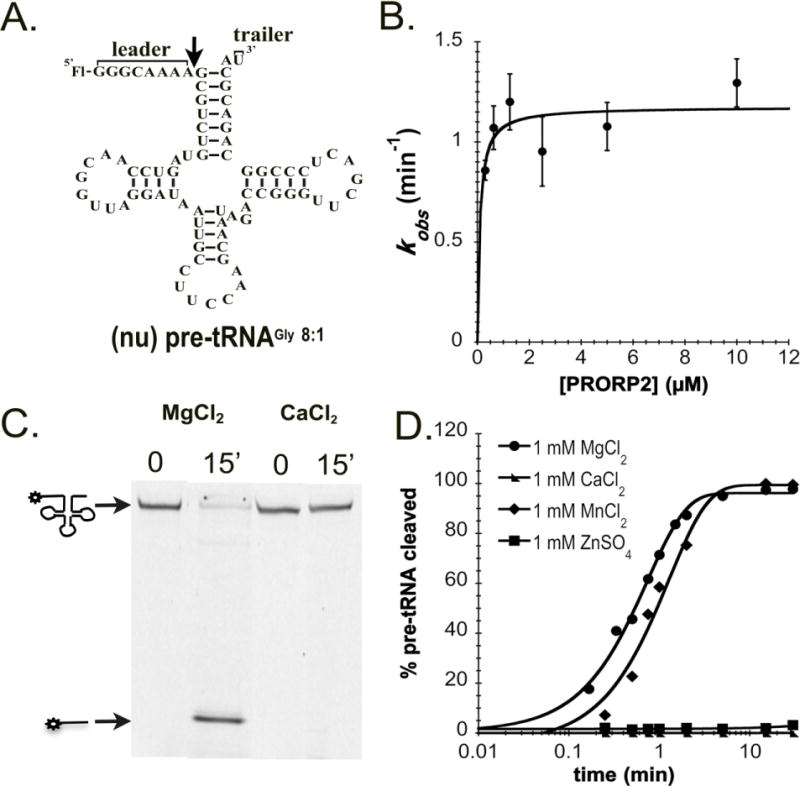
A. Proposed secondary structure of nuclear pre-tRNAGly 8:1 used in our in vitro assays. Pre-tRNAs are 5′ fluorescently labeled by fluorescein (Fl). The 5′ leader and 3′ trailer are noted, and the black arrow indicates the cleavage site. B. The dependence of the single turnover cleavage rate constant (kobs) on the PRORP2 concentration for (nu)pre-tRNAGly 8:1. C. Representative denaturing PAGE gel displaying the cleavage activity of PRORP2 for (nu)pre-tRNAGly 8:1 after a 15 min incubation with buffer containing 1 mM MgCl2 or CaCl2. Cleavage is observed in the presence of MgCl2 but not CaCl2 under standard reaction conditions. D. Representative timecourses for the cleavage of a 5′ “fluorescein-labeled” (nu)pre-tRNAGly 8:1 substrate catalyzed by PRORP2 under standard single turnover conditions in the presence of MgCl2 (circle), MnCl2 (diamond), ZnCl2 (square) and CaCl2 (triangle).
Scheme 1. Minimal kinetic mechanism for PRORP2.

PRORP2 (E) binds pre-tRNA to form an enzyme-substrate complex. The substrate is then cleaved (k2), resulting in an enzyme-tRNA-5′ leader complex. The products of the reaction (tRNA and 5′ leader) are subsequently released.
Since all PRORPs contain a conserved metallonuclease domain we first asked if PRORP2, like PRORP1, uses a divalent metal ion as its catalytic co-factor. We measured the single-turnover rate constants for PRORP2 cleaving a nuclear pre-tRNAGly with an 8 nucleotide long 5′ leader and a 1 nucleotide long 3′ trailer ((nu)pre-tRNAGly 8:1) (Figure 1A) in the presence of four distinct divalent metal ions commonly found in metallonuclease active sites (Mg2+, Mn2+, Ca2+, Zn2+)8; 17; 18; 19. PRORP2 is active in the presence of Mg2+ and Mn2+ but not Ca2+ or Zn2+ (Figure 1C, 1D, Supplemental Figure 1) similar to PRORP18. Given that PRORP2 cleaves substrates with comparable single turnover rate constants in the presence of Mg2+ and Mn2+ (kobs,Mg2 = 1.4 ± 0.1 min−1, kobs,Mn2+ = 0.8 ± 0.1 min−1, Figure 1D) and Mg2+ is at least 5-fold more abundant in the cell20 our data suggest that Mg2+ is the likely catalytic metal ion in vivo.
PRORP2 preferentially binds pre-tRNA substrates with short 5′ leader and 3′ trailer sequences
The traditional ribozyme form of RNase P has a strong preference for binding and cleaving pre-tRNAs with particular 5′ leader and 3′ trailer lengths21; 22. However, it is unknown if the length of the 5′ leader and 3′ trailer are also important determinants of PRORPs binding pre-tRNAs. To investigate this question we measured the dissociation constants (KD) for PRORP2 binding 5′ fluorescently labeled nuclear pre-tRNAGly substrates with varying 5′ leader and 3′ trailer lengths ((nu)pre-tRNAGly 23:1, (nu)pre-tRNAGly 13:1, (nu)pre-tRNAGly 8:1, (nu)pre-tRNAGly 23:10, (nu)pre-tRNAGly 23:5, (nu)pre-tRNAGly 23:1) (Table 1). KD values were determined using a previously described fluorescence polarization assay developed for PRORP1 in which the concentration of 5′ “fluorescein-labeled” substrates is held constant (20 nM) and the enzyme is titrated (0–1000 nM)8. Since PRORP2 is incapable of cleaving pre-tRNA in the presence of Ca2+ (Figure 1C, 1D) and previous studies demonstrated that PRORP1 binds pre-tRNAs in the presence of Ca2+ 8, our KD values were measured in the presence of Ca2+ instead of Mg2+.
Table 1.
Dissociation (KD) and single-turnover rate constants (kobs) for PRORP2 with (nu)pre-tRNAGly substrates containing varying 5′ leader and 3′ trailer lengths. The data were measured in buffers containing 30 mM MOPS pH 7.8, 150 mM NaCl, 1 mM DTT and 1 mM MgCl2 (kobs) or CaCl2 (KD). The standard error values displayed were obtained from at least three independent experiments.
| Nuclear Substrate | 5′ leader sequence | 3′ trailer sequence | KD (nM) | kobs (min−1) |
|---|---|---|---|---|
| pre-tRNAGly 23:10 | GGGUAUUUGCUCGAUAUGCAAAA | UUUAUAUUUU | 118 ± 26 | 0.7 ±0.1 |
| pre-tRNAGly 23:5 | GGGUAUUUGCUCGAUAUGCAAAA | UUUAU | 52 ± 12 | 1.0 ±0.1 |
| pre-tRNAGly 23:1 | GGGUAUUUGCUCGAUAUGCAAAA | U | 17 ± 5 | 0.7 ±0.1 |
| pre-tRNAGly 13:1 | GGGAUAUGCAAA | U | 6 ± 1 | 0.7 ±0.1 |
| pre-tRNAGly 8:1 | GGGCAAAA | U | 3 ± 1 | 1.1 ±0.1 |
We found that PRORP2 preferentially binds substrates with short 3′ trailers. The affinity of PRORP2 for a (nu)pre-tRNAGly substrate containing a 23 nucleotide long 5′ leader is enhanced by 7-fold when the 3′ trailer length is shortened from 10 to 1 nucleotides (Figure 2A, Table 1). Additionally, PRORP2 prefers to bind substrates with shorter (< 23 nucleotide) 5′ leaders. When the 5′ leader length of (nu)pre-tRNAGly substrate with a 1 nucleotide 3′ trailer is reduced from 23 to 8 nucleotides, the KD decreases by 5-fold (Figure 2A, Table 1). Thus, PRORP2 has higher affinity for substrates with short 3′ trailer and 5′ leader.
Figure 2. PRORP2 discriminates betweeen binding, but not cleaving, substrates with varied 5′ leader and 3′ trailer lengths.
