Abstract
For pathogens that infect multiple species the distinction between reservoir hosts and spillover hosts is often difficult. In Alaska, three variants of the arctic rabies virus exist with distinct spatial distributions. We test the hypothesis that rabies virus variant distribution corresponds to the population structure of the primary rabies hosts in Alaska, arctic foxes (Vulpes lagopus) and red foxes (V. vulpes) in order to possibly distinguish reservoir and spill over hosts. We used mitochondrial DNA (mtDNA) sequence and nine microsatellites to assess population structure in those two species. mtDNA structure did not correspond to rabies virus variant structure in either species. Microsatellite analyses gave varying results. Bayesian clustering found 2 groups of arctic foxes in the coastal tundra region, but for red foxes it identified tundra and boreal types. Spatial Bayesian clustering and spatial principal components analysis identified 3 and 4 groups of arctic foxes, respectively, closely matching the distribution of rabies virus variants in the state. Red foxes, conversely, showed eight clusters comprising 2 regions (boreal and tundra) with much admixture. These results run contrary to previous beliefs that arctic fox show no fine-scale spatial population structure. While we cannot rule out that the red fox is part of the maintenance host community for rabies in Alaska, the distribution of virus variants appears to be driven primarily by the artic fox Therefore we show that host population genetics can be utilized to distinguish between maintenance and spillover hosts when used in conjunction with other approaches.
Keywords: Boreal, Disease Transmission, Gene flow, Tundra, Vulpes lagopus, Vulpes vulpes
Introduction
Infectious agents can be isolated from wildlife reservoir or spillover hosts, which can have drastically different roles in long-term disease maintenance and dynamics of epizootics. Maintenance hosts can perpetuate a pathogen and as a reservoir host can serve as a source for the infection of spillover host populations that cannot maintain the infectious agents beyond short-lived chains of pathogen transmission (Haydon et al. 2002). Prevalence and incidence rates in suspected reservoir and target species is often used to distinguish between these types of hosts in a system (Lembo et al.2008, Viana et al. 2014). The specific role of a species as a wildlife reservoir host or spillover host is not easily distinguished as can be seen in the case of Ebola virus in the tropical rain forest where fruit bats have been implicated as reservoir hosts (Leroy et al. 2009) but the role of intermediate spillover hosts in human infection is still not clear (Dhama et al. 2015). Rabies is another example of a disease where the agents are isolated from multiple hosts (Dyer et al. 2014).
Here we demonstrate how host population genetic structure may be used to further our insight into the role of two different host species in disease transmission dynamic, using rabies in Alaska as a model system. This model system also addresses infectious disease dynamics in the face of environmental change, as it involves a disease that has potentially spread with its host species, posing a significant public health concern (Hueffer et al. 2013). Disease spread between species and disease maintenance by highly mobile species such as arctic foxes (Vulpes lagopus) can have far-reaching consequences as can be seen by the incursion of arctic rabies variant 1 into southern Ontario and its establishment in red fox (V. vulpes) populations as well as other host species (Nadin-Davis et al. 2006). Another example is phocine distemper outbreaks in European harbor seals (Phoca vitulina). These very sedentary animals were likely infected from more mobile grey seals (Halichoerus grypus), or this species could serve as a subclinical wildlife reservoir (Härkönen et al 2006).
Rabies is a zoonotic disease of global public health importance. The causative virus normally is maintained in a primary mammalian reservoir host (Rupprecht et al. 2011). Host species vary regionally and different genetic variants of rabies virus are associated with each host (Rupprecht et al. 2002). During an epizootic outbreak, the disease can spillover into other mammalian species that can also spread infection temporally, although these spillover hosts do not maintain the virus over long periods in their populations (Dyer et al. 2014).
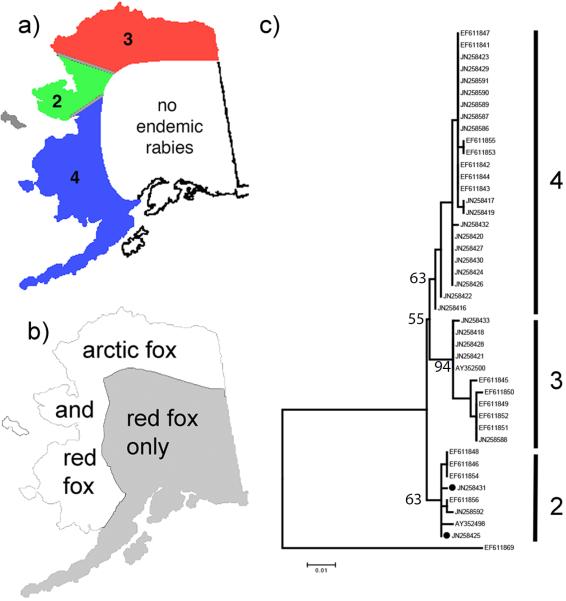
The rabies virus is common in Alaska along the northern and western coasts, an area that is characterized by continuous tundra habitat. On the other hand, the Alaska Interior, southcentral Alaska, and southeastern Alaska are currently devoid of endemic rabies in terrestrial mammals (Fig. 1a). Kuzmin et al. (2008) and Nadin-Davis et al. (2012) identified three different variants of arctic rabies virus among Alaska rabies isolates. Arctic rabies virus variant 2 is generally restricted in Alaska to the Seward Peninsula and regions on the mainland in close proximity with only two isolations described from the North Slope region. Arctic rabies virus variant 3 is found on the northern coast of Alaska, whereas arctic rabies virus variant 4 is found in southwestern Alaska (Fig. 1a). However, the precise boundaries cannot be easily determined given the limited numbers of virus isolated over the large area of Alaska. Currently, arctic rabies variant 4 has only been isolated in Alaska (Kuzmin et al. 2008). The distributions of isolates have been geographically stable at least between 1989 until 2008 with only the two exceptions mentioned above, suggesting that these variants are locally maintained between epizootic outbreaks (Kuzmin et al. 2008; Nadin-Davis et al. 2012). How the distinct spatial structure of the virus is maintained over multiple epizootics in Alaska is currently not well understood. However, geographic stability clearly indicates that the virus is transmitted in a spatially limited fashion, which could be explained by limited contact of infected hosts with susceptible hosts. Alternative explanations could be local genetic adaptations of the virus or hosts that favor transmission in a given ecological or genetic context.
Fig. 1.
Approximate a) rabies virus variant distribution and b) fox ranges by species in Alaska as well as c) phylogenetic relationship of Alaska rabies variants. Circles at two branch tips of the tree represent variants collected in region 3 that are more closely aligned genetically with those from region 2. Labeling of clades refers to the previously described arctic rabies variants 2, 3, and 4. Bootstrap support for the organization of the major clades is provided. Adapted from Kuzmin et al. (2008), MacDonald and Reynolds (2008), Nadin-Davis et al. 2012 Alaska Department of Health and Social Services (2014), and Angerbjörn and Tannerfeldt (2014).
