Abstract
Purpose
Like all viruses, Human immunodeficiency virus type 1 (HIV-1) requires host cellular factors for productive replication. Identification of these factors may lead to the development of novel cell-based inhibitors.
Experimental design
A Strep-tag was inserted into the C-terminus of the Matrix region of the HIV-1 gag gene. The resultant virus was replication competent and used to infect Jurkat T-cells. Matrix complexes were affinity purified with Strep-Tactin agarose. Protein quantification was performed using SWATH mass spectrometry, data was log-2 transformed, and student t-tests with Bonferroni correction used to determine statistical significance. Several candidate proteins were validated by immunoblot and investigated for their role in virus infection by siRNA knockdown assays.
Results
A total of 17 proteins were found to be statistically different between the infected versus uninfected and untagged control samples. Ku70, Ku80 and YB-1 were confirmed to interact with Matrix by immunoblot. Knockdown of two candidates, EZRIN and YB-1, enhanced HIV infection in vitro.
Conclusions and clinical relevance
The Strep-tag allowed for the capture of viral protein complexes in the context of virus replication. Several previously described factors were identified and at least two candidate proteins were found to play a role in HIV-1 infection. These data further increase our understanding of HIV-host cell interactions.
Keywords: HIV, Matrix, SWATH, Ku70, Ku80, EZRIN, YB-1
Introduction
Acquired Immune Deficiency Syndrome (AIDS) is a pandemic disease that has resulted in the death of more than 30 million people since the discovery of the infectious agent human immunodeficiency virus type 1 (HIV-1) [1, 2]. HIV-infected patients can be treated with a triple combination therapy to control virus replication and indefinitely extend their lives, but no cure is available [3–5]. Importantly, resistance to existing drugs is on the rise [6], therefore new inhibitors are still needed to treat infected patients and prevent spread of the virus. HIV, like all viruses, requires host cellular factors to facilitate its replication, which include fusion and entry of the virus, reverse transcription of the viral RNA into DNA, integration of the DNA into the host chromosome, viral protein expression, virion assembly, and budding and maturation. Identification of the host-cell interactions that mediate these steps and delineation of how HIV interacts with these factors may lead to discovery of new targets for HIV inhibitor development.
As a member of the retrovirus family, HIV has 3 major genes: gag, pol and env. The gag gene contains the structural proteins of the virus. It is synthesized as a single polyprotein and cleaved by the viral protease during maturation into four proteins: Matrix, Capsid, Nucleocapsid, and p6 [7, 8]. The Matrix protein is critical for both virus entry and release in the HIV life cycle. During the early stage of virus replication, Matrix is associated with the reverse transcription and preintegration complex (RTC and PIC, respectively) [9–11]. Matrix binds both viral RNA and DNA, but also binds ribosomal RNA (rRNA) and heterologous DNA nonspecifically [12–15]. Importantly, the exact role of Matrix in these complexes and throughout the early steps of replication remains unclear [16]. During the late steps of replication Matrix facilitates targeting of the Gag polyprotein to the plasma membrane and incorporation of viral envelope proteins into virus particles [17, 18].
In an effort to further delineate the role of Matrix in virus replication we employed a proteomic approach to investigate the interactome of Matrix during virus replication. Several other genetic and proteomic studies have been performed on Matrix utilizing individually expressed Matrix or Gag proteins [19–22]. To identify Matrix interactions in the context of replicating virus we constructed a molecular clone (MA-Strep) with the Strep-tag sequence interjected into the C-terminus of Matrix at a site previously shown to be amenable to insertions [23, 24]. The Strep-tag is eight amino acids which should not impact protein folding or secretion [25]. The resulting virus was replication competent. Matrix protein complexes were isolated from infected Jurkat T-cells and proteins identified and quantified using the Sequential Window Acquisition of all Theoretical fragment-ion spectra (SWATH) method of MS. Statistical analyses identified 17 proteins to be differentially present in the MA-Strep samples compared to both uninfected and infected-untagged control cells. These studies present several novel Matrix-interacting proteins that may be important for HIV replication.
Materials and methods
DNA constructs
The TEV protease and Strep-tag sequences, as well as linker sequences (Figure 1) were inserted by overlap PCR mutagenesis into pNLX clone of HIV-1 [26] using the overlapping primers NLX_117_REV (5’-TGCGGATGGCTCCAGCTCTGAAAATACAGGTTTTCGCCTCCTATCTGGCTGTTGTTTCC-3’) and NLX_1171_FOR (5’-TTCAGAGCTGGAGCCATCCGCAGTTTGAAAAAGGGGGGAGCATAGTCAGCCAAAATTAC-3’) and flanking primers that overlap unique BssHII and SpeI restriction sites in pNLX (NLX_708 (5’-GAAGCGCGCACGGCAAGAGGC-3’) and NLX_1630 (5’-TCCAGAATGCTGGTAGGGC-3’)). The HIV firefly luciferase reporter virus plasmid, pNL-luc-R+E−, was obtained from NIH AIDS Research and Reference Reagent Program (Germantown, MD) [27, 28]. The VSVg envelope expression vector pMD2.G was obtained from the Addgene Plasmid Repository.