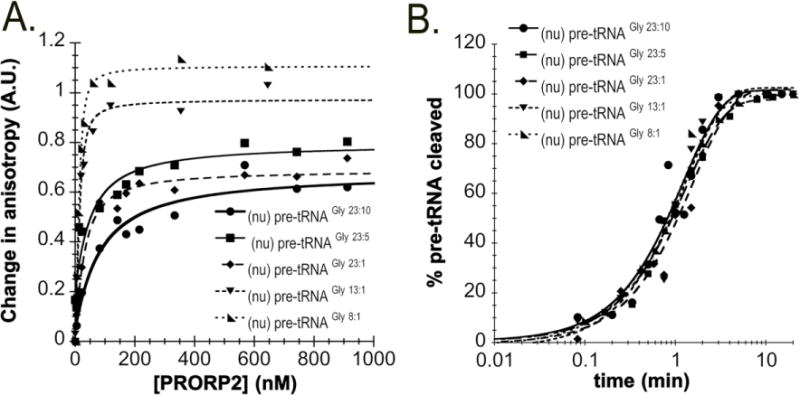
A. Representative plots of fluorescent polarization assays performed at 25 °C in 30 mM MOPS pH 7.8, 150 mM NaCl, 1mM DTT and 6 mM CaCl2 with varied PRORP2 (0–1000 nM) and 20 nM 5′ fluorescently labeled: (nu)pre-tRNAGly 23:10 (closed circle), (nu)pre-tRNAGly 23:5 (closed square), (nu)pre-tRNAGly 23:1 (diamond), (nu)pre-tRNAGly 13:1 (closed upside down triangle), and (nu)pre-tRNAGly 8:1 (right triangle). B. Representative single turnover assays using standard assay conditions with 5 μM PRORP2 and 50 nM nuclear fluorescently labeled (nu)pre-tRNAGly 23:10 (closed circle), (nu)pre-tRNAGly 23:5 (closed square), (nu)pre-tRNAGly 23:1 (diamond), (nu)pre-tRNAGly 13:1 (closed upside down triangle), and (nu)pre-tRNAGly 8:1 (right triangle).
We also measured the kobs values in single turnover assays for nuclear pre-tRNAGly substrates with varied 5′ leader and 3′ trailer lengths ((nu)pre-tRNAGly 23:1, (nu)pre-tRNAGly 13:1, (nu)pre-tRNAGly 8:1, (nu)pre-tRNAGly 23:10, (nu)pre-tRNAGly 23:5, (nu)pre-tRNAGly 23:1). The kobs values are comparable for all five of these substrates (kobs = 0.7–1 min −1) (Figure 2B, Table 1) suggesting that the leader and trailer lengths do not affect stabilization of the transition state. Additionally, we measured the kobs values for PRORP1 cleaving the (nu)pre-tRNAGly 23:1 and (nu)pre-tRNAGly 8:1 substrates (kobs = 1.2 ± 0.06 min−1 and 1.2 ± 0.1 min−1, respectively); PRORP1 and PRORP2 cleave both substrates with comparable rate constants.
PRORP2 inefficiently cleaves mitochondrial encoded nad6 t-element
Next, we assessed if PRORP2 can recognize and cleave other substrates encoded in the mitochondria using single turnover kinetic assays. For this purpose, we measured the kobs values for PRORP1 and PRORP2 cleaving the mitochondria specific nad6 t-element possessing a 14 nucleotide long 5′ leader and 6 nucleotide long 3′ trailer sequence ((mt) pre- nad6 t-element14:6, Figure 3A); PRORP1 was previously shown to cleave this substrate3. T-elements are encoded near, and in the same transcriptional orientation as, the 5′ or 3′ ends of many mRNAs and rRNAs in the mitochondrial DNA of plants23. These elements assume tRNA-like secondary structures and have been proposed to act as signals for endonucleic cleavage catalyzed by PRORPs3. The (mt) pre-nad6 t-element14:6 is proposed to fold into a structure much like a tRNA, however it differs significantly from a canonical pre-tRNA substrate because it lacks an anti-codon stem loop (Figure 3A). PRORP1 cleaves the (mt) pre-nad6 t-element14:6 with rate constants similar to other substrates (kobs = 1.5 ± 0.2 min−1)8; 12 in contrast to PRORP2, which exhibited a ~10-fold slower single-turnover rate constant (kobs = 0.13 ± 0.02 min−1, Figure 3B). Furthermore, PRORP2 binds the precursor nad6 t-element significantly less tightly (KD= 615 ± 100) than the nuclear encoded precursor tRNAGly substrates. This suggests that differentially localized A. thaliana PRORPs exhibit some specificity for binding and cleaving a subset of non-canonical substrates.
Figure 3. PRORP2 cleaves mitochondrial pre-nad6 t-element.
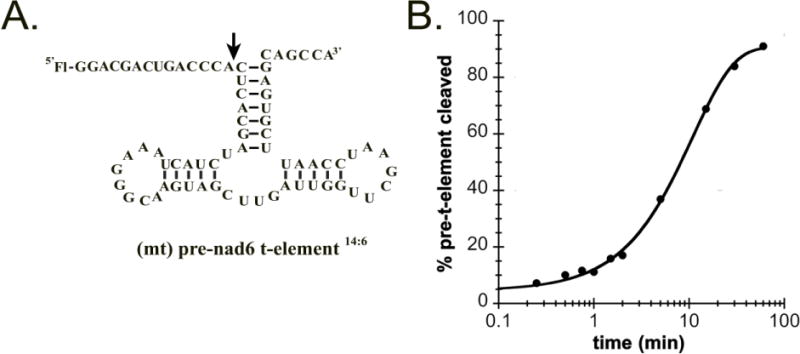
A. Proposed secondary structure of the (mt)pre-nad6 t-element14:6 used in our in vitro assays. B. Percentage of the (mt)pre-nad6 t-element14:6 cleaved as a function of time under single turnover conditions (50 nM 5′ fluorescein end labeled (mt)pre-nad6 t-element14:6, 5 μM PRORP2, 25°C, 30 mM MOPS pH 7.8, 150 mM NaCl, 1 mM DTT and 1 mM MgCl2).
PRORP1 and PRORP2 share overall structural characteristics
The existence of three different PRORPs in A. thaliana that localize to distinct cellular compartments provides a unique opportunity to investigate general PRORP characteristics by comparing their structural differences and similarities. We solved a 3.2 Å crystal structure of full-length nuclear PRORP2 (Figure 4A, PDB ID 5DIZ) by molecular replacement based on our previously determined 1.98 Å structure of the mitochondrial and chloroplast Δ76 PRORP18 (Figure 4B, PDB ID 4G23, Table 2). In PRORP1, where the N-terminal region is longer than that of PRORP2 (Supplemental Figure 3) due to the presence of an organellar localization sequence, electron density is visible from the 95th residue. This density corresponds to the beginning of the first helix of the PPR domain. Similarly, in PRORP2 electron density is seen from the 28th residue in the beginning of the PPR domain. Since we could map neither the first 28 residues of PRORP2 nor the residues between the localization signal and the 94th residue in PRORP1, we predict that these regions may be structurally disordered. It was previously suggested that this region might be functionally important based on reduced activity of truncated versions of PRORP114. To test if this region is indeed important for the catalytic activity of PRORP2, we measured the single-turnover rate constants for PRORP2 containing truncations at the N- tand C-termini (residues 21–515). This mutant PRORP2 correlates well with Δ76 PRORP1 in sequence length since it starts at approximately the same residue and lacks the C-terminal extension of PRORP2 (Supplemental Figure 2.). We find that truncated 21–515 PRORP2 cleaves the (nu)pre-tRNAGly 8:1 substrate (data not shown) with a single-turnover rate constant similar to that of wild type full-length PRORP2. This suggests that the truncated residues (including the N-terminal disordered region and the PRORP2 specific extension) are not important for the substrate binding and/or cleavage in vitro (Scheme 1).
Figure 4. Comparison of PRORP1 and PRORP2 crystal structures.
