Abstract
The flavoprotein L-hydroxynicotine oxidase (LHNO) catalyzes an early step in the bacterial catabolism of nicotine. Although, the structure of the enzyme establishes that it is a member of the monoamine oxidase family, LHNO is generally accepted to oxidize a carbon-carbon bond in the pyrrolidine ring of the substrate and has been proposed to catalyze the subsequent tautomerization and hydrolysis of the initial oxidation product to yield 6-hydroxypseudooxynicotine (Kachalova et al. (2011) Proc. Natl. Acad. Sci. USA 108, 4800–4805). Analysis of the product of the enzyme from Arthrobacter nicotinovorans by NMR and continuous-flow mass spectrometry establishes that the enzyme catalyzes the oxidation of the pyrrolidine carbon-nitrogen bond, the expected reaction for a monoamine oxidase, and that hydrolysis of the amine to form 6-hydroxypseudooxynicotine is nonenzymatic. Based on the kcat/Km and kred values for (S)-hydroxynicotine and several analogs, the methyl group contributes only marginally (~0.5 kcal/mol) to transition state stabilization, while the hydroxyl oxygen and pyridyl nitrogen each contribute ~4 kcal/mol. The small effects on activity of mutagenesis of His187, Glu300, or Tyr407 rule out catalytic roles for all three of these active-site residues.
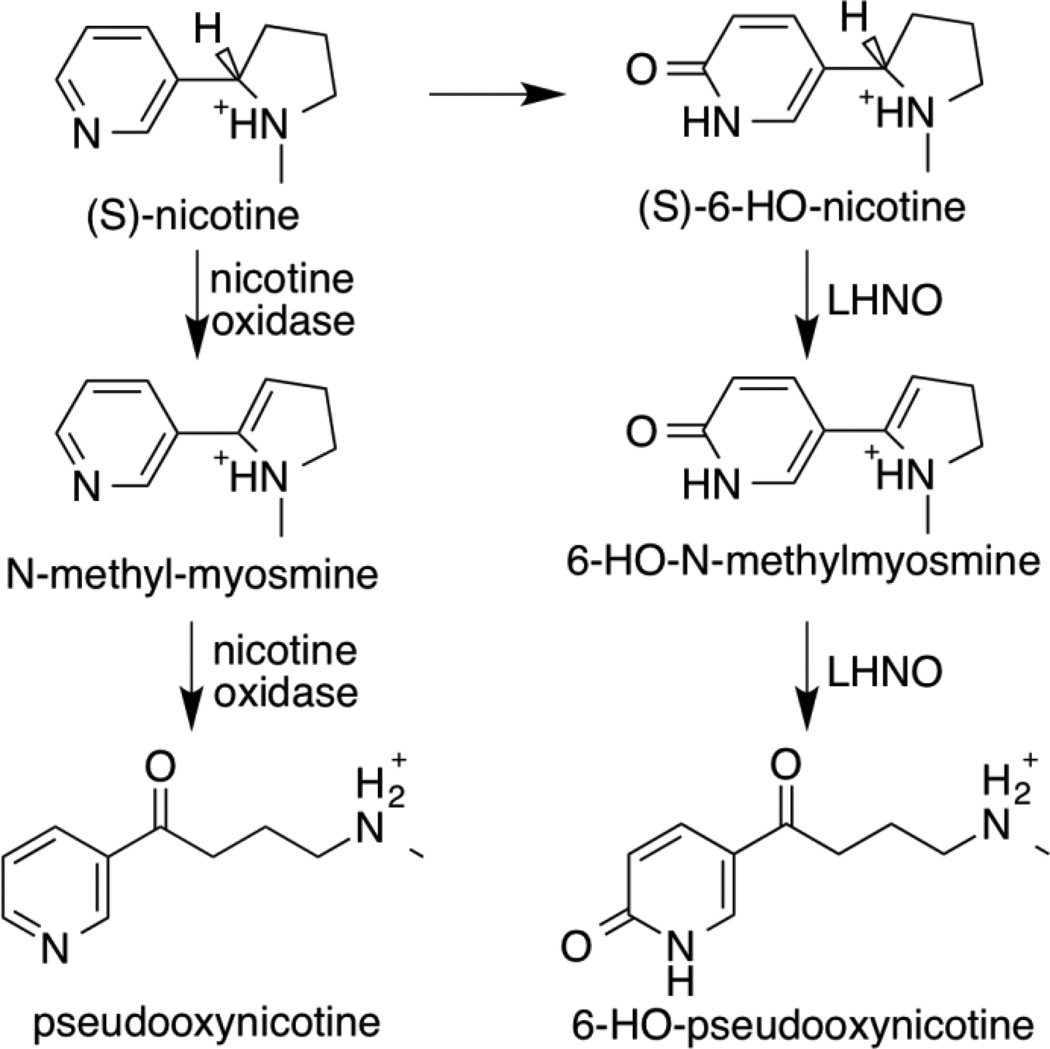
A variety of microorganisms are able to grow on nicotine, the predominant alkaloid in tobacco plants. The best-characterized pathways are in Arthrobacter nicotinovorans and several pseudomonads.1–4 In Arthrobacter the pathway (Scheme 1) begins with the hydroxylation of the pyridyl ring of nicotine by the molybdopterin nicotine dehydrogenase to yield (S)-6-hydroxynicotine.5 The flavoprotein L-6-hydroxy-nicotine oxidase (LHNO) is then proposed to oxidize a carbon-carbon bond in the pyrrolidine ring of (S)-6-hydroxynicotine to yield 6-hydroxy-N-methylmyosmine, which is hydrolyzed to 6-hydroxypseudooxynicotine.6, 7 In the pseudomonad pathway, the first step is proposed to be oxidation of a carbon-carbon bond in the pyrrolidine ring of nicotine by the flavoprotein nicotine oxidase8 to yield N-methylmyosmine, which is hydrolyzed to pseudooxynicotine. Both LHNO and nicotine oxidase are proposed to catalyze essentially identical reactions, oxidation of a carbon-carbon bond in the pyrrolidine ring of the substrate. With both enzymes, the ring-opened pseudooxynicotine rather than the initially formed methylmyosmine has been identified as the product of the enzyme-catalyzed reaction.6–8
Scheme 1.