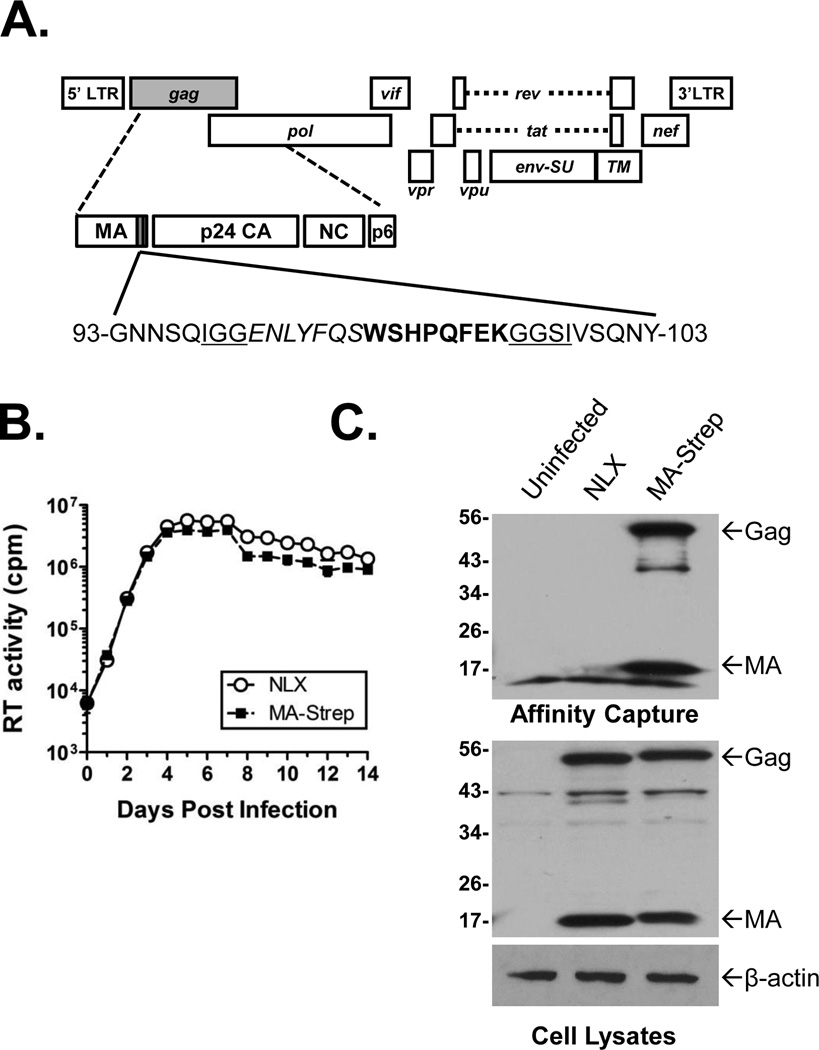
Figure 1.
HIV-1 MA-Strep Tag clone (A). Approximate location of insertion into C-terminus of Matrix is indicated by shaded box. The insert contained the Strep-tag (bold), a TEV cleavage site (italics), and linker sequences (underlined). (B) Replication of MA-Strep. Jurkat cells were inoculated overnight with normalized levels of the indicated viruses, the cells washed, and propagated. Supernatants were collected and clarified by centrifugation on the days indicated. At the end of the experiment the samples were analyzed for exogenous RT activity using a [32P]TTP incorporation assay. (C) Expression and affinity purification of tagged Matrix. Jurkat cells were infected as indicated above blots. Bottom two panels show immunoblots of Matrix and β-actin (as a control) in cell lysates prior to affinity capture. Upper panel shows anti-Matrix immunoblot of samples affinity purified with Strep-Tactin beads. Location of HIV-1 Gag and Matrix are indicated.
Cell lines and virus production
293T and Jurkat cells were grown in DMEM and RPMI 1640 medium, respectively, supplemented with 10% Fetal Clone III Hyclone (Logan, UT USA). All cells were grown at 37 °C with a 5% CO2 atmosphere. Virus stocks were produced by transfection of 1×107 293T cells with 5 µg of viral molecular clone using Polyjet transfection reagent (SignaGen Laboratories, Rockville, MD). HIV-Luc reporter viruses were generated by co-transfection of 5 µg of pNL-luc-R+E− and 1 µg of pMD2.G VSVg vector.
Affinity purifications
For each biological replicate 1×108 Jurkat-T4 cells were infected with 1 ml virus overnight, washed, and incubated for an additional 2–3 days until the appearance of syncitia. Uninfected samples were prepared in parallel as negative controls. Cells were pelleted, washed once with PBS, and lysed in 1 ml 25 mM Tris-HCl pH 7.4/150 mM NaCl/1 mM EDTA/1% Triton X-100 for 15 min. at 4°C with rotation. Samples were clarified by centrifugation and incubated with Sepharose 6B (Sigma-Aldrich, St. Louis, MO) at 4°C with rotation to reduce background. Samples were transferred to a new tube and rotated overnight with 50 µl Strep-Tactin agarose beads (Qiagen, Venlo, Netherlands) at 4°C. Beads were pelleted and washed thrice for 15 minutes with cell lysate buffer, resuspended in 10 mM NH4HCO3 and stored in −80°C until processed for MS.