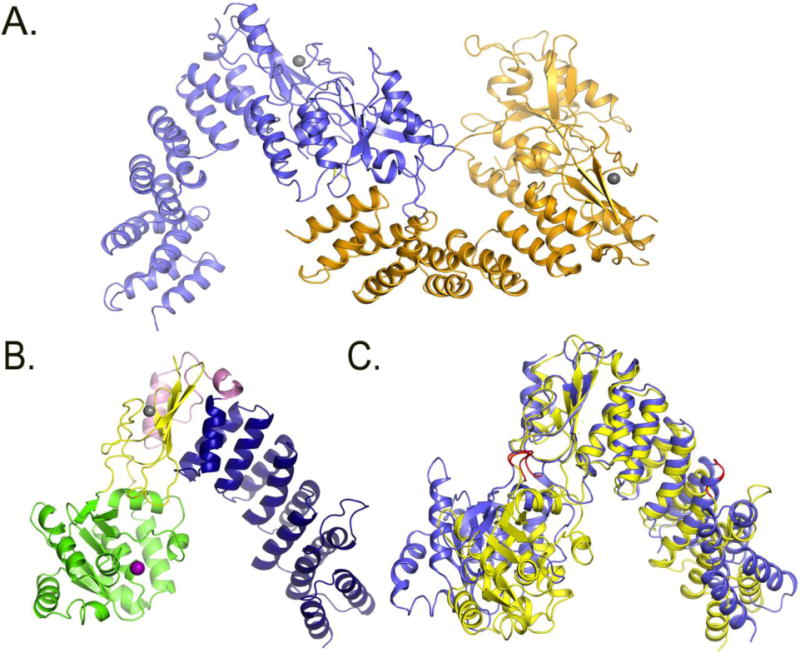
A. Crystal structure of the two monomers of PRORP2 in the asymmetric unit. B. Crystal structure of PRORP1. blue: PPR domain, yellow: central domain, Pink: A. thaliana specific insertion, green: metallonuclease domain, Grey: zink ion, Purple: modeled magnesium ion. C. PRORP1 and PRORP2 superimposition based on the central domains. The structure of PRORP2 (blue) is in an “open” conformation, while PRORP1 (yellow) is in a “closed” conformation. The predicted PRORP2 mechanical hinges are shown in red in the metallonuclease domain and the 3rd PPR motif.
Table 2. Crystallographic data.
Data processing and refinement statistics.
| Protein | PRORP2 |
|---|---|
| Data Collection | |
| Space Group | P1 |
| Cell Dimensions | |
| a, b, c (Å) | 70.0, 77.0, 80.1 |
| α, β, γ (°) | 72.7, 64.1, 77,8 |
| Wavelength (Å) | 1.033 |
| Resolution (Å) | 50–3.20, (3.31–3.20) |
| Rsym (%) | 5.9 (65.4) |
| I/σI | 8.7 (1.4) |
| Completeness (%) | 99.1 (98.6) |
| Redundancy | 2.2 (2.2) |
|
| |
| Refinement | |
| Resolution (Å) | 45.6–3.2 |
| No. reflections | 23,241 |
| Rwork/Rfree | 0.228/0.272 |
| No. atoms | |
| Protein | 7493 |
| Water | 5 |
| Zn | 2 |
| B-factors | |
| Protein | 123.5 |
| Water | 66.8 |
| Zn | 127.8 |
| R.m.s Deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 0.970 |
| Ramachandran plot (%) | |
| Favored/allowed/outliers | 93.70/5.66/0.64 |
| MolProbity Score | 1.23 (100th percentile) |
| Protein Data Bank code | 5DIZ |
PRORP2 shares the overall V-shaped structure of PRORP1 but has an “open” conformation relative to PRORP1 (Figure 4C). Analysis of PRORP1 and PRORP2 structures by the DynDom protein motion server24; 25 predicts that both proteins form three rigid bodies: the metallonuclease domain, the first five PPR α-helices, and the central domain with the C-terminal portion of the PPR domain. When the central domains of PRORP1 and PRORP2 are superimposed, the first 5 N-terminal helices are rotated by 34.8° and the metallonuclease domain is rotated by 44.5° in PRORP2 relative to PRORP1. Potential flexible hinges are present between the metallonuclease and central domain (PRORP2 residues 291–293 and 480–481, and PRORP1 residues 354–56 and 534–35) and in the intra-motif loop of the third PPR motif in the PPR domain (PRORP2 residues 122–123, and PRORP1 residues 189–190) (Figure 4C).
Since macromolecules can undergo many different conformational changes we performed normal mode analysis for PRORP1 and PRORP2 to predict the most probable large-scale functional motions using the ElNemo26 and iMods27 normal mode analysis servers. Normal mode analysis further indicates the presence of flexible regions in the PPR domain and between the metallonuclease and central domain. We propose that these potential flexible hinges could facilitate the motion of the metallonuclease domain and the PPR domain towards each other, potentially playing a role in accommodating substrates.
PRORP2 is a dimer in crystallo and monomeric in solution
PPR motif containing proteins can exist as homodimers, exemplified by HCF15228 and PPR1029; 30, or monomers, such as PPR4 and PPR529; 30; 31; 32. In our crystal structure PRORP2 appears as a homodimer. This is in contrast to PRORP1, which is a monomer in both the crystal and solution. This disparity raises the question whether PRORP2 forms a dimer in the crystal as a consequence of its packing or if its dimerization is physiologically relevant. The PRORP2 homodimer in the crystal has an unusual inter-domain interaction in which the first PPR motif is inserted into the nuclease active site (Figure 4A). We performed interdomain surface analysis to provide insight into the physiological relevance of this interaction using the PISA webserver33. The dimerization interface surface for PRORP2 was 790 Å2 however the Complex Formation Significance Score (CFSS) of zero indicated that PRORP2 dimerization is unlikely in vivo.
Nonetheless, we tested if residues involved in dimerization in the crystal structure are important for catalysis. Our structure of PRORP2 suggests that dimerization may be mediated via an interaction between the K42 residue in the first PPR domain of one PRORP2 monomer and the metallonuclease domain active site residues D422 and D421 in an adjacent PRORP2 monomer (Figure 5A). We speculated that if dimerization is important for PRORP2 function then disruption of this putative interaction might alter the activity of PRORP2. However, the K42A mutation does not change the cleavage rate constant of PRORP2 for (nu)pre-tRNAGly 8:1 (kobs,wild-type = 0.8 ± 0.1 min−1 and kobs,K42A = 0.8 ± 0.03 min−1) (Figure 5B). Lastly, we directly tested the dimerization state of PRORP2 in solution using Analytical Ultracentrifugation (AUC) with 1 and 10 mg/mL PRORP2. Even at high concentrations of PRORP2 only a single peak representing the monomeric form of PRORP2 is observed at 3.6S (Figure 5C). Analysis of this peak suggests the presence of a 58 kDa protein in solution, which correlates well with the 60 kDa molecular weight of the PRORP2. Our results indicate that PRORP2 is monomeric in solution and that the observed dimerization of PRORP2 in the asymmetric unit is due to crystal packing without in vivo relevance.
Figure 5. PRORP2 is a monomer in solution.
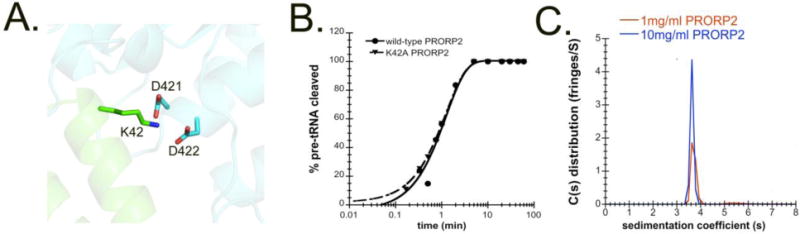
A. Residue K42 in the PPR domain and active site residues of two adjacent in crystallo PRORPs (residues D421 and D422). B. Representative plot of 5 μM K42A PRORP2 cleaving 50 nM (nu)tRNAGly8:1 in single turnover cleavage assays under standard reaction conditions. C. Analytical Ultracentrifugation experiment with 1 and 10 mg/ml PRORP2.
