Abstract
Preadult determinants of adult fitness and behavior have been documented in a variety of organisms with complex life cycles, but little is known about expression patterns of genes underlying these adult traits. We explored the effects of differences in egg to adult development time on adult transcriptome and cuticular hydrocarbon variation in order to understand the nature of the genetic correlation between preadult development time and premating isolation between populations of Drosophila mojavensis reared in different host cactus environments. Transcriptome variation was analyzed separately in flies reared on each host and revealed that hundreds of genes in adults were differentially expressed (FDR P < 0.05) due to development time differences. For flies reared on pitaya agria cactus, longer preadult development times caused increased expression of genes in adults enriched for ribosome production, protein metabolism, chromatin remodeling, and regulation of alternate splicing and transcription. Baja California flies reared on organ pipe cactus showed fewer differentially expressed genes in adults due to longer preadult development time, but these were enriched for ATP synthesis and the TCA cycle. Mainland flies reared on organ pipe cactus with shorter development times showed increased transcription of genes enriched for mitochondria and energy production, protein synthesis, and glucose metabolism: adults with longer development times had increased expression of genes enriched for adult life span, cuticle proteins and ion binding, although most differentially expressed genes were unannotated. Differences due to population, sex, mating status, and their interactions were also assessed. Adult cuticular hydrocarbon profiles also showed shifts due to egg to adult development time, and were influenced by population and mating status. These results help to explain why preadult life history variation determines subsequent expression of the adult transcriptome along with traits involved with reproductive isolation and revealed previously undocumented connections between genetic and environmental influences over the entire life cycle in this desert insect.
Keywords: Development time, life cycle, gene expression, microarrays, cuticular hydrocarbons, cactus, transcriptome
Introduction
Classical notions of life history evolution typically depend on the existence of genetic variation and covariation in functionally related components of fitness allowing populations to respond to patterns of environmental variation over the life cycle (Coulson et al. 2010; Gadgil & Bossert 1970; Istock 1981; Martin 1995; Reznick et al. 1996; Rose & Charlesworth 1981; Stearns 1977; Steiner et al. 2014). Identifying mortality risk at different stages/ages, particularly in stochastic environments (Orzack et al. 2011; Orzack & Tuljapurkar 1989; Steiner & Tuljapurkar 2012; Tuljapurkar et al. 2009), has furthered understanding of the causes for the wide diversity of observed life histories and causes of demographic heterogeneity and aging (Jones et al. 2014). However, suites of genes and their patterns of stage/age expression forming the functional basis of evolving life histories are not well understood for most organisms. Work in the genetics and physiology of development has identified candidate genes controlling preadult ontogenetic shifts (Tennessen & Thummel 2011), and studies of aging adults have categorized genes influencing life span (de Magalhães et al. 2009; Paaby & Schmidt 2009), but few studies have identified genes and their expression patterns responsible for the evolution of correlated life history traits (but see Flatt et al. 2005; Paaby et al. 2014). In general, if the genetic basis of these correlations is often polygenic, then whole genome studies are required in order to identify genes and characterize genome regions contributing to the expression of quantitative genetic variation in life histories (e.g. Bochdanovits & de Jong 2004) and other traits correlated with them.
Preadult determinants of adult fitness have been hypothesized to influence adult mortality in fish (Reznick et al. 2004), birds and mammals (Martin et al. 2015; Ricklefs 2006; Rollo 2002), and lizards (Olsson & Shine 2002). In humans, longer gestation times and higher birth weights are correlated with increased child cognitive abilities (Figlio et al. 2014). Environmental influences in embryonic and preadult stages on later life history stages have also been widely studied in a diversity of organisms, some described as carry-over effects (reviewed in Pechenik 2006). Thus, preadult influences on adult phenotypes over the life cycle seem widespread, including holometabolous insects (Doyon & Boivin 2005; Tu & Tatar 2003; Valtonen et al. 2012). Functional characterization of those genes that are differentially expressed over the life cycle due to such carry over effects has not been well studied.
We set out to directly assess the carry-over effects of variation in a preadult life history trait, egg to adult development time (DEVT), on determinants of adult mate choice, epicuticular hydrocarbon composition and whole genome transcription levels in Drosophila mojavensis. Assessing carry-over effects in D. mojavensis was motivated by previous evidence documenting influences of variation in DEVT and preadult conditions on adult mating preferences and cuticular hydrocarbon composition, including; 1) mainland and Baja California populations cultured on fermenting cactus showed significantly reduced sexual isolation between populations as well as male mating speed of Baja California flies vs. those reared on laboratory media (Brazner & Etges 1993), 2) bidirectional artificial selection revealed realized heritabilities of 8–16 percent for DEVT and correlated responses in premating isolation between populations (Etges 1998), 3) four cuticular hydrocarbon QTLs associated with male mating success were correlated with variation in DEVT (r = 0.59, P = 0.02; Etges et al. 2010), 4) offspring of females allowed to choose mates had shorter DEVT than those not allowed to choose mates (Havens et al. 2011), and 5) mainland adults with longer DEVT cultured on fermenting cactus had more cuticular hydrocarbons than adults eclosing earlier (Etges 2014) . Thus, within-population variation in DEVT is genetically correlated with determinants of adult mate choice behaviors responsible for sexual selection and sexual isolation (Etges & Tripodi 2008; Havens & Etges 2013). Understanding this correlation and identifying the causative genes involved should reveal how reproductive isolation is driven by genetic differences in life histories, and may suggest how variation in the adult transcriptome explains the basis of genetic variation in reproductive isolation (reviewed in Mullen & Shaw 2014; Seehausen et al. 2014) and ecological speciation (Arnegard et al. 2014; Funk 1998; Funk et al. 2006; McKinnon et al. 2004; Nosil 2012; Schluter & Conte 2009).
Natural history and biogeography of D. mojavensis
Of the four endemic drosophilids in the Sonoran and Mojave Deserts, D. mojavensis uses different host cacti across its range and is characterized by geographic differentiation and population structuring across desert regions (Etges et al. 1999; Heed 1982; Markow et al. 2002). In Baja California, agria cactus, Stenocereus gummosus, is the preferred host plant with occasional use of Myrtillocactus cochal. In northwestern Mexico and Arizona, organ pipe cactus, S. thurberi, is the major host with some use of sina cactus, S. alamosensis, it shares with its sibling species, D. arizonae. In the Mojave Desert, California barrel cactus, Ferocactus cylindraceus, is the principal host, and prickly pears, Opuntia littoralis, O. oricola, and O. demissa (O. oricola X O. ficus-indica hybrids) are the only known host plants used by Santa Catalina Island populations near Los Angeles, California (Barbour et al. 2007). Ancestral populations of D. mojavensis became isolated in Baja California after splitting from what is now D. arizonae on the mainland ca 1.3 mya, but gene flow continued until ca 270 kya. At about the same time, Baja California populations of D. mojavensis invaded mainland Sonora and are now partially sympatric with D. arizonae. Switching from agria to organ pipe cactus was accompanied by the evolution of life history differences including longer egg to adult development times and lower viabilities in mainland populations (Etges 1990, 1993; Etges et al. 2010) associated the slower fermentation rates of organ pipe cactus tissues (Etges 1989). Mojave Desert populations of D. mojavensis diverged from mainland Mexico and Arizona populations ca 117 kya (Lohse et al. 2015; Smith et al. 2012). Analysis of mtDNA COI sequence variation (Richmond et al. 2013) and chromosomal inversions (Delprat et al. 2014) suggested that Santa Catalina Island populations are derived from Baja California.
Here, we focused on Baja California and mainland populations by culturing egg to imago stages on fermenting tissues of two different cacti and quantified whole genome transcriptome variation in aged male and female D. mojavensis grouped by egg to adult DEVT that were either unmated or exposed to members of the opposite sex. We were most interested in discovering if adults with different egg to adult DEVT had different gene expression profiles more than a week after eclosion when adults attain sexual maturity. We predicted that these patterns of adult differential gene expression should reveal why phenotypes including mating behavior and cuticular hydrocarbons are affected by preadult rearing conditions and variation in DEVT. Thus, we tested the null hypothesis that there should be no significant differences in gene expression in same age, sexually mature adult flies reared in either of two host cacti due to differences in egg to adult DEVT.
Materials and Methods
Origin of Stocks
A Baja California population of D. mojavensis originated from Punta Prieta in January 2008, and a Sonora population was collected in Las Bocas in March 2009. All flies were netted over fermented bananas or by collecting adults emerged from cactus rots returned to the lab. A total of 465 baited adults were collected in Punta Prieta, and 1264 baited adults plus 9 adults that emerged from sina cactus, S. alamosensis, rots collected in Las Bocas. For site locations, see Etges et al. (2010). Once returned to the lab, each population was cultured on banana food (Brazner & Etges 1993) in 35 ml shell vials at room temperature until the experiments began in September 2009.
Cactus culture conditions
Each population was introduced into a separate cage (12,720 cm3) for 7–10 days to allow random mating in an incubator programmed for a 14:10 LD photoperiod and 27:17° C. Eggs collected from food cups attached to these cages were reared to eclosion at moderate larval densities in 250 ml bottles containing banana food. Eclosed adults were separated in small same sex groups and transferred to 35 ml shell vials containing banana food until they were sexually mature (8–10 days). Approximately 200 females and 200 males from each population were introduced into 2–3 separate oviposition chambers and allowed to mate and oviposit for 10 h each day. Eggs were collected from a 5.5 cm diameter petri dish attached to each oviposition chamber containing a 1% agar-cactus media, and washed in sterile deionized water, 70% ethanol, and again in deionized water. Eggs were counted into groups of 200 onto a 1 cm2 piece of sterilized filter paper, and placed in bottles containing 75 g of fermenting cactus tissue and cultured in the incubator described above. Each bottle contained 75 g of aquarium gravel and a 5.5 cm diameter filter paper circle that was autoclaved before adding cactus tissues. Each culture was inoculated with 1 ml of a pectolytic bacterium, Erwinia cacticida (Alcorn et al. 1991), and a mixture of seven cactophilic yeasts: Dipodascus starmeri, Candida sonorensis, C. valida, Starmera amethionina, Pichia cactophila, P. mexicana, and Sporopachydermia cereana. All unhatched eggs were counted to allow calculation of egg to adult viability. Eclosed adults from each replicate culture were counted daily allowing determination of egg-to adult development time. Because of the large numbers of adults required for each day of development time, we cultured each population on each cactus 14 times for a total n = 56 cactus cultures.
Upon emergence, flies were sexed and stored in groups separated by day of development time in shell vials containing lab food in the incubator. Development time was based on mid-morning aspiration of adults from each culture bottle each day because emergence is diurnal, i.e. eclosion occurs entirely in the morning insuring all imagoes had time to harden before handling. Because we were most interested in within population fast-slow differences in DEVT, we grouped flies based on egg to emergence time of the first adults to eclose as emergence day 1, adults eclosing the next day as emergence day 2, etc. In order to compare same-aged virgin (unmated) and mated adults, groups of 24 virgin females were aged for 9 days and groups of 24 virgin males were aged to 13 days in 35 ml vials containing lab food in the incubator described above (Fig. 1). Sexual maturity in D. mojavensis under laboratory conditions is 3–6 da for females and 8–10 da for males (Etges & Klassen 1989; Markow 1982). For mated adults, groups of 24 females 8 days old were combined with 24 males that were 12 days old in 35 ml vials, held for 24 hr in the incubator, separated by sex, snap frozen in liquid nitrogen, and stored at -80° C for RNA extraction or frozen at −20° C for CHC analysis. While we labeled these flies as mated, we realize that not all adults may have mated, but all were exposed to members of the opposite sex for 24 hr similar to the design in Etges et al. (2009) that we replicated here. In one-hour multiple mating trials with cactus-reared D. mojavensis, almost all copulations occur in the first 20–30 minutes (Brazner & Etges 1993; Havens et al. 2011). All flies were frozen ca 2 hr after lights on in the morning, i.e. 8:00 AM.
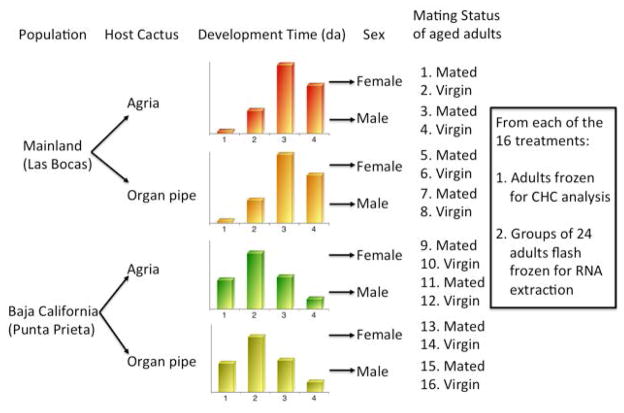
Figure 1.
