Abstract
Evidence links aryl hydrocarbon receptor (AHR) activation to rheumatoid arthritis (RA) pathogenesis, although results are inconsistent. AHR agonists inhibit pro-inflammatory cytokine expression in macrophages, pivotal cells in RA aetiopathogenesis, which hints at specific circuits that regulate the AHR pathway in RA macrophages. We compared microRNA (miR) expression in CD14+ cells from patients with active RA or with osteoarthritis (OA). Seven miR were downregulated and one (miR-223) upregulated in RA compared to OA cells. miR-223 upregulation correlated with reduced Notch3 and Notch effector expression in RA patients. Overexpression of the Notch-induced repressor HEY-1 and co-culture of healthy donor monocytes with Notch ligand-expressing cells showed direct Notch-mediated downregulation of miR-223. Bioinformatics predicted the AHR regulator ARNT (AHR nuclear translocator) as a miR-223 target. Pre-miR-223 overexpression silenced ARNT 3’UTR-driven reporter expression, reduced ARNT (but not AHR) protein levels and prevented AHR/ARNT-mediated inhibition of pro-inflammatory cytokine expression. miR-223 counteracted AHR/ARNT-induced Notch3 upregulation in monocytes. Levels of ARNT and of CYP1B1, an AHR/ARNT signalling effector, were reduced in RA compared to OA synovial tissue, which correlated with miR-223 levels. Our results associate Notch signalling to miR-223 downregulation in RA macrophages, and identify miR-223 as a negative regulator of the AHR/ARNT pathway through ARNT targeting.
Macrophages are an integral part of the innate immune system, with important functions in the onset and development of chronic inflammatory diseases1. Many pathological alterations in RA result from aberrant macrophage hyperactivation in the inflamed synovial membrane2. These activated macrophages are the main producers of pro-inflammatory factors, including tumor necrosis factor (TNF)-α, interleukin (IL)-1β and IL-6, targets of current therapies3. Synovial macrophage accumulation correlates well with disease activity and joint destruction, and is the most sensitive biomarker of a positive response to therapy4.
MicroRNA (miR), small noncoding RNA molecules of ~15–25 nucleotides, act as posttranscriptional negative regulators that fine-tune protein expression by binding to the 3′-untranslated regions (UTR) of their mRNA targets; this causes their degradation or translation repression depending on the degree of base-pair complementarity between the miR and its target mRNA5. Specific miR can also positively regulate gene expression5. miR participate in immune cell differentiation, maturation and function6, and their aberrant expression is associated with pathogenesis in several osteoarticular diseases. Synovial fibroblasts from RA patients upregulate miR-155, miR-203, miR-221, miR-222 and miR-323-3p; immune cells from these patients also show altered miR-16, miR-132, miR-146a, miR-155 and miR-223 levels7,8. The miR profile in RA patients differs from that in patients with OA9, a non-autoimmune disease characterised by low inflammation levels. The relevance of these miR in RA aetiopathogenesis is poorly understood.
The aryl hydrocarbon receptor (AHR), or dioxin receptor is a cytosolic sensor of xenobiotics such as benzo[a]pyrene (BaP), found in large amounts in tobacco smoke10, a known environmental risk