PPR domains of PRORP1 and PRORP2 exhibit structural differences
PPR motifs are found in all plants and are comprised of ~35 amino acid long sequences forming a helix-turn-helix arrangement (α-hairpin). These motifs can vary in length and their sequences are highly degenerate. The PPR domain of PRORP2 consists of five and a half PPR motifs corresponding to eleven consecutive α-helices similar to PRORP1. Based on the available structures of PPR motif containing proteins8; 29; 30; 34; 35; 36 sequential PPR motifs packing in a parallel fashion results in a spiral of anti-parallel α-helices that form a right-handed superhelical structure. The two helices in a PPR motif are referred to as helix A and helix B, with all odd-numbered helices A in a superhelical PPR arrangement forming an inner concave surface while all even-numbered helices B are facing outwards and form the convex surface. The PPR domains of PRORP1 and PRORP2 share high amino acid identity (~45 %, Supplemental Figure 3) and structural homology. As expected with paralogous proteins37 most of the differences between PRORP1 and PRORP2 are localized in the loop regions connecting the well-conserved structural elements. Example differences include the observation that the intra-domain loop of the second PPR motif in A. thaliana PRORPs is eight amino acids longer than in a canonical PPR repeat (Supplemental Figure 4A), and the presence of a two amino acid insertion in the third motif of the PRORP2 intradomain loop that is missing in PRORP1.
Superimposition of each individual equivalent PPR motif from PRORP1 and PRORP2 shows little variation in their structures (RMSD values = 0.58–0.68 Å) and in the absolute values of the angles between the two antiparallel α-helices (intra-PPR motif A-B angles) as calculated by PROMOTIF38 (Supplemental Table 1). However, superimposition of the PPR domains of PRORP1 and PRORP2 in their entirety (and not just as individual motifs) no longer demonstrates a high degree of overlap between the structures (RMSD = 2.94 Å). While there is little variation in the angles between sequential PPR motifs (inter-PPR motif angles A-A′), closer examination reveals that the inter-PPR motif angle between helices 5 and 6 in PRORP1 is 153.1°, in contrast to the corresponding angle in PRORP2 of −166.5°. Furthermore, PRORP2 has a greater degree of curvature compared to PRORP1 (Figure 6A, Supplemental Table 1) when only the 5th helix of PRORP1 and PRORP2 PPR domains are aligned. The global rearrangement observed in the PPR domains of the two PRORPs is not due to differences between the intra-PPR and inter-PPR motif angles since these are equally and globally distributed among all PPR motifs, but rather results from differences between the orientation of helices 5 and 6 in the third PPR motif. This difference in orientation between helices 5 and 6 in the two proteins is shown in Figure 5A, which displays how PRORP2 helices 4 and 6 on the convex surface have moved away from each other by more than 2 Å compared to the equivalent helices in PRORP1. We speculate that this rearrangement between helices 5 and 6 may be indicative of a flexible connecting loop.
Figure 6. Comparison of the PPR, central and metallonuclease domains of PRORP1 and PRORP2.
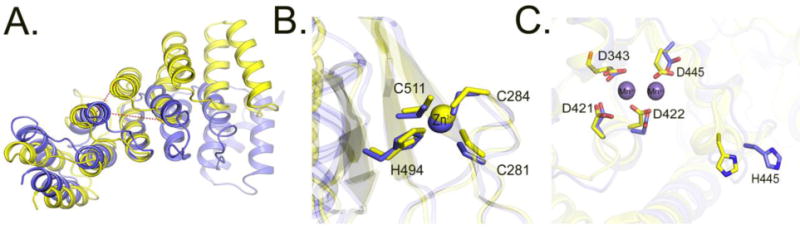
A. Superimposition of the 5th PPR motif of PRORP1 and PRORP2. Dotted red lines indicate differences in distance between the same residues in the 4th and 6th PPR helices in PRORP1 and PRORP2. B. Superimposition of the central domains of PRORP1 and PRORP2. Residues numbering correspond to the conserved amino acids in PRORP2. C. Superimposition of the active sites of PRORP1 (yellow) and PRORP2 (blue). Residue numbers correspond to conserved amino acids in PRORP2. Positions of Mn2+ ions are depicted based on the structure of PRORP1.
We performed normal mode analysis of PRORP1 and PRORP2 independently using the iMod and DynDom servers24; 25; 27. Our analyses indicate that the PPR domain displays a potential mechanical hinge between the two helices 5 and 6 of the PPR3 motif in both PRORPs (PRORP2 residues 122–123, and PRORP1 residues 190–191). This would permit motion between two rigid domains (where the first domain is defined by the 5 first helices and second domain is comprised of the subsequent 6 helices) and is consistent with our observed structural differences between helices 5 and 6. We hypothesized that such a motion might be relevant to catalysis and tested the potential contribution of the first three PPR motifs to PRORP function by measuring the catalytic activity of an N-terminal truncated version of PRORP2 (Δ141) lacking the first three PPR motifs. This mutant cleaves (nu)pre-tRNAGly 8:1 with a single turnover rate constant (kobs < 0.001 min −1) that is over 100-fold slower than wild-type PRORP2 (Table 3) under conditions where wild-type PRORP2 is saturating. To test if Δ141 PRORP2 cleaves slowly under these conditions due, at least in part, to an inability to bind substrate tightly we measured the KD of this mutant for (nu)pre-tRNAGly 8:1. We find that the affinity of the mutant for (nu)pre-tRNAGly 8:1 is over 58-fold weaker (KD > 1 μM) than the wild-type enzyme. Our data indicate that the first three PPR motifs of PRORP2 contribute significantly to substrate binding.
Table 3. Single turnover rate constants for PRORP2 cleaving (nu)pre-tRNAGly 8:1.
The reactions were perfomed at 25°C in the presence of 30 mM MOPS pH 7.8, 150 mM NaCl, 1 mM DTT and 1 mM MgCl2 with 50 nM 5′ “fluorescein-labeled” (nu)pre-tRNAGly 8:1 and 5 μM PRORP2. The kobs values and standard errors reflect at least three independent experiments.
| PRORP2 | kobs (min−1) |
|---|---|
| Wild type | 1.1 ± 0.1 |
| D393A | < 0.001 |
| D421A | < 0.001 |
| D422A | < 0.001 |
| D440A | < 0.001 |
| H445A | 0.02 ± 0.004 |
| Δ141 | < 0.001 |
Central domain of PRORP2 harbors a conserved structural zinc binding site
The central domain of PRORP2 is sandwiched between the PPR and metallonuclease domain and harbors a zinc-binding motif that is a conserved in all of the PRORPs. The Zn2+ binding motif observed in our PRORP2 structure is comprised of four conserved residues (C281, C284, H494 and C511) that coordinate a Zn2+ ion. As in PRORP1, Zn2+ binding appears to be important for structure, as the metal is distal from the enzyme active site and the Zn2+ does not directly participate in substrate cleavage. The central domain interacts with the PPR domain through a four-stranded antiparallel β-sheet and is anchored to the metallonuclease domain through two long loops similar to that of PRORP1 (Figure 6B). Overall, the structure of the central domain is highly similar between PRORP1 and PRORP2 (RMSD = 0.644 Å).
Active sites in the metallonuclease domains of A. thaliana PRORPs are conserved
The metallonuclease domains of PRORP1 and PRORP2 are nearly structurally identical (overall RMSD= 0.956 Å, Figure 4C and 6C) based on their superposition. The few differences between PRORP1 and PRORP2 metallonuclease domains occur primarily in loop regions (e.g. the connecting loop between α-helices (PRORP2 residues 349–358, PRORP1 residues 403–411)) that possess different conformations (Supplemental Figure 4B)). In the structure of PRORP1, two Mn2+ ions were observed in the active site8 when Mn2+ ions were soaked into PRORP1 crystals. In the absence of any added heavy metals, the electron density of water and Mg2+ ions cannot be unambiguously discerned. This is especially true for our PRORP2 crystal structure, where due to the limited resolution and the absence of any added heavy metals, we were unable map Mg2+ ions in the active site. The core of PRORP1 and PRORP2 active sites share very high amino acid identity (~80 %) and structure (RMSD < 0.8 Å). The conserved aspartate residues of the active site (D343, D421, D422, D440 in PRORP2 and D399, D474, D475, D493 in PRORP1) overlap when superimposed (Figure 5C). The histidine residue in the active site (H445 in PRORP2, H498 in PRORP1) is found in a different position in the structures of PRORP2 and PRORP1 (Figure 6C) suggesting flexibility in this region. This residue was initially proposed to protonate the leaving product8 but was later shown not to participate in acid-base chemistry12.