Diagram of the design of this experiment. Each population was reared on fermenting host cacti, and eclosed adults were grouped by day of development time. Males and females were aged to sexual maturity, 9 days for females and 13 days for males, in vials containing laboratory food. They were either stored separately or combined with an equal number of members of the opposite sex for 24 hours. From each of the 16 treatment groups, adults were frozen for CHC analysis or flash frozen in liquid nitrogen for RNA extraction in groups of 24 adults. Some replicates of organ pipe cactus-reared, mated, mainland adults were missing, see text for details.
Epicuticular hydrocarbon analysis
Total epicuticular hydrocarbons were extracted by immersing each adult in hexane for 20 min in a 300 μL glass vial insert (Microliter Analytical Supplies, Suwanee, GA), evaporating off all hexane in a 40°C heating block, and freezing each sample at −20° C until analysis. Individual CHC extracts were redissolved in 5 μl of heptane containing a known amount of docosane (C22) as an internal standard. Each sample was analyzed by capillary gas-liquid chromatography using an automated Shimadzu GC-17A (Shimadzu Scientific Instruments, Columbia, MD) fitted with a flame ion detector (FID) and a 15 m (ID = 0.22 mm) Rtx-5 fused-silica column (Restek Corporation, Bellefont, PA). Injector and detector temperatures were set at 290° C and 345° C, respectively, with the injector port in split mode (3:1 ratio), and the column was heated from 200° C to 345° C at 15° C/min holding at 345° C for 4 min. We quantified amounts of 31 CHC components (Etges & Ahrens 2001; Etges & Jackson 2001; Stennett & Etges 1997) in all flies by analysis of peak integrations using Class VP 4.2 software provided by Shimadzu, quantified by using C as an internal standard, and expressed as nanograms/fly.22
cDNA synthesis, hybridization and visualization
We used the same protocols described in Rajpurohit et al. (2013). Briefly, total RNA was isolated from each group of 24 adults using RNeasy mini-kits (Qiagen, Valencia, California USA) and stored at −80° C. Double-stranded cDNA was synthesized with Invitrogen Superscript Double-Stranded cDNA Synthesis kits, and cDNA concentrations assessed using a NanoDrop spectrophotometer (NanoDrop Technologies) to insure that all cDNA samples were ≥ 100ng/ul, A260/A280 ≥ 1.8, and A260/A230 ≥ 1.8. All cDNA samples were Cy3 labeled using a NimbleGen One Color DNA Labeling kit.
We used Roche NimbleGen 12-plex microarrays with each array designed to include 14,528 unique transcripts based on the D. mojavensis genome (ver 1.3 released on 4/14/2009) with nine probes per transcript yielding 130,705 probes (each microarray included 135K probes; see Gene Expression Omnibus entry GSE43220 for details). A NimbleGen Hybridization System (Hybridization System 4, BioMicro Systems, Inc.) was used for sample hybridizations and spot intensities were scanned with a GenePix 4000B scanner (Molecular Devices) and GenePix Pro software. All spot intensities were normalized using quantiles (Bolstad et al. 2003) with NimbleScan v2.5 software. Gene call files were produced with the Robust Multichip Average (RMA) algorithm (Irizarry et al. 2003).
Ortholog search and functional annotation clustering
Of the 14,528 D. mojavensis transcripts submitted to Flybase (Tweedie et al. 2009), only 9117 were orthologous to D. melanogaster genes, i.e. only ~ 63 percent of predicted D. mojavensis genes could be functionally analyzed. We performed gene ontology analyses (GO) using DAVID Bioinformatics Resources 6.7 (Huang et al. 2009) by submitting lists of D. mojavensis transcripts of interest after determining the subset of those transcripts that had D. melanogaster orthologs. We used the corresponding D. melanogaster genes in our GO analyses. Gene annotation clusters were determined by DAVID’s clustering algorithm with initial classification stringencies set to ‘Moderate’. Further inspection of annotated gene function was performed by identifying KEGG pathways (Kanehisa & Goto 2000).
Statistical analyses
All CHC data were log10 transformed to improve normality. We sampled 4–8 adults for each combination of population, cactus, sex, mating status, and DEVT, and performed MANCOVA. Principal Components Analysis (PCA) was used to identify different combinations of correlated CHC amounts and these PCs were used in ANOVAs to assess overall sources of variation. Regression analysis of CHC amounts with day of development time was also performed for comparisons with the results from Etges et al. (2010) for F2 male CHC-development time associations. All analyses were performed with SAS (SAS-Institute 2004). GC-MS identification of most of these CHCs was described in Toolson et al. (1990) and Etges and Jackson (2001).
Our cuticular hydrocarbon and microarray experimental design was planned to include 4 replicates for each combination of two populations, two sexes, mating status (mated or unmated), two host cacti (AG and OP), and day of development time. Both the data and the normalized fluorescence for each microarray probe set was subjected to the same replicated 5-way mixed model ANOVA in SAS using PROC MIXED (SAS-Institute 2004):
where μ is the grand mean, Pj is the effect of population, Hk is the effect of host cactus, Sl is the effect of sex, Mm is the effect of mating status, Dn is development time day, IPxH is the interaction between population and cactus, IPxS is the interaction between population and sex, IPxM is the interaction between population and mating status, etc., and Eijkmn is the error term. Population was considered a random effect. For the CHC data we also analyzed this full model with development time as a covariate rather than a random effect, but this had little effect on the results. We used PROC NLMIXED in SAS implemented for microarray data as it does not assume normally distributed data (Allison et al. 2002). We calculated least-squares means with the LSMEANS statement to assess the significance of each term using the DIFFS option. To correct for multiple comparisons, we calculated false-discovery rates (FDR) for the overall ANOVA data and for all pair-wise comparisons between treatments (Benjamini & Hochberg 1995) and further filtered these lists by concentrating on expression differences with > 1.5 X fold changes.
Results
A total of 9641 D. mojavensis were reared on both cactus substrates. Egg to adult development times and viability were consistent with previous experiments where mainland, Las Bocas flies expressed longer egg to adult development times (DEVT) than Punta Prieta, Baja California flies (LSMEAN X̄ ± 1 SE da, Las Bocas = 16.23 ± 0.024; Punta Prieta = 15.27 ± 0.024; Suppl. Fig. 1), and organ pipe cactus caused longer DEVT than agria with a significant Population x Cactus interaction (Suppl. Table 1). There were no differences in egg to adult viability between populations (overall LSMEAN X̄ ± 1 SE, 77.9 ± 1.78) but a significant difference due to cactus was observed (Suppl. Table 1).
Epicuticular hydrocarbon variation
MANCOVA revealed most model effects and interactions between them had significant effects on CHC variation, especially population, mating status, and development time (Table 1). The number of significant interaction terms revealed the sensitivity of CHC expression due to multiple sources of variation, particularly the degree to which DEVT and its interaction with other factors influenced variation in adult CHCs. To identify which groups of CHCs were influenced by these treatment effects, PCA revealed 8 PCs that explained 89.3 % of the total variation in the data (Suppl. Table 2). PC 1 loadings were all positive as expected as PC 1 included total CHC variation among individuals and sources of experimental error (Etges et al. 2009). The first six PCs were then subjected to ANOVA using the complete factorial model above in order to identify sources of variation represented by each PC. PC 2 explained 12 % of the variation and was influenced by variation due to population, mating status, sex x mating status interaction, and development time (Table 2). Since we expected large geographical differences (Etges & Ahrens 2001) and effects of exposure to the opposite sex (Etges et al. 2009) on CHC variation, covariation in CHCs represented by PC 2 (and PC 5) was of special interest because variation along this axis was associated with DEVT differences (Table 2). Changes in total CHC amounts per fly with increasing development time was population specific and shifted significantly due to exposure to flies of the opposite sex causing a DEVT x Mate interaction, F = 6.02, P = 0.015 (Table 2, Fig. 2). Mated flies had significantly more CHCs than unmated flies (Table 1, Fig. 2). There was also a significant Sex x Mate interaction although the main effect of sex was not significant (F = 10.6, P = 0.001).
Table 1.
MANCOVA results for variation in CHCs for adult D. mojavensis due to population, cactus, mating status (Mate; virgin vs mated), sex, egg to adult development time as the covariate, and all interactions. n = 399, all df = 31,277.
| Source of variation | Wilks' λ | F | Pr > F |
|---|---|---|---|
| Cactus | 0.7008 | 3.81 | < 0.0001 |
| Sex | 0.7277 | 3.34 | < 0.0001 |
| Cactus x Sex | 0.8226 | 1.93 | 0.003 |
| Population | 0.3522 | 16.44 | < 0.0001 |
| Cactus x Population | 0.8390 | 1.71 | 0.013 |
| Sex x Population | 0.8395 | 1.71 | 0.014 |
| Cactus x Sex x Population | 0.8442 | 1.65 | 0.0196 |
| Mate | 0.6712 | 4.38 | < 0.0001 |
| Cactus x Mate | 0.7406 | 3.13 | < 0.0001 |
| Sex x Mate | 0.7322 | 3.27 | < 0.0001 |
| Cactus x Sex x Mate | 0.8158 | 2.02 | 0.002 |
| Population x Mate | 0.7895 | 2.38 | 0.0001 |
| Cactus x Population x Mate | 0.8882 | 1.12 | 0.303 |
| Sex x Population x Mate | 0.8450 | 1.64 | 0.021 |
| Cactus x Sex x Population x Mate | 0.8364 | 1.75 | 0.011 |
| Development time | 0.6019 | 5.91 | < 0.0001 |
| DEVT x Cactus | 0.6871 | 4.07 | < 0.0001 |
| DEVT x Sex | 0.8036 | 2.18 | 0.0005 |
| DEVT x Population | 0.7281 | 3.34 | < 0.0001 |
| DEVT x Cactus x Sex | 0.8314 | 1.81 | 0.007 |
| DEVT x Cactus x Population | 0.8468 | 1.62 | 0.024 |
| DEVT x Sex x Population | 0.8635 | 1.41 | 0.078 |
| DEVT x Cactus x Sex x Population | 0.8472 | 1.61 | 0.025 |
| DEVT x Mate | 0.7044 | 3.75 | < 0.0001 |
| DEVT x Cactus x Mate | 0.7428 | 3.09 | < 0.0001 |
| DEVT x Sex x Mate | 0.8151 | 2.03 | 0.002 |
| DEVT x Cactus x Sex x Mate | 0.8342 | 1.78 | 0.009 |
| DEVT x Population x Mate | 0.8145 | 2.04 | 0.001 |
| DEVT x Cactus x Population x Mate | 0.8904 | 1.10 | 0.333 |
| DEVT x Sex x Population x Mate | 0.8483 | 1.60 | 0.027 |
| DEVT x Cactus x Sex x Population x Mate | 0.8520 | 1.55 | 0.036 |
Table 2.