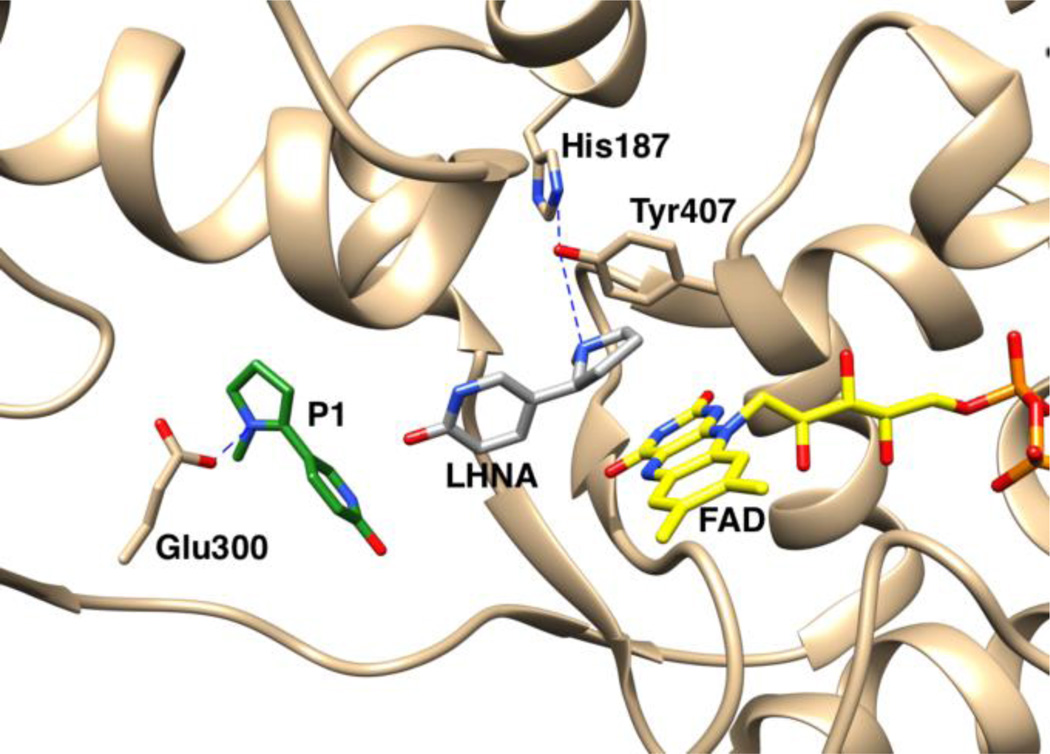
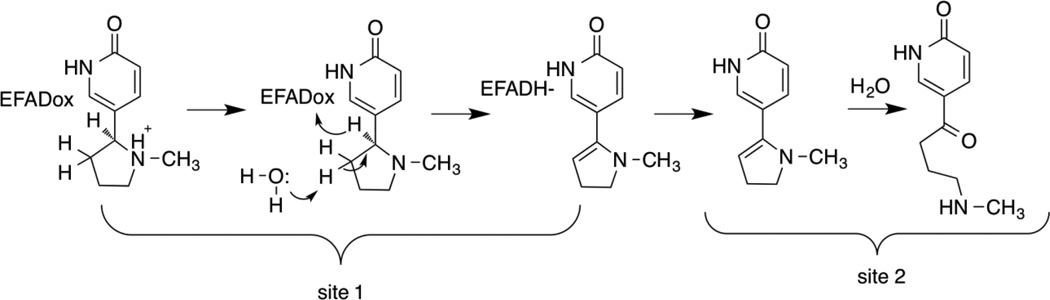
A number of structures of LHNO are available with substrates and/or inhibitors bound9, 10. These establish that the enzyme is a flavoprotein in the monoamine oxidase (MAO) family11. The structure of nicotine oxidase has not been reported, but its sequence is 30% identical to that of LHNO8, establishing that it is also a flavoprotein oxidase. Figure 1 shows the structure of LHNO with (S)-6-hydroxynicotine bound in the active site and the putative oxidized amine bound in a separate site. Based on this and related structures that show ligands can bind at two separate sites, a detailed catalytic mechanism has been proposed for LHNO that involves discrete reactions in two different active sites (Scheme 2).9 In the first part of the reaction, after binding and deprotonation of the cationic substrate, the carbon-carbon bond of 6-hydroxynicotine is oxidized by hydride transfer to the flavin. Tyr407 and His187 have been proposed to act as part of a proton relay that removes the proton from the pyrrolidine nitrogen prior to hydride transfer. The resulting 6-hydroxymyosmine is then hydrolyzed in a second site to form 6-hydroxypseudooxynicotine, the product released by the enzyme. Glu300 has been proposed to act as an active site base in this step by activating an active-site water molecule for the hydrolysis.
Figure 1.
Active site of LHNO with (S)-6-hydroxynicotine (LHNA) and the proposed oxidized amine (P1) bound, from PDB file 3NGC.
Scheme 2.
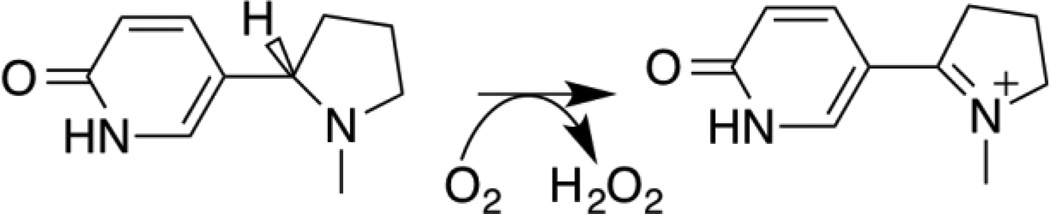
The active site of LHNO is homologous to that of MAO, and the substrates bind in identical fashion in both enzymes.9, 12 All well-characterized flavin amine oxidases in the MAO family catalyze oxidation of the carbon-nitrogen bond in the substrate, with any subsequent hydrolysis of the oxidized amine occuring nonenzymatically.13 Thus, the proposed reactions for both LHNO and nicotine oxidase and the proposed mechanism for the former would make these enzyme unique members of the family. Oxidation of a carbon-carbon bond would also represent a significant mechanistic divergence within a common flavoprotein fold. Based on precedent with other flavoprotein amine oxidases, an alternative is that LHNO and nicotine oxidase bind the neutral form of the substrate and catalyze the oxidation of the substrate carbon-nitrogen bond (Scheme 3), and that the initial oxidation product is hydrolyzed non-enzymatically after release from the enzyme. The experiments described here were designed to evaluate these mechanistic proposals for LHNO.
Scheme 3.