SWATH MS, statistics, and bioinformatic analyses
Samples were digested with 0.1 µg/µL modified trypsin (Promega, Madison, WI USA) overnight at 37°C. Beads were pelleted by centrifugation and the supernatants transferred to a new tube. The digested peptides were extracted with three washes of 0.1% Trifluroacetic acid in 60% acetonitrile with shaking for 60 minutes at room temperature. The washes were combined and peptides purified using C18 Zip Tips® according to manufacturer's procedure (Millipore, Billerica, MA USA). Samples were dried by vacuum centrifugation and resuspended in 0.1% formic acid in HPLC-grade water for LC-MS/MS and SWATH-MS analysis. Protein identifications were made by ProteinPilot using a UniProt Swiss-Prot database containing human and HIV-1 proteins; common laboratory contaminants were excluded. The FDR cutoff threshold was 1%. PeakView v.2.0 (AB Sciex) was used for spectral alignment and targeted data extraction of SWATH-MS samples using a previously established library for human monocytes [29] using the following parameters: extraction window of 5 min, 8 peptides, 5 transitions, peptide confidence of >99%, exclusion of shared peptides, and XIC width set at 50 ppm. Each SWATH-MS experiment was log-2 transformed independently of other experiments before statistical analysis. Student t-tests were performed with Bonferroni correction for three conditions: MA-Strep vs. uninfected, NLX vs. uninfected, and MA-Strep vs. NLX. The proteins that were significantly different (p<0.05) between the MA-Strep condition vs. both the uninfected and NLX controls were identified as candidate MA interactors. PANTHER v. 9.0 (www.pantherdb.org) was used for functional analysis of the candidate proteins using gene ontology (GO) molecular function terms. Hierarchical analysis of GO terms was performed using the Gene Ontology Consortium (http://geneontology.org).
Immunoblot Analyses
Cell lysates were resolved by SDS-PAGE and transferred to a PVDF membrane. The following primary antibodies were used for immunoblot: Anti-Ku70 (C-19), anti Ku80 (C-20), and anti-NCL (MS-3) were from Santa Cruz Biotechnology (Dallas, TX); and anti YB-1 (ab76149) from Abcam. Primary antibodies were detected by incubation for 1 h with species-specific HRP-conjugated secondary antibodies (Santa Cruz Biotechnology, Dallas, Texas). Proteins were visualized using SuperSignal West Pico substrate (Pierce Biotechnology, Rockford, IL) and exposure to autoradiography film.
Viral assays
For viral infectivity assays, 1×105 293T were seeded a 6-well plate. The following day each well was transfected with siRNAs (Santa Cruz Biotechnology, Dallas, Texas). At 24 h post transfection, the cells were inoculated with HIV-Luc overnight, the media changed and the cells grown an additional 24 h. Cells were lysed with M-PER solution (Pierce Biotechnology, Rockford, IL) and clarified by centrifugation. Luciferase activity was measured using One-glo luciferase reagent (Promega, Madison, WI) and protein concentrations determined by BCA assay (Pierce Biotechnology, Rockford, IL). Luciferase values were normalized to protein concentration. Statistics were performed using GraphPad Prism software (version 5.04).
Results
Production of HIV-1 with a Strep-tag Matrix protein (MA-Strep)
Previous studies established that the C-terminus of Matrix was amenable to the insertion of heterologous tags [23, 24], including a virus clone we had made with a biotin acceptor sequence fused in this region. That virus was replication competent, but the biotin-streptavidin capture system proved ineffective due to a large amount of background capture of biotin utilizing proteins [30]. To address that issue, we constructed a new virus with a TEV protease recognition site and the Strep-tag sequence flanked by two linker sequences inserted at amino acid 97 of Matrix (Fig. 1A). The TEV site was intended to allow for the release of bound proteins from the beads, but was not efficiently cleaved by the protease (data not shown). The linker sequences were necessary for Gag expression and virus release (data not shown).
To assess the viability of MA-Strep, virus stocks of it and the parental NLX virus were made by transient transfection of 293T cells. Jurkat cells were inoculated overnight with normalized amounts of virus, washed, and the cells propagated for 14 days. The supernatants were sampled at various days post infection and virus replication measured by an in vitro [32P]TTP incorporation reverse transcriptase (RT) assay. Overall, the MA-Strep virus replicated with kinetics similar to wild-type virus (Fig 1B), demonstrating that the introduction of the tag did not affect virus infectivity in vitro.
Next we confirmed the expression and capture of the Strep-tagged MA protein. To do this, cell lysates were collected from infected cells at 48 hpi and protein complexes captured by incubation with Strep-tactin beads. Both the cell lysates and affinity purified samples were analyzed by immunoblot using an anti-Matrix antibody. As shown in Figure 1C, Strep-tagged Matrix was expressed at similar levels to NLX Matrix in the lysates of infected Jurkat cells. As expected, a slightly larger sized band was detected in the MA-Strep samples due to the addition of the insert. Notably, no additional smaller sized bands were observed, suggesting that the insertion did not destabilize the protein. Anti-actin immunoblots were performed as controls (bottom panel). Affinity capture of the whole cell lysates using the Strep-Tactin beads demonstrated efficient capture of Matrix as well as full-length Gag polyprotein from the samples (Fig. 1C, top blot).
SWATH-MS analysis of Matrix protein complexes
To identify cellular proteins that interact with Matrix during virus replication, we performed three replicate experiments in which we infected Jurkat cells until syncitia formation was observed (typically 2–3 days). Two controls were used for these experiments- uninfected (-) cells and cells infected with the parental, untagged NLX virus. Cells were collected, lysed and Matrix complexes captured using Strep-Tactin beads. The captured protein complexes from each group were digested on the beads with trypsin, the peptides eluted, purified, and analyzed by SWATH-MS using a previously established library for human monocytes [29]. The exported results contain quantitative output for the area under the curve for intensity of individual ions and peptides, as well as the summed intensity of the peptides for each identified protein, which was used for our analyses. A total of 27,611 spectral peptide counts were captured for all three experiments (9477 for MA-Strep, 7333 for the NLX control, and 10,801 for the uninfected control). Supplemental dataset 1 contains the summed intensity of the proteins identified in all three biological replicates. A total of 3276 proteins were identified in all replicates. The protein intensities were log2 transformed for parametric statistics. Student t-tests with Bonferroni correction were calculated for three conditions: MA-Strep vs. uninfected, NLX vs. uninfected, and MA-Strep vs. NLX. Ninety eight proteins were significantly different (p<0.05) between the MA-Strep condition vs. the uninfected cells; and 62 were significantly different between MA-Strep and the untagged NLX control group (supplemental dataset 2). Seventeen proteins overlapped between both datasets, these were considered to be candidate MA-interacting factors. The proteins in that dataset that were validated in this study are presented in Table 1.