ANCOVA results for the first 6 CHC Principal Components for D. mojavensis compared by day of eclosion (covariate) from two populations reared on agria and organ pipe cactus that were either allowed to mate or were virgins. Effects significant at P < 0.01 are italicized in bold.
| PC 1 | PC 2 | PC 3 | PC 4 | PC 5 | PC 6 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Source of variation | F | Pr | F | Pr | F | Pr | F | Pr | F | Pr | F | Pr |
| Model | 3.90 | < 0.0001 | 29.85 | < 0.0001 | 24.96 | < 0.0001 | 4.74 | < 0.0001 | 7.55 | < 0.0001 | 0.07 | 0.785 |
| Cactus | 0.79 | 0.374 | 1.44 | 0.231 | 0.65 | 0.421 | 1.09 | 0.298 | 2.59 | 0.109 | 0.65 | 0.421 |
| Sex | 1.12 | 0.292 | 2.23 | 0.136 | 19.17 | < 0.0001 | 4.63 | 0.032 | 2.47 | 0.117 | 0 | 0.949 |
| Cactus x Sex | 12.02 | 0.001 | 6.24 | 0.013 | 0.28 | 0.597 | 4.91 | 0.028 | 0.26 | 0.607 | 12.9 | 0.0004 |
| Population | 21.17 | < 0.0001 | 62.12 | < 0.0001 | 0.1 | 0.756 | 10.86 | 0.001 | 7.86 | 0.005 | 0.39 | 0.533 |
| Cactus x Population | 0.2 | 0.658 | 0.72 | 0.397 | 2.22 | 0.137 | 2.4 | 0.122 | 0.12 | 0.731 | 6.44 | 0.012 |
| Sex x Population | 3.02 | 0.083 | 0.72 | 0.397 | 1.72 | 0.191 | 0.89 | 0.347 | 4.52 | 0.034 | 1.43 | 0.233 |
| Cactus x Sex x Population | 10.58 | 0.001 | 6.68 | 0.010 | 1.99 | 0.160 | 1.06 | 0.305 | 1.87 | 0.173 | 24.2 | < 0.0001 |
| Mate | 1.28 | 0.260 | 6.98 | 0.009 | 3.81 | 0.052 | 1.52 | 0.219 | 3.31 | 0.070 | 2.33 | 0.128 |
| Cactus x Mate | 0 | 0.950 | 0.68 | 0.409 | 1.28 | 0.258 | 1.07 | 0.302 | 3.81 | 0.052 | 0.38 | 0.538 |
| Sex x Mate | 8.62 | 0.004 | 10.6 | 0.001 | 1.57 | 0.212 | 0.21 | 0.650 | 6.63 | 0.011 | 3.31 | 0.070 |
| Cactus x Sex x Mate | 8.35 | 0.004 | 4.98 | 0.026 | 0.16 | 0.693 | 1.37 | 0.243 | 0.23 | 0.630 | 3.15 | 0.077 |
| Population x Mate | 5.24 | 0.023 | 0 | 0.996 | 5.66 | 0.018 | 2.47 | 0.117 | 4.04 | 0.045 | 1.06 | 0.304 |
| Cactus x Population x Mate | 0.04 | 0.842 | 0.13 | 0.716 | 0.01 | 0.936 | 0.54 | 0.465 | 0.89 | 0.346 | 0.22 | 0.641 |
| Sex x Population x Mate | 0.62 | 0.431 | 1.05 | 0.306 | 0.4 | 0.527 | 14.25 | 0.0002 | 0.66 | 0.419 | 0.56 | 0.454 |
| Cactus x Sex x Population x Mate | 0.73 | 0.394 | 0.43 | 0.511 | 0.76 | 0.384 | 5.47 | 0.020 | 2.3 | 0.131 | 21.05 | < 0.0001 |
| Development time | 0.89 | 0.346 | 13.6 | 0.0003 | 1.07 | 0.301 | 2.27 | 0.133 | 27.51 | < 0.0001 | 0.18 | 0.676 |
| DEVT x Cactus | 1.34 | 0.248 | 1.77 | 0.184 | 0.12 | 0.728 | 4.15 | 0.043 | 2.72 | 0.100 | 0.05 | 0.816 |
| DEVT x Sex | 4.04 | 0.045 | 0.93 | 0.335 | 0.88 | 0.349 | 3.38 | 0.067 | 0.04 | 0.837 | 0.4 | 0.529 |
| DEVT x Cactus x Sex | 11.76 | 0.001 | 6.06 | 0.014 | 0.36 | 0.550 | 3.81 | 0.052 | 0.01 | 0.934 | 6.4 | 0.012 |
| DEVT x Population | 6.93 | 0.009 | 0.09 | 0.767 | 4.58 | 0.033 | 5.63 | 0.018 | 10.28 | 0.002 | 0.28 | 0.594 |
| DEVT x Cactus x Population | 0.36 | 0.549 | 1.24 | 0.267 | 1.24 | 0.266 | 3.73 | 0.054 | 0 | 0.962 | 10.21 | 0.002 |
| DEVT x Sex x Population | 2.81 | 0.095 | 0.06 | 0.811 | 1.37 | 0.243 | 0.22 | 0.636 | 1.13 | 0.289 | 2.16 | 0.143 |
| DEVT x Cactus x Sex x Population | 10.48 | 0.001 | 4.96 | 0.027 | 2.94 | 0.087 | 1.40 | 0.237 | 1.07 | 0.301 | 16.23 | < 0.0001 |
| DEVT x Mate | 0.79 | 0.376 | 6.02 | 0.015 | 2.35 | 0.126 | 1.06 | 0.304 | 4.24 | 0.040 | 0 | 0.948 |
| DEVT x Cactus x Mate | 0.48 | 0.487 | 0.51 | 0.474 | 0.93 | 0.337 | 1.77 | 0.184 | 3.21 | 0.074 | 0.59 | 0.441 |
| DEVT x Sex x Mate | 6.06 | 0.014 | 4.84 | 0.029 | 0.2 | 0.654 | 0.03 | 0.863 | 3.66 | 0.057 | 2.34 | 0.127 |
| DEVT x Cactus x Sex x Mate | 7.46 | 0.007 | 2.98 | 0.085 | 0.02 | 0.895 | 0.29 | 0.591 | 0.83 | 0.363 | 2.04 | 0.154 |
| DEVT x Population x Mate | 3.29 | 0.071 | 0.31 | 0.581 | 5.58 | 0.019 | 2.87 | 0.091 | 1.26 | 0.263 | 0 | 0.988 |
| DEVT x Cactus x Population x Mate | 0.47 | 0.4935 | 0.47 | 0.494 | 0.03 | 0.867 | 1.92 | 0.167 | 0.79 | 0.375 | 0.03 | 0.863 |
| DEVT x Sex x Population x Mate | 0.67 | 0.412 | 2.71 | 0.101 | 0.26 | 0.612 | 10.01 | 0.002 | 0.02 | 0.899 | 0.23 | 0.633 |
| DEVT x Cactus x Sex x Population x Mate | 1.50 | 0.222 | 2.08 | 0.150 | 0.22 | 0.638 | 8.59 | 0.004 | 2.24 | 0.135 | 0.07 | 0.785 |
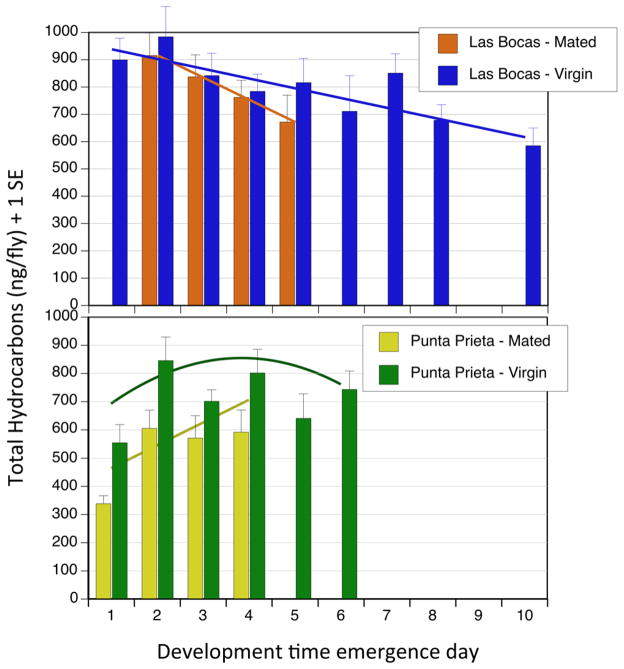
Figure 2.
Change in total CHCs per aged adult fly with egg to adult development time. Regression equations for mainland, Las Bocas adults were: mated (NS) y = − 79.5x + 1075.9, r2 = 0.08; virgin (P < 0.05) y = −33.6x + 967.3, r2 = 0.05; Baja California, Punta Prieta: mated (NS) y = 58.1x + 405.5, r2 = 0.07; virgin (P < 0.05) y = 188.5x* −25.9x2* + 435.7, r2= 0.05. Terms in parentheses indicate regression model significance, NS =- not significant, * P < 0.05 for individual regression terms.
Within population variation in egg to adult development had population-specific effects on adult CHC variation. Regression analysis of pooled males and females revealed linear decreases in total CHC amounts per fly with increasing development times in the mainland, Las Bocas population for both mated and virgin flies (Fig. 2). CHC amounts increased with DEVT in mated and virgin Punta Prieta adults, with a significant curvilinear relationship (P < 0.05) in virgin Punta Prieta adults similar to the results in Etges et al. (2010). All regressions were sequentially fitted with linear, squared, and cubic terms for total CHCs: all higher order terms were not significant except for the latter case. Comparisons were somewhat limited by the absence of more mated flies, but they were needed for microarray analysis. PC 2 and PC 5 showed population-specific shifts in CHC variation with day of emergence with parallel increases up to day 4–5, and then decreases in PC 2 (Suppl. Fig. 2). A Population X DEVT interaction for PC 5 resulted from Las Bocas CHCs shifting downwards with emergence day, but Punta Prieta CHCs showed little change from emergence day 1 to 6 (Suppl. Fig. 2).
Sex differences in CHCs were the only major source of variation for PC 3, and population differences along with a number of higher order interactions with populations were major sources of variation in PC 4. PC 6 reflected variation due to cactus substrates and their interactions with sex, population and development time (Table 2). Therefore, there were large, significant changes in covarying groups of CHCs in aged, adult D. mojavensis that were in part caused by differences in egg to adult development time consistent with previous observations (Etges 2014; Etges et al. 2010).
Transcriptome variation
In all cases we were able to detect the effects of development time differences on transcriptional variation in same age, sexually mature adults. For the complete model with all interactions, PROC NLMIXED (SAS-Institute 2004) did not converge on a result suggesting that the error distribution of the data did not fit a simple mixture of uniform and beta distributions (Allison et al. 2002). This was likely due to the unbalanced nature of the data because of the contrasting distributions of development times of Baja California and mainland populations of D. mojavensis, and some missing replicates due to insufficient numbers of flies to make replicate groups of 24 adults required for RNA extraction and analysis. There was a total of 137 whole genome hybridizations. In an effort to balance the design, we analyzed agria-reared adults from eclosion day 1, 2, 3, organ pipe-reared Baja adults on days 1, 2, 3, 4, and organ pipe-reared mainland adults on days 2, 3, and 4. Here, eclosion day 1 corresponded to a DEVT of 14 days, eclosion day 2 to DEVT of 15 days, etc. Thus, we divided the data into balanced subsets so that we could calculate least square means for all genes showing significant log2 fluorescence differences after FDR correction. For agria-reared flies, the data were balanced so we assessed the full factorial model including population, mating status, sex, and development time in PROC MIXED (SAS-Institute 2004). For organ pipe cactus-reared flies, there were insufficient numbers of mated mainland adults and no emergence day 1 flies owing to the longer DEVT of mainland populations, so we analyzed the Baja California population separately using a complete factorial model including mating status, sex and development time. Unmated mainland Las Bocas adults were assessed for the effects of sex, development time, and sex X development time interactions on levels of gene expression.
Agria cactus-reared flies
Differences in gene expression revealed by the full linear model were observed for all treatment effects and their interactions for 9295 different genes after correction with FDR < 0.01 and at least 1.5 X fold changes (Table 3). A total of 5695 of these genes had D. melanogaster orthologs, but clearly, most of the most significant effects on gene expression were apparent in the interaction terms between the effects of population, sex, mating status, and development time.
Table 3.
Gene expression summary for agria-reared male and female D. mojavensis from populations in mainland Mexico and Baja California. For all model main effects and interactions, numbers of differentially expressed genes are shown at FDR P> 0.01 together with a 1.5 fold change cutoff. For development time, FDR P<0.05 was also used.
| Treatments | No. pairwise differences; FDR < 0.01 | No. unique genes | No. genes with Dmel orthologs | No. pairwise differences; 1.5 x fold change cut-off | No. unique genes; 1.5 x cutoff | No. genes with Dmel orthologs |
|---|---|---|---|---|---|---|
| Main effects | ||||||
| Population (P) | 206 | 206 | 89 | 78 | 78 | 21 |
| Mating status (M) | 58 | 58 | 47 | 17 | 17 | 11 |
| Sex (S) | 8959 | 8959 | 5606 | 6851 | 6851 | 3982 |
| Development time (D) | 31 (297)* | 31 (281)* | 18 (199)* | 12 | 10 | 0 |
| Interaction effects | ||||||
| 5. P X M | 532 | 311 | 182 | 295 | 158 | 69 |
| 6. P X S | 29010 | 8829 | 5522 | 25085 | 7714 | 4591 |
| 7. P X D | 428 | 147 | 66 | 324 | 102 | 33 |
| 8. M X S | 28326 | 8578 | 5523 | 24070 | 7030 | 4276 |
| 9. M X D | 86 | 55 | 40 | 44 | 25 | 19 |
| 10. S X D | 55023 | 7735 | 4949 | 49830 | 6950 | 4272 |
| 11. M X S X D | 172360 | 7909 | 5064 | 163950 | 7419 | 4628 |
| 12. P X M X D | 1203 | 314 | 189 | 999 | 249 | 132 |
| 13. P X M X S | 91910 | 9023 | 5710 | 85168 | 8303 | 5078 |
| 14. P X S X D | 168314 | 7607 | 4811 | 162009 | 7295 | 4534 |
| 15. P X M X S D | 492160 | 7765 | 4950 | 484552 | 7604 | 4799 |
| Column totals | 1048606 | 67527 | 42766 | 1003272 | 59805 | 36445 |
| Number of unique genes | 10050 | 6327 | 9295 | 5695 | ||
Number in parentheses refers differentially expressed genes with FDR P < 0.05.