To verify that the putative active site residues are essential for PRORP2 catalysis, we created a series of mutants where the active site aspartate and histidine residues are substituted with alanine (D343A, D421A, D422A, D440A, H445A). We found that all of the aspartate to alanine mutants are unable to cleave (nu)pre-tRNAGly 8:1 in our single turnover assays (Table 3), and some of these mutants have significantly weaker substrate affinity than the wild type enzyme (Supplemental Table 2)6. The H445A PRORP2 mutant reduced the kobs value measured in cleavage assays by approximately 50-fold relative to the wild type protein. Since we expect that mutation of active site residues that directly participate in the pre-tRNA cleavage reaction to lower activity by more than 50-fold, we tested if H455 plays a role in pre-tRNA binding and recognition by measuring the KD of the PRORP2 H455A mutant for (nu)pre-tRNAGly 8:1. We find that this mutant does not alter substrate binding relative to the wild-type enzyme (Supplemental Table 2). Given that H455A binds substrates well and, in contrast to the aspartate active site mutants, does not render the enzyme entirely inactive, we suggest that H455 might play an indirect role in catalysis such as orienting the substrate for cleavage following binding.
Discussion
While protein only RNase Ps are found in many eukaryotic organisms (including humans) how this new class of RNA processing enzymes generally functions remains poorly understood. In some Eukaryotes, such as land plants, PRORPs have entirely taken over the role of “ribozyme-based” RNase Ps4; 5; 6; 9. Despite their essential nature9 to date only a single PRORP, the A. thaliana PRORP1 localized to mitochondria and chloroplast, has been characterized biochemically and structurally4; 6; 8; 12; 15. To broadly understand how PRORPs function, our work here is the first structure and function study of a nuclear localized PRORP, A. thaliana PRORP2.
Our 3.2 Å crystal structure of A. thaliana PRORP2 demonstrates that the overall structure of single enzyme PRORPs, including the active site and structural zinc site, are conserved. We were unable to visualize the active site metal(s) previously seen in the PRORP1 structure8 in our PRORP2 crystal structure. However, our biochemical data indicate that PRORP2, like PRORP1, uses a catalytic Mg2+ cofactor (Figure 1C, 1D). Interestingly, we discovered that PRORP2 exhibits a more open conformation than PRORP1; this difference cannot be explained simply by crystal contacts and suggests that despite a high degree of sequence identity (~48%) there can be significant variation even between PRORP paralogues from a single organism. The structural differences that we observe are localized to the PPR domain and the region connecting the central and metallonuclease domains. Within the PPR domain we found that the different rotation angles of the A-B helices of the third PPR motif in PRORP1 and PRORP2 account for much of the difference in protein conformations. Additionally, normal mode analysis of PRORP1 and PRORP2 revealed the presence of putative mechanical hinges in the intra-motif loop of the third PPR and between the metallonuclease domain and central domain in both PRORPs. These crystal structures combined with the normal mode analysis are the first evidence of potential flexibility within the PPR motifs. Kinetic assays with truncated PRORP2 indicate that the first three PPR helices, forming the first rigid group in the PRORP2 PPR domain, are relevant to PRORP2 function since truncation leads to reduced activity. The N-terminal helices may be important for both providing residues that mediate protein-nucleic acid interactions and helping to accommodate conformational changes within the PPR domain necessary for substrate binding. Our structural analysis suggests that the relative motion of the two observed rigid group of helices within the PPR domain could permit two different superhelical arrangements of the PPR domain. These arrangements potentially represent different PPR domain conformations during the PRORP catalytic cycle and reveal how the PPR domain might adopt different conformations in response to RNA binding.
We find that PRORP2 has high affinity for pre-tRNA (KD values in low nanomolar range) consistent with dissociation constants reported for PRORP18; 12 and other PPR proteins (eg. PPR 10)29. Furthermore, the single turnover rate constants that we measure for PRORP2 cleaving (nu)pre-tRNAGly substrates are comparable with previously published rate constants for PRORP1 catalyzing cleavage of (mt)pre-tRNAs under the same conditions8; 12. However these activities are at least 10-fold lower than that of bacterial and yeast RNA based RNase P6.
“Ribozyme-based” RNase P typically recognizes pre-tRNA substrates using a variety of sequence and secondary structural characteristics (e.g. D and T-stem loops, 5′ leader length) based on in vitro kinetic experiments and the co-crystal structure of RNase P with tRNAPhe 21; 39; 40; 41; 42; 43; 44; 45; 46; 47; 48. Activity assays and footprinting experiments indicate that PRORP1 recognizes individual residues in the pre-tRNA D- and T-loops49 but it is unknown if, like RNA-based RNase P, the pre-tRNA 5′ leader and immature 3′ trailer length are also determinants of PRORP substrate recognition. Our binding studies (Figure 2A, Table 1) indicate that PRORP2 preferentially associates with pre-tRNA substrates containing short 3′ trailers and 5′ leaders. This is in contrast to the bacterial RNA-based form of the enzyme, which does not discriminate between binding pre-tRNAs with leaders longer than 5 nucleotides21; 22; 39. PRORP2 demonstrates a preference towards binding pre-tRNAs with short 3′ trailers, making it unlikely that 3′ trailers of pre-tRNAs are important in substrate recognition. The preference for pre-tRNAs with short 3′ trailers differs from the eukaryotic nuclear RNA-based RNase P found in yeast39, indicating that there are potential differences between how eukaryotic RNA based RNase-P and PRORPs recognize their substrates. It is possible that PRORP2 prefers to bind pre-tRNAs with short 5′ leader and 3′ trailer sequences because extended single stranded RNA regions have the potential to base pair and/or form secondary structures, thereby altering the substrate structure. While pre-tRNA 5′ end processing by RNase P is conserved, tRNA 3′ end maturation is not; depending on the organism 3′ maturation is catalyzed by different endo- (e.g. RNase Z, RNase PH) and exonucleases (e.g Rex1.), and even sometimes requires multiple enzymes7. Furthermore, the order of 3′ and 5′ pre-tRNA maturation is not conserved7. While in human mitochondria 5′ end processing is proposed to be the first step50, given the substrate preferences that we observe we speculate that nuclear pre-tRNAs in A. thaliana may be first processed by 3′ end tRNA processing enzymes before 5′ end maturation by PRORP2 in vivo.
Whether or not differentially localized PRORPs in a single organism display specificity for substrates localized to their compartments is mostly unknown51. Answering this question is of interest because it might help us to rationalize why some organisms evolved multiple PRORP paralogues. We first approached this by measuring the single-turnover rate constants for PRORP2 cleaving a mitochondria specific t-element and nuclear encoded pre-tRNAs. Our in vitro studies demonstrate that PRORP1 single turnover activity for the mitochondrial t-element is 10-fold higher than that of PRORP2, suggesting that some differences in cleavage specificity amongst PRORPs exist. Moreover, despite the similarity of PPR domains in PRORP proteins, there are not only sequence differences but also structural dissimilarities within PRORP PPR domains. These observed subtle variances in the PPRs may account for differences in substrate recognition strategies between PRORPs of different organellar localization. This observation can help to explain why PRORPs with different sequences and slight structural variations may be required within a single organism.