Table 1.
Candidate Proteins validated in this study.
| UNIPROT | Protein names | Gene name |
NCBI HIV DB1 |
Ratio MA/Cont2 |
MA v Cont. p-value3 |
NLX v Cont. p-value3 |
Ratio MA/NLX2 |
MA v NLX p-value3 |
|---|---|---|---|---|---|---|---|---|
| P67809 | Nuclease-sensitive element- binding protein 1 |
YBX1 | Y | 3.5857 | 0.0002 | 0.0641 | 2.1596 | 0.0189 |
| P13010 | X-ray repair cross- complementing protein 5 |
XRCC5 | Y | 4.0628 | 0.0006 | 1.1429 | 3.1297 | 0.0787 |
| P15311 | Ezrin | EZR | Y | 4.4379 | 0.0011 | 2.2313 | 4.7015 | 0.0110 |
| Q9NTK5 | Obg-like ATPase 1 | OLA1 | Y | 5.1634 | 0.0098 | 1.5512 | 5.7393 | 0.0054 |
| P12956 | X-ray repair cross- complementing protein 6 |
XRCC6 | Y | 2.7185 | 0.0275 | 1.7390 | 2.1573 | 0.1463 |
Protein is present in NIH NCBI HIV interaction database.
Ratio of SWATH count after log2 transformation of data.
Calculated by student t-test.
Bioinformatic analysis
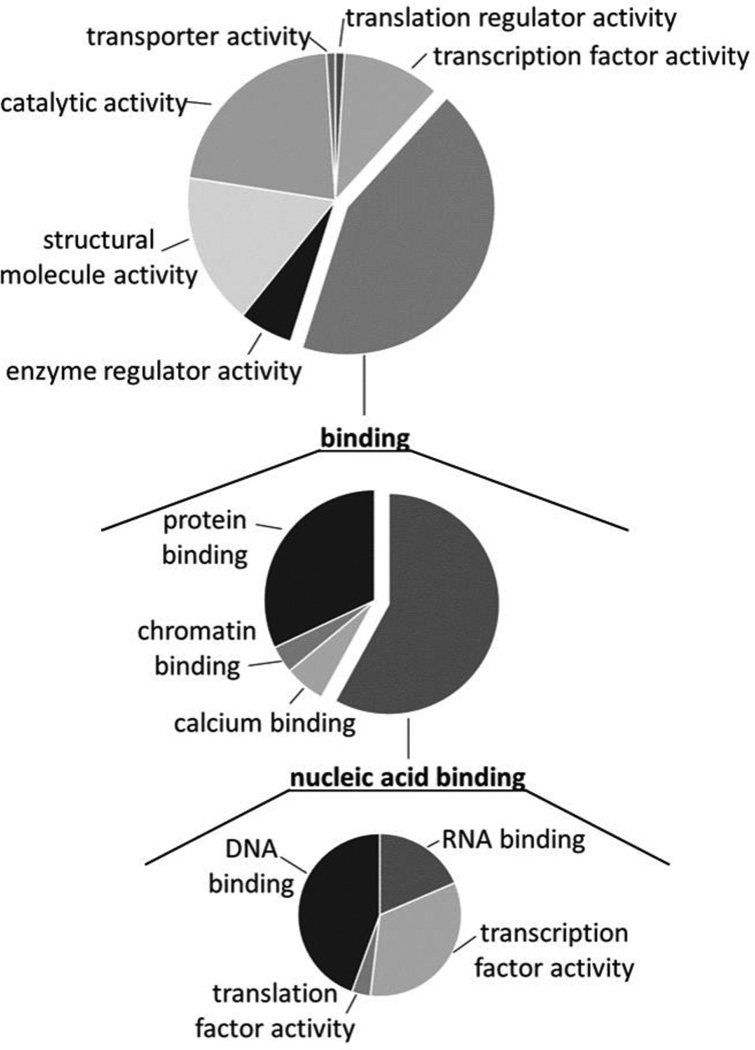
Next, the molecular functions for the 98 proteins identified in the comparison between the MA-Strep and uninfected groups were analyzed using PANTHER (Fig. 2). Four major categories were identified and the largest subset of the proteins were categorized as “binding.” Secondary refinement of the “binding” family revealed an enrichment for nucleic acid binding proteins (middle chart). Further refinement showed enrichment of both RNA and DNA binding proteins, as well as proteins associated with transcription factor activity (bottom chart). These data suggested that a major area of Matrix interaction during virus replication is with nucleic acid-binding proteins, including transcription factors and transcription regulators.
Figure 2.
Bioinformatic analyses reveal alterations in nucleic acid binding proteins. PANTHER analysis of molecular function GO terms revealed enrichment for binding proteins (top). Expansion of this family of proteins (center) identified nucleic acid binding proteins as substantially enriched and was further expanded (bottom).