Development time
Expression levels of genes involved with both transcription and translation in adults were determined by length of egg to adult development time in flies reared on fermenting agria cactus tissues. There were 31 genes (Suppl. Table 3) that were differentially expressed due to day of emergence (day 1, 2, or 3) in agria-reared flies (FDR P < 0.01), 19 of which were annotated. Of these, 12 genes also showed 1.5 X fold differences, but none were annotated. Of the differentially transcribed 19 genes, there were two weakly supported functional groups associated with DNA binding and gastrulation (Suppl. Table 3). In order to fully explore development time-related transcription differences, we relaxed our FDR cutoff to P < 0.05 (Suppl. Table 3), and found 281 unique differentially expressed genes, 199 of which had D. melanogaster orthologs. Functionally enriched gene clusters were associated with DNA binding, regulation of transcription, and ribosome biogenesis (Enrich scores = 2.74, 1.81, 1.6, respectively). Increasing stringency to include only those genes with 1.5 X fold change resulted in a smaller group of 10 unique genes, none of which had D. melanogaster orthologs (Suppl. Table 3).
Overall differences in adult gene expression were further inspected in all binary comparisons between each day of emergence (i.e., day 1 vs 2, 1 vs 3, 2 vs 3) to identify which genes were up or down-regulated between emergence days (Table 4). These comparisons were not independent, but inspection of each contrast revealed the nature of transcriptome variation due to development time that persisted through adulthood in 9 day old females and 13 day old males. There were four genes that showed increased expression (FDR P < 0.01) in day 2 vs day 1 eclosed adults, three of which had D. melanogaster orthologs. These included Dmoj\GI1606, GI19815, and GI22216 that are associated with regulation of transcription, folic acid transport, and phospholipid biosynthesis, respectively (Suppl. Table 3). For FDR P < 0.05, there were 22 of 26 differentially expressed genes with D. melanogaster annotation with a variety of metabolic functions including ion binding and transcriptional regulation (Table 4). Only two genes showed greater expression in day 1 vs day 2 adults (FDR P < 0.01), but neither was annotated. There were 19 genes with greater expression levels (FDR P < 0.05), but just 5 were annotated, one, Dmoj\GI1097, with neurotransmitter secretion function and the other, Dmoj\GI21297, involved with phagocytosis. Thus, a small number genes associated with general metabolic processes differed in expression levels between aged adults that eclosed on the first vs. those on the second day.
Table 4.
Development time differences in gene expression for agria-reared flies from both populations comparing up/down expression differences for day 1, 2, and 3 development times, all FDR P < 0.05. Comparisons indicate greater (>) and lesser (<) transcript amounts for each comparison and numbers of genes involved.
| Emergence day comparison | ||||||||
|---|---|---|---|---|---|---|---|---|
| Day 1 vs 2 | Day 1 vs 3 | Day 2 vs 3 | ||||||
| Up/down comparison+ |
GOterm | Enrich score |
Up/down comparison+ |
GOterm | Enrich score |
Up/down comparison+ |
GOterm | Enrich score |
| Day 1 > 2 (19, 5) + | none | Day 1 > 3 (50, 20) |
|
0.81 | Day 2 > 3 (16, 13) |
|
2.32** 1.35* |
|
| Day 1 < 2 (26, 22) |
|
0.13 - |
Day 1 < 3 (161, 122) |
|
2.18** 1.85* 1.84* 1.27* |
Day 2 < 3 (24, 17) |
|
|
number of genes, number of genes with D. melanogaster orthologs
The largest numbers of differentially expressed genes in agria-reared flies were observed in emergence day 1 vs. 3 comparisons (Table 4, Suppl. Table 3). For adults eclosing on day 3, 15 genes were upregulated compared with day 1 adults at FDR P < 0.01, but just one had > 1.5 X fold change, FBtr0172291, an unannotated transcript. At FDR P < 0.05, there were 122 of 161 genes of diverse metabolic function that have D. melanogaster orthologs and were enriched for rRNA processing/ribosome manufacture, calponin-like actin-binding in flight muscle (Winder & Walsh 1993), cyclin-dependent protein kinase regulation, and chromatin remodeling (Table 4). So, 9 day old female and 13 day old male D. mojavensis reared on agria cactus with preadult development times just two days longer than those that eclosed on the first day had significantly higher rates of gene expression for 161 different genes, including those associated with regulation of transcription, protein synthesis, cell cycle regulation, and regulation of actin in flight muscle.
One gene annotated in D. mojavensis, FBtr0172018 (Dmoj\GI21293, FBgn0144023), was expressed at significantly higher levels in flies that eclosed on the first day vs the third day, 1.52 X fold change, FDR P = 5.57 X 10−6). This gene encodes a GTP binding factor associated with ribosomal protein S5 (St. Pierre et al. 2014) suggesting early eclosing adults have higher rates of polypeptide elongation than flies eclosing on day 3. Dmoj\GI21293 has no orthologs in D. melanogaster, but has 84.1 % BLASTn sequence similarity with Dvir\GJ20895 indicating this gene is virilis-repleta group-specific. Two other genes, Dmoj\GI15831 and GI14896, were also upregulated in adults that eclosed on day 1 vs 3, the former orthologous to rho-6 in D. melanogaster that functions in serine-type endopeptidase activity. The latter gene is unannotated. At FDR P < 0.05, there were 50 genes up-regulated in day 1 vs 3 adults, 20 of which with D. melanogaster orthologs (Table 4). Three of these were weakly enriched for transcription regulation, and the others were of diverse functions including imaginal disc development, signal transduction, and microsome-related electron transport (Suppl. Table 3). Flies emerging on day 2 were enriched for increased expression of DNA binding and neuron development genes vs day 3 adults. Attempts to assess population X development time interactions were unsuccessful because of the lack of annotation of most orthologs involved (Suppl. Table 4).
Population, sex, and mating status
Gender differences in gene expression for agria-reared flies were observed for 61.2 % (8959/14,528) of all genes on our microarray (Table 3). Large sex-specific differences in gene expression were expected (Brown et al. 2014; Chen et al. 2014; Graveley et al. 2011), so we filtered this gene list to only those showing greater the 1.5 X fold changes, and found 4310 genes were at higher expression levels in females vs. males, with another 2541 genes more highly expressed in males than females. Of the latter, 1088 had D. melanogaster orthologs and were enriched for microtubule-dynein function and mitochondrion membrane gene clusters. For genes overexpressed in females, 2894 were annotated and highly enriched for nuclear lumen, chromosome, gene regulation, chromatin splicing, and DNA replication (Table 5; Suppl. Table 5). A more detailed analysis of gender-specific expression differences is provided below given the large interaction effects including sex (Table 3).
Table 5.
Gene ontology and enrichment for population, mating status, population X mating status, and sex x mating status interactions for agria-reared flies. Punta Prieta = PP, Las Bocas = LB. All functional clustering was based on genes with FDR P < 0.01 and > 1.5 fold change for each treatment effect and interaction.
| Comparison | No. Genes (No. Annotated) | GOTerm | Enrich score |
|---|---|---|---|
| Population | |||
| 1. Punta Prieta > Las Bocas | 114 (48) | 1. proteolysis, peptidase activity, hydrolase | 1.7 |
| 2. protease, glycoprotein | 1.1 | ||
| 2. Las Bocas > Punta Prieta | 92 (41) | 1. oxidation reduction, P450 | 1.5 |
|
| |||
| Mating status | |||
| 3. Mated > Virgin | 55 (44) | 1. proteolysis, peptidase activity | 1.4 |
| 2. intracellular transport, Golgi | 0.9 | ||
| 4. Mated < Virgin | 3 (3) | Dmoj\GI23785; glycine catabolic process | |
| Dmoj\GI23785; chorion | |||
| Dmoj\GI16601; Insulin-like peptide 8, chorion formation, ovary | |||
|
| |||
| Sex | |||
| 5. Female < Male | 2541 (1088) | 1. microtubule-based movement | 3.5 **** |
| 2. cytoskeleton | 3.3 **** | ||
| 3. mitochondria | 2.8 **** | ||
| 4. cellular retinaldehyde-binding | 2.8 * | ||
| 5. glycolysis | 2.3 ** | ||
|
| |||
| 6. Female > Male | 4310 (2894) | 1. nuclear lumen | 37.8 **** |
| 2. chromosome | 26.9 **** | ||
| 3. regulation of transcription | 22.1 **** | ||
| 4. DNA replication | 18.6 **** | ||
| 5. chromatin modification, histones | 16.6 **** | ||
| 6. ribosome function | 15.5 **** | ||
|
| |||
| Population X Mating status | |||
|
| |||
| 7. PP mated > PP virgin | 24 (21) | 1. intracellular transport, Golgi | 2.0 |
| 2. peptidase activity, hydrolase | 0.9 | ||
|
| |||
| 8. LB virgin > PP virgin | 116 (78) | 1. oxidation reduction, P450 | 4.0 **** |
|
| |||
| 9. LB virgin < PP virgin | 77 (29) | 1. proteolysis, peptidase activity, hydrolase | - |
|
| |||
| 10. LB mated > PP mated | 23 (8) | none | |
|
| |||
| 11. LB mated < PP mated | 30 (12) | 1. ion binding, oxidation reduction | - |
|
| |||
| 12. LB virgin > PP mated | 27 (12) | 1. heme, ion binding | 1.4 |
|
| |||
| 13. LB virgin < PP mated | 99 (53) | 1. proteolysis, peptidase activity, hydrolase | 4.4 **** |
| 2. endopeptidase activity | 2.5 | ||
|
| |||
| Mating status X Sex | |||
|
| |||
| 14. Mated ♀ > Virgin ♀ | 46 (29) | 1. egg production | 2.0 * |
| 2. regulation of transcription | 1.3 | ||
|
| |||
| 15. Mated ♀ < Virgin ♀ | 10 (7) | 1. female meiosis | - |
|
| |||
| 16. Mated ♂ > Virgin ♂ | 21 (17) | 1. membrane, Golgi redox | 0.7 |
|
| |||
| 17. Mated ♂ < Virgin ♂ | 0 | ||
|
| |||
| 18. Mated ♂ < Virgin ♀ | 2416 (1684) | 1. nuclear lumen | 21.3 **** |
| 2. transcription regulation | 15.0 **** | ||
| 3. chromatin modulation | 9.6 **** | ||
|
| |||
| 19. Mated ♂ > Virgin ♀ | 1323 (611) | 1. transmembrane | 2.6 |
| 2. microtubule movement | 1.9 | ||
| 3. cilium assembly | 1.8 | ||
| 4. pigmentation | 1.5 | ||
|
| |||
| 20. Virgin ♂ < Virgin ♀ | 1104 (886) | 1. chromosome | 19.8 **** |
| 2. DNA replication | 17.9 **** | ||
| 3. nuclear lumen | 16.6 **** | ||
| 4. nucleotide binding | 14.9 **** | ||
|
| |||
| 21. Virgin ♂ > Virgin ♀ | 1128 (437) | 1. Cellular retinaldehyde-binding | 2.2 * |
| 2. microtubule cytoskeleton | 2.0 * | ||
| 3. mitochondrial membrane | 1.9 ** | ||
|
| |||
| 22. Mated ♀ < Mated ♂ | 1219 (553) | 1. exonuclease activity | 1.5 |
| 2. endonuclease activity | 1.5 | ||
| 3. microtubule cytoskeleton | 1.4 | ||
|
| |||
| 23. Mated ♀ > Mated ♂ | 2358 (1700) | 1. nuclear lumen | 22.7 **** |
| 2. chromosome | 15.8 **** | ||
| 3. transcription regulation | 13.5 **** | ||
| 4. mRNA processing | 10.0 **** | ||
|
| |||
| 24. Virgin ♀ < Mated ♂ | 1085 (417) | 1. mitochondrial membrane | 3.5 ** |
| 2. microtubule cytoskeleton | 2.7 ** | ||
| 3. tubulin-tyrosine ligase activity | 2.4 | ||
| 4. metallopeptidase activity | 2.4 | ||
|
| |||
| 25. Virgin ♀ > Mated ♂ | 672 (521) | 1. ribosome biogenesis | 12.8 **** |
| 2. chromosome | 12.1 **** | ||
| 3. nuclear lumen | 10.4 **** | ||
| 4. nucleotide binding | 8.7 **** | ||
|
| |||
| 26. Mated ♀ < Virgin ♂ | 1783 (779) | 1. tubulin-tyrosine ligase activity | 2.7 |
| 2. Cellular retinaldehyde-binding | 1.9 | ||
| 3. mitochondrial membrane | 1.9 | ||
|
| |||
| 27. Mated ♀ > Virgin ♂ | 3420 (2461) | 1. nuclear lumen | 20.2 **** |
| 2. chromosome | 15.1 **** | ||
| 3. transcription regulation | 13.1 **** | ||
| 4. mRNA processing | 8.9 **** | ||
|
| |||
| 28. Virgin ♀ < Virgin ♂ | 3207 (1328) | 1. mitochondrial membrane | 2.9 ** |
| 2. microtubule cytoskeleton | 2.4 **** | ||
| 3. Cellular retinaldehyde-binding | 2.0 | ||
| 4. glycolysis | 1.9 | ||
|
| |||
| 29. Virgin ♀ > Virgin ♂ | 4306 (3076) | 1. nuclear lumen | 38.3 **** |
| 2. chromosome | 21.9 **** | ||
| 3. transcription regulation | 19.8 **** | ||
| 4. DNA replication | 15.4 **** | ||
Population-specific differences in aged adult D. mojavensis involved contrasting patterns of gene expression associated with protein catabolism, ion binding related P450 cytochrome activity, and several genes involved with courtship behavior when reared on agria cactus. There were 206 genes that differed in expression between populations (FDR < 0.01), but only 89 (43.2%) had D. melanogaster orthologs, and of the 78 genes showing 1.5 X fold change differences in expression, only 21 (26.9%) had D. melanogaster orthologs (Table 3). GO clustering of the 89 orthologs (Table 5) showed that genes with higher expression in Punta Prieta flies (PP > LB) were enriched for proteolysis and peptidase genes (Enrich score = 1.7). Las Bocas flies (LB > PP) were enriched for ion binding and P450 gene expression (Enrich score = 1.6) consistent with previous observations (Etges 2014), with increased expression of candidate behavioral genes including takeout involved with adult feeding and courtship behavior, Pbprp5 or odorant-binding protein 28a involved with pheromone perception, and Gr2a, a taste receptor gene. Similar results were observed for the genes with at least 1.5 X fold changes where the same two gene clusters were recovered (Suppl. Table 5).