Experimental Procedures
Materials
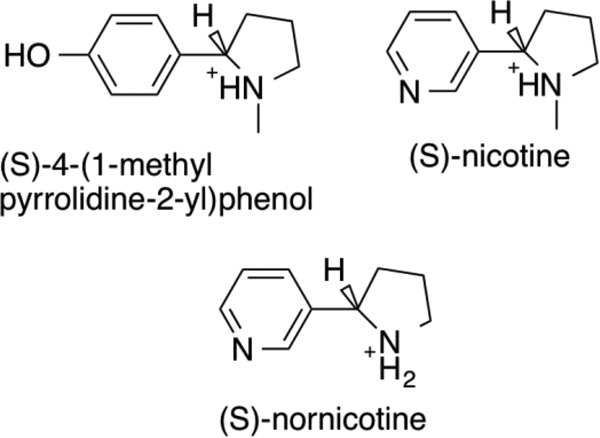
(R,S)-6-Hydroxynicotine was from Princeton Biomolecular Research (Princeton, NJ). (S)-Nicotine was from Sigma Aldrich. (R,S)-4-(1-Methyl pyrrolidine-2-yl)phenol was from Aurora Fine Chemicals LLC (San Diego, CA). (S)-6-Hydroxynornicotine was from Asiba Pharmatech Inc (Milltown, NJ). (R,S)-Nornicotine was from Ark Pharm (Libertyville, IL).
Protein expression and purification
A synthetic gene for wild-type LHNO from A. nitotinovorans optimized for expression in Escherichia coli was obtained from DNA2.0 (Menlo Park, CA). The gene, which contains a histidine tag at the C-terminus, was extracted from the commercial vector by digestion with the restriction enzymes NcoI and BamHI and cloned into the expression vector pET23d cut with the same enzymes. The resulting construct (pETHLNO) was transformed into BL21 (DE3) competent cells for protein expression. To generate expression plasmids for mutant proteins, site-directed mutagenesis of pETHLNO was carried out using the QuikChange protocol (Stratagene). The DNA sequences of the genes in the resulting plasmids were determined to confirm the mutations and rule out adventitious mutations.
For purification of wild-type and mutant LHNO, BL21(DE3) cells containing the desired version of pETHLNO were grown in 2–6 L LB medium containing 100 µg/l ampicillin at 37 °C to an absorbance at 600 nm of 0.7; the temperature was then lowered to 18 °C, and IPTG was added to a final concentration of 0.5 mM. The cells were harvested by centrifugation after growing overnight at 18 °C. The cell paste was resuspended in a lysis buffer of 50 mM HEPES (pH 8.0), 0.1 M NaCl, 10 mM imidazole, 2 µM pepstatin A, 2 µM leupeptin, and 100 µg/mL lysozyme, and lysed by sonication. The resulting lysate was centrifuged at 4 °C at 22,400g for 20 min. Streptomycin sulfate was added to a final concentration of 1.5%, and nucleic acids were precipitated by centrifugation at 22,400g for 20 min. The resulting supernatant was loaded onto a 5 ml HisTrap HP column (GE Healthcare) equilibrated with the lysis buffer. The column was washed with 50 ml lysis buffer; the protein was eluted with 100 ml of the same buffer using a 0 to 300 mM imidazole gradient. Fractions containing LHNO were pooled and concentrated by the addition of solid ammonium sulfate to 45% saturation. The pellet was redissolved in 50 mM HEPES (pH 8.0), 0.1 M NaCl, and dialyzed against the same buffer before being stored at −80 °C. The purity of all enzyme preparations was greater than 95% based on polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate.
Assays
Initial rates of amine oxidation were routinely measured by monitoring oxygen consumption in 0.1 M Hepes (pH 8.0), 0.1 M NaCl, at 25 °C using an oxygen electrode (Model 5300A, Yellow Springs Instruments, Yellow Springs, OH). With (R,S)-6-hydroxynicotine and (S)-6-hydroxynornicotine, the concentration of enzyme was typically 0.1 µM; this was increased to 4 µM for (S)-nicotine and (R,S)-4-(1-methyl pyrrolidine-2-yl)phenol. The concentration of oxygen was varied by bubbling the desired concentration of oxygen (62 µM - 1.25 mM) into the oxygen electrode cell for ~10 min. kcat/Km values for amine substrates were determined either by varying the concentration of the amine substrate at 250 µM oxygen and fitting the data to the Michaelis-Menten equation or varying the concentrations of both the amine and oxygen and fitting the data to eq 1, where A is the concentration of the amine substrate. Values of kcat and kcat /KO2 were determined by varying the concentration of both the amine and oxygen and fitting the data to eq 1 or by varying the concentration of oxygen at a saturating concentration of the amine (1–2 mM) and fitting the data to the Michaelis-Menten equation. Ki values were determined by varying the concentration of inhibitor and amine in 250 µM oxygen and fitting the data to the equation for competitive inhibition. Data were fit to the relevant equations using the program KaleidaGraph (Synergy Software). Kinetic parameters for racemic substrates were corrected to the concentration of the active stereoisomer.
| (1) |
Stopped-flow experiments were carried out using an Applied Photophysics (United Kingdom) SX-20 MV stopped-flow spectrophotometer. The conditions were ~ 20 µM LHNO, and 60–2000 µM substrate in 0.1 M Hepes (pH 8.0), 0.1 M NaCl, at 25 °C. Oxygen was removed from the instrument and reagents as previously described.14 Spectra of intermediates in the reductive half reaction were obtained by monitoring the reduction of LHNO with 1 mM amine using an Applied Photophysics diode array detector and fitting the data to a two step model with SFit (Bio-Logic SAS).
Continuous flow mass spectrometry
Products of the reaction of LHNO with (R,S)-6-hydroxynicotine and (S)-6-hydroxynornicotine were detected by high-resolution mass spectrometry using a Thermo Scientific (Waltham, MA) LTQ OrbiTrap Discovery and a New Objectives (Woburn, MA) PicoView nanospray source with an incorporated custom peltier-controlled nano-volume continuous-flow mixer (Eksigent, Dublin, CA).15 Reactions were performed at 25 °C by mixing a solution of 70 µM LHNO in 25 mM ethylenediamine/acetate, 25 mM ammonium chloride (pH 8.5) with an equal volume of 70 µM amine substrate in the same buffer containing 10% methanol. Flow-rates of 5.0 and 0.50 µL/min gave reaction times of 1.2 and 12 s, respectively. Reactions were monitored in spectral mode over the range m/z 110 – 400. Substrates and products were identified by their [M + H]+ ions with a mass error of less than 1 ppm.
NMR spectroscopy
1H-NMR spectra were collected at 300 K on a Bruker Avance 600 spectrometer using a 5 mm TXI (1H/13C/15N) CryoProbe with z-axis pulsed field gradients. To determine the product of the enzymatic reaction, the 1D-1H spectrum of 200 µM (S)-6-hydroxynornicotine was obtained with water suppression before and after incubation for 10 min at 22 °C with 0.5 µM LHNO in 50 mM phosphate (pD 8.0), 0.1 M NaCl, in D2O.