It has been proposed that PPR domains interact with RNAs via both the RNA sugar-phosphate backbone and/or base specific contacts14; 29; 49. These types of contacts are more likely to be made between the PRORP PPR domains and pre-tRNA D- and T-arms, and the leader sequences49 than with pre-tRNA aniticodon loops. This is reinforced by the observation that PRORPs process precursor forms of t-elements that completely lack the anticodon loop (Figure 3A). Bioinformatic analyses of PPR proteins suggest that amino acids that potentially interact with the RNA substrates are typically found in the first, third, sixth, tenth and thirteenth position of a given PPR motif (when the first residue of a PPR motif is defined as the first amino acid of the first α-helix of a PPR motif)29; 52; 53. The crystal structure of maize PPR10 PPR protein with its cognate RNA substrate provided hints as to the potential importance of amino acids in the third and the sixth position of PPR motifs in substrate binding. Mutagenesis studies have shown that amino acids in the 6th position of the second and third PPR motif of PRORP1 might play a role in recognition of the tRNA substrate14. We mapped the positions of all the residues thus far identified as potentially important for A. thaliana PRORPs binding pre-tRNAs onto the PRORP1 and PRORP2 crystal structures. The positions of these residues on the two structures are similar, consistent with our findings suggesting that the enzymes typically have similar reactivity toward pre-tRNAs. We expect that this will not be the case for non-canonical substrates, such as the mitochondrial t-elements. Further studies, including foot-printing, crosslinking, and crystal structures of PRORPs bound to their substrates will be necessary to tease out the precise contributions of individual PRORP structures to substrate recognition.
The work presented here provides the first biochemical and structural insight into a nuclear localized PRORP. This work demonstrates that there are subtle, but potentially important, differences between the structures of PRORPs and identifies a number of structural features likely important for substrate recognition. Furthermore, we reveal that differentially localized PRORPs exhibit some preference for cleaving a subset of localized non-canonical substrates, partially explaining the need for multiple PRORP paralogues in a single organism. In summary, our findings further our knowledge of a new class of RNA processing enzymes by providing mechanistic insight into how they recognize and cleave their substrates, and uncovering a number of conserved and non-conserved features of their structure and function.
Materials and methods
Protein expression
Δ75 PRORP1 was prepared according to previously published protocols8; 12. Full length PRORP2 was cloned into pMCSG7 vector54. Mutants were generated by Quickchange mutagenesis. Expression of full-length wild type PRORP2 was induced upon addition of 200 μM IPTG to BL21(DE3) E. coli at 18 °C. Bacterial cells were collected by centrifugation (10.000 × g, 15 minutes), resuspended in lysis buffer (20 mM MOPS pH 7.8, 1 M NaCl, 1 mM tris(2-carboxyethyl)phosphine (TCEP), 0.2 % Tween, 0.1 mg/ml lysozyme and 0.1 mg/ml phenylmethylsulfonyl fluoride (PMSF), and incubated for 1 hour at 4°C. The cells were subsequently subjected to sonication, and the resulting suspension was centrifuged at 34.000 × g at 4°C for 1 hour. The supernatant was loaded onto a HisTrap (GE Healthcare) nickel column and protein was eluted with 500 mM imidazole. The purified His6-PRORP were dialyzed into 20 mM MOPS pH 7.8, 150 mM NaCl, 1 mM TCEP and 5 % glycerol overnight at 4°C and incubated with TEV protease (1:20 molar protein:TEV protease ratio) to remove the His tag. His6-TEV protease was removed by running the sample through the HisTrap nickel column. For in vitro assays PRORP2 was dialyzed into 20 mM MOPS pH 7.8, 1 mM TCEP, 150 mM NaCl and 2 mM EDTA overnight at 4 °C to remove any possible metal contamination. Then, buffer was exchanged for 20 mM MOPS pH 7.8, 1 mM TCEP, 150 mM NaCl using a desalting column (BioRad). For crystallization purposes, PRORP2 was further fractionated using a size exclusion column before setting up crystallization trays.
Transcription and 5′ labeling of precursor tRNAs
Pre-tRNA sequences were commercially synthesized and cloned into pCR vector (Zero Blunt Topo PCR cloning kit, Invitrogen) and templates were amplified by PCR for in vitro transcription. The Nad6 t-element sequence was cloned into a pUC18 vector; the final vector contains a T7 promoter and a Bstn1 restriction site at the 5′ and 3′ of the template, respectively. Bstn1 was used for linearizing the template for runoff transcription reactions. Reactions were carried out in 50 mM Tris-HCl pH 8.0, 4 mM (for pre-tRNA) or 25 mM (pre- nad6 t-element) MgCl2, 1 mM spermidine, 5 mM DTT, 4 mM ATP, 4 mM CTP, 4 mM UTP, 1 mM GTP, 4 mM guanosine- 5′-O-monophosphorothioate (GMPS), 350 μg/ml purified T7 RNA polymerase, 12.5 μM purified DNA template containing T7 promoter and 4 U/μl SUPERaseIn. After stopping the transcription by addition of 50 mM EDTA and 500 mM NaCl, the pre-tRNA was washed with degassed TE pH 7.2 three times using an Amicon spin column (10 kDa MWCO). The washed pre-tRNA (~200 μl) was incubated with 20 μl 45 mM fluorescein overnight at 37 °C to label the 5′end. The reaction was stopped by addition of an equal volume of 2X loading dye (0.05 % Bromo-Phenol-Blue, 0.05 % Xylene Cyanol dye, 50 % m/v urea, 0.1 M EDTA) and run on a 12% urea-polyacrylamide gel. The pre-tRNA was eluted into crush-soak buffer (TE, 0.1 % SDS and 0.5 M NaCl) overnight at 4°C. The next day the mixture was filtered, concentrated and washed with degassed TE using an Amicon spin column (10 kDa MWCO). The pre-tRNA was ethanol precipitated and the resulting pellet was re-suspended in RNase free H2O. The concentrations of total and labeled pre-tRNA were measured from absorbance using a Nanodrop spectrophotometer.
Crystallization of PRORP2
Protein samples were concentrated to 8 mg ml−1 in 50 mM MOPS buffer at pH 7.8, 100 mM NaCl, 1 mM TCEP. The PRORP2 crystals were grown under oil (mixture containing 1:2 paraffin to silicone oil) in microbatch trays at 4 °C by mixing 0.3 μl of protein solution with 0.6 μl of reservoir solution, which contained 19.9 % (w/v) PEG3000, and 0.21 M sodium citrate. Crystals were cryoprotected for a few minutes before being flash frozen in liquid N2, by transfer to a solution of 20 % glycerol, 20% PEG3000 (w/v), and 0.2 M sodium citrate, in 20 mM MOPS pH 7.8. Crystals of PRORP2 were of space group P1 (a= 70.0 Å, b= 77.0 Å, c= 80.1 Å, α= 72.6°, β= 64.1°, γ= 77.8°) with 2 monomers in the asymmetric unit (Matthews’ coefficient, VM = 3.1 Å3/Da for 2 PRORP2 monomers, 60.6% solvent content).
Data collection, structure determination and refinement
Diffraction data were collected at GM/CA-CAT 23-IDB (Advanced Photon Source, Argonne National Laboratory) on a Mar 300 detector and processed with HKL200055 to 3.2 Å resolution. Initial phases were obtained by the molecular replacement (MR) method with Phaser56 using the three domains of PRORP1 (PDB accession number 4G26) separately as search models. Specifically a molecular replacement strategy was employed to search firstly for two excised PRORP1 metallonuclease domains, followed by two PPR and then finally by two central domains. It has to be noted that all loops and areas with high B-factors were omitted from the search models. Phaser successfully obtained a solution with all 6 individual domains The PRORP2 structure was refined originally with PHENIX57 including rigid body refinement of the individual domains followed by simulated annealing in torsional and Cartesian space, coordination minimization, and restrained individual B-factor adjustment with maximum-likelihood targets. Refmac558 in the CCP4i suite59 was subsequently employed for restrained refinement of using isotropic individual B-factors with maximum-likelihood targets using a simple model for bulk solvent scaling, followed by model building and modification with Coot60. Several iterative rounds of refinement followed by model building/modifications were performed. In the early rounds of refinement, restraints and ideal targets for tetrahedral geometry at zinc were added. Crystallographic information as well as refinement statistics are provided in the Table 3. The geometric quality of the model and its agreement with the structure factors were assessed with MolProbity For PRORP2, MolProbity61 reported a clash and a molprobity score of 1.62 (100th perecentile) and1.48 (100th percentile) respectively, while 92.52 % of the residues were in the favored Ramachandran plot regions with 0.96% residues in outlier regions. Figures showing crystal structures were generated with PyMOL62.