Validation of Matrix interactors
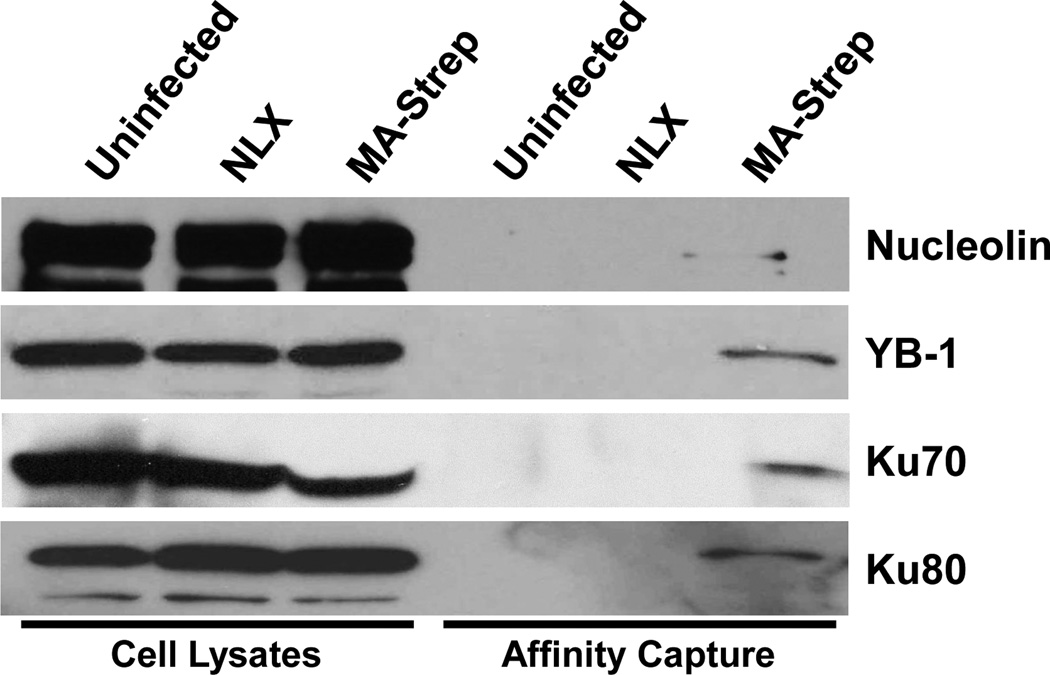
Next we sought to validate the interactions of several candidate proteins with Matrix by western blot. We focused on several proteins that we identified in previous proteomic studies of PICs and HIV infected nuclei, including Ku70, Ku80 (XRCC6 and XRCC5, respectively), and Nuclease-sensitive element-binding protein (YB-1) [31, 32]. We also included a previously described MA binding factor nucleolin (NCL) as a control [33, 34]. Each factor was detected in whole cell lysate input controls, and we were able to detect the presence of Ku70, Ku80, NCL, and YB-1 in the AP samples (Fig. 3), confirming the interaction of each factor with Matrix. Ku70 and Ku80 are components of the Ku heterodimer that is a component of the nonhomologous end-joining (NHEJ) pathway [35]. A previous study showed that Ku80 is present in the core of HIV particles [36] and that depletion of Ku80 inhibits HIV integration [37]. Ku70 has been shown to interact with HIV-1 integrase and protects it from the Lys48-linked polyubiquitination proteasomal pathway [38]. Knockdown of Ku70 in C8166 cells inhibits 2-LTR circles formation and reduces the level of integrated DNA [38]. YB-1 has been found in other mass spectrometry analysis that interacts with HIV gag. YB-1 participates varies of mRNA pathway, including pre-mRNA transcription and splicing, mRNA packaging, and regulation of mRNA stability and translation [39]. NCL is high abundance protein in nucleus and regulates various aspects of DNA and RNA metabolism [40], and has been reported co-localized with Matrix [33], as well as upregulated in the nuclei of HIV-infected cells [32]. Several other factors from the dataset, including ezrin, MED6 and OLA1, were tested in these assays and were not validated, possibly due to the lack of sensitivity of the western-blot approach (data not shown).
Figure 3.
Candidate protein interactions with Matrix. Cell lysates (left lanes) and samples purified with Strep-Tactin agarose were prepared from uninfected cells and cells infected with the indicated viruses. Samples were separated by SDS-PAGE and immunoblotted for the factors indicated at right.