Results
Kinetic mechanism
The steady-state kinetic parameters for oxidation by LHNO of 6-hydroxynicotine and several analogs (Scheme 4) were determined as probes of the kinetic mechanism and substrate specificity of the enzyme. Assays in which the concentrations of both (S)-6-hydroxynicotine and oxygen were varied, either independently or in fixed ratio16, established that the kinetic data with this substrate are well-fit by eq 1 (results not shown), consistent with LHNO exhibiting a typical flavoprotein oxidase kinetic mechanism. The steady-state kinetic parameters for (S)-6-hydroxynicotine as a substrate for LHNO at pH 8 are given in Table 1. The kinetic parameters for (S)-hydroxynornicotine in air-saturated buffer were similar to those for (S)-hydroxynicotine, so a more complete kinetic characterization was carried out with this substrate. As with (S)-6-hydroxynicotine, the data when the concentrations of both the amine and oxygen were varied established eq 1 as the appropriate kinetic mechanism. The kinetic parameters for (S)-hydroxynornicotine, given in Table 1, establish that the methyl group is not a significant determinant of specificity. In contrast, both (S)-nicotine, which lacks the oxygen on the pyridyl ring, and (S)-4-(1-methyl-pyrrolidine-2-yl)phenol, in which the pyridyl dienone is replaced by phenol, are very slow substrates; because of their low activity, the kinetics were only determined in air-saturated buffer (Table 1). No activity could be detected with (R,S)-nornicotine, which acted as a competitive inhibitor versus the amine substrate with a Ki value of 3.5 ± 0.5 mM at pH 8.
Scheme 4.
Table 1.
Steady-State Kinetic Parameters for LHNO*
| Substrate | kcat, s−1 |
kcat/Kamine, mM−1s−1 |
kcat/KO2, mM−1s−1 |
Kamine, mM |
KO2, mM |
|---|---|---|---|---|---|
| (S)-6-hydroxynicotine | 78 ± 10 (30 ± 1) a |
600 ± 260 (730 ± 100)a |
270 ± 60 | 0.13 ± 0.03 (0.042 ± 0.007)a |
0.29 ± 0.10 |
| (S)-6-hydroxynornicotine | 70 ± 4 (16 ± 1)a |
370 ± 50 (480 ± 90)a |
150 ± 10 | 0.20 ± 0.03 (0.064 ± 0.014)a |
0.46 ± 0.07 |
| (S)-nicotinea | 0.033 ± 0.002a | 0.042 ± 0.007 | NDb | 0.80 ± 0.16a | ND |
| (S)-4-(1-methyl pyrrolidine-2-yl)phenola | 0.044 ± 0.002a | 0.069± 0.008 | ND | 0.64 ± 0.10a | ND |
Conditions: 0.1 M Hepes (pH 8.0), 0.1 M NaCl, 25 °C.
Apparent value determined by varying the concentration of the amine with 250 µM oxygen and fitting the data to the Michaelis-Menten equation.
ND, not determined
Rapid-reaction kinetics
The kinetics of reduction of LHNO by amine substrates were determined using anaerobic stopped-flow spectroscopy. With both (S)-6-hydroxynicotine and (S)-6-hydroxynornicotine, mixing enzyme and amine resulted in a biphasic decrease in the visible absorbance of the flavin, with the rapid phase having an amplitude at 450 nm ~10 times that of the slower. The rate constants for the two phases were obtained by fitting the data to eq 2; here, ΔA1 and ΔA2 are the amplitudes of the two phases, kobs1 and kobs2 are the respective rate constants, and A∞ is the absorbance at the end of the reaction. In addition, a diode-array detector was used to follow the reduction of LHNO by 1 mM (S)-6-hydroxynicotine and by 1 mM (S)-6-hydroxynornicotine. The data at all wavelengths from 330–750 nm could be fit with a two-step kinetic model, with an intermediate having a spectrum very similar to reduced flavin and no spectral features consistent with the presence of a radical intermediate (results not shown). With both substrates, the rate constant for the first phase varied with the concentration of the amine, while the rate constant for the slower phase did not. The effect of the concentration of (6)-hydroxynicotine or (6)-hydroxynornicotine on the rate constant for the first phase was fit to eq 3 to obtain the values of kred, the first-order rate constant for reduction of the flavin when the amine substrate is bound, and the apparent Kd values for binding of the amine to the oxidized enzyme; these values and the rate constants for the slow second phase (kobs2) are given in Table 2. Reduction by (S)-nicotine and (S)-4-(1-methyl pyrrolidine-2-yl)phenol was ~1000-fold slower; for these two substrates only a single phase was observed.
| (2) |
| (3) |
Table 2.
Rapid-Reaction Kinetic Parameters for LHNO*
| Substrate | kred, s−1 | Kd, mM | kobs2, s−1 |
|---|---|---|---|
| (S)-6-hydroxynicotine | 450 ± 50 | 0.10 ± 0.05 | 41 ± 24 |
| (S)-6-hydroxynornicotine | 160 ± 5 | 0.30 ± 0.03 | 3.0 ± 1.0 |
| (S)-nicotine | 0.17 ± 0.05 | 4.6 ± 1.6 | - |
| (S)-4-(1-methyl pyrrolidine-2-yl)phenol | 0.34 ± 0.06 | 2.0 ± 1.2 | - |
Conditions: 0.1 M Hepes (pH 8.0), 0.1 M NaCl, 25 °C.
Characterization of reaction products
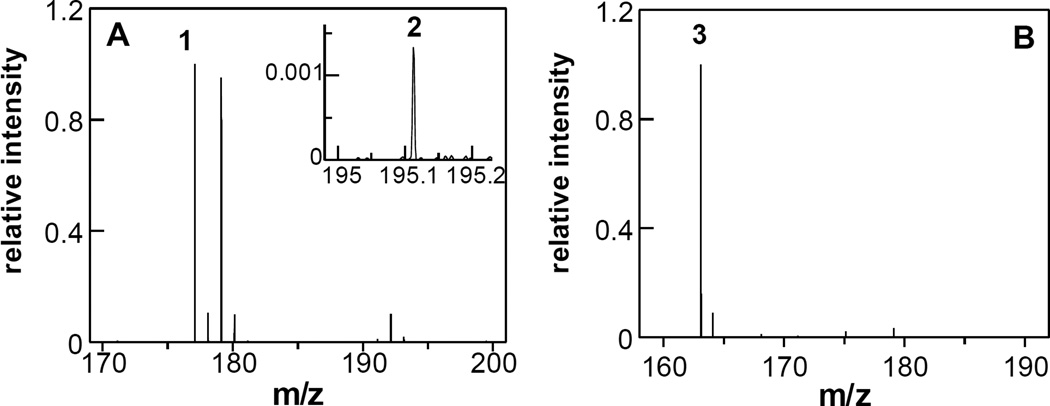
Continuous-flow high-resolution mass spectrometry was used to identify the products of the reaction catalyzed by LHNO. This method allows analysis of potentially unstable products such as enamines by injecting an entire reaction mixture into the mass spectrometer within seconds after mixing enzyme and substrate15. LHNO was mixed with (R,S)-6-hydroxynicotine in air-saturated buffer and the reaction injected into the mass spectrometer after 1.2 or 12 s. At both times the mass spectra showed significant ions at m/z 177.1023 (1) and 179.1179 (Figure 2A). The ion at m/z 179.1179 corresponds to the [M + H]+ ion of 6-hydroxynicotine (m/z 179.1023). Ion 1 is precisely two hydrogen atoms lower in mass, confirming that LHNO catalyzes dehydrogenation of (S)-6-hydroxynicotine. At 1.2 s, the intensities of the ions for 1 and 6-hydroxynicotine were nearly equivalent, with relative intensities of 100% and 95%, respectively, consistent with the use of the racemic substrate. In addition to the signal for 1, an additional product was detected at very low levels at m/z 195.1120 (2) (inset, Figure 2A). This is exactly one water larger than the oxidation product, consistent with 2 being 6-hydroxypseudooxynicotine. The intensity of 2 increased 10-fold (from 0.13% to 1.6%) between 1.2 and 12 s.