Analytical Ultracentrifugation
All the AUC sedimentation velocity experiments were carried out in 20 mM MOPS (pH 7.8), 100 mM NaCl, and 1 mM TCEP buffer using a Beckman ProteomeLab XL-1 instrument with An-50 Ti rotor at 2000×g and 20 °C using 1 and 10 mg/mL of protein concentrations. The values for buffer density, viscosity and protein partial specific volume were calculated using Sednterp63. During the runs, changes in the concentration gradient were monitored by absorbance at 280 nm. The data were analyzed by the SEDFIT using the continuous C(s) distribution model.
Single turnover assays
All single turnover reactions were conducted in buffer containing 30 mM MOPS pH 7.8, 150 mM NaCl, 1 mM MgCl2 (or CaCl2, MnCl2, ZnCl2 for data displayed in Figure 1C,D), 1 mM DTT at 25 °C. To initiate the reaction 20–100 nM fluorescently labeled substrate was added to 5 μM of recombinant purified PRORP2. Reactions were incubated for up to 60 minutes at 25 °C, time points were obtained by stopping the reactions with 0.05 % Bromo-Phenol-Blue, 0.05 % Xylene Cyanol dye, 50 % m/v urea, 0.1 M EDTA. Samples were visualized on a polyacrylamide-urea gel (8–20%) by direct scanning on a Storm 860 imager. Densitometry was performed with ImageQuant and data from at least three independent experiments were analyzed using Kaleidagraph 4.1.3. Eq 1. was fit to the data, as described in Howard et al, 20128 to calculate the observed rate constant (kobs) and the standard error.
| Eq 1 |
Fluorescent anisotropy binding assay
The binding assays were performed in 30 mM MOPS pH 7.8, 150 mM NaCl, 6 mM CaCl2, 1 mM DTT at 25 °C. For all assays, 20 nM of 5′ fluorescein labeled pre-tRNAGly was incubated with increasing concentrations (0–1000 nM) of PRORP2 for 5 minutes before changes in anisotropy was measured. Data were corrected with the anisotropy measured in the absence of the protein. The quadratic form of a binding isotherm (Eq. 2)22 was fit to the fluorescence anisotropy data from at least three independent experiments using Kaleidagraph 4.1.3 software to calculate the dissociation constant (KD) and the standard error.
| Eq 2 |
ΔFP= Observed enhancement in fluorescent anisotropy
ΔFP0= Fluorescent anisotropy in the absence of enzyme
ΔFPmax= Fluorescent anisotropy at enzyme saturation
Supplementary Material
Highlights.
Proteinaceous only RNase Ps (PRORPs) were recently discovered in Eukaryotes
In vitro activity of nuclear PRORP2 is comparable to that of organellar PRORP1
Nuclear PRORP2 processes the mitochondrial pre-t-element slower
PRORP1 & 2 have similar structures but are trapped in two different conformations
Nuclear and organellar PRORPs have conserved structural and functional properties
Acknowledgments
We would like to thank the National Institute of General Medical Sciences and National Cancer Institute Collaborative Access Team (GM/CA CAT) at the Advanced Light Source for beam time. We also thank Dr. Kristin S. Koutmou for her advice in setting up kinetic experiments and reading this manuscript. This work was supported by the National Institutes of Health [R01 GM055387 to C.A.F]; in part by the Molecular Biophysics Training Grant [T32 GM8270-25 to M.J.H.] and by Uniformed Services University of the Health Sciences start-up funds (R071KB to M.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS: Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 5DIZ.
References
- 1.Altman S. Ribonuclease P. Philos Trans R Soc Lond B Biol Sci. 2011;366:2936–41. doi: 10.1098/rstb.2011.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClain WH, Lai LB, Gopalan V. Trials, travails and triumphs: an account of RNA catalysis in RNase P. J Mol Biol. 2010;397:627–46. doi: 10.1016/j.jmb.2010.01.038. [DOI] [PubMed] [Google Scholar]
- 3.Gobert A, Gutmann B, Taschner A, Gossringer M, Holzmann J, Hartmann RK, Rossmanith W, Giege P. A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol. 2010;17:740–4. doi: 10.1038/nsmb.1812. [DOI] [PubMed] [Google Scholar]
- 4.Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–74. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Taschner A, Weber C, Buzet A, Hartmann RK, Hartig A, Rossmanith W. Nuclear RNase P of Trypanosoma brucei: a single protein in place of the multicomponent RNA-protein complex. Cell Rep. 2012;2:19–25. doi: 10.1016/j.celrep.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howard MJ, Liu X, Lim WH, Klemm BP, Fierke CA, Koutmos M, Engelke DR. RNase P enzymes: Divergent scaffolds for a conserved biological reaction. RNA Biol. 2013;10:909–14. doi: 10.4161/rna.24513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossmanith W. Of P and Z: mitochondrial tRNA processing enzymes. Biochim Biophys Acta. 2012;1819:1017–26. doi: 10.1016/j.bbagrm.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howard MJ, Lim WH, Fierke CA, Koutmos M. Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5′ processing. Proc Natl Acad Sci U S A. 2012;109:16149–54. doi: 10.1073/pnas.1209062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutmann B, Gobert A, Giege P. PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis. Genes Dev. 2012;26:1022–7. doi: 10.1101/gad.189514.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, Liu X, Zhou W, Yang X, Shen Y. Auto-inhibitory Mechanism of the Human Mitochondrial RNase P Protein Complex. Sci Rep. 2015;5:9878. doi: 10.1038/srep09878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinhard L, Sridhara S, Hallberg BM. Structure of the nuclease subunit of human mitochondrial RNase P. Nucleic Acids Res. 2015;43:5664–72. doi: 10.1093/nar/gkv481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard MJ, Klemm BP, Fierke CA. Mechanistic Studies Reveal Similar Catalytic Strategies for Phosphodiester Bond Hydrolysis by Protein-only and RNA-dependent Ribonuclease P. J Biol Chem. 2015 doi: 10.1074/jbc.M115.644831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Small ID, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–7. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 14.Imai T, Nakamura T, Maeda T, Nakayama K, Gao X, Nakashima T, Kakuta Y, Kimura M. Pentatricopeptide repeat motifs in the processing enzyme PRORP1 in Arabidopsis thaliana play a crucial role in recognition of nucleotide bases at TpsiC loop in precursor tRNAs. Biochem Biophys Res Commun. 2014;450:1541–6. doi: 10.1016/j.bbrc.2014.07.030. [DOI] [PubMed] [Google Scholar]
- 15.Pavlova LV, Gossringer M, Weber C, Buzet A, Rossmanith W, Hartmann RK. tRNA processing by protein-only versus RNA-based RNase P: kinetic analysis reveals mechanistic differences. Chembiochem. 2012;13:2270–6. doi: 10.1002/cbic.201200434. [DOI] [PubMed] [Google Scholar]
- 16.Zhou W, Karcher D, Fischer A, Maximova E, Walther D, Bock R. Multiple RNA processing defects and impaired chloroplast function in plants deficient in the organellar protein-only RNase P enzyme. PLoS One. 2015;10:e0120533. doi: 10.1371/journal.pone.0120533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Németh, E., Kožíšek. M., Schilli, G. K. & Gyurcsik, B.