Functional validation of the candidate host factors
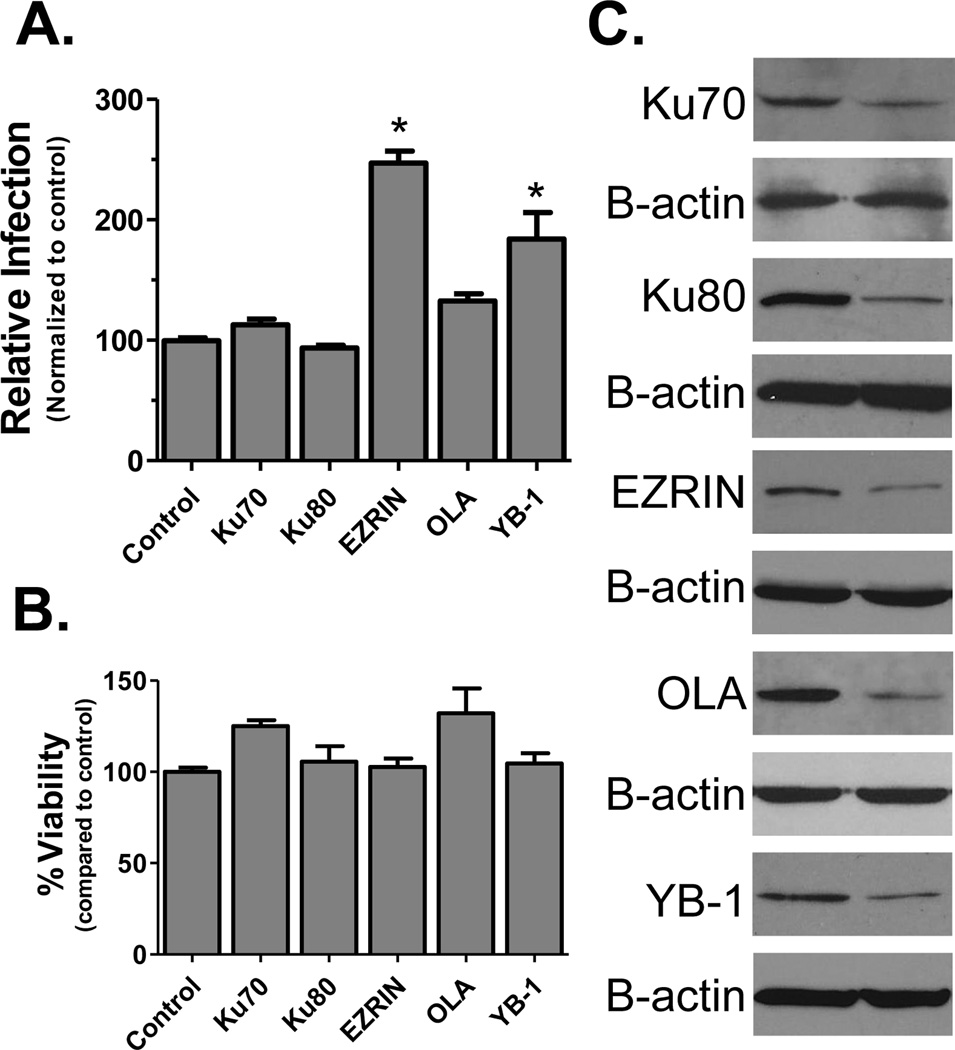
To further investigate the importance of the candidate factors in HIV biology, we tested the effect of depletion of factor expression on HIV infection by RNA interference. Cells were pre-transfected with siRNAs targeting the candidate factors and infected with a HIV luciferase reporter virus (Fig. 4A). Cell health was monitored by MTT assay to verify the effects were not due to changes in cell viability (Fig 4B). Knockdown of each protein was confirmed by immunoblot using β-actin as a control (Fig. 4C). The reduction of Ezrin and YB-1 box were found to enhance HIV replication. Ezrin is a member of the Ezrin-Radixin-Moesin family which regulates microtubule formation and has been shown to limit retroviral infection when over-expressed [41]. Our results were consistent with previous studies showing that knockdown of Ezrin enhances HIV replication and HIV-1-induced cell-cell fusion [42, 43]. YB-1 has been reported as a HIV Tat and TAR RNA binding factor that can modulate viral promoter activity [44, 45]. Reduced expression of Ku70 and OLA also slightly promoted HIV infection.
Figure 4.
siRNA screens for candidate factors. (A) Infection assays. Cells were transfected with siRNAs to indicated proteins 24 h prior to infection with a VSVg-pseudotyped HIV-Luciferase reporter virus. At 24 hpi cells were harvested and infection quantified by luciferase assay and normalized for protein concentration (* = p<0.001 by unpaired 2-tailed t-test). Control represents cells transfected with scrambled siRNAs. (B) Cell viability upon siRNA treatment was tested by MTT assay. Results in both graphs are presented relative to the control group and error bars represent standard error of the mean. (C) Knock-down of proteins was checked by immunoblot of transfected cells lysates using β-actin as a loading control (bottom panel for each protein).
Discussion
Combination antiviral treatment (cART) is successful at controlling HIV-1 replication and the development of disease but does not eliminate infection. The development of drug resistance is a major concern during life-long treatment of infected individuals. As an obligatory parasite, HIV-1 utilizes host cell factors and processes for viral propagation and dissemination. Defining virus-host interactions and altered pathways could lead to development of novel anti-viral treatments. This is a challenging task due to individual response of infected individuals and multistep complexity of viral life cycle. In this study we used a novel affinity purification approach to capture HIV-1 Matrix complexes during virus replication of T-cells. A SWATH-MS pipeline identified 3276 proteins across three biological replicates. Statistical analysis of protein abundance with Bonferroni correction identified 17 proteins in the MA-Strep samples compared to both the uninfected and infected-untagged control cells.
These data contribute to an evolving picture of HIV-host interactions in the cell. Over half of the identified proteins (11 of 17) are members of the NCBI HIV interaction database (supplemental dataset 2). Three (OLA1, Ezrin, YB-1) are present in the dataset of a broad study of the interacome of all HIV proteins [20]. Two of the proteins (IsoC2, HistH2A) were also identified previously in our study that captured Matrix complexes with an in vivo biotinylation system [30]. Previous studies have also shown the interaction of Matrix with LRPPRC and EEF1A1 [20, 30]. Notably, those factors that have not been reported previously may represent novel observations and warrant further characterization. In this study we confirmed the interaction of Ku70, Ku80 and YB-1. Functional validation studies also demonstrated that the knock-down of several factors modulated HIV infection. Combined, these data support the efficacy of our proteomic approach to study viral-host interactions and identify factors crucial for virus replication.
Global proteomics is an excellent approach to study virus-host interactions as it can direct us to intertwining cellular processes that are altered by virus infection. However, there is a dilemma to how focused a proteomic study should be without losing its power of global analysis. Using too global of an approach may pose difficulties in data interpretation; whereas too focused of an approach may lose crucial information. Previous AP-MS studies of Matrix interactions examine protein interactomes using singly transfected expression vectors. While a widely used strategy, this approach may not be advantageous for a number of reasons. First, factors such as exposure to transfection reagents or the non-physiological over-expression of a protein may alter the host cell functions or induce stress responses which skew the proteome of the cell [20, 46]. Second, certain virus-host interactions may involve multiple viral components and will be missed if only a single protein is expressed [47]. Third, it is very difficult to perform single viral protein AP-MS studies in primary cells due to low transfection efficiencies. The tagging of Matrix at a location that maintained HIV replication overcomes these challenges since the protein is expressed at physiological levels in the context of replicating virus. This study only demonstrated the feasibility of investigating the Matrix interactome in a T-cell line, but future studies will utilize this system to study Matrix interactions in infected primary T-cells and macrophages.