Figure 2.
Identification of the LHNO product by continuous flow mass spectrometry. A: Mass spectrum of the reaction of 35 µM LHNO with 35 µM (R,S)-6-hydroxynicotine at a reaction time of 1.2 s at 25 °C. Ion signals are normalized to the most intense ion (1). Inset: A close-up of the mass spectrum showing the second reaction product (2). B: Mass spectrum of the reaction of 35 µM LHNO with 35 µM (S)-6-hydroxynornicotine at a reaction time of 0.5 s at 25 °C. Ion signals are normalized to the most intense ion (3).The spectra are averages of 200 scans.
A similar reaction was carried out with (S)-6-hydroxynornicotine (m/z 165.1022). At a reaction time of 0.5 s, a single ion at m/z 163.0866 (3) was dominant (Figure 2B), with only trace amounts of substrate observed. The signal for 3 corresponds to the dehydrogenated substrate. No signal could be detected for the hydrolyzed product (m/z 181.0972).
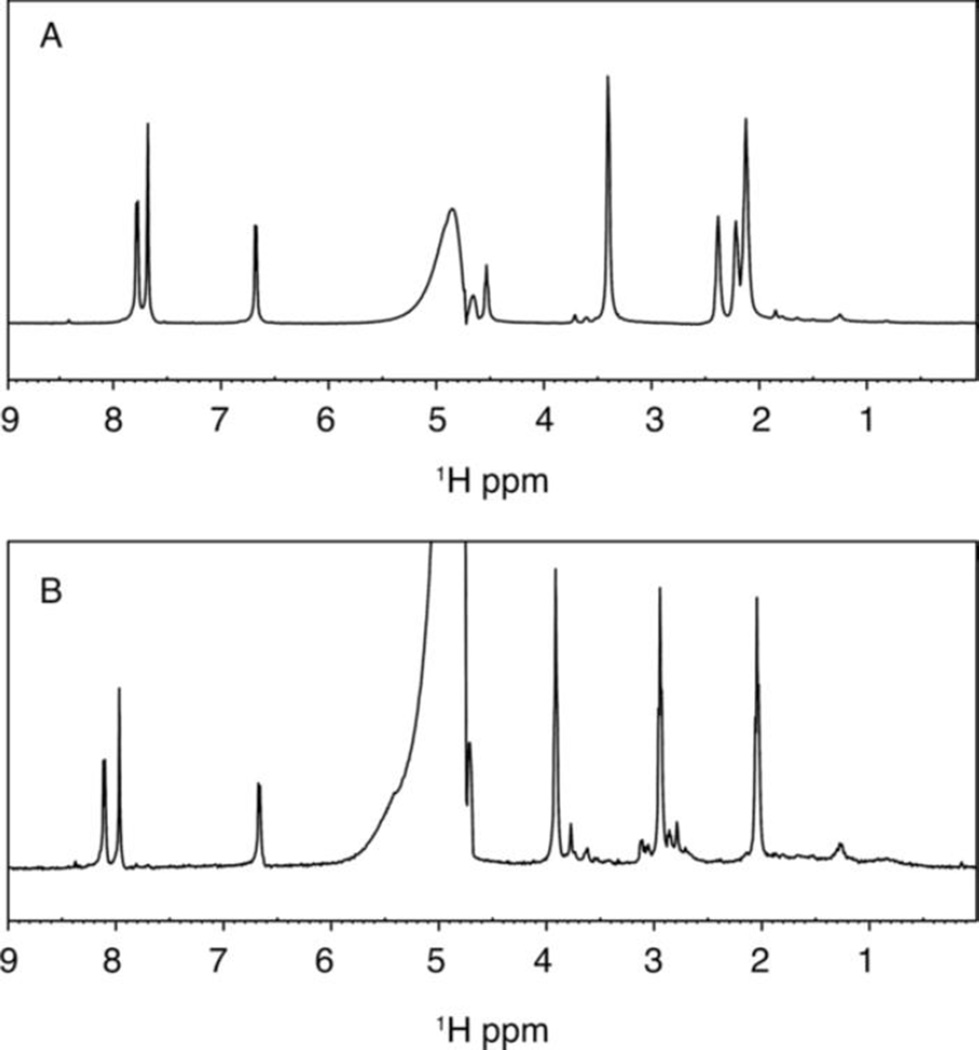
NMR spectroscopy was used to further characterize the product of the LHNO reaction. (S)-6-Hydroxynornicotine was used for this analysis because of its simpler NMR spectrum. Figure 3A shows the 1H-NMR spectrum of 200 µM (S)-6-hydroxynornicotine in D2O in 50 mM sodium phosphate (pD 8.0). Figure 3B shows the spectrum 10 min after the addition of LHNO at a final concentration of 0.5 µM. The product spectrum shows the most dramatic change between 2 and 4 ppm, with three peaks of equal intensity. All three integrate as two protons, consistent with the oxidation product shown in Scheme 3. There is no evidence in the NMR spectrum for significant formation of the hydrolysis product 6-hydroxypseudooxynornicotine.
Figure 3.
Proton NMR spectra of 0.2 mM (S)-6-hydroxynornicotine before (A) and after (B) incubation for 10 min at 22 °C with 0.5 µM LHNO in 50 mM sodium phosphate, 100 mM NaCl in D2O (pD 8.0).