Arctic and red foxes both occur in Alaska and both species can function as wildlife reservoirs of endemic rabies infections in the arctic and temperate regions respectively. (Rupprecht et al. 2002). In fact, all three arctic virus variants from Alaska have been found in both species of fox (Kuzmin et al. 2008; Nadin Davis et al. 2012). However, in Alaska, endemic rabies infections are maintained only in areas that closely match the range of the arctic fox, whereas areas inhabited solely by red fox are considered free of endemic rabies (Fig. 1a and b). This distribution suggests that red foxes represent a spillover host rather than a true reservoir in Alaska. However, in the spring of 2013 rabies virus was detected in two wolves (Canis lupus) in the northern part of Interior Alaska (Alaska Department of Fish and Game 2013), an area solely inhabited by red foxes. Although the red fox clearly represents a significant portion of rabid wildlife in Alaska (Kim et al. 2013), it is not clear if this species is a maintanance or spillover host for arctic rabies virus in the state.
Differences in movements between the two host species could greatly affect disease transmission and the distribution of rabies virus variants. Arctic foxes travel long distances in search of food, particularly during the winter months (Pamperin et al. 2008; Tarroux et al. 2010). Red foxes are comparatively sedentary, with home ranges in tundra habitats of around 16 km2 (Jones & Theberge 1982).
Factors decreasing gene flow in carnivores include geographic features (McRae et al. 2005), sea ice patterns (Paetkau et al. 1999), and human development (Proctor et al. 2005). In arctic foxes, population structure is influenced by seasonal variation in movements (Norén et al. 2011b), habitat fragmentation (Dalén et al. 2006), kin structure (Ehrich et al. 2012), and variability in food resources (Norén et al. 2011b). Throughout the circumpolar range of the arctic fox, sea ice greatly influences gene flow and plays an important role in the connectivity of different populations (Dalén et al. 2007, Geffen et al. 2007; Norén et al. 2011a). In addition, a recent modeling study identified a correlation between sea ice extent and reported rabies cases in foxes (both red and arctic) in Alaska (Kim et al. 2013), though a causal relationship for this correlation cannot necessarily be assumed. In Alaska, population genetics studies on arctic foxes have been limited to island populations (Geffen et al. 2007) and animals from the northern coastal plain of Alaska (Carmichael et al. 2007).
Climate change in the Arctic has been more pronounced than at lower latitudes (Meehl et al. 2007). Movement of red foxes into more northern areas of Alaska and other regions has been documented and is expected to continue as the Arctic's climate warms ((Hersteinsson et al. 1989, Stickney et al. 2014, Foden & Stuart 2009). A replacement of arctic foxes by red foxes would have unknown consequences on the distribution of rabies and other diseases based on differences in the behavior and ecology of these two fox species (Hueffer et al. 2011). Therefore rabies virus ecology in a rapidly changing Arctic can serve as a model to better understand the effects of climate change on the dynamics of host-pathogen interactions. In this study we explored if population structure of the two main hosts (red and arctic foxes) of rabies virus in Alaska can be used to further our understanding of their respective role in rabies ecology in Alaska as reservoir or spillover hosts.
Methods
Sample collection
Fox tissue samples or extracted DNA samples from across Alaska were donated by a variety of agencies and organizations including Alaska Department of Fish and Game, Centers for Disease Control and Prevention - Rabies Program, Alaska Department of Health and Social Services – Public Health Laboratory, the Alaska Trappers Association, the University of Alaska Museum of the North, and by individual trappers across the state (Fig. S1). For mitochondrial DNA (mtDNA) we analyzed 294 samples of red fox and 93 samples of arctic fox. For microsatellites, we analyzed 257 red fox samples and 78 arctic fox samples. Sample distribution by region and year for both analyses is included in Tables 2–5 and Supplemental Table 1. DNA was extracted from the tissue samples using the DNEasy Blood and Tissue Kit (QIAGEN, Germantown, Maryland, USA). When possible, tissue samples paired with skulls were deposited into the curated tissue bank at the University of Alaska Museum of the North. The Arctos Database contains data associated with specific samples such as location and date of collection as well as links to data deposited in Genbank and Dryad.
Table 2.
Genetic differentiation between pairs of arctic fox populations based on mtDNA sequences. ΦST values are below the diagonal and their significance levels are above the diagonal. Negative ΦST values have been changed to equal zero.
| Southwest (n=43) | Seward Peninsula (n=8) | North Slope (n=42) | |
|---|---|---|---|
| Southwest | 0.47 | 0.06 | |
| Seward Peninsula | 0.000 | 0.71 | |
| North Slope | 0.031 | 0.000 |
Table 5.
Genetic differentiation between pairs of red fox populations based on microsatellites. FST values are below the diagonal and their significance levels are above the diagonal.
| South-west (n=91) | Seward peninsula (n=74) | North Slope (n=38) | Interior (n=66) | South-central (n=25) | |
|---|---|---|---|---|---|
| Southwest | <0.001 | 0.003 | <0.001 | <0.001 | |
| Seward Peninsula | 0.010 | 0.040 | <0.001 | <0.001 | |
| North Slope | 0.012 | 0.006 | <0.001 | <0.001 | |
| Interior | 0.041 | 0.031 | 0.017 | <0.001 | |
| Southcentral | 0.059 | 0.050 | 0.040 | 0.032 |
For some analyses, samples from both species were assigned a geographic location within the state of Alaska corresponding to the distribution of arctic rabies virus variants described by Kuzmin et al. (2008) and Nadin-Davis et al. (2012), referred to as Seward Peninsula (arctic rabies virus variant 2), North Slope (arctic rabies virus variant 3), or Southwest (arctic rabies virus variant 4), or a geographic area in Alaska without endemic rabies, the Interior or Southcentral (Fig. 1a, Fig. 2). The Interior is the central area of Alaska, north of the Alaska Range and south of the Brooks Range. Southcentral is the area of the state south of the Alaska Range; these are referred to as sampling areas. Finally, the red fox samples were also split into tundra foxes (Seward Peninsula, North Slope, and Southwest) and boreal foxes (Interior and Southcentral) according to the dominant habitat type of the region for some analyses; these are referred to as regions.
Fig. 2.
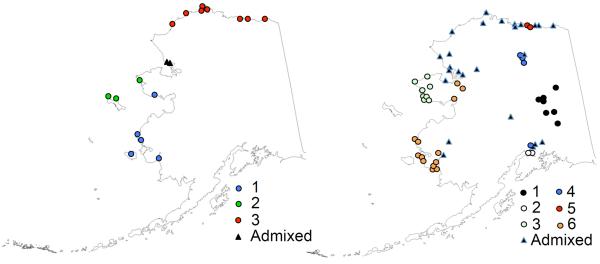
Distribution of mitochondrial haplotypes in a) arctic foxes and b) red foxes in regions defined by rabies strain distribution - North Slope region (arctic rabies variant 3) in red, Seward Peninsula region (arctic rabies variant 2) in green, and the Souwestern region (arctic rabies variant 4) in blue. The area of the pie charts are scaled to sample size. In red foxes only, samples were collected from two additional regions in Figure 3b – the Interior in yellow and Southcentral in orange. Major regional hub cities are marked with black stars, from west to east - Nome, Bethel, Barrow, Anchorage and Fairbanks.