Remarkably few genes showed expression changes due to exposure to the opposite sex for 24 hr consistent with previous observations (Smith et al. 2013), but most of the significant differences in gene expression involving mating status were due to interactions with the other main effects (Table 3, 5). All but 3 of the 58 genes differentially expressed due to mating status were upregulated in adults exposed to the opposite sex vs. virgins (Suppl. Table 5). Several weakly supported clusters identified with DAVID (Huang et al. 2009) indicated mated adults had higher levels of gene expression in proteolysis and peptidase, intracellular protein transport, and regulation of transcription (Table 5). Three candidate CHC gene orthologs, CG2781 (fatty acid elongase), CG6271 (triglyceride lipase), and bond (fatty acid elongase) were upregulated in mated flies (Supple. Table 5) suggesting exposure to the opposite sex increased CHC production. In D. melanogaster, bond is required for male sex pheromone synthesis (J. Yew, pers. comm.). While there were 532 genes showing significant Population x Mating status (P x M) interactions (FDR P < 0.01), 295 genes also passed the 1.5 X fold filter. Examination of individual P x M contrasts revealed increased expression of genes with significant functional clustering for redox, P450 function in Las Bocas vs. Punta Prieta virgin adults and proteolysis, peptidase activity, and hydrolase activities in mated Punta Prieta adults vs virgin Las Bocas adults (Table 5) similar to the main effects of population (see above). While increased expression of protease and hydrolase gene clusters in mated vs. virgin adults suggests these genes may function in female reproductive tract interactions with male ejaculates, none of the orthologs here overlapped with those in previous studies (Kelleher et al. 2007).
By breaking down Sex X Mating status interactions into all possible pairwise comparisons we hoped to reveal the sex-specific nature of mating status differences, but these interactions revealed little more than the main effects of sex on gene expression (Table 5). For same-sex comparisons (Table 5, rows 12–15), mated females showed enrichment for genes upregulated for egg production and transcription regulation vs. virgin females while expression of genes associated with meiosis was greater in virgin females. Mated males showed increased expression of membrane bound Golgi apparatus genes vs. virgin males, and no genes in virgin males were detected with greater expression levels than in mated males. The remaining pairwise comparisons all involved males vs. females where females showed significantly higher levels of expression for gene clusters involved in DNA replication and regulation of transcription than males (Table 5; rows 16, 18, 21, 23, 25, 27) and males showed increased expression for a wider spectrum of genes with less significant GO clustering, including microtubule cytoskeleton, mitochondrial membrane, and exo- and endonuclease activity (Table 5; rows 17, 19, 20, 22, 24, 26). Thus, in agria cactus-reared adults, most sex-specific differences in gene expression were unaffected by exposure to the opposite sex. We did not attempt to interpret any of the three or four way interactions given the numbers of genes involved, but underscore how gender specific differences in gene expression were influenced by significant interactions with all of the main effects in this experiment (Table 3).
Organ pipe cactus-reared flies
Each population of D. mojavensis was analyzed separately because of the unbalanced design (Tables 6, 7). Expression of 70.5 % (10,237/14,528) of all genes in Punta Prieta, Baja California adults was influenced by sex with mating status and development time influencing expression of far fewer orthologs. For mainland, Las Bocas adults, 59 % (8570) of all genes were differentially transcribed in males and females with expression of 20 different genes influenced by development time (FDR P < 0.01), and 194 genes at FDR P < 0.05. The vast majority of significant gene expression differences were observed as interactions between main model effects (Table 6, 7).
Table 6.
Gene expression summary for organ pipe-reared Punta Prieta, Baja California flies only. OP1b
| Treatments | No. pairwise differences; FDR P < 0.01 | No. unique genes | No. genes with Dmel orthologs | No. pairwise differences; 1.5 x fold change cut-off | No. unique genes; 1.5 x cutoff | No. genes with Dmel orthologs |
|---|---|---|---|---|---|---|
| Main effects | ||||||
| Mating status (M) | 420 | 420 | 344 | 46 | 46 | 27 |
| Sex (S) | 10237 | 10237 | 6415 | 6778 | 6778 | 3801 |
| Development time (D) 1 | 32 (305) | 28 (254) | 20 (147) | 54 | 47 | 14 |
| Interaction effects | ||||||
| M X S | 18241 | 10135 | 6300 | 9543 | 7065 | 3784 |
| M X D | 1537 | 864 | 650 | 5134 | 2229 | 829 |
| S X D | 84033 | 9475 | 5944 | 84079 | 9175 | 5175 |
| M X S X D | 296984 | 10206 | 6532 | 235710 | 10690 | 6159 |
| Column totals | 411484 | 41365 | 26205 | 341344 | 36030 | 19789 |
| Number of unique genes | 11622 | 7295 | 11027 | 6367 | ||
Numbers in parentheses correspond to FDR P < 0.05.
Table 7.
Gene expression summary for organ pipe-reared Las Bocas, mainland, unmated flies only. OP2b
| Treatments | No. pairwise differences; FDR P < 0.01 | No. unique genes | No. genes with Dmel orthologs | No. pairwise differences; 1.5 x fold change cut-off | No. unique genes; 1.5 x cutoff | No. genes with Dmel orthologs |
|---|---|---|---|---|---|---|
| Sex | 8570 | 8570 | 5513 | 7037 | 7037 | 4079 |
| Development time 1 | 21 (208) | 20 (194) | 11 (123) | 55 | 47 | 5 |
| Sex X Development time | 56956 | 7951 | 5148 | 51674 | 7243 | 1691 |
| Column totals | 65547 | 16541 | 10673 | 58766 | 14354 | 5775 |
| Number of unique genes | 8709 | 5615 | 7874 | 4557 |
Numbers in parentheses correspond to FDR P < 0.05.
Development time
At FDR P < 0.01, 32 orthologs in organ pipe-reared Punta Prieta, Baja California adults showed differences in expression due to development time, 20 had orthologs in D. melanogaster, and 12 were annotated (Table 6). DAVID identified four clusters of genes enriched for mitochondrial iron ion binding, oxidative phosphorylation, oxidation reduction, and membrane function (enrich scores = 1.97, 1.33, 1.02, 0.46, respectively; Supple. Table 6). So, adults from this Baja California population reared on organ pipe cactus showed significant differences in transcript abundance determined by differences in preadult DEVT for genes associated with mitochondrial ATP production and electron transfer.
In order to assess +/− directional differences due to DEVT in adult transcript levels, we assessed all pairwise comparisons of DEVT day at FDR P < 0.05 as for agria-reared flies (Table 8). Because organ pipe cactus caused longer DEVT than agria (Supple. Table 1, Supple Fig. 1), there were enough flies to form emergence day samples from 1 to 4. Adults emerging on the day 1 vs. day 2 showed few differences in gene expression, with day 2 adults expressing higher transcript levels of aconitate hydratase, Dmoj\GI18654, and two unannotated genes than day one adults. Dmoj\GI18654 is orthologous to pAbp that has pleiotropic effects on synaptic transmission, regulation of translation, oogenesis, and dorsal/ventral pattern formation. One annotated gene, Gadd45, which is part of a protein kinase cascade associated with the regulation of oviposition was expressed at higher levels in day 1 adults than day 3 emerged adults. Two annotated orthologs were expressed at higher levels in day 3 than day 1 adults, CG31559 and pAbp, that are associated with cellular electron transport and synaptic transmission, respectively (Table 8).
Table 8.
Development time differences in gene expression for organ pipe-reared Punta Prieta, Baja California flies comparing up/down expression differences for day 1, 2, 3, and 4 development times, all FDR P < 0.05. Comparisons indicate greater (>) and lesser (<) transcript amounts for each comparison and numbers of genes involved.
| Emergence day comparison | ||||||||
|---|---|---|---|---|---|---|---|---|
| Day 1 vs 2 | Day 1 vs 3 | Day 1 vs 4 | ||||||
| Up/down comparison+ | GOterm/ID | Enrich score | Up/down comparison+ | GOterm/ID | Enrich score | Up/down comparison+ | GOterm/ID | Enrich score |
| Day 1 > 2 (1, 0) | none | Day 1 > 3 (1, 1) | Dmoj\GI19497 Gadd45; regulation of oviposition | Day 1 > 4 (4, 1) | 1. Dmoj\GI24142, endosome transport | |||
| Day 1 < 2 (3, 2) | 1. Dmoj\GI21890 aconitate hydratase 2. Dmoj\GI18654 pAbp; poly(A) RNA binding |
Day 1 < 3 (5,2) | 1. CG31559; electron carrier 2. Dmoj\GI18654 pAbp; poly(A) RNA binding |
Day 1 < 4 (40, 35) | 1. Mitochondria, ATP synthesis 2. TCA cycle 3. endopeptidase activity |
4.2**** 2.7* 0.2 |
||
| Day 2 vs 3 | Day 2 vs 4 | Day 3 vs 4 | ||||||
| Up/down comparison+ | GOterm/ID | Enrich score | Up/down comparison+ | GOterm/ID | Enrich score | Up/down comparison+ | GOterm/ID | Enrich score |
| Day 2 > 3 (6, 4) | 1. Dmoj\GI19844; exu, bicoid mRNA localization 2. desat2 3. Dmoj\GI23403; triglyceride lipase 4. Dmoj\GI18075; ATP synthesis |
Day 2 > 4 (1, 0) | FBtr0162777 (not annotated) | Day 3 > 4 (31, 14) | 1. nucleotide binding | 1.5 | ||
| Day 3 > 2 (2, 2) | unknown | Day 4 > 2 (65,49) | 1. mitochondria 2. mitochondria, phosphorylation 2. metal ion binding |
1.4 1.0 0.2 |
Day 4 > 3 (95, 59) | 1. ATP synthesis 2. signal peptide |
1.3 0.5 |
|
numbers in parentheses denote (number of genes, number of genes with D. melanogaster orthologs)
Most transcriptional variation due to longer DEVT in organ pipe cactus-reared Baja California adults was associated with increased expression of genes involved in ATP synthesis and energy production. By far, the largest numbers of genes showing expression differences were those including day 4 emergence adults, i.e., adults with longer egg to adult DEVT. There were 40 orthologs with increased expression in day 4 vs day 1 adults, FDR P < 0.05. Of these 35 were annotated and were enriched for gene clusters involving ATP synthesis, the TCA cycle and endopeptidase activity. Similar, overlapping patterns were observed for day 4 > 2 and 4 > 3 comparisons (Table 8). A candidate cuticular hydrocarbon gene, desat2, was one of four annotated genes at higher expression in day 2 vs day 3 adults suggesting increased adult CHC processing in adults with somewhat shorter DEVT.