Characterization of active site mutants
His187, Glu300, and Tyr407 have all been proposed to be involved in proton transfers during the LHNO-catalyzed reaction.9 Consequently, each was mutated to a residue incapable of acting as an active site acid or base. The steady-state kinetic parameters at pH 8 with (S)-6-hydroxynicotine as substrate for the mutant proteins are given in Table 3. The only mutation that alters the value of a steady-state kinetic parameter more than 3-fold is H187N; the smaller effects of the H187Q mutation suggest that the effects of the H187N mutation are due to introduction of a smaller residue rather than the loss of an active site acid or base.
Table 3.
Steady-state kinetic parameters with (S)-6-hydroxynicotine for site-specific variants of LHNO*
| Mutants | kcata, s−1 |
kcat/Kamine,b mM−1s−1 |
kcat/KO2a, mM−1s−1 |
Kamineb, µM |
KO2a, mM |
|---|---|---|---|---|---|
| H187Q | 75 ± 8 | 320 ± 40 | 122 ± 17 | 32 ± 5 | 0.61 ± 0.14 |
| H187N | 28 ± 2 | 410 ± 60 | 50 ± 5 | 58 ± 10 | 0.56 ± 0.10 |
| E300Q | 104 ± 8 | 660 ± 80 | 120 ± 8 | 44 ± 7 | 0.88 ± 0.13 |
| Y407F | 75 ± 3 | 490 ± 90 | 110 ± 10 | 48 ± 10 | 0.69 ± 0.12 |
Conditions: 0.1 M Hepes (pH 8.0), 0.1 M NaCl, 25 °C.
Determined by varying the concentration of oxygen at 2 mM amine.
Determined by varying the concentration of the amine in air-saturated buffer.
Discussion
The isoalloxazine ring in flavin cofactors is capable of accepting one or two electrons reversibly and the reduced cofactor reacts readily with oxygen.17 While this allows flavoproteins to catalyze a wide range of metabolic reactions, the versatility of this cofactor means that individual enzymes must enhance one reactivity while suppressing the others. Efforts to understand how this is done necessarily assume that flavoenzymes with similar structures catalyze chemically similar reactions. Consistent with such a model, most flavoenzymes that catalyze oxidation of amines can be grouped into one of two major structural families, the MAO family and the D-amino acid/sarcosine oxidase (DAAO) family, with a few such as berberine bridge enzyme (BBE) and trimethylamine dehydrogenase making up much smaller families.13 A growing set of mechanistic, structural, and computational data support the conclusion that the MAO and the DAAO families and BBE utilize a common mechanism for amine oxidation, hydride transfer from the neutral amine.13, 18 The structures of members of the larger MAO family exhibit two structural domains.11 There is a highly conserved FAD-binding domain and a much less conserved substrate binding domain. This suggests that this family of enzymes evolved from an ancestral amine oxidase by conserving the residues that tune the reactivity of the flavin for amine oxidation while varying the residues involved in substrate binding. If this is correct, finding that a protein has a structure that places it in the MAO family makes it very likely that the protein will catalyze amine oxidation. The structure of LHNO places it in the MAO family of flavoproteins,10 and (S)-6-hydroxynicotine binds with the carbon-nitrogen bond of its pyrrolidine ring oriented to the flavin in a fashion identical to the way that the carbon-nitrogen bond in substrates oxidized by MAO bind that enzyme. The assignment of LHNO and the related enzyme nicotine oxidase as enzymes that catalyze carbon-carbon bond oxidation raises the possibility that the core MAO FAD domain can be readily adapted to catalyze carbon-carbon bond oxidation. The results presented here establish that LHNO, and presumably nicotine oxidase, indeed catalyzes a typical MAO reaction, oxidizing a carbon-nitrogen bond in the substrate and releasing the iminium product to solution. This result strongly suggests that any protein with an MAO FAD-binding domain has evolved to oxidize carbon-nitrogen bonds and has no meaningful activity on carbon-carbon bonds.
The product of the oxidation of (S)-6-hydronicotine by LHNO was initially identified as 6-hydroxypseudooxynicotine by analysis of the product formed in cells and by the isolated enzyme after extensive incubation.7, 19 The reaction was proposed to occur in two steps: initial oxidation of a carbon-carbon bond in the pyrrolidine ring followed by hydrolysis of the iminium carbon-nitrogen bond. While the hydrolysis was initially proposed to be spontaneous rather than enzyme-catalyzed,7 it has more recently been proposed to occur while the iminium ion is still bound to the enzyme.9 The product analyses described here establish that LHNO catalyzes the oxidation of a carbon-nitrogen bond as shown in Scheme 3. The mechanism in Scheme 2 proposes that the enzyme oxidizes the carbon-carbon bond to form an enamine and then catalyzes tautomerization to form the iminium product. The NMR experiment shown in Figure 3 was carried out in D2O; under these conditions tautomerization of the proposed enamine formed by carbon-carbon bond oxidation would result in incorporation of deuterium at C2’. An upper limit can be set on the extent of any deuterium incorporation at this carbon of 5%, making this mechanism unlikely. The results from both mass and NMR spectrometry establish that the iminium product is hydrolyzed much more slowly than the enzyme turns over and thus must be non-enzymatic.
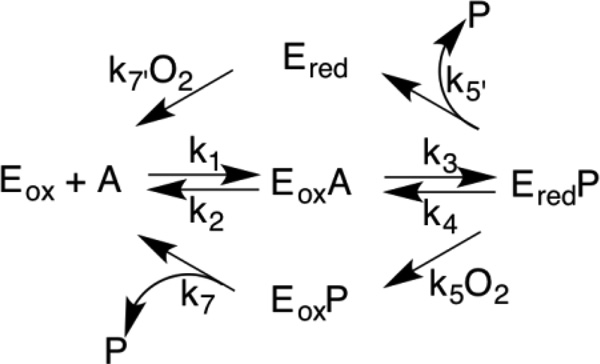
The kinetic mechanism for most flavoprotein oxidases is shown in Scheme 5. Whether the lower or upper path for flavin oxidation occurs depends on the relative values of the rates of product release from the reduced enzyme (k5’) and the rate of oxidation of the reduced enzyme-product complex by oxygen.20 Irrespective of the details of flavin oxidation, this mechanism can result in one of two steady-state kinetic equations depending upon whether oxidation of the amine is significantly reversible. If k4 is effectively zero due either to the thermodynamics of the reaction or to rapid release of the oxidized amine from the reduced enzyme, the kinetic mechanism is given by eq 1 and is identical to the common equation for bi bi ping pong kinetics. A kinetically significant value for k4 yields eq 4, which is identical to the common equation for bi bi sequential kinetics. The two possibilities are distinguished by the presence of the additional term in eq 3.20 This term has the effect that the kcat/Km value for each substrate depends on the concentration of the other substrate for eq 3 and is independent of the concentration of the other substrate for eq 1. The observation that the kinetics of LHNO are well fit by eq 1 establish eq 1 as valid for LHNO.
