Abstract
Nutritional guidelines for maintaining healthy blood glucose levels are commonly portrayed as universally applicable. A new study however, demonstrates that the impact of each food on blood glucose varies dramatically across individuals and largely depends on personal characteristics and gut microbiota composition, laying the foundations for broad implementation of personalized nutrition.
The prevalence of diabetes among U.S. adults has increased dramatically in recent years, from 3.5% in 1980 to 9% in 2011 (CDC). Obesity rates have likewise increased from 13% in 1962 to 36% in 2010 (NIH). These epidemics in turn, have met a flood of entrepreneurial diet books and TV shows promoting the latest and greatest food fad, with a constant flow of overhyped news articles describing new research, vilifying yesterday’s nutritional golden boy and exonerating a previously convicted food item (think eggs, coffee, wine, grains). The resulting public sentiment is epitomized by one Internet commenter who stated recently: “Every day is April Fool’s in nutrition” [1].
One feature though, seems to remain constant in this quagmire: the notion that certain types of food are either universally good (and should be a part of any healthy diet) or bad (and should be avoided or consumed in moderation). This view underappreciates a potentially substantial metabolic variation in individuals responding to identical diets; one person’s miracle diet may fail miserably for another. For instance, a 2005 clinical trial randomly assigned individuals to one of four contrasting popular diets, and found that although on average, each group’s participants lost a small amount of weight, the variability in response within each group was high, with some individuals in every diet group gaining weight over the one-year study [2]. Similar variability has been observed across individuals’ blood glucose levels in response to identical single food products; the post-prandial glucose responses (PPGR), which represent an important risk factor in the development of type II diabetes [3].
The variability in people’s responses to diet may result from a range of factors, from genetics and lifestyle, to various environmental parameters, including toxin exposure. Indeed, factors beyond diet and exercise are increasingly seen as important contributors to obesity, as they may influence fat storage and glycemic regulation [4]. One such factor is the gut microbiome, whose composition varies widely across individuals and has been associated with a wide range of health outcomes, including diabetes, obesity, and other metabolic disorders [5]. Until recently however, a systematic, large-scale assessment of interpersonal variability in response to food has been lacking, and the contribution of various personal and environmental factors to such variability, unclear.
To address this challenge, a recent study by Zeevi et al. [6] in Cell, characterized in detail the interpersonal variability of human glycemic responses, charting its determinants (Figure 1). The blood glucose levels of 800 individuals were continuously monitored for seven days, in conjunction with detailed record keeping on diet and lifestyle, using a smartphone-adjusted website. Lifestyle and medical background data (including sleep/wake cycles and physical activity), blood parameters, anthropometric measures, and stool samples were also collected. The stool samples were important in assessing the composition of both species and genes in each individual’s gut bacterial microbiome. Participants were instructed to follow their usual diet, except for small, standardized meals at the beginning of each day. The process resulted in a large-scale dataset, including ~47,000 real-life meals, ~5,000 standardized meals, and detailed continuous glucose monitoring data.