Supplementary Material
Clinical Relevance.
Viruses are obligate intracellular parasites that utilize host cell interactions to replicate. Proteomics can map these interactions on a global scale and lead to the identification of critical factors for infection and/or pathogenesis. Here, we constructed a replication competent clone of HIV-1 with an affinity tag in the Matrix protein of the virus. This virus was used to investigate the interactome of Matrix in HIV-infected T-cells. A SWATH-MS protocol identified numerous novel Matrix binding proteins. These data give insight into the function of Matrix during virus replication and may provide targets for the future development of novel, cell-based HIV inhibitors.
Acknowledgments
This work was funded by Grant AI080348 from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health.
Abbreviations
- AP
affinity purification
- AIDS
acquired immune deficiency syndrome
- Ku70
X-ray repair cross-complementing protein 6
- Ku80
X-ray repair cross-complementing protein 5
- cART
Combination antiviral treatment
- HIV
human immunodeficiency virus
- MA
matrix
- RPMI
Roswell Park Memorial Institute medium
- YB-1
Y-box binding protein 1
- rRNA
Ribosomal RNA
- RTC
reverse transcription complex
- PIC
preintegration complex
Footnotes
The authors declare no conflict of interest.
References
- 1.Gallo RC. A reflection on HIV/AIDS research after 25 years. Retrovirology. 2006;3:72. doi: 10.1186/1742-4690-3-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pneumocystis pneumonia--Los Angeles. MMWR Morb Mortal Wkly Rep. 1981;30:250–252. [PubMed] [Google Scholar]
- 3.World Health Organization. World Health Organization. Department of HIV/AIDS. Antiretroviral therapy for HIV infection in adults and adolescents : recommendations for a public health approach. [PubMed]
- 4.World Health Organization. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection : recommendations for a public health approach
- 5.Sax PE, Baden LR. When to start antiretroviral therapy--ready when you are? N Engl J Med. 2009;360:1897–1899. doi: 10.1056/NEJMe0902713. [DOI] [PubMed] [Google Scholar]
- 6.Vogel M, Schwarze-Zander C, Wasmuth JC, Spengler U, et al. The treatment of patients with HIV. Dtsch Arztebl Int. 2010;107:507–515. doi: 10.3238/arztebl.2010.0507. quiz 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanstrom R, Wills JW. Synthesis, Assembly, and Processing of Viral Proteins. 1997 [PubMed] [Google Scholar]
- 8.Freed EO. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 9.Bukrinsky MI, Sharova N, McDonald TL, Pushkarskaya T, et al. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci U S A. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karageorgos L, Li P, Burrell C. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res Hum Retroviruses. 1993;9:817–823. doi: 10.1089/aid.1993.9.817. [DOI] [PubMed] [Google Scholar]
- 11.Iordanskiy S, Berro R, Altieri M, Kashanchi F, Bukrinsky M. Intracytoplasmic maturation of the human immunodeficiency virus type 1 reverse transcription complexes determines their capacity to integrate into chromatin. Retrovirology. 2006;3:4. doi: 10.1186/1742-4690-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meric C, Spahr PF. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J Virol. 1986;60:450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parent LJ, Gudleski N. Beyond plasma membrane targeting: role of the MA domain of Gag in retroviral genome encapsidation. J Mol Biol. 2011;410:553–564. doi: 10.1016/j.jmb.2011.04.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alfadhli A, McNett H, Tsagli S, Bachinger HP, et al. HIV-1 matrix protein binding to RNA. J Mol Biol. 2011;410:653–666. doi: 10.1016/j.jmb.2011.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai M, Huang Y, Craigie R, Clore GM. Structural basis of the association of HIV-1 matrix protein with DNA. PLoS One. 2010;5:e15675. doi: 10.1371/journal.pone.0015675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alfadhli A, Barklis E. The roles of lipids and nucleic acids in HIV-1 assembly. Front Microbiol. 2014;5:253. doi: 10.3389/fmicb.2014.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Checkley MA, Luttge BG, Freed EO. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol. 2011;410:582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta K, Ott D, Hope TJ, Siliciano RF, Boeke JD. A human nuclear shuttling protein that interacts with human immunodeficiency virus type 1 matrix is packaged into virions. Journal of Virology. 2000;74:11811–11824. doi: 10.1128/jvi.74.24.11811-11824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jager S, Cimermancic P, Gulbahce N, Johnson JR, et al. Global landscape of HIV-human protein complexes. Nature. 2011;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lama J, Trono D. Human immunodeficiency virus type 1 matrix protein interacts with cellular protein HO3. Journal of Virology. 1998;72:1671–1676. doi: 10.1128/jvi.72.2.1671-1676.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peytavi R, Hong SS, Gay B, d'Angeac AD, et al. HEED, the product of the human homolog of the murine eed gene, binds to the matrix protein of HIV-1. The Journal of biological chemistry. 1999;274:1635–1645. doi: 10.1074/jbc.274.3.1635. [DOI] [PubMed] [Google Scholar]