Analysis of rabies isolate sequences
We used a 500-base pair (bp) segment of the rabies virus nucleoprotein gene to construct an Alaska-specific phylogeny of rabies isolates. Sequences of isolates collected in Alaska were obtained from Genbank and stem from studies by Kuzmin et al. (2008) and Nadin-Davis et al. (2012), both of which provide location information but often only at a coarse level. Kuzmin et al. (2008) determined that variants were segregated geographically so we assumed geographic location based on variant identification. Nadin-Davis et al. (2012) provided community names for collection sites along with variant identification. A maximum likelihood phylogeny was created in Mega 6.06 (Tamura et al. 2013). A Kimura 2-parameter model (Kimura 1980) with gamma-distributed mutation rates (Γ = 0.22) was determined by the software to be the most appropriate for the data. The tree was rooted by an arctic-like sequence obtained from a dog in Iraq (Kuzmin et al. 2008).
Mitochondrial DNA analysis
We sequenced 342 bp of the mitochondrial control region of red foxes and 366 bp of arctic foxes using the primer pair VVDL1 and VVDL6 (Aubry et al. 2009). Individuals were sequenced in both directions and a consensus sequence was determined. DNA sequencing was performed by Elim Biopharm (Hayward, CA, USA). Chromatograms were edited using Ridom TraceEdit (Ridom GmbH, Germany) before being imported into SeaView 4.5 (Guoy et al. 2010) for alignment.
We estimated indices of molecular diversity and conducted an analysis of molecular variance (AMOVA) using Arlequin 3.5 (Lischer and Excoffier 2010) for the three arctic fox sampling areas and for the five red fox sampling areas; for red foxes we added a regional level to the analysis of boreal versus tundra. We computed ΦST values using pairwise differences as the distance measure and tested them for significance using 10,000 permutations in Arlequin.
Microsatellite analysis
We selected nine previously published microsatellite loci that amplified in both arctic fox and red fox: CPH3, CPH9, CPH15 (Fredholm & Winterø 1995), AHT121, AHTh171, Co4.140, Co1.424, REN105L03, and REN247M23 (Molecular Ecology Resources Primer Development Consortium et al. 2010). We amplified the microsatellite loci using fluorescently labeled primers and the QIAGEN Multiplex PCR kit (QIAGEN, Germantown, Maryland, USA) in the following three multiplexes, grouped according to the web program MultiPLX 2.1 (Kaplinski et al. 2005): Co4.140, REN105L03, and AHTh171; CPH3, AHT121, and REN247M23; CPH9, Co1.424, and CPH15. DNA extracted from arctic fox tissues was diluted 1:10 prior to PCR amplification whereas DNA from red fox was diluted 1:20 prior to PCR amplification. Multiplexed PCR products were analyzed using the fragment analysis protocol with the LIZ-500 size standard at the Yale University DNA Analysis Facility. We scored the fragment analysis output with GeneMapper ver. 3.7 (Life Technologies, Grand Island, NY, USA).
We used Micro-Checker (van Oosterhout et al. 2004) to identify potential null alleles, large allele dropout, and binning errors while scoring microsatellites. Hardy-Weinberg equilibrium (HWE) at both local and statewide scales, linkage disequilibrium, and extent of differentiation among sampling units using allelic composition were assessed using Genepop ver. 4.2 (Raymond & Rousset, 1995; Rousset 2008) for both fox species. Comparisons using allelic composition were assessed using the default G test in Genepop. The sequential Bonferroni correction (Rice 1989) was applied to alpha values when conducting multiple testing. AMOVA, pairwise FST values, and associated P-values were estimated using Arlequin.
We analyzed population structure with two different Bayesian clustering approaches. First, we used the program Structure ver. 2.3.4 (Pritchard et al. 2000; Falush et al. 2003; Hubisz et al. 2009) to determine the number of homogeneous clusters of individuals (K), assuming correlated allele frequencies with admixture. We used a burn-in of 100,000 replicates before running 400,000 replicates for data analysis, varying K from 1-10 for red foxes and from 1-5 for arctic foxes, with 3 iterations for each value of K. We used the locprior option, which uses the sampling area as a prior and is better at detecting subtle structure (Hubisz et al. 2009). We considered an individual as a member of a cluster if its probability of membership (q) was ≥0.8. Any individual with q <0.8 for its assigned cluster was considered admixed. Output summaries were submitted to the Structure Harvester website (Earl and vonHoldt 2012) for determination of the most likely value of K using both Pritchard's ad hoc method (Pritchard et al. 2000) and Evanno's ΔK method (Evanno et al. 2005). We also visually inspected bar plots from all levels of K to determine if biologically meaningful solutions existed that were not detected by the prior methods because Structure is known to be biased toward selecting solutions with higher-level structure (Meirmans 2015). Finally, we used the programs CLUMPP (Jakobsson and Rosenberg 2007) and Distruct (Rosenberg 2004) to average replicate runs and to generate bar graphs of Structure results, respectively.
We used Geneland ver. 4.0.3 (Guillot et al. 2005a; Guillot et al. 2005b; Guillot 2008; Guillot et al. 2008; Guedi & Guillot 2011) to conduct spatially explicit structuring. Fox locations were expressed in Universal Transverse Mercator (UTM) coordinates to avoid bias introduced by expressing locations as latitude and longitude measurements close to the North Pole. We specified an uncertainty value of 10 km for locations. This action allowed different foxes collected at the same location to be considered members of different populations. We used 1,000,000 iterations of a model, thinning every 1000 to yield 1000 data points for analysis, the first 500 of which were discarded as burn-in. We ran the program using the model for uncorrelated allele frequencies and followed that by running the program with the model for correlated allele frequencies. The correlated allele frequencies model assumes that all populations can be traced to a single ancestral population from which they diverged and that they have experienced drift since the divergence (Guillot 2008), which we believe is a more realistic model of our system than the uncorrelated model. Again, we varied K from 1-10 for red foxes and 1-5 for arctic foxes. The most likely value of K was determined by comparing log-likelihood scores of different runs. To determine probability of membership in each cluster identified by Geneland, we ran a second series of 10 iterations with K fixed for the value found most likely for each species. As with Structure, we considered an individual as a member of a cluster if its probability of membership (q) was ≥0.8.
Spatial Bayesian clustering algorithms may sometimes overestimate the number of clusters due to the presence of isolation by distance in the data (Frantz et al. 2009). The Geneland manual notes that this may be true for the correlated allele frequencies model (The Geneland Development Group 2012). To test for isolation by distance in our data we performed Mantel tests for each species using the software GenAlEx ver. 6.5 (Peakall and Smouse 2012).
We used a second spatial approach, spatial principal components analysis (sPCA; Jombart et al. 2008), that was independent of the assumptions of HWE and linkage equilibrium common to Structure and Geneland, and, unlike those two packages, assumes autocorrelation in the data. First, we tested for positive spatial autocorrelation using the test described by Jombart et al. (2008). If autocorrelation existed, we conducted sPCA by starting with a Delauney triangulation network. The number of eigenvalues to be retained for analysis was determined by examination of a screeplot. The first 3 lagged PCA scores were plotted on a map of the study area with the PCA scores represented as values along the red, green, and blue color spectra. Similar colors of individuals indicate similar genetic make-up and changes in color over the landscape indicate either clinal variation or the presence of barriers to gene flow, depending upon the degree of color change.