Adult flies from each population of D. mojavensis reared on organ pipe cactus showed contrasting patterns of expression for gene clusters associated with oxidative phosphorylation and energy production due to preadult DEVT. For Las Bocas, mainland adults, a large fraction of expression differences due to DEVT involved unannotated genes (Table 7). Only 20 genes were differentially expressed at FDR P < 0.01, and 11 were annotated (Supple. Table 9). Most of these genes were involved with amino acid, carbohydrate, and glycogen metabolism, as well as mitochondrial redox homeostasis and one gene, ade5, associated with male-male behavior. In order to explore these data further, we relaxed our cutoff to FRD P < 0.05. This resulted in a total of 123 differentially expressed genes due to DEVT differences that were annotated. DAVID identified a few weakly enriched gene clusters associated with vitamin binding, NADP binding, mitochondrial function, and tRNA aminoacylation. Relaxing the cutoff to FDR P < 0.1 resulted in DEVT day comparisons for 797 transcripts, of which 538 were annotated. Functional enrichment clustering yielded similar results with those at FDR P < 0.05, and in contrast to Baja California adults, mainland adults with shorter DEVT (day 2 > 3, 2 > 4) were functionally enriched for genes with higher transcription rates for mitochondrial ATP synthesis, phosphorylation, and protein synthesis (Table 10). There were 168/196 annotated genes with increased expression in development day 3 vs 4 adults that were enriched for protein stacking, charged tRNA synthesis, Golgi body formation, and ATP binding all suggesting increased protein metabolism. However, there were also a large number of other genes with increased expression of diverse metabolic functions (Supple. Table 9).
Table 10.
Development time differences in gene expression for organ pipe-reared Las Bocas flies comparing up/down expression differences for day 2, 3, and 4 development times1, all FDR P < 0.05. Comparisons indicate greater (>) and lesser (<) transcript amounts for each comparison and genes involved. See Table 7 for gene summary.
| Emergence day comparison | ||||||||
|---|---|---|---|---|---|---|---|---|
| Day 2 vs 3 | Day 2 vs 4 | Day 3 vs 4 | ||||||
| Up/down comparison+ | GOterm/ID | Enrich score | Up/down comparison+ | GOterm/ID | Enrich score | Up/down comparison+ | GOterm/ID | Enrich score |
| Day 2 > 3 (79,32) | 1. mitochondria | 2.4 * | Day 2 > 4 (308,260) | 1. mitochondria 2. cofactor binding, protein synthesis 3. ATP binding 4. RNA synthesis 5. Aldehyde dehydrogenase 6. energy production 7. glucose metabolism |
4.7 * 3.4 **** 2.4 2.3 2.1 1.8 *** 1.7 * |
Day 3 > 4 (196,168) | 1. Ankyrin repeat 2. tRNA aminoacylation 3. Golgi apparatus 4. ATP binding 5. proteolysis |
2.0 1.8 1.6 1.6 1.3 |
| Day 3 > 2 (25,10) | 1. adult life span | - | Day 4 > 2 (37,13) | 1. zinc ion binding | 0.34 | Day 4 > 3 (152,55) | 1. cuticle protein 2. ion binding |
1.3 0.5 |
Insufficient numbers of day 1 organ pipe cactus reared flies were caused by longer development times of mainland flies.
numbers in parentheses denote (number of genes, number of genes with D. melanogaster orthologs)
Mating status, sex, and mating status X sex interactions
More genes showed differences in expression due to mating status in organ pipe-reared, Baja California adults (Table 9) than when reared on agria cactus (Table 5). There were 315 genes of which 269 were annotated that were overexpressed in adults exposed to the opposite sex than in virgin flies. DAVID identified significantly enriched gene clusters in mated adults associated with Golgi associated protein transport and proteolysis as with agria-reared flies (Table 5). Of 105 genes, 75 were annotated that were overexpressed in virgin adults associated with amino acid metabolism including both carboxylic acid biosynthesis and carboxylesterase activity (Table 9; Supple. Table 7). Differences due to sex were consistent in all three data sets (Tables 5, 9, Supple. Table 10) where females showed greater expression in thousands of genes enriched for nucleus, DNA replication, regulation of transcription, ribosome activity and chromatin organization associated with egg and embryo formation (Etges et al. 2015). However males showed significantly greater expression of thousands of genes highly enriched for microtubule, cytoskeleton, mitochondrial membrane, glycolysis/alcohol metabolism, and intercellular transport function. Similar to agria-reared flies Table 5), evaluation of interaction terms for organ pipe-reared flies, i.e. Mating status X Sex and Sex X Development time, revealed little insights into functional gene enrichment other than the effects of sex differences (Table 9, Supple. Table 10).
Table 9.
Gene ontology and enrichment for mating status, sex, mating status X sex, and sex x mating status interactions for organ pipe cactus-reared, Baja California flies. All functional clustering was based on genes with FDR P < 0.01 and > 1.5 fold change for each treatment effect and interaction - see Table 6.
| Comparison | No. Genes (No. Annotated) | GOTerm | Enrich score |
|---|---|---|---|
| Mating status | |||
| 1. Mated > Virgin | 315 (269) | 1. protein transport | 3.4* |
| 2. vesicle-mediated transport | 3.0* | ||
| 3. Golgi transport | 2.6* | ||
| 4. proteolysis | 2.5* | ||
| 2. Mated < Virgin | 105 (75) | 1. carboxylic acid biosynthesis | 2.4 |
| 2. carboxylesterase activity | 1.8 | ||
| 3. amino acid metabolism | 1.8 | ||
|
| |||
| Sex | |||
| 3. Female < Male | 2441 (954) | 1. microtubule, cytoskeleton | 4.0 **** |
| 2. mitochondrial membrane | 3.8** | ||
| 3. Tubulin-tyrosine ligase | 3.3 ** | ||
| 4. Glutathione metabolism | 2.1 | ||
|
| |||
| 4. Female > Male | 4247 (2847) | 1. nucleus | 31.9 **** |
| 2. chromosome | 25.6 **** | ||
| 3. transcription regulation | 24.7 **** | ||
| 4. DNA replication | 18.8 **** | ||
| 5. ribosome biogenesis | 14.8 **** | ||
| 6. mRNA processing | 12.8 **** | ||
|
| |||
| Mating status X Sex | |||
|
| |||
| 5. Mated ♂ > Virgin ♂ | 70 (35) | 1. peptidase, hydrolase, zinc ion binding | 1.5 |
|
| |||
| 6. Mated ♂ < Virgin ♂ | 91 (67) | 1. glycine metabolic pathway | 3.0 |
| 2. vitamin binding | 2.0 | ||
| 3. amino acid catabolism | 1.8 | ||
|
| |||
| 7. Mated ♂ < Virgin ♀ | 4056 (2602) | 1. transcription regulation | 31.5 **** |
| 2. nuclear lumen | 29.7 **** | ||
| 3. chromosome | 20.6 **** | ||
| 4. DNA replication | 19.4 **** | ||
| 5. chromatin modification | 15.0 **** | ||
| 6. ribosome biogenesis | 13.5 **** | ||
|
| |||
| 8. Mated ♂ > Virgin ♀ | 2428 (975) | 1. microtubule movement | 4.1 **** |
| 2. mitochondrial membrane | 3.5 ** | ||
| 3. microtubule motor | 3.1 **** | ||
| 4. pyruvate metabolism | 2.2 * | ||
|
| |||
| 9. Virgin ♂ < Virgin ♀ | 4089 (2813) | 1. nuclear lumen | 31.4 **** |
| 2. transcription regulation | 24.2 **** | ||
| 3. chromosome | 22.5 **** | ||
| 4. nucleotide binding | 20.1 **** | ||
| 5. DNA replication | 19.5 **** | ||
|
| |||
| 10. Virgin ♂ > Virgin ♀ | 2519 (1008) | 1. mitochondrial membrane | 3.9 **** |
| 2. microtubule cytoskeleton | 3.7 **** | ||
| 3. glutathione metabolism | 2.1 * | ||
| 4. glycolysis | 2.0 * | ||
Discussion
Differences in preadult development time were associated with variation in expression of genes and CHCs in adult D. mojavensis, revealing developmental ties between disparate parts of the life history and determinants of adult mating success. In addition to other sources of variation including sex, population, host cactus, age, mating status, and adult rearing conditions (Etges & de Oliveira 2014; Etges et al. 2015; Pletcher et al. 2002; Smith et al. 2013), variation in DEVT had carry-over effects that influenced transcriptome variation into early adulthood. Egg to adult development time is a key life history character that varies genetically between Baja California and mainland Mexico and Arizona populations of D. mojavensis and its expression is influenced by cactus rearing substrates (Etges 1990). While we could not evaluate the full range of DEVT variation and its effects on adult transcriptome variation (see results), we did uncover hundreds of genes that were differentially expressed in mature mated and unmated adults of different development times. Organ pipe cactus tissues ferment slower than agria tissues that lengthens DEVT (Etges 1989), particularly in mainland populations that use organ pipe cactus in nature. Thus, agria-reared flies show less rearing substrate induced DEVT variation, usually emerging over a span of 3–4 days, yet there were significant differences in expression of genes enriched for regulation of transcription, DNA binding, RNA splicing, ribosome biogenesis, protein synthesis, cell cycle regulation, neuron development, and regulation of actin in flight muscle (Table 4). Most of these differences were apparent for flies with longer DEVT suggesting that flies that spend more time as larvae/pupae have higher rates of gene expression and somatic tissue development as young, but sexually mature adults.
For organ pipe-reared flies, development time differences had quite contrasting effects on patterns of adult gene expression that were population specific. Baja California adults that took longer to eclose showed higher expression of genes enriched for ATP synthesis, the TCA cycle, and endopeptidase activity indicating higher metabolic rates in flies with longer DEVT. Few candidate CHC genes showed any effects of DEVT except desat2 (Table 8), a stearoyl-coA 9-desaturase encoding gene responsible for adding double bonds to growing hydrocarbon chains in oenocytes (Chung & Carroll 2015; Gleason et al. 2009). As 30–50% of adult CHCs are alkadienes (Etges & Jackson 2001; Toolson et al. 1990), and desat2 is very near a QTL that influences male mating success and a number of covarying CHCs (Etges et al. 2007; Etges et al. 2009), how DEVT influences desat2 expression may be a focal link in understanding the correlation between DEVT differences and mating success in D. mojavensis.
In contrast to Baja California flies reared on organ pipe cacti, mainland adults with shorter DEVT had higher expression of genes associated with mitochondrial ATP synthesis, phosphorylation, energy production, and protein synthesis (Table 10). As organ pipe cactus is the host plant used by most mainland populations of D. mojavensis, this may reflect a subtle form of host plant adaptation, but the phenotypic consequences of this shift in gene expression need to be evaluated. Mainland populations have significantly longer DEVT than Baja California populations and agria is the preferred host used almost exclusively in Baja California. Significant region X cactus and population X cactus interaction terms from common garden experiments for DEVT, as well as egg to adult viability, have suggested regional life history differences are due to host plant adaptation (Etges et al. 2010).
Patterns of preadult gene expression showed stage specific differences in the expression of hundreds of genes that differed due to host cactus and population (Etges et al. 2015), but differences in gene expression causing DEVT differences are not yet known. Variation in eclosion times approximates left skewed normal distributions with higher frequencies of longer DEVT in mainland populations, particularly when reared on organ pipe cactus (Fig. 2, Supple. Fig. 3). Studies of the genetic basis of DEVT variation have revealed effects of at least 8 QTL, 8 G(QTL) X E(cactus) interactions, and several cases of cactus influenced multiple QTL epistasis across all chromosomes. Line cross analyses revealed cactus, autosomal, X chromosome, cytoplasmic, and cactus interaction effects on DEVT, with some effects cross-specific (Etges et al. 2010). Thus, the genetic basis of regional DEVT differences in D. mojavensis is multigenic, significantly influenced by rearing substrates, and is clearly in need of further resolution through whole genome mapping.
Observation of direct effects of DEVT variation on adult gene expression suggests genetic and environmental variation in preadult stages can persist through somatic remodeling in the pupal stage of holometabolous insects like Drosophila. In agria-reared flies, several genes were over-expressed in emergence day 2 vs day 3 adults involved with neuronal development (Table 4) including homologs of derailed (drl) and ladybird early (lbe) involved in axon guidance, and Ets21C involved in dendrite morphogenesis. This is not a novel observation: some larval neurons are maintained into adulthood in the mushroom body of D. melanogaster (Lee et al. 1999), while some are reduced before addition of adult-specific neuronal projections (Marin et al. 2005). Larval experience can influence adult behavior in a number of insects, including learned behaviors in larval lepidopterans that persist through metamorphosis (Blackiston et al. 2008), and larval learning of adult nest mate recognition in ants (Signorotti et al. 2014). Further, there is evidence that some larval somatic tissues that could reflect larval diets survive into adulthood, e.g., larval fat body in D. melanogaster (Aguila et al. 2007). Differences in alternate splicing patterns in expression of ion channel and nervous system development genes were influenced by preadult cactus rearing conditions and mating success in male D. mojavensis (Smith et al. 2013) reinforcing the view that preadult rearing conditions can directly influence patterns of gene expression that alter adult courtship behavior.