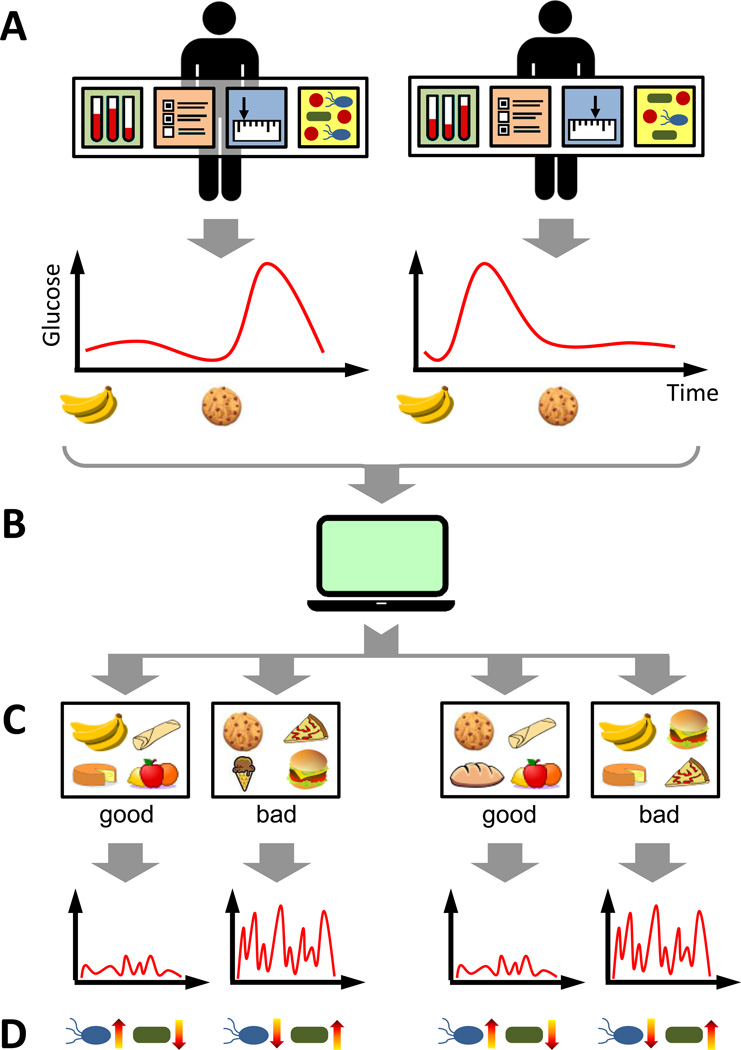
Figure 1. The Impact of Diet on Blood Glucose Is Highly Variable, but Predictable.
(A) Personal and microbiome properties are analyzed in response to diet (including clinical and anthropometric measures, lifestyle, medical background, and the functional pathways and taxonomic composition/species of the gut microbiome). These properties markedly affect the glycemic response over time to various food items for each individual. One person, for example, may have a high postprandial (post-meal) glycemic response to bananas and a low response to cookies, while this ordering may be reversed for another person. (B) Using large-scale data on such personal and microbiome properties, along with continuous glucose monitoring and detailed dietary logs, Zeevi et al., (2015) developed an algorithm to predict with high accuracy, individual glycemic responses for each meal (computer symbol). (C) With this algorithm, it is possible to generate personalized intervention diets designed to regulate glycemic responses, promoting either low (“good” diet) or high responses (“bad” diet). (D) “Good” intervention diets can promote microbiome shifts towards a healthier composition, lowering the abundance of specific bacteria previously associated with diabetes and/or obesity (“blue” coded), while increasing the abundance of bacteria associated with good health (“green” coded).
Linking blood glucose measurements to dietary logs and nutritional values, the research group found a dramatic variation in glycemic responses to the same food items between individuals (Figure 1A). To explore whether this variation was predictable, they developed a machine-learning algorithm to predict an individual’s PPGR to each meal, based on the meal’s nutritional content, and on each individual’s personal and microbiome data (Figure 1B). They then demonstrated that this algorithm successfully predicted person-specific PPGR for each meal. In fact, the algorithm approached a high accuracy level, providing predictions based on the same individual’s previous response to an identical meal. Importantly, this algorithm performed equally well on an independent 100-person validation cohort.
To further examine whether these predictions could be used to promote healthy blood glucose levels, Zeevi and colleagues applied their algorithm to generate comprehensive intervention diets, designed to regulate PPGRs (Figure 1C). They recruited 26 additional subjects and after collecting a week of data using the same format, assigned each participant two diets predicted by the algorithm; one to promote low PPGRs (the “good” diet) and one to promote high PPGRs (the “bad” diet). Paradoxically, certain foods including pizza, hummus, and potatoes, were included in the “good” diet for some participants and in the “bad” diet for others. Results showed that PPGRs were indeed significantly lower in the “good” diet group, with fewer glucose spikes, when compared to PPGRs in the “bad” diet group.