Results
Rabies isolate phylogeny
The data yielded a shallow phylogeny with 3 clades; each clade contained only isolates collected in one of the 3 rabies isolate areas with the exception of 2 isolates, representing 2 individuals, collected in area 3 but with sequences falling within those from area 2 (Fig. 1c). Clades representing areas 3 (North Slope) and 4 (Southwest) were more closely related to each other than either was to the clade containing isolates from area 2 (Seward Peninsula).
Mitochondrial data
We detected 18 haplotypes in arctic foxes, and 20 haplotypes in red foxes. Our results align with a previous study as all but one red fox were included within the Holarctic clade, the most prevalent clade in Alaska (Aubry et al. 2009); a single fox from Delta Junction in the Interior sampling area was part of the Nearctic clade, which is found less frequently in Alaska (Aubry et al. 2009). For red foxes, haplotype diversity was highest in the Southwest sampling area, but the number of pairwise differences and nucleotide diversity were greatest in the Interior. For arctic fox, all diversity indices were greatest in the North Slope sampling area (Fig. 2, Table 1).
Table 1.
Indices of mitochondrial sequence diversity for arctic and red foxes.
| Southwest | Seward Peninsula | North Slope | Interior | Southcentral | |
|---|---|---|---|---|---|
| Red foxes | |||||
| N | 91 | 74 | 38 | 66 | 25 |
| Number of haplotypes | 11 | 8 | 9 | 11 | 5 |
| Haplotype diversity | 0.86 | 0.72 | 0.82 | 0.75 | 0.64 |
| Mean number of pairwise differences | 3.1 | 2.6 | 2.9 | 3.3 | 2.4 |
| Nucleotide diversity | 0.0092 | 0.0076 | 0.0086 | 0.0098 | 0.0069 |
| Arctic foxes | |||||
| N | 26 | 8 | 59 | ||
| Number of haplotypes | 7 | 4 | 16 | ||
| Haplotype diversity | 0.83 | 0.82 | 0.89 | ||
| Mean number of pairwise differences | 3.2 | 2.4 | 4.5 | ||
| Nucleotide diversity | 0.010 | 0.0081 | 0.015 |
For arctic foxes, AMOVA indicated that variance among the 3 sampling areas was not significant (ΦST = 0.005, P = 0.32). Nonetheless, the North Slope and Southwest sampling areas differed marginally in pairwise comparison for ΦST (Table 2).
Based on sampling areas, genetic structure of red fox populations could not be explained by habitat characteristics, i.e., boreal (populations with no endemic rabies) and tundra (populations in rabies virus variant areas). AMOVA based on mtDNA variation revealed an insignificant level of total variance explained by that regional classification (ΦCT = 0.0004, P = 0.60) whereas variance at the among-sampling area level within region (ΦSC = 0.038, P = 0.0035) and at the among-sampling area level (ΦST = 0.038, P = 0.0008) were significant. For pairwise ΦST values, Southwest differed from all other areas but no other significant differences existed (Table 3).
Table 3.
Genetic differentiation between pairs of red fox populations based on mtDNA sequences. ΦST values are below the diagonal and their significance levels are above the diagonal. A negative estimate of ΦST has been changed to zero.
| Southwest (n=56) | Seward peninsula (n=77) | North Slope (n=32) | Interior (n=67) | Southcentral (n=25) | |
|---|---|---|---|---|---|
| Southwest | 0.005 | 0.019 | 0.001 | 0.014 | |
| Seward Peninsula | 0.005 | 0.22 | 0.17 | 0.17 | |
| North Slope | 0.018 | 0.007 | 0.36 | 0.31 | |
| Interior | 0.068 | 0.008 | 0.000 | 0.11 | |
| Southcentral | 0.069 | 0.018 | 0.003 | 0.027 |
Microsatellite data
For red foxes, no null alleles were identified by Micro-Checker in the 5 areas sampled. For arctic foxes, Micro-Checker identified one locus with potential null alleles in the Seward Peninsula (REN247M23) and 2 loci for the North Slope (Co1.424 and CPH15). No loci were identified as having null alleles in all areas; therefore, we chose not to eliminate those loci. For red foxes, no loci were out of Hardy Weinberg equilibrium (HWE) when considered as a single population when the Bonferroni correction for multiple comparisons was used. When split into tundra and boreal regions, no loci were out of HWE. For arctic foxes, one locus (Co1.424) was out of HWE on an entire population basis after Bonferroni correction. Nonetheless, no loci were out of HWE for more than a single area after Bonferroni correction. Considering that no loci were out of HWE for all 3 areas, all 9 loci were kept in the analysis. Five of 36 pairwise comparisons were in linkage disequilibrium for red foxes and two of 36 were in disequilibrium for Arctic foxes, but none of those comparisons were significant after Bonferroni correction.
Considering differences in allelic composition in red foxes on the basis of 5 sampling areas, all areas differed from all other areas (P < 0.0008) with the exceptions of the Seward Peninsula and North Slope (P = 0.17). When considered as boreal or tundra, the 2 regions differed highly significantly (P < 0.00001). For arctic fox, the Seward Peninsula and Southwest areas (P = 0.0006) and the North Slope and Southwest areas (P < 0.00001) differed significantly in allelic composition but not the North Slope and Seward Peninsula areas (P = 0.15).
For the arctic fox, AMOVA indicated significant differentiation among the three rabies variant regions (FST = 0.02, P = 0.001). The only significant difference between areas based on FST, however, was between Southwest and North Slope areas although the difference between Southwest and Seward Peninsula was marginal (Table 4). For red foxes, AMOVA indicated the lack of existence of a regional structure of boreal versus tundra when using the 5 a priori sampling areas as populations (FCT = 0.02, P = 0.09) but significant structure among populations within regions (FSC = 0.015, P < 0.0001) and among populations (FST = 0.035, P < 0.0001). For pairwise FST values, all areas differed significantly from all other areas (Table 5).
Table 4.
Genetic differentiation between pairs of arctic fox populations based on microsatellites. FST values are below the diagonal and their significance levels are above the diagonal.
| Southwest (n=26) | Seward Peninsula (n=8) | North Slope (n=44) | |
|---|---|---|---|
| Southwest | 0.06 | <0.001 | |
| Seward Peninsula | 0.017 | 0.31 | |
| North Slope | 0.023 | 0.004 |
Results of microsatellite data from Structure indicated K = 2 for both species (Figs. S2 and S3, Supporting Information; Fig. 3). For arctic foxes, the North Slope and Southwest areas were distinct from each other, with the Seward Peninsula showing some admixture but a closer fit with the North Slope than Southwest. For red foxes, those in Southcentral and Interior comprised one group and North Slope, Seward Peninsula, and Southwest comprised a second group although the North Slope area was somewhat admixed with the Interior group. There seemed to be a south-north cline of variation from Southcentral through Interior to the North Slope area within the proportion of genomes belonging to the tundra region. This suggests north-south directional gene flow in both tundra and boreal regions but little gene flow from east to west. Solutions of K = 3 and 4 for red fox also were biologically meaningful. For K = 3, all tundra sampling areas clustered into a single unit whereas Southcentral and the Interior were assigned to their own clusters. Except for Southcentral and Southwest, however, most individuals were admixed. For K = 4, results were similar to those for K = 3 except Southwest was assigned to its own cluster and most individuals were admixed in all clusters.