Scheme 5.
The slow phase seen in the anaerobic oxidation of (S)-6-hydroxynicotine and (S)-6-hydroxynornicotine can be attributed to dissociation of the iminium product from the reduced enzyme, so that the kobs2 values in Table 2 yield the k5’ values for these two substrates. With (S)-6-hydroxynicotine as substrate, the value of k5’ suggests that the enzyme can partition between the upper and lower pathway, with the relative flux depending on the concentration of oxygen. In the absence of the upper pathway, the kcat/KO2 value equals k5. In air-saturated buffer at 25 °C ([O2] = 250 µM), the kcat/KO2 value with (S)-6-hydroxynicotine (Table 1) yields a value of 68 s−1 for oxidation of the reduced flavin in the enzyme-product complex. This is comparable to the value of kobs2 of 45 s−1 for this substrate. Similar behavior was recently described for the flavoprotein spermine oxidase.16 Analyses with that enzyme demonstrated that this partitioning results in the measured kcat/KO2 value being intermediate between the values of k5 and k7’, but still results in a hyperbolic dependence of the rate of oxidation on the concentration of oxygen. The much lower value of kobs2 for (S)-6-hydroxynornicotine suggests that the bottom pathway in Scheme 5 predominates with this substrate.
| (4) |
With both (S)-6-HO-nicotine and (S)-6-HO-nornicotine, the intrinsic kcat values were determined. Comparison of these kcat values (Table 1) with the kred values (Table 2) shows that the former are significantly smaller. For these substrates a step other than amine oxidation must therefore limit kcat. The most likely candidate for such a step is product release. Since the lower pathway will dominate for both substrates at high oxygen concentrations, kcat = k3k7/(k3+k7). The value of k3 is given by kred, so the k7 values for (S)-6-HO-nicotine and (S)-6-HO-nornicotine can be calculated to be 94 ± 15s−1 and 124 ± 8 s−1, respectively. For the slower substrates (S)-nicotine and (S)-4-(1-methyl pyrrolidine-2-yl)phenol only the apparent kcat values were determined. In both cases these are smaller than the kred values, but we cannot rule out the possibility that this difference is due to oxygen not being saturating. It is likely that amine oxidation is rate-limiting for kcat with these very slow substrates.
The kinetic parameters in Table 1 provide insight into the substrate specificity of LHNO. Comparison of the kcat/Km values for (S)-6-hydroxynicotine and (S)-6-hydroxynornicotine establishes that the presence of the methyl group has only a small effect on the catalytic efficiency of the enzyme. This is confirmed by comparing the kred values for amine oxidation in Table 2 for these two substrates. Both sets of kinetic parameters set the contribution of the methyl group to transition state stabilization at ~0.5 kcal/mol. The structure of LHNO with (S)-6-hydroxynicotine bound9 shows that the side chains of Tyr407 and Leu198, the FAD, and two water molecules are at distances of 3.5–4 Å from the substrate methyl group, with a third water at 4.3 Å. This arrangement would be expected to readily allow a water molecule to take the place of the methyl group when (S)-6-hydroxynornicotine is the substrate and provides a rationale for the small effect of loss of the substrate methyl group. In contrast, loss of the oxygen atom from the pyridyl ring or replacement of the hydroxypyridyl ring with phenol decreases the kcat/Kamine and the kred values by ~1000-fold, establishing that these two atoms contribute ~4 kcal/mol to proper binding of the substrate and transition state stabilization.
His187 and Tyr407 have been proposed to assist in removal of the proton from the positively-charged nitrogen in the pyrrolidyl ring of the substrate prior to hydride transfer.9 The lack of a significant change in the kcat/Km value for (S)-6-hydroxynicotine when either residue is mutated makes it unlikely that either is involved in proton transfer during the catalytic reaction. While the substrate nitrogen will be predominantly protonated at pH 8 and oxidation of the carbon-nitrogen bond is expected to require the neutral nitrogen, flavin amine oxidases generally bind substrate with this nitrogen unprotonated.21 There is no other amino acid residue in the active site of LHNO with its side chain appropriately placed to accept a proton from the substrate amine. This suggests that the active form of the substrate for LHNO is uncharged.
Members of the monoamine oxidase family of flavoproteins generally contain two aromatic residues on opposite sides of the substrate amine.22, 23 The two residues in LHNO are Trp371 and Tyr407. Mutation to phenylalanine of the tyrosine in MAO A or B that corresponds to Tyr407 in LHNO decreases the kcat/Km value for the amine ~4-fold for MAO A and 4 to 40-fold for MAO B, depending upon the substrate.24 In mouse polyamine oxidase the decrease is ~3-fold.25 In other members of the family this residue is replaced with phenylalanine, tryptophan or nonaromatic residues in the wild-type enzyme. This variability and the frequent observation of small effects when the tyrosine is mutated to phenylalanine suggests that the major role of this residue in this enzyme family is to maintain the active site shape, with the size of the residue being the most important feature. Consistent with such a proposal, the largest effect of mutating this residue in MAO B is seen with tryptophan.22
The lack of an effect of the E300Q mutation on steady-state kinetic parameters is consistent with the observation that hydrolysis of the enamine is not enzyme-catalyzed. A number of LHNO structures show ligands bound at a site other than the active site, near Glu300 (Figure 1).9 MAO B contains a similar site between the protein surface and the active site,12 and the structure of Calloselasma rhodostoma L-amino acid oxidase, another member of the MAO family, shows several substrate molecules bound in a channel leading to the active site.26 In neither case has a specific role in catalysis been assigned to the cavity.
Conclusions
The present results establish that LHNO catalyzes carbon-nitrogen bond oxidation, consistent with its structural similarities to other flavin amine oxidases, and that the product released from the enzyme is the enamine rather than its hydrolysis product. The pyridyl dienone ring is the major determinant for substrate specificity with (S)-nicotine analogs.
Acknowledgments
Support for the Center for NMR Spectroscopy from the Office of the Vice President for Research at UTHSCSA is gratefully acknowledged.
Funding: This work was supported in part by the NIH (grant R01 GM058698) and The Welch Foundation (AQ-1245). The NMR Spectroscopy Core at UTHSCSA is supported in part by NIH grant P30 NCI/CA054174.