- 23.Belshan M, Matthews JM, Madson CJ. Replication of biotinylated human immunodeficiency viruses. Journal of Virological Methods. 2011;171:299–302. doi: 10.1016/j.jviromet.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller B, Daecke J, Fackler OT, Dittmar MT, et al. Construction and Characterization of a Fluorescently Labeled Infectious Human Immunodeficiency Virus Type 1 Derivative. J. Virol. 2004;78:10803–10813. doi: 10.1128/JVI.78.19.10803-10813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt TG, Skerra A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc. 2007;2:1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- 26.Brown HE, Chen H, Engelman A. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J Virol. 1999;73:9011–9020. doi: 10.1128/jvi.73.11.9011-9020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J, Choe S, Walker R, Di Marzio P, et al. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 29.Haverland NA, Fox HS, Ciborowski P. Quantitative proteomics by SWATH-MS reveals altered expression of nucleic acid binding and regulatory proteins in HIV-1-infected macrophages. Journal of Proteome Research. 2014;13:2109–2119. doi: 10.1021/pr4012602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schweitzer CJ, Matthews JM, Madson CJ, Donnellan MR, et al. Knockdown of the Cellular Protein LRPPRC Attenuates HIV-1 Infection. PLoS One. 2012;7:e40537. doi: 10.1371/journal.pone.0040537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schweitzer CJ, Jagadish T, Haverland N, Ciborowski P, Belshan M. Proteomic analysis of early HIV-1 nucleoprotein complexes. Journal of Proteome Research. 2013;12:559–572. doi: 10.1021/pr300869h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeBoer J, Jagadish T, Haverland NA, Madson CJ, et al. Alterations in the nuclear proteome of HIV-1 infected T-cells. Virology. 2014;468–470:409–420. doi: 10.1016/j.virol.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao W, Li M, Zhang J. Tandem immunoprecipitation approach to identify HIV-1 Gag associated host factors. J Virol Methods. 2014;203:116–119. doi: 10.1016/j.jviromet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 34.Ueno T, Tokunaga K, Sawa H, Maeda M, et al. Nucleolin and the packaging signal, psi, promote the budding of human immunodeficiency virus type-1 (HIV-1) Microbiol Immunol. 2004;48:111–118. doi: 10.1111/j.1348-0421.2004.tb03496.x. [DOI] [PubMed] [Google Scholar]
- 35.Fell VL, Schild-Poulter C. Ku regulates signaling to DNA damage response pathways through the Ku70 von Willebrand A domain. Mol Cell Biol. 2012;32:76–87. doi: 10.1128/MCB.05661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos S, Obukhov Y, Nekhai S, Bukrinsky M, Iordanskiy S. Virus-producing cells determine the host protein profiles of HIV-1 virion cores. Retrovirology. 2012;9:65. doi: 10.1186/1742-4690-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeanson L, Subra F, Vaganay S, Hervy M, et al. Effect of Ku80 depletion on the preintegrative steps of HIV-1 replication in human cells. Virology. 2002;300:100–108. doi: 10.1006/viro.2002.1515. [DOI] [PubMed] [Google Scholar]
- 38.Zheng Y, Ao Z, Wang B, Jayappa KD, Yao X. Host Protein Ku70 Binds and Protects HIV-1 Integrase from Proteasomal Degradation and Is Required for HIV Replication. J Biol Chem. 2011;286:17722–17735. doi: 10.1074/jbc.M110.184739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB-1 protein: functions and regulation. Wiley Interdiscip Rev RNA. 2014;5:95–110. doi: 10.1002/wrna.1200. [DOI] [PubMed] [Google Scholar]
- 40.Tajrishi MM, Tuteja R, Tuteja N. Nucleolin: The most abundant multifunctional phosphoprotein of nucleolus. Commun Integr Biol. 2011;4:267–275. doi: 10.4161/cib.4.3.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naghavi MH, Valente S, Hatziioannou T, de Los Santos K, et al. Moesin regulates stable microtubule formation and limits retroviral infection in cultured cells. Embo J. 2007;26:41–52. doi: 10.1038/sj.emboj.7601475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy NH, Lambele M, Chan J, Symeonides M, Thali M. Ezrin is a component of the HIV-1 virological presynapse and contributes to the inhibition of cell-cell fusion. J Virol. 2014;88:7645–7658. doi: 10.1128/JVI.00550-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haedicke J, de Los Santos K, Goff SP, Naghavi MH. The Ezrin-radixin-moesin family member ezrin regulates stable microtubule formation and retroviral infection. J Virol. 2008;82:4665–4670. doi: 10.1128/JVI.02403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ansari SA, Safak M, Gallia GL, Sawaya BE, et al. Interaction of YB-1 with human immunodeficiency virus type 1 Tat and TAR RNA modulates viral promoter activity. J Gen Virol. 1999;80(Pt 10):2629–2638. doi: 10.1099/0022-1317-80-10-2629. [DOI] [PubMed] [Google Scholar]
- 45.Gautier VW, Gu L, O'Donoghue N, Pennington S, et al. In vitro nuclear interactome of the HIV-1 Tat protein. Retrovirology. 2009;6:47. doi: 10.1186/1742-4690-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engeland CE, Brown NP, Borner K, Schumann M, et al. Proteome analysis of the HIV-1 Gag interactome. Virology. 2014;460–461:194–206. doi: 10.1016/j.virol.2014.04.038. [DOI] [PubMed] [Google Scholar]
- 47.Greco TM, Diner BA, Cristea IM. The Impact of Mass Spectrometry–Based Proteomics on Fundamental Discoveries in Virology. Annual Review of Virology. 2014;1:581–604. doi: 10.1146/annurev-virology-031413-085527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.