Fig. 3.
Results from Structure microsatellite analysis of a) arctic fox and b) red fox from Alaska. Vertical bars represent individuals and color indicates the proportion of the genome assignable to a specific cluster. NS = North Slope, SP = Seward Peninsula, SW = Southwest, SC = Southcentral, and INT = Interior.
Geneland, when using the uncorrelated model, mirrored the results from Structure for red fox (i.e., K = 2; Fig. 3), with the exception that 3 individuals from Interior and 10 from the North Slope were considered admixed. For arctic fox, Geneland found K = 1 when using the uncorrelated model. When using the correlated model, however, arctic foxes were divided into three clusters (Fig. 4a) that corresponded closely to the three regions for rabies strains described by Kuzmin et al. (2008) and Nadin-Davis et al. (2012). Of the 8 arctic foxes collected in the Seward Peninsula area, five were assigned to a single cluster, 1 was assigned to the southwestern cluster and the 2 closest to the North Slope cluster were considered admixed. All individuals in the North Slope and Southwest areas were assigned to separate clusters and none were admixed (Table 6). For red foxes, the correlated model yielded K = 8. However, when admixed animals were removed from the analysis, only 6 populations remained (Fig. 4b). This result remained consistent even if we lowered our threshold for membership to q ≥0.6. The 6 clusters were assigned within the boreal or tundra regions, with populations 1, 2, and 4 assigned to the boreal region and populations 3, 5, and 6 assigned to the tundra region (Table 6). Admixed individuals were mostly found in the Seward Peninsula and North Slope areas, and admixed individuals made up a larger proportion of total individuals in red foxes (27%) than in arctic foxes (2.6%).
Fig. 4.
Results from a Geneland analysis of a) arctic fox and b) red fox in Alaska. Locations sharing a colored symbol indicate membership in the same cluster. Sample locations are denoted with single symbols regardless of the number of samples from that location. Black triangles indicate admixed individuals.
Table 6.
Proportions of arctic foxes and red foxes that were assigned to clusters using the correlated allele frequencies model in Geneland comprising representatives from each sampling area. Membership in a cluster was assigned if q ≥0.8. Individuals with assignments <0.8 were considered admixed.
| Cluster | Red fox | Arctic fox | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | Admixed | 1 | 2 | 3 | Admixed | |
| N | 59 | 17 | 37 | 9 | 6 | 58 | 71 | 29 | 5 | 42 | 2 |
| Southwest | 0.88 | 0.01 | 1.00 | ||||||||
| Seward Pen. | 1.00 | 0.12 | 0.53 | 1.00 | |||||||
| North Slope | 1.00 | 0.35 | 1.00 | 1.00 | |||||||
| Interior | 1.00 | 0.67 | 0.04 | ||||||||
| Southcentral | 1.00 | 0.33 | 0.07 | ||||||||
Isolation by distance among arctic foxes was weak (r = 0.055, P = 0.06). The extent of isolation by distance among the 5 a priori red fox sampling areas varied, however. Values for individuals within boreal (r = 0.066, P = 0.052) and tundra (r = 0.068, P = 0.063) regions were weak and similar to that seen for arctic foxes, but relationships between boreal and tundra regions were much stronger (e.g., Southcentral-North Slope: r = 0.134, P = 0.002; Interior-North Slope: r = 0.134, P = 0.004; Interior-Seward Peninsula: r = 0.138, P < 0.001; Interior-Southwest: r = 0.146, P < 0.001). Therefore, the overall extent for red fox isolation by distance seems to be driven by inter-regional differences (barriers to gene flow) rather than differences within regions that would be indicative of limited local dispersal (isolation by distance).
The tests for positive autocorrelation in the data were significant for both red fox (max(t) = 0.015, P = 0.009) and arctic fox (max(t) = 0.023, P = 0.003); therefore, we went forward with sPCA. Three positive eigenvalues were retained for the analysis of red fox and 6 were retained for arctic fox, although only the first 3 were used in the color plot analysis; no negative eigenvalues were retained. Color for arctic foxes (Fig. 5a) indicated four separate populations in Southwestern, the Seward Peninsula, Northwestern, and the eastern North Slope. The Southwestern and the Seward Peninsula populations coincided with the areas where rabies virus variants 4 and 2 occurred, respectively. The Northwestern and eastern North Slope populations both occurred in the area where rabies virus variant 3 occurred. Color variation for red fox samples (Fig. 5a) indicated that separate populations exist in Southcentral, Interior, Southwest, the Seward Peninsula, and the eastern North Slope. Color variation between the eastern North Slope and the Seward Peninsula indicate a zone of admixture in northwestern Alaska. This is the same area where Geneland found many admixed individuals (Fig. 5b). Although red foxes in tundra habitats seemingly exhibited structure consistent with rabies virus variants, the high degree of admixture observed is inconsistent with the spatially distinct rabies structure.
Fig. 5.
Clustering of Alaskan a) arctic and b) red foxes as determined by spatial principal components analysis.
Discussion
Fox population structure and rabies variant distribution
As both red and arctic foxes are reported as rabies positive in about equal numbers in Alaska the roles of these two species as maintenance or spillover hosts are not clear. Furthermore, the virus could circulate in Alaska in a multi-host system (Fenton and Pedersen, 2005) in which more than one species are necessary for disease maintenance. To investigate if host population genetics could serve as a tool to further elucidate the role of two hosts in maintaining a pathogen we took a host population genetics approach to test if correlation between host population genetics and pathogen strain distribution in the landscape can provide information to distinguish the roles of the two hosts in rabies maintenance in Alaska. Spatially based methods reveal structure in arctic foxes that mirror that of rabies virus variants. Although the spatial structure of tundra red foxes is also similar to that of rabies virus variants, the high degree of admixture shown within groups indicates high levels of gene flow that are inconsistent with the spatial structure of rabies. This suggests, therefore, that the arctic fox likely is more important in long–term disease ecology of rabies in Alaska compared to the red fox. Nonetheless, we recognize that different methods provided conflicting views of population structure in the two hosts and that these differences require explanation.
Although our analysis of mtDNA in red foxes clearly supports several populations with distinct genetic makeup in Alaska, the relatively low ΦST values from the mtDNA analysis for both the arctic fox and red fox suggest that there is possibly substantial gene flow among these various populations. The lack of differentiation in mtDNA, however, may be indicative of historical processes of population colonization and expansion in eastern Beringia following the last glacial maximum (Kutschera et al. 2013) rather than contemporary population processes, and therefore might have no effect on rabies virus variant distribution.
Analyses with microsatellite-only data indicated little structure in arctic fox, although AMOVA results indicated structure among the three sampling areas. This result likely was driven by the difference between Southwest and North Slope sampling areas as indicated by allelic composition data and FST values. Results from Structure also indicated two groups of arctic foxes, with Southwest foxes differing from Seward Peninsula and North Slope foxes. Thus we can conclude that Southwest foxes comprise a distinct group but that Seward Peninsula and North Slope foxes may exhibit admixture to some degree, particularly in northwestern Alaska. Some admixture in this area also seems reflected in virus variant distribution as two individuals with variant 2 haplotypes were found in the North Slope area (Nadin-Davis et al. 2012).