- 18.Chinami M, Shingu M. Hydrogen-1 nuclear magnetic resonance studies of staphylococcal nuclease variant H124L: pH dependence of histidines and tyrosines. Arch Biochem Biophys. 1989;270:126–36. doi: 10.1016/0003-9861(89)90014-3. [DOI] [PubMed] [Google Scholar]
- 19.Hsiao YY, Nakagawa A, Shi Z, Mitani S, Xue D, Yuan HS. Crystal structure of CRN-4: implications for domain function in apoptotic DNA degradation. Mol Cell Biol. 2009;29:448–57. doi: 10.1128/MCB.01006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finney LA, O’Halloran TV. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science. 2003;300:931–6. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 21.Crary SM, Niranjanakumari S, Fierke CA. The protein component of Bacillus subtilis ribonuclease P increases catalytic efficiency by enhancing interactions with the 5′ leader sequence of pre-tRNAAsp. Biochemistry. 1998;37:9409–16. doi: 10.1021/bi980613c. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh J, Fierke CA. Conformational change in the Bacillus subtilis RNase P holoenzyme–pre-tRNA complex enhances substrate affinity and limits cleavage rate. RNA. 2009;15:1565–77. doi: 10.1261/rna.1639409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forner J, Weber B, Thuss S, Wildum S, Binder S. Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res. 2007;35:3676–92. doi: 10.1093/nar/gkm270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–54. [PubMed] [Google Scholar]
- 25.Hayward S, Kitao A, Berendsen HJ. Model-free methods of analyzing domain motions in proteins from simulation: a comparison of normal mode analysis and molecular dynamics simulation of lysozyme. Proteins. 1997;27:425–37. doi: 10.1002/(sici)1097-0134(199703)27:3<425::aid-prot10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 26.Suhre K, Sanejouand YH. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32:W610–4. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Blanco JR, Aliaga JI, Quintana-Orti ES, Chacon P. iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42:W271–6. doi: 10.1093/nar/gku339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura T, Meierhoff K, Westhoff P, Schuster G. RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur J Biochem. 2003;270:4070–81. doi: 10.1046/j.1432-1033.2003.03796.x. [DOI] [PubMed] [Google Scholar]
- 29.Yin P, Li Q, Yan C, Liu Y, Liu J, Yu F, Wang Z, Long J, He J, Wang HW, Wang J, Zhu JK, Shi Y, Yan N. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013;504:168–71. doi: 10.1038/nature12651. [DOI] [PubMed] [Google Scholar]
- 30.Li Q, Yan C, Xu H, Wang Z, Long J, Li W, Wu J, Yin P, Yan N. Examination of the Dimerization States of the Single-stranded RNA-recognition Protein PPR10. J Biol Chem. 2014 doi: 10.1074/jbc.M114.575472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–95. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barkan A, Small I. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 2014;65:415–42. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- 33.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Ban T, Ke J, Chen R, Gu X, Tan MH, Zhou XE, Kang Y, Melcher K, Zhu JK, Xu HE. Structure of a PLS-class pentatricopeptide repeat protein provides insights into mechanism of RNA recognition. J Biol Chem. 2013;288:31540–8. doi: 10.1074/jbc.M113.496828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gully BS, Cowieson N, Stanley WA, Shearston K, Small ID, Barkan A, Bond CS. The solution structure of the pentatricopeptide repeat protein PPR10 upon binding atpH RNA. Nucleic Acids Res. 2015;43:1918–26. doi: 10.1093/nar/gkv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwinghammer K, Cheung AC, Morozov YI, Agaronyan K, Temiakov D, Cramer P. Structure of human mitochondrial RNA polymerase elongation complex. Nat Struct Mol Biol. 2013;20:1298–303. doi: 10.1038/nsmb.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joseph AP, Valadie H, Srinivasan N, de Brevern AG. Local structural differences in homologous proteins: specificities in different SCOP classes. PLoS One. 2012;7:e38805. doi: 10.1371/journal.pone.0038805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hutchinson EG, Thornton JM. PROMOTIF–a program to identify and analyze structural motifs in proteins. Protein Sci. 1996;5:212–20. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziehler WA, Day JJ, Fierke CA, Engelke DR. Effects of 5′ leader and 3′ trailer structures on pre-tRNA processing by nuclear RNase P. Biochemistry. 2000;39:9909–16. doi: 10.1021/bi000603n. [DOI] [PubMed] [Google Scholar]
- 40.Loria A, Niranjanakumari S, Fierke CA, Pan T. Recognition of a pre-tRNA substrate by the Bacillus subtilis RNase P holoenzyme. Biochemistry. 1998;37:15466–73. doi: 10.1021/bi9816507. [DOI] [PubMed] [Google Scholar]
- 41.Loria A, Pan T. Recognition of the T stem-loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1997;36:6317–25. doi: 10.1021/bi970115o. [DOI] [PubMed] [Google Scholar]
- 42.Loria A, Pan T. Recognition of the 5′ leader and the acceptor stem of a pre-tRNA substrate by the ribozyme from Bacillus subtilis RNase P. Biochemistry. 1998;37:10126–33. doi: 10.1021/bi980220d. [DOI] [PubMed] [Google Scholar]
- 43.Loria A, Pan T. The cleavage step of ribonuclease P catalysis is determined by ribozyme-substrate interactions both distal and proximal to the cleavage site. Biochemistry. 1999;38:8612–20. doi: 10.1021/bi990691f. [DOI] [PubMed] [Google Scholar]
- 44.Koutmou KS, Day-Storms JJ, Fierke CA. The RNR motif of B. subtilis RNase P protein interacts with both PRNA and pre-tRNA to stabilize an active conformer. RNA. 2011;17:1225–35. doi: 10.1261/rna.2742511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zahler NH, Christian EL, Harris ME. Recognition of the 5′ leader of pre-tRNA substrates by the active site of ribonuclease P. RNA. 2003;9:734–45. doi: 10.1261/rna.5220703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christian EL, McPheeters DS, Harris ME. Identification of individual nucleotides in the bacterial ribonuclease P ribozyme adjacent to the pre-tRNA cleavage site by short-range photo-cross-linking. Biochemistry. 1998;37:17618–28. doi: 10.1021/bi982050a. [DOI] [PubMed] [Google Scholar]
- 47.Brannvall M, Mattsson JG, Svard SG, Kirsebom LA. RNase P RNA structure and cleavage reflect the primary structure of tRNA genes. J Mol Biol. 1998;283:771–83. doi: 10.1006/jmbi.1998.2135. [DOI] [PubMed] [Google Scholar]
- 48.Lustbader JW, Cirilli M, Lin C, Xu HW, Takuma K, Wang N, Caspersen C, Chen X, Pollak S, Chaney M, Trinchese F, Liu S, Gunn-Moore F, Lue LF, Walker DG, Kuppusamy P, Zewier ZL, Arancio O, Stern D, Yan SS, Wu H. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer’s disease. Science. 2004;304:448–52. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- 49.Gobert A, Pinker F, Fuchsbauer O, Gutmann B, Boutin R, Roblin P, Sauter C, Giege P. Structural insights into protein-only RNase P complexed with tRNA. Nat Commun. 2013;4:1353. doi: 10.1038/ncomms2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brzezniak LK, Bijata M, Szczesny RJ, Stepien PP. Involvement of human ELAC2 gene product in 3′ end processing of mitochondrial tRNAs. RNA Biol. 2011;8:616–26. doi: 10.4161/rna.8.4.15393. [DOI] [PubMed] [Google Scholar]
- 51.Sugita C, Komura Y, Tanaka K, Kometani K, Satoh H, Sugita M. Molecular Characterization of Three PRORP Proteins in the Moss Physcomitrella patens: Nuclear PRORP Protein Is Not Essential for Moss Viability. PLoS One. 2014;9:e108962. doi: 10.1371/journal.pone.0108962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi K, Kawabata M, Hisano K, Kazama T, Matsuoka K, Sugita M, Nakamura T. Identification and characterization of the RNA binding surface of the pentatricopeptide repeat protein. Nucleic Acids Res. 2012;40:2712–23. doi: 10.1093/nar/gkr1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 55.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 56.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 58.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–7. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 60.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schrodinger LLC. The PyMOL Molecular Graphics System. Version. 2010;1:4. [Google Scholar]
- 63.Cole JL, Lary JW, T PM, Laue TM. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 2008;84:143–79. doi: 10.1016/S0091-679X(07)84006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