Other potential causes of preadult carry-over effects are epigenetic modifications over the life cycle and hormonal shifts controlling development (Etges et al. 2015; Flatt et al. 2005; Smith et al. 2013; Snell-Rood et al. 2013). A significant component of gene expression over the life cycle of D. mojavensis involved groups of covarying genes in early development and late in adult life that were enriched for epigenetic regulation of gene expression (Etges et al. 2015). An enriched group of 15 genes was identified by DAVID (Huang et al. 2009) including Pimet, Bka, Sce, egg, mof, Sirt6, Su(z)12, and Mt2 that are involved with RNA, DNA and histone methylation (see Table S3 in Etges et al. 2015). Smith et al. (2013) observed that genes involved in DNA, RNA, and protein methylation in mated males were consistently upregulated vs. unmated males under rearing conditions similar to those used here. Thus, epigenetic modification occurs throughout the life cycle and is also associated with adult mating success in D. mojavensis.
A role of major genes controlling integrated life histories includes the pleiotropic consequences of steroid hormones such as juvenile hormone (JH) and ecdysone on Drosophila physiology, development and life history. Because so much is known about the antagonistic effects of JH and ecdysone as regulators of life cycle timing, body size, reproduction, and life span (Riddiford et al. 2003; Thummel 2001), expression of JH and genes downstream from it could explain much about life history evolution (reviewed in Flatt et al. 2005). Integrating expression of ecdysteroid biosynthesis over the life cycle and life history evolution in D. mojavensis is incomplete, but observed stage and age specific gene expression patterns involved pupal stages and young adult flies (Etges et al. 2015). Of 3506 genes down-regulated between early pupae and late pupae (288 and 384 hr old, respectively), 1796 were annotated including 10 weakly enriched for hormone regulation (Supple. Table 12). Included were homologs for Jhe, Jheh2, and Jheh3 that catabolize JH and sad associated with ecdysone synthesis. In 6 day old adults, there were 279 genes at their maximum levels of expression over the life cycle, and of the 213 annotated orthologs, 8 were significantly enriched for steroid metabolism (P < 0.0001), including CG1513, CG9205, Eo, phm, and Cyp18a1, all involved with ecdysone biosynthesis or steroid metabolism (Supple. Table 12). How these orthologs influence observed life history variation in D. mojavensis, and how rearing substrates influence the expression of these genes in different populations remains to be explored.
Cuticular hydrocarbons, egg to adult development time, and reproductive isolation
Variation in eclosion time caused CHC variation in adult D. mojavensis, a remarkable life history carry over effect that has direct relevance to understanding the genetic and ecological basis of ecological speciation (Etges 1998; Etges et al. 2010; Etges et al. 2009). While the within population shifts in adult CHC amounts associated with DEVT (Fig. 2) were more complex than previously observed, this was the first attempt to include the effects of population, sex, and mating status in one experiment. In a previous analysis, virgin F2 males showed consistent curvilinear shifts in CHC amounts with DEVT, while F2 males exposed to females showed linear decreases in CHC amounts over 6 days of eclosion (n = 1650) (see Figure S2 and S3 in Etges et al. 2010). These observations were also consistent with observed increases in CHCs in mainland populations between virgin adults separated into those that emerged on the first two days, “fast”, vs. all the remaining flies labeled “slow” (Etges 2014). Total hydrocarbons per fly were in greater abundance in the “slow” groups than in the “fast” groups (t = 2.38, P < 0.022). These patterns were similar to the observed CHC shifts for Baja virgins and mainland flies, respectively, in the present study (Figure 1), but the degree to which CHC amounts were influenced by most effects interacting with DEVT (Table 1) made clear that effects of preadult DEVT were underestimated. Sex, region, population, preadult rearing substrates, adult rearing substrates, adult age, exposure to desiccation, and adult temperature conditions all influence CHC composition in D. mojavensis (Etges & Ahrens 2001; Etges & de Oliveira 2014; Rajpurohit et al. 2013; Etges et al., unpublished data).
Both host plant differences and climatic variation have shaped life history variation in D. mojavensis (Etges 1989, 1990). Here, we revealed hundreds of genes differentially expressed in mature adults due to development time differences. Major categories included genes involved with adult energy metabolism, gene regulation, rates of protein synthesis, neural development, and others including a candidate behavioral isolation gene, desat2 (Table 8). The forces that shaped life history evolution in D. mojavensis have provided a window into the genomic basis of adaptation to different environments and resulting genetically based physiological and behavioral systems underlying the early stages of reproductive isolation. These functional connections across the life cycle suggest that transcriptomic approaches to understanding classical genetic correlations between life history characters should provide a deeper understanding of the genetics of life history evolution.
Supplementary Material
Acknowledgments
We thank A. J. Marlon and G. Almeida for excellent technical assistance and C. Ross for statistical help. Funding was provided by National Science Foundation grants DEB-0211125 to WJE and EF-0723930 to AGG and WJE, and a grant from the Center on the Economics and Demography of Aging (CEDA) – UC Berkeley to S. Tuljapurkar and WJE. The UNLV Genomics Core Facility was supported by grants from the National Center for Research Resources (5P20RR016464-11) and the National Institute of General Medical Sciences (8P20GM103440-11). We also thank the Secretaria de Medio Ambiente y Recursos Naturales in Mexico City for issuing a CITES permit, and M. A. Armella for helping us obtain it.
Footnotes
Data archiving
MIAME compliant microarray data are available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSM1858214. All cuticular hydrocarbon data are archived at Dryad doi:10.5061/dryad.483hr.
Contributor Information
William J. Etges, Email: wetges@uark.edu.
Cássia de Oliveira, Email: cassia.oliveira@lyon.edu.
Subhash Rajpurohit, Email: subhashrajpurohit@gmail.com.
Allen G. Gibbs, Email: allen.gibbs@unlv.edu.
Literature Cited
- Aguila JR, Suszko J, Gibbs AG, Hoshizaki DK. The role of larval fat cells in adult Drosophila melanogaster. Journal of Experimental Biology. 2007;210:956–963. doi: 10.1242/jeb.001586. [DOI] [PubMed] [Google Scholar]
- Alcorn SM, Orum TV, Steigerwalt AG, et al. Taxonomy and pathogenicity of Erwinia cacticida sp. nov. International Journal of Systematic Bacteriology. 1991;41:197–212. doi: 10.1099/00207713-41-2-197. [DOI] [PubMed] [Google Scholar]
- Allison D, Gadbury G, Heo M, et al. A mixture model approach for the analysis of microarray gene expression data. Computational Statistics and Data Analysis. 2002;39:1–20. [Google Scholar]
- Arnegard ME, McGee MD, Matthews B, et al. Genetics of ecological divergence during speciation. Nature. 2014;511:307–311. doi: 10.1038/nature13301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour M, Keeler-Wolf T, Schoenherr AA. Terrestrial Vegetation of California. 3. University of California Press; Berkeley, CA: 2007. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- Blackiston DJ, Silva Casey E, Weiss MR. Retention of memory through metamorphosis: Can a moth remember what it learned as a caterpillar? PLoS ONE. 2008;3:e1736. doi: 10.1371/journal.pone.0001736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochdanovits Z, de Jong G. Antagonistic pleiotropy for life-history traits at the gene expression level. Proceedings of the Royal Society of London. Series B: Biological Sciences. 2004;271:S75–S78. doi: 10.1098/rsbl.2003.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Brazner JC, Etges WJ. Pre-mating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. II. Effects of larval substrates on time to copulation, mate choice, and mating propensity. Evolutionary Ecology. 1993;7:605–624. [Google Scholar]
- Brown JB, Boley N, Eisman R, et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512:393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z-X, Sturgill D, Qu J, et al. Comparative validation of the D. melanogaster modENCODE transcriptome annotation. Genome Research. 2014;24:1209–1223. doi: 10.1101/gr.159384.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Carroll SB. Wax, sex and the origin of species: Dual roles of insect cuticular hydrocarbons in adaptation and mating. BioEssays. 2015;37:822–830. doi: 10.1002/bies.201500014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson T, Tuljapurkar S, Childs D. Using evolutionary demography to link life history theory, quantitative genetics and population ecology. Journal of Animal Ecology. 2010;79:1226–1240. doi: 10.1111/j.1365-2656.2010.01734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhães JP, Budovsky A, Lehmann G, et al. The Human Ageing Genomic Resources: online databases and tools for biogerontologists. Aging Cell. 2009;8:65–72. doi: 10.1111/j.1474-9726.2008.00442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delprat A, Etges WJ, Ruiz A. Reanalysis of polytene chromosomes in Drosophila mojavensis populations from Santa Catalina Island, California, USA. Drosophila Information Service. 2014;97:53–57. [Google Scholar]
- Doyon J, Boivin G. The effect of development time on the fitness of female Trichogramma evanescens. Journal of Insect Science. 2005;5:4. doi: 10.1093/jis/5.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etges WJ. Evolution of developmental homeostasis in Drosophila mojavensis. Evolutionary Ecology. 1989;3:189–201. [Google Scholar]
- Etges WJ. Direction of life history evolution in Drosophila mojavensis. In: Barker JSF, Starmer WT, MacIntyre RJ, editors. Ecological and Evolutionary Genetics of Drosophila. Plenum; New York: 1990. pp. 37–56. [Google Scholar]
- Etges WJ. Genetics of host-cactus response and life-history evolution among ancestral and derived populations of cactophilic Drosophila mojavensis. Evolution. 1993;47:750–767. doi: 10.1111/j.1558-5646.1993.tb01231.x. [DOI] [PubMed] [Google Scholar]
- Etges WJ. Premating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. IV. Correlated responses in behavioral isolation to artificial selection on a life history trait. The American Naturalist. 1998;152:129–144. doi: 10.1086/286154. [DOI] [PubMed] [Google Scholar]
- Etges WJ. No boundaries: genomes, organisms, and ecological interactions responsible for divergence and reproductive isolation. Journal of Heredity. 2014;105:756–770. doi: 10.1093/jhered/esu039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etges WJ, Ahrens MA. Premating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. V. Deep geographic variation in epicuticular hydrocarbons among isolated populations. The American Naturalist. 2001;158:585–598. doi: 10.1086/323587. [DOI] [PubMed] [Google Scholar]
- Etges WJ, de Oliveira CC. Premating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. X. Age-specific dynamics of adult epicuticular hydrocarbon expression in response to different host plants. Ecology and Evolution. 2014;4:2033–2045. doi: 10.1002/ece3.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etges WJ, de Oliveira CC, Gragg E, et al. Genetics of incipient speciation in Drosophila mojavensis. I. Male courtship song, mating success, and genotype x environment interactions. Evolution. 2007;61:1106–1119. doi: 10.1111/j.1558-5646.2007.00104.x. [DOI] [PubMed] [Google Scholar]
- Etges WJ, de Oliveira CC, Noor MAF, Ritchie MG. Genetics of incipient speciation in Drosophila mojavensis. III. Life history divergence and reproductive isolation. Evolution. 2010;64:3549–3569. doi: 10.1111/j.1558-5646.2010.01096.x. [DOI] [PubMed] [Google Scholar]
- Etges WJ, de Oliveira CC, Ritchie MG, Noor MAF. Genetics of incipient speciation in Drosophila mojavensis. II. Host plants and mating status influence cuticular hydrocarbon QTL expression and G x E interactions. Evolution. 2009;63:1712–1730. doi: 10.1111/j.1558-5646.2009.00661.x. [DOI] [PubMed] [Google Scholar]
- Etges WJ, Jackson LL. Premating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. VI. Epicuticular hydrocarbon variation in Drosophila mojavensis cluster species. Journal of Chemical Ecology. 2001;27:2125–2149. doi: 10.1023/a:1012203222876. [DOI] [PubMed] [Google Scholar]
- Etges WJ, Johnson WR, Duncan GA, Huckins G, Heed WB. Ecological genetics of cactophilic Drosophila. In: Robichaux R, editor. Ecology of Sonoran Desert Plants and Plant Communities. University of Arizona Press; Tucson: 1999. pp. 164–214. [Google Scholar]