Adding spatial data to the analysis separated Seward Peninsula arctic foxes from North Slope foxes when using the correlated allele frequencies approach in Geneland. This pattern of structure was identical to that of rabies virus variants. We believe that the correlated model is superior to the uncorrelated model in Geneland because current evidence shows that arctic foxes historically have exhibited high levels of gene flow around the Arctic (Dalen et al. 2005, Norén et al. 2011) and therefore we would expect to find correlated allele frequencies among foxes in different areas. Spatial Bayesian analyses of structure must be interpreted with caution, however, if isolation by distance occurs in the data because they can overestimate population structure (Frantz et al. 2009). Although isolation by distance was weak in our arctic fox data, we conducted the sPCA analysis because it assumes spatial autocorrelation in the data (Jombart et al. 2008), thus providing an independent estimate of structure that still incorporates spatial variation into the analysis. The four populations identified by sPCA were identical to those identified by Geneland with the exception that the northwestern Alaska foxes differed from eastern North Slope foxes and together those two populations were consistent with the zone of occurrence of rabies virus variant 3.
For red fox, differences among the five sampling areas indicated regional structure although AMOVA results did not identify separate boreal and tundra population segments. Structure results, however, did show differences between boreal and tundra areas, as did the correlated allele frequencies model in Geneland. Finally, sPCA results identified five separate populations with extensive admixture in northwestern Alaska, a finding similar to that demonstrated by Geneland.
We recognize that our sample size for Seward Peninsula arctic foxes is small (n = 8) and that this may affect our results. We note, however, that Landguth et al. (2012) examined the effect of sample size on the ability to detect spatial genetic patterns via partial Mantel tests and found that results from samples of 10 and 1000 were similar although results from the smaller sample size yielded lower precision. Therefore, although it may be less than ideal, we consider our sample from the Seward Peninsula as minimally sufficient.
Of interest is the fact that the rabies-endemic area in Alaska is coincident with the tundra red fox population observed in the Structure analysis and the tundra population structure exhibited in the Geneland and sPCA analyses. If red foxes are effective maintenance hosts of rabies virus, we would expect to see more mixing of rabies variants within this region, particularly in northwestern Alaska where red fox admixture is high. Nevertheless, the only mixing of virus variants observed was between areas 2 and 3 (Nadin-Davis et al. 2012), which come into contact in the northwest.
The fact that many red foxes are detected as rabid could result from continuous spillover infections or short-term epizootics in that species, whereas long-term maintenance, and hence distribution, of variants is more reliant on arctic foxes as true reservoir hosts. This view is further supported by an apparent correlation of areas with endemic rabies and arctic fox range, whereas areas solely inhabited by the red fox are considered free of endemic rabies in Alaska.
Alternatively a true multi-host system of pathogen maintenance (Fenton and Pederson, 2005) is consistent with our results of conflicting host population structure. The relative importance of red and arctic fox in this system could be explained by their similar population distributions in the tundra region of our study. Such a multi-host system could also explain the absence of endemic rabies in areas solely inhabited by the red fox, if both species are necessary for rabies virus maintenance. Further studies on rabies ecology and a more thorough understanding of fox ecology and behavior in different ecoregions in Alaska will be necessary to further solidify conclusions about the respective roles of the two fox species in rabies strain distribution in Alaska. The respective roles of these hosts in rabies could change with continued environmental change in the far North, including climate change. Although the specific consequences of that change are not well understood (Hueffer, 2011), the consequences would likely be different if rabies was maintained only by the arctic fox or within a true multi-host system.
The distinct distribution of rabies strains could alternatively be explained through the distribution or movement patterns of other potential host species such as coyotes (Canis latrans), wolves or dogs (C. familiaris). Although the low number of diagnosed rabies cases in wolves and coyotes and the vaccination of dogs and human related movement of dogs makes this alternative explanation less likely. Furthermore, adaptation of rabies virus to local conditions (such as different host species) or host species ecology as well as the fast evolutionary adaptation of viruses compared to their hosts could explain the rabies virus distribution. However, no data exist to support these alternatives for Alaska at this time.
Arctic fox population structure
Previous studies on the population structure of mainland Alaska arctic foxes used the North Slope to represent arctic fox populations throughout Alaska (Carmichael et al. 2007, Norén et al. 2011a), which may be why no population structure had been found. Carmichael et al. (2007) studied microsatellite variation among arctic foxes throughout North America and found an average FST of 0.002, which is an order of magnitude lower than our average of 0.015 for the a priori groups of this study. Norén et al. (2011), also using microsatellites, found a large-scale, pan-Arctic population that included all North American arctic foxes as one group. Geffen et al. (2007) and Dalén et al. (2005), on the other hand, found ΦST values among artic fox populations throughout the Arctic that were greater than what we observed among our sampling areas but their geographic scale of sampling was much greater than ours.
Arctic foxes were considered to exhibit little population structure except over vast geographic scales or between island and mainland populations (Carmichael et al. 2007, Norén et al. 2011a), although kin structure in arctic foxes in Svalbard has been suggested (Ehrich et al. 2012). Arctic foxes, especially those without access to year-round marine resources, are known to travel long distances over sea ice in search of food in winter (Pamperin et al. 2008), and this scale of movement is often cited as a reason for genetically homogeneous populations over large geographic scales (Dalen et al. 2005, Carmichael et al. 2007, Norén et al. 2011) as the presence of sea ice is important for population connectivity (Geffen et al. 2007). Our findings of fine-scale population structure in arctic foxes in Alaska seemingly contradicts those findings but actually may not. Southwest Alaska, the Yukon-Kuskokwim Delta, has more inconsistent sea ice than the two more northern areas (Seward Peninsula and North Slope). The lack of long-term sea ice may result in a subpopulation of arctic foxes in Southwest Alaska maintaining a different strain of rabies virus (arctic rabies variant 4). Especially as this rabies virus variant is found only in Alaska, the lack of movement of arctic foxes due to lack of sea ice could explain the maintenance of an Alaska-specific rabies strain and a distinct population of foxes similar to the genetic isolation of Iceland arctic foxes due to lack of sea ice connectivity (Dalen et al. 2005). Nonetheless, if rabid foxes disperse to neighboring populations, even if they die before getting an opportunity to breed, they would spread their genetic variant of rabies virus within the population. In Alaska, this is obviously not the case on a consistent basis, and we hypothesize that rabid foxes likely die before being able to travel long distances, thus maintaining the genetic structure of rabies virus variants.
In summary, the population structure of the arctic fox matches that of rabies strains in Alaska more so than the population structure of red foxes. This supports the hypothesis that the population structure of the arctic fox maintains rabies strains with distinct spatial distribution in Alaska Alternatively a multi-host system could explain our results with a stronger role of arctic foxes in maintaining the virus in Alaska. We also provide evidence for fine-scale population structure in highly mobile arctic foxes in Alaska.