Abbreviations used
- LHNO
L-6-hydroxy- nicotine oxidase
- MAO
monoamine oxidase
- DAAO
D-amino acid oxidase
- BBE
berberine bridge enzyme
References
- 1.Brandsch R. Microbiology and biochemistry of nicotine degradation. Appl. Microbiol. Biotechnol. 2006;69:493–498. doi: 10.1007/s00253-005-0226-0. [DOI] [PubMed] [Google Scholar]
- 2.Thacker R, Rørvig O, Kahlon P, Gunsalus IC. NIC, a conjugative nicotine-nicotinate degradative plasmid in Pseudomonas convexa. J. Bacteriol. 1978;135:289–290. doi: 10.1128/jb.135.1.289-290.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang SN, Liu Z, Tang HZ, Meng J, Xu P. Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype A strain S16. Microbiology. 2007;153:1556–1565. doi: 10.1099/mic.0.2006/005223-0. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Li X, Duan Y, Zhang KQ, Yang J. Biotransformation of nicotine by microorganism: the case of Pseudomonas spp. Appl Microbiol Biotechnol. 2010;86:11–17. doi: 10.1007/s00253-009-2427-4. [DOI] [PubMed] [Google Scholar]
- 5.Grether-Beck S, Igloi GL, Pust S, Schilz E, Decker K, Brandsch R. Structural analysis and molybdenum-dependent expression of the pAO1-encoded nicotine dehydrogenase genes of Arthrobacter nicotinovorans. Mol. Microbiol. 1994;13:929–936. doi: 10.1111/j.1365-2958.1994.tb00484.x. [DOI] [PubMed] [Google Scholar]
- 6.Gries FA, Decker K, Bruehmueller M. Decomposition of nicotine by bacterial enzymes. V. The oxidation of L-6-hydroxynicotine to γ-methylaminopropyl 6-hydroxy-3-pyridyl ketone. Hoppe-Seyler's Z. Physiol. Chem. 1961;325:229–241. [Google Scholar]
- 7.Decker K, Dai VD. Mechanism and specificity of L-and D-6-hydroxynicotine oxidase. Eur. J. Biochem. 1967;3:132–138. doi: 10.1111/j.1432-1033.1967.tb19507.x. [DOI] [PubMed] [Google Scholar]
- 8.Qiu J, Ma Y, Zhang J, Wen Y, Liu W. Cloning of a novel nicotine oxidase gene from Pseudomonas sp. strain HZN6 whose product nonenantioselectively degrades nicotine to pseudooxynicotine. Appl. Environ. Microbiol. 2013;79:2164–2171. doi: 10.1128/AEM.03824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kachalova G, Decker K, Holt A, Bartunik HD. Crystallographic snapshots of the complete reaction cycle of nicotine degradation by an amine oxidase of the monoamine oxidase (MAO) family. Proc. Natl. Acad. Sci. USA. 2011;108:4800–4805. doi: 10.1073/pnas.1016684108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kachalova GS, Bourenkov GP, Mengesdorf T, Schenk S, Maun HR, Burghammer M, Riekel C, Decker K, Bartunik HD. Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans. J. Mol. Biol. 2010;396:785–799. doi: 10.1016/j.jmb.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Gaweska H, Fitzpatrick PF. Structures and mechanism of the monoamine oxidase family. BioMol. Concepts. 2011;2:365–377. doi: 10.1515/BMC.2011.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edmondson DE, Binda C, Mattevi A. Structural insights into the mechanism of amine oxidation by monoamine oxidases A and B. Arch. Biochem. Biophys. 2007;464:269–276. doi: 10.1016/j.abb.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzpatrick PF. Oxidation of amines by flavoproteins. Arch. Biochem. Biophys. 2010;493:13–25. doi: 10.1016/j.abb.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denu JM, Fitzpatrick PF. pH and kinetic isotope effects on the reductive half-reaction of D-amino acid oxidase. Biochemistry. 1992;31:8207–8215. doi: 10.1021/bi00150a013. [DOI] [PubMed] [Google Scholar]
- 15.Roberts KM, Tormos JR, Fitzpatrick PF. Characterization of unstable products of flavin- and pterin-dependent enzymes by continuous-flow mass spectrometry. Biochemistry. 2014;53:2672–2679. doi: 10.1021/bi500267c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adachi MS, Juarez PR, Fitzpatrick PF. Mechanistic studies of human spermine oxidase: Kinetic mechanism and pH effects. Biochemistry. 2010;49:386–392. doi: 10.1021/bi9017945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massey V. The chemical and biological versatility of riboflavin. Biochem. Soc. Trans. 2000;28:283–296. [PubMed] [Google Scholar]
- 18.Gaweska HM, Roberts KM, Fitzpatrick PF. Isotope effects suggest a stepwise mechanism for berberine bridge enzyme. Biochemistry. 2012;51:7342–7347. doi: 10.1021/bi300887m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gries FA, Decker K, Eberwein H, Bruehmueller M. Decomposition of nicotine by bacterial enzymes. VI. Enzymic rearrangement of γ-methylaminopropyl 6-hydroxy-3-pyridyl ketone. Biochem. Zeit. 1961;335:285–302. [Google Scholar]
- 20.Palmer G, Massey V. Mechanisms of flavoprotein catalysis. In: Singer TP, editor. Biological Oxidation. New York: John Wiley and Sons; 1968. pp. 263–300. [Google Scholar]
- 21.Fitzpatrick PF. Combining solvent isotope effects with substrate isotope effects in mechanistic studies of alcohol and amine oxidation by enzymes. Biochim. Biophys. Acta. 2015;1854:1746–1755. doi: 10.1016/j.bbapap.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akyüz MA, Erdem SS, Edmondson DE. The aromatic cage in the active site of monoamine oxidase B: effect on the structural and electronic properties of bound benzylamine and p-nitrobenzylamine. J. Neural Transm. 2007;114:693–698. doi: 10.1007/s00702-007-0670-3. [DOI] [PubMed] [Google Scholar]
- 23.Fitzpatrick PF. Amine and amino acid oxidases and dehydrogenases. In: Miller S, Hille R, Palfey BA, editors. Handbook of Flavoproteins. Berlin: Walter de Gruyter; 2013. pp. 119–137. [Google Scholar]
- 24.Li M, Binda C, Mattevi A, Edmondson DE. Functional role of the "aromatic cage" in human monoamine oxidase B: structures and catalytic properties of Tyr435 mutant proteins. Biochemistry. 2006;45:4775–4784. doi: 10.1021/bi051847g. [DOI] [PubMed] [Google Scholar]
- 25.Royo M, Fitzpatrick PF. Mechanistic studies of mouse polyamine oxidase with N1,N12-bisethylspermine as a substrate. Biochemistry. 2005;44:7079–7084. doi: 10.1021/bi050347k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pawelek PD, Cheah J, Coulombe R, Macheroux P, Ghisla S, Vrielink A. The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site. EMBO J. 2000;19:4204–4215. doi: 10.1093/emboj/19.16.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]