- Etges WJ, Klassen CS. Influences of atmospheric ethanol on adult Drosophila mojavensis: Altered metabolic rates and increases in fitness among populations. Physiological Zoology. 1989;62:170–193. [Google Scholar]
- Etges WJ, Tripodi AD. Premating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. VIII. Mating success mediated by epicuticular hydrocarbons within and between isolated populations. Journal of Evolutionary Biology. 2008;21:1641–1652. doi: 10.1111/j.1420-9101.2008.01601.x. [DOI] [PubMed] [Google Scholar]
- Etges WJ, Trotter MV, de Oliveira CC, et al. Deciphering life history transcriptomes in different environments. Molecular Ecology. 2015;24:151–179. doi: 10.1111/mec.13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figlio DN, Guryan J, Karbownik K, Roth J. The effects of poor neonatal health on children's cognitive development. American Economic Review. 2014;104:3921–3955. doi: 10.1257/aer.104.12.3921. [DOI] [PubMed] [Google Scholar]
- Flatt T, Tu M-P, Tatar M. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. BioEssays. 2005;27:999–1010. doi: 10.1002/bies.20290. [DOI] [PubMed] [Google Scholar]
- Funk DJ. Isolating a role for natural selection in speciation: host adaptation and sexual isolation in Neochlamisus bebbianae leaf beetles. Evolution. 1998;52:1744–1759. doi: 10.1111/j.1558-5646.1998.tb02254.x. [DOI] [PubMed] [Google Scholar]
- Funk DJ, Nosil P, Etges WJ. Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proceedings of the National Academy of Sciences. 2006;103:3209–3213. doi: 10.1073/pnas.0508653103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadgil M, Bossert WH. Life historical consequences of natural selection. American Naturalist. 1970;104:1–24. [Google Scholar]
- Gleason JM, James RA, Wicker-Thomas C, Ritchie MG. Identification of quantitative trait loci function through analysis of multiple cuticular hydrocarbons differing between Drosophila simulans and Drosophila sechellia females. Heredity. 2009;103:416–424. doi: 10.1038/hdy.2009.79. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens JA, Etges WJ. Premating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. IX. Host plant and population specific epicuticular hydrocarbon expression influences mate choice and sexual selection. Journal of Evolutionary Biology. 2013;26:562–576. doi: 10.1111/jeb.12073. [DOI] [PubMed] [Google Scholar]
- Havens JA, Orzack SH, Etges WJ. Mate choice opportunity leads to shorter offspring development time in a desert insect. Journal of Evolutionary Biology. 2011;24:1317–1324. doi: 10.1111/j.1420-9101.2011.02265.x. [DOI] [PubMed] [Google Scholar]
- Heed WB. The origin of Drosophila in the Sonoran Desert. In: Barker JSF, Starmer WT, editors. Ecological Genetics and Evolution: The Cactus-Yeast-Drosophila Model System. Academic Press; Sydney: 1982. pp. 65–80. [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protocols. 2009;4(1):44–45. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Istock CA. Natural selection and life history variation: Theory plus lessons from a mosquito. In: Denno RF, Dingle H, editors. Insect life history patterns: habitat and geographic variation. Springer-Verlag; New York: 1981. pp. 113–127. [Google Scholar]
- Jones OR, Scheuerlein A, Salguero-Gomez R, et al. Diversity of ageing across the tree of life. Nature. 2014;505:169–173. doi: 10.1038/nature12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Swanson WJ, Markow TA. Gene duplication and adaptive evolution of digestive proteases in Drosophila arizonae female reproductive tracts. PLoS Genetics. 2007;3:e148. doi: 10.1371/journal.pgen.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo LQ. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lohse K, Clarke M, Ritchie MG, Etges WJ. Genome-wide tests for introgression between cactophilic Drosophila implicate a role of inversions during speciation. Evolution. 2015;69:1178–1190. doi: 10.1111/evo.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Watts R, Tanaka N, Ito K, Luo L. Developmentally programmed remodeling of the Drosophila olfactory circuit. Development. 2005;132:725–737. doi: 10.1242/dev.01614. [DOI] [PubMed] [Google Scholar]
- Markow TA. Mating systems of cactophilic Drosophila. In: Barker JSF, Starmer WT, editors. Ecological Genetics and Evolution: The Cactus-Yeast-Drosophila Model System. Academic Press; Sydney: 1982. pp. 273–287. [Google Scholar]
- Markow TA, Castrezana S, Pfeiler E. Flies across the water: genetic differentiation and reproductive isolation in allopatric desert Drosophila. Evolution. 2002;56:546–552. doi: 10.1111/j.0014-3820.2002.tb01365.x. [DOI] [PubMed] [Google Scholar]
- Martin TE. Avian life history evolution in relation to nest sites, nest predation and food. Ecological Monographs. 1995;65:101–127. [Google Scholar]
- Martin TE, Oteyza JC, Mitchell AE, Potticary AL, Lloyd P. Postnatal growth rates covary weakly with embryonic development rates and do not explain adult mortality probability among songbirds on four continents. The American Naturalist. 2015;185:380–389. doi: 10.1086/679612. [DOI] [PubMed] [Google Scholar]
- McKinnon JS, Mori S, Blackman BK, et al. Evidence for ecology's role in speciation. Nature. 2004;429:294–298. doi: 10.1038/nature02556. [DOI] [PubMed] [Google Scholar]
- Mullen SP, Shaw KL. Insect speciation rules: Unifying concepts in speciation research. Annual Review of Entomology. 2014;59:339–361. doi: 10.1146/annurev-ento-120710-100621. [DOI] [PubMed] [Google Scholar]
- Nosil P. Ecological Speciation. Oxford University Press; Oxford: 2012. [Google Scholar]
- Olsson M, Shine R. Growth to death in lizards. Evolution. 2002;56:1867–1870. doi: 10.1111/j.0014-3820.2002.tb00202.x. [DOI] [PubMed] [Google Scholar]
- Orzack SH, Steiner UK, Tuljapurkar S, Thompson P. Static and dynamic expression of life history traits in the northern fulmar Fulmarus glacialis. Oikos. 2011;120:369–380. doi: 10.1111/j.1600-0706.2010.17996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzack SH, Tuljapurkar S. Population dynamics in variable environments. VII. The demography and evolution of iteroparity. American Naturalist. 1989;133:901–923. [Google Scholar]
- Paaby AB, Bergland AO, Behrman EL, Schmidt PS. A highly pleiotropic amino acid polymorphism in the Drosophila insulin receptor contributes to life-history adaptation. Evolution. 2014;68:3395–3409. doi: 10.1111/evo.12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby AB, Schmidt PS. Dissecting the genetics of longevity in Drosophila melanogaster. Fly. 2009;3:29–38. doi: 10.4161/fly.3.1.7771. [DOI] [PubMed] [Google Scholar]
- Pechenik JA. Larval experience and latent effects: metamorphosis is not a new beginning. Integrative and Comparative Biology. 2006;46:323–333. doi: 10.1093/icb/icj028. [DOI] [PubMed] [Google Scholar]
- Pletcher SD, Macdonald SJ, Marguerie R, et al. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Current Biology. 2002;12:712–723. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- Rajpurohit S, Oliveira CC, Etges WJ, Gibbs AG. Functional genomic and phenotypic responses to desiccation in natural populations of a desert drosophilid. Molecular Ecology. 2013;22:2698–2715. doi: 10.1111/mec.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick DN, Bryant MJ, Roff D, Ghalambor CK, Ghalambor DE. Effect of extrinsic mortality on the evolution of senescence in guppies. Nature. 2004;431:1095–1099. doi: 10.1038/nature02936. [DOI] [PubMed] [Google Scholar]
- Reznick DN, Butler MJI, Rodd FH, Ross P. Life history evolution in guppies (Poecilia reticulata). 6. Differential mortality as a mechanism for natural selection. Evolution. 1996;50:1651–1660. doi: 10.1111/j.1558-5646.1996.tb03937.x. [DOI] [PubMed] [Google Scholar]
- Richmond MP, Johnson S, Haselkorn TS, et al. Genetic differentiation of island populations: geographical barrier or a host switch? Biological Journal of the Linnean Society. 2013;108:68–78. [Google Scholar]
- Ricklefs RE. Embryo development and ageing in birds and mammals. Proceedings of the Royal Society B: Biological Sciences. 2006;273:2077–2082. doi: 10.1098/rspb.2006.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford LM, Hiruma K, Zhou X, Nelson CA. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochemistry and Molecular Biology. 2003;33:1327–1338. doi: 10.1016/j.ibmb.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Rollo CD. Growth negatively impacts the life span of mammals. Evolution and Development. 2002;4:55–61. doi: 10.1046/j.1525-142x.2002.01053.x. [DOI] [PubMed] [Google Scholar]
- Rose MR, Charlesworth B. Genetics of life history in Drosophila melanogaster. I. Sib analysis of adult females. Genetics. 1981;97:173–186. doi: 10.1093/genetics/97.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS-Institute. SAS/STAT 9.1.2. SAS Institute, Inc; Cary, NC: 2004. [Google Scholar]
- Schluter D, Conte GL. Genetics and ecological speciation. Proceedings of the National Academy of Sciences (USA) 2009;106:9955–9962. doi: 10.1073/pnas.0901264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O, Butlin RK, Keller I, et al. Genomics and the origin of species. Nature Reviews Genetics. 2014;15:176–192. doi: 10.1038/nrg3644. [DOI] [PubMed] [Google Scholar]
- Signorotti L, Jaisson P, d'Ettorre P. Larval memory affects adult nest-mate recognition in the ant Aphaenogaster senilis. Proceedings of the Royal Society B: Biological Sciences. 2014;281 doi: 10.1098/rspb.2013.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G, Fang Y, Liu X, et al. Transcriptome-wide expression variation associated with environmental plasticity and mating success in cactophilic Drosophila mojavensis. Evolution. 2013;67:1950–1963. doi: 10.1111/evo.12082. [DOI] [PubMed] [Google Scholar]
- Smith G, Lohse K, Etges WJ, Ritchie MG. Model-based comparisons of phylogeographic scenarios resolve the intraspecific divergence of cactophilic Drosophila mojavensis. Molecular Ecology. 2012;21:3293–3307. doi: 10.1111/j.1365-294X.2012.05604.x. [DOI] [PubMed] [Google Scholar]
- Snell-Rood EC, Troth A, Moczek AP. DNA methylation as a mechanism of nutritional plasticity: Limited support from horned beetles. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2013;320:22–34. doi: 10.1002/jez.b.22479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Pierre SE, Ponting L, Stefancsik R, McQuilton P the-FlyBase-Consortium. FlyBase 102 - advanced approaches to interrogating FlyBase. Nucleic Acids Research. 2014;42:D780–D788. doi: 10.1093/nar/gkt1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns SC. The evolution of life history traits: A critique of the theory and a review of the data. Annual Review of Ecology and Systematics. 1977;8:145–171. [Google Scholar]
- Steiner UK, Tuljapurkar S. Neutral theory for life histories and individual variability in fitness components. Proceedings of the National Academy of Sciences (USA) 2012;109:4684–4689. doi: 10.1073/pnas.1018096109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner UK, Tuljapurkar S, Coulson T. Generation time, net reproductive rate, and growth in stage-age-structured populations. The American Naturalist. 2014;183:771–783. doi: 10.1086/675894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennett MD, Etges WJ. Pre-mating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. III. Epicuticular hydrocarbon variation is determined by use of different host plants in Drosophila mojavensis and Drosophila arizonae. Journal of Chemical Ecology. 1997;23:2803–2824. [Google Scholar]
- Tennessen JM, Thummel C. Coordinating growth and maturation - Insights from Drosophila. Current Biology. 2011;21:R750–R757. doi: 10.1016/j.cub.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Developmental Cell. 2001;1:453–465. doi: 10.1016/s1534-5807(01)00060-0. [DOI] [PubMed] [Google Scholar]
- Toolson EC, Markow TA, Jackson LL, Howard RW. Epicuticular hydrocarbon composition of wild and laboratory-reared Drosophila mojavensis Patterson and Crow (Diptera: Drosophilidae) Annals of the Entomological Society of America. 1990;83:1165–1176. [Google Scholar]
- Tu M-P, Tatar M. Juvenile diet restriction and the aging and reproduction of adult Drosophila melanogaster. Aging Cell. 2003;2:327–333. doi: 10.1046/j.1474-9728.2003.00064.x. [DOI] [PubMed] [Google Scholar]
- Tuljapurkar S, Gaillard J, Coulson T. From stochastic environments to life histories and back. Philosophical Transactions of the Royal Society of London. Series B, Biological sciences. 2009;364:1499–1509. doi: 10.1098/rstb.2009.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Research. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtonen TM, Kangassalo K, Pölkki M, Rantala MJ. Transgenerational effects of parental larval diet on offspring development time, adult body size and pathogen resistance in Drosophila melanogaster. PLoS ONE. 2012;7:e31611. doi: 10.1371/journal.pone.0031611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder SJ, Walsh MP. Calponin: thin filament-linked regulation of smooth muscle contraction. Cellular signalling. 1993;5:677–686. doi: 10.1016/0898-6568(93)90029-l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.