This study shows that host population genetics can be used to determine the likely role of different host species as wildlife reservoirs versus spillover hosts in the dynamic of infectious agents. However, our study shows this approach is best utilized together with other techniques to help determine these roles in other systems and could improve our understanding of multi-host disease dynamics and management of those diseases.
Supplementary Material
Acknowledgements
We thank all contributors of samples and Link Olson at the University of Alaska Museum of the North for curating samples. This work was supported by the UAF Office of Undergraduate Research and Scholarly Activities and the Alaska INBRE program through grants from the National Center for Research Resources (5P20RR016466) and the National Institute of General Medical Sciences (8P20GM103395-12). In addition support was provided by the National Institute of General Medical Sciences under three linked awards number RL5MD009600, TL4MD009628 and 1UL1MD009610. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health.
Footnotes
Data Accessibility All mitochondrial DNA sequences were deposited to GenBank (accession numbers KJ642756 - KJ643139). When possible (as determined by submitter) samples were deposited in the UA Museum of the North and sample associated data can be accessed via the ARCTOS database (http://arctos.database.museum). Museum catalogue numbers are available via GenBank. Microsatellite and sequence data used in this study has been deposited in Dryad (http://dx.doi.org/10.5061/dryad.dc1q8).
Author Contributions KH and KJH designed the study, BR, CJC, EAH, KJH, and KH performed mitochondrial sequence analysis, EWG and KJH performed microsatellite analysis. EWG, KJH and KH wrote the manuscript.
References
- Alaska Department of Fish & Game [Accessed 8/5/2014];Rabies Detected in New Area of State. Alaska Department of Fish and Game Press Release. 2013 4/23/2013. Available from URL: http://www.adfg.alaska.gov/index.cfm?adfg=pressreleases.pr04232013.
- Alaska Department of Health and Social Services [Accessed 8/5/2014];Regions of Alaska where rabies is considered enzootic (always present at a certain level) among foxes. 2014 Available from URL: http://www.epi.hss.state.ak.us/id/rabies/regions.gif.
- Angerbjörn A, Tannerfeldt M. [Accessed 5/8/2014];Vulpes lagopus. The IUCN Red List of Threatened Species. Version 2014.2. 2014 Available from URL: http://www.iucnredlist.org.
- Aubry KB, Statham MJ, Sacks BN, et al. Phylogeography of the North American red fox: vicariance in Pleistocene forest refugia. Molecular Ecology. 2009;18:2668–2686. doi: 10.1111/j.1365-294X.2009.04222.x. [DOI] [PubMed] [Google Scholar]
- Carmichael LE, Krizan J, Nagy JA, et al. Historical and ecological determinants of genetic structure in arctic canids. Molecular Ecology. 2007;16:3466–3483. doi: 10.1111/j.1365-294X.2007.03381.x. [DOI] [PubMed] [Google Scholar]
- Dalén L, Fuglei E, Hersteinsson P, et al. Population history and genetic structure of a circumpolar species: the arctic fox. Biological Journal of the Linnean Society. 2005;84:79–89. [Google Scholar]
- Dalén L, Kvaløy K, Linnell JDC, et al. Population structure in a critically endangered arctic fox population: does genetics matter? Molecular Ecology. 2006;15:2809–2819. doi: 10.1111/j.1365-294X.2006.02983.x. [DOI] [PubMed] [Google Scholar]
- Dhama K, Malik YS, Malik SV, Singh RK. Ebola from emergence to epidemic: the virus and the disease, global preparedness and perspectives. Journal of Infection in Developing Countries. 2015;9:441–455. doi: 10.3855/jidc.6197. [DOI] [PubMed] [Google Scholar]
- Dyer JL, Yager P, Orciari L, et al. Rabies surveillance in the United States during 2013. Journal of the American Veterinary Medical Association. 2014;245:1111–1123. doi: 10.2460/javma.245.10.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and programming for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Ehrich D, Carmichael L, Fuglei E. Age-dependent genetic structure of arctic foxes in Svalbard. Polar Biology. 2012;35:53–62. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analysis under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton A, Pedersen AB. Community epidemiology framework for classifying disease threats. Emerging Infectious Diseases. 2005;11:1815–1821. doi: 10.3201/eid1112.050306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foden W, Stuart SN. Species and Climate Change: More than just the Polar Bear. International Union for Conservation of Nature Species Survival Commission. 2009 Retrieved from https://portals.iucn.org/library/efiles/documents/2009-051.pdf.
- Frantz AC, Cellina S, Krier A, et al. Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance? Journal of Applied Ecology. 2009;46:493–505. [Google Scholar]
- Fredholm M, Winterø AK. Variation of short tandem repeats within and between species belonging to the Canidae family. Mammalian Genome. 1995;6:11–18. doi: 10.1007/BF00350887. [DOI] [PubMed] [Google Scholar]
- Geffen E, Waidyaratne S, Dalen L, et al. Sea ice occurrence predicts genetic isolation in the Arctic fox. Molecular Ecology. 2007;16:4241–4255. doi: 10.1111/j.1365-294X.2007.03507.x. [DOI] [PubMed] [Google Scholar]
- Guedj B, Guillot G. Estimating the location and shape of hybrid zones. Molecular Ecology Resources. 2011;11:1119–1123. doi: 10.1111/j.1755-0998.2011.03045.x. [DOI] [PubMed] [Google Scholar]
- Guillot G, Estoup A, Mortier AF, Cosson JF. A spatial statistical model for landscape genetics. Genetics. 2005a;170:1261–1280. doi: 10.1534/genetics.104.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot G, Mortier F, Estoup A. Geneland: A program for landscape genetics. Molecular Ecology Notes. 2005b;5:712–715. [Google Scholar]
- Guillot G, Santos F, Estoup A. Analysing georeferenced population genetics data with Geneland: a new algorithm to deal with null alleles and a friendly graphical user interface. Bioinformatics. 2008;24:1406–1407. doi: 10.1093/bioinformatics/btn136. [DOI] [PubMed] [Google Scholar]
- Guillot G. Inference of structure in subdivided populations at low levels of genetic differentiation. The correlated allele frequencies model revisited. Bioinformatics. 2008;24:2222–2228. doi: 10.1093/bioinformatics/btn419. [DOI] [PubMed] [Google Scholar]
- Guoy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Härkönen T, Dietz R, Reijnders P, et al. The 1988 and 2002 phocine distemper virus epidemics in European harbour seals. Diseases of Aquatic Organisms. 2006;68:115–130. doi: 10.3354/dao068115. [DOI] [PubMed] [Google Scholar]