Yet, perhaps the most intriguing finding of this study was the observation that the “good” intervention diet not only resulted in healthier PPGRs, but could also promote gut microbiome shifts toward microbiota compositions previously associated with health (though this was not 100% the case) (Figure 1D). Clearly, the impact of a specific microbiome composition on health is in many cases mediated by microbiome metabolism of dietary compounds, which can subsequently modulate both immune and/or metabolic pathways in the host [7]. The microbiome’s composition itself can in turn be modulated by a range of diet components, selecting certain microbial species (bacteria, viruses) and hindering the growth of others [8]. Nevertheless, there is no a priori reason to assume that diet-induced shifts in gut microbiome composition regulate in concert, both positive and negative metabolic effects on heath. But yet, a diet that beneficially impacts one aspect of health via the metabolic activity of an individual’s current microbiome composition (such as short-term PPGR), could in theory, promote the long-term growth of bacteria with negative effects on health. The findings from this study therefore imply that microbiome activity and selection are strongly and positively intertwined, and furthermore, that the effect of the microbiome on health may be amplified by a concordant selection towards microbiome compositions that reinforce the same effect (be it beneficial or harmful). Moreover, these observations suggest that diet may be an important trigger governing the direction of molecular cascades associated with gut microbiota composition and determining whether these cascades ultimately spiral towards a healthy or unhealthy outcome for the host.
Undoubtedly, tailoring medical and nutritional decisions based on detailed personal information is a promising opportunity. The study by Zeevi et al. therefore represents an exciting first step towards translating such high-dimensional personal health data into actionable personalized recommendations with clinical relevance. With the rise in accuracy and popularity of health and lifestyle monitoring technologies as well as the drop in DNA sequencing costs, generating data such as these will become easier and more accessible. Similar machine-learning approaches could be applied to tailor other medical recommendations, such as drug dosage administration. In addition, this study highlights the need for personalized nutrition, but it also suggests that this type of analysis can be used to inform diet recommendations globally, indicating the extent of variation of food-specific responses, and the associated benefits versus damages across a given population.
Of note, while this study underscores the potential promise of personalized nutrition, it also reveals that many questions remain unknown, particularly those concerning the complex interactions between diet and the microbiome. For example, it is unclear which spatio-temporal scales or taxonomic designations matter for an accurate prediction of microbial associations [9]. Moreover, the extent to which such personalized interventions prevent disease, promote wellness long-term, and are able to generate a stable microbiome composition, remain to be determined. Importantly, this predictive algorithm, though powerful, is a black box that reveals complex statistical associations, but not the mechanisms underlying such associations. A better systems-level understanding of the mechanisms by which the gut microbiome functions, metabolizes the host diet, and ultimately contributes to host health will be essential for the development of microbiome-based therapies in the future [10].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bohannon J. I fooled millions into thinking chocolate helps weight loss. Here’s how., io9. 2015 May 27; [Google Scholar]
- 2.Dansinger ML, et al. Comparison of the Atkins, Ornish, Weight Watchers, and Zone Diets for weight loss and heart disease risk reduction: a randomized trial. JAMA. 2005;293:43. doi: 10.1001/jama.293.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Vrolix R, Mensink RP. Variability of the glycemic response to single food products in healthy subjects. Contemp. Clin. Trials. 2010;31:5–11. doi: 10.1016/j.cct.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 4.McAllister EJ, et al. Ten putative contributors to the obesity epidemic. Crit. Rev. Food Sci. Nutr. 2009;49:868–913. doi: 10.1080/10408390903372599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nieuwdorp M, et al. Role of the microbiome in energy regulation and metabolism. Gastroenterology. 2014;146:1525–1533. doi: 10.1053/j.gastro.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Zeevi D, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Donia MS, Fischbach MA. Small molecules from the human microbiota. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roopchand DE, et al. Dietary polyphenols promote growth of the gut bacterium Akkermansia muciniphila and attenuate high-fat diet–induced metabolic syndrome. Diabetes. 2015;64:2847–2858. doi: 10.2337/db14-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenblum S, et al. Extensive strain-level copy-number variation across human gut microbiome species. Cell. 2015;160:583–594. doi: 10.1016/j.cell.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waldor MK, et al. Where next for microbiome research? PLOS Biol. 2015;13:e1002050. doi: 10.1371/journal.pbio.1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]