- Haydon D, Cleaveland S, Taylor LH, Laurenson MK. Identifying reservoirs of infection: A conceptual and practical challenge. Emerging Infectious Diseases. 2002;8:1468–1473. doi: 10.3201/eid0812.010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersteinsson P, Angerbjörn A, Frafjord K, Karikusalo A. The Arctic Fox in Fennoscandia and Iceland: Management Problems. Biological Conservation. 1989;49:67–81. [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueffer K, O'Hara TM, Follmann EH. Adaptation of mammalian host-pathogen interactions in a changing arctic environment. Acta Veterinaria Scandinavica. 2011;53 doi: 10.1186/1751-0147-53-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueffer K, Parkinson AJ, Gerlach R, Berner J. Zoonotic infections in Alaska: disease prevalence, potential impact of climate change and recommended actions for earlier disease detection, research, prevention and control. International Journal of Circumpolar Health. 2013;72 doi: 10.3402/ijch.v72i0.19562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Dufour A-B, Pontier D. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity. 2008;101:92–103. doi: 10.1038/hdy.2008.34. [DOI] [PubMed] [Google Scholar]
- Jones DB, Theberge JB. Summer home range and habitat utilization of the red fox (Vulpes vulpes) in a tundra habitat, northwest British Columbia. Canadian Journal of Zoology. 1982;60:807–812. [Google Scholar]
- Kaplinski L, Andreson R, Puurand T, Remm M. MultiPLX: automatic grouping and evaluation of PCR primers. Bioinformatics. 2005;21:1701–1702. doi: 10.1093/bioinformatics/bti219. [DOI] [PubMed] [Google Scholar]
- Kim BI, Blanton JD, Gilbert A, et al. A Conceptual Model for the Impact of Climate Change on Fox Rabies in Alaska, 1980–2010. Zoonoses and Public Health. 2013;61:72–80. doi: 10.1111/zph.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kutschera VE, Lecomte N, Janke A, et al. A range-wide synthesis and timeline for phylogeographic events in the red fox (Vulpes vulpes) BMC Evolutionary Biology. 2013;13:114. doi: 10.1186/1471-2148-13-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin IV, Hughs GJ, Botvinkin AD, et al. Arctic and Arctic-like rabies virus: distribution, phylogeny, and evolutionary history. Epidemiology and Infection. 2008;136:509–519. doi: 10.1017/S095026880700903X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landguth EL, Fedy BC, Oyler-McCance SJ, et al. Effects of sample size, number of markers, and allelic richness on the detection of spatial genetic pattern. Molecular Ecology. 2012;12:276–284. [Google Scholar]
- Lembo T, Hampson K, Haydon DT, et al. Exploring reservoir dynamics: a case study of rabies in the Serengeti ecosystem. J Appl Ecol. 2008;45(4):1246–1257. doi: 10.1111/j.1365-2664.2008.01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy EM, Epelboin A, Mondonge V, et al. Human Ebola outbreak resulting from direct exposure to fruit bats in Luebo, Democratic Republic of Congo, 2007. Vector Borne Zoonotic Dis. 2009;9:723–728. doi: 10.1089/vbz.2008.0167. [DOI] [PubMed] [Google Scholar]
- MacDonald DW, Reynolds JC. [Accessed 8/5/2014];Vulpes vulpes. The IUCN Red List of Threatened Species. Version 2014.2. 2008 Available from URL: http://www.iucnredlist.org.
- McRae BH, Beier P, Dewald LE, et al. Habitat barriers limit gene flow and illuminate historical events in a wide-ranging carnivore, the American puma. Molecular Ecology. 1999;14:1965–1977. doi: 10.1111/j.1365-294x.2005.02571.x. [DOI] [PubMed] [Google Scholar]
- Meehl GA, Stocker TF, Collins WD, et al. Global Climate Projections. In: Solomon S, Qin D, Manning M, Chen Z, Marquis M, Averyt KB, Tignor M, Miller HL, editors. Climate Change 2007: The Physical Science Basis. Cambridge University Press; New York: 2007. pp. 747–846. [Google Scholar]
- Meirmans PG. Seven common mistakes in population genetics and how to avoid them. Molecular Ecology. 2015 doi: 10.1111/mec.13243. In Press. [DOI] [PubMed] [Google Scholar]
- Molecular Ecology Resources Primer Development Consortium. An J, Bechet A, et al. Permanent Genetic Resources added to Molecular Ecology Resources Database 1 October 2009–30 November 2009. Molecular Ecology Resources. 2010;10:404–408. doi: 10.1111/j.1755-0998.2009.02827.x. [DOI] [PubMed] [Google Scholar]
- Mørk T, Prestrud P. Arctic Rabies – A Review. Acta Veterinaria Scandinavica. 2004;45:1–9. doi: 10.1186/1751-0147-45-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadin-Davis SA, Muldoon F, Wandeler AI. Persistence of genetic variants of the arctic fox strain of Rabies virus in southern Ontario. The Canadian Journal of Veterinary Research. 2006;70:11–19. [PMC free article] [PubMed] [Google Scholar]
- Nadin-Davis SA, Sheen M, Wandeler AI. Recent emergence of the Arctic rabies virus lineage. Virus Research. 2012;163:352–362. doi: 10.1016/j.virusres.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Norén K, Carmichael L, Dalén L, et al. Arctic fox Vulpes lagopus population structure: circumpolar patterns and processes. Oikos. 2011a;120:873–885. [Google Scholar]
- Norén K, Carmichael L, Fuglei E, et al. Pulses of movement across the sea ice: population connectivity and temporal genetic structure in the arctic fox. Oecologia. 2011b;166:973–984. doi: 10.1007/s00442-011-1939-7. [DOI] [PubMed] [Google Scholar]
- Paetkau D, Amstrup SC, Born EW, et al. Genetic structure of the world's polar bear populations. Molecular Ecology. 1999;8:1571–1584. doi: 10.1046/j.1365-294x.1999.00733.x. [DOI] [PubMed] [Google Scholar]
- Pamperin NJ, Follmann EH, Person BT. Sea-ice use by arctic foxes in northern Alaska. Polar Biology. 2008;31:1421–1426. [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor MF, McLellan BN, Strobeck C, Barclay RMR. Genetic analysis reveals demographic fragmentation of grizzly bears yielding vulnerably small populations. Proceedings of the Royal Society B. 2005;272:2409–2416. doi: 10.1098/rspb.2005.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. Genepop (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. Distruct: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4:137–138. [Google Scholar]
- Rousset F. Genepop'007: a complete reimplementation of the Genepop software for Windows and Linux. Molecular Ecology Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Rupprecht CE, Hanlon CA, Hemachudha T. Rabies re-examined. The Lancet Infectious Diseases. 2002;2:327–343. doi: 10.1016/s1473-3099(02)00287-6. [DOI] [PubMed] [Google Scholar]
- Rupprecht CE1, Turmelle A, Kuzmin IV. A perspective on lyssavirus emergence and perpetuation. Current Opinion in Virolology. 2011;6:662–70. doi: 10.1016/j.coviro.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Stickney AA, Obritschkewitsch T, Burgess RM. Shifts in Fox Den Occupancy in the Greater Prudhoe Bay Area, Alaska. Arctic. 2014;67(2) [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetic analysis version 6. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarroux A, Berteaux D, Bety J. Northern nomads: ability for extensive movements in adult arctic foxes. Polar Biology. 2010;33:1021–1026. [Google Scholar]
- The Geneland Development Group [Accessed 21 November 2014];Population genetic and morphometric data analysis using R and the GENELAND program. 2012 http://www2.imm.dtu.dk/~gigu/Geneland/
- Van Oosterhout C, Hutchison WF, Wills DPM, Shipley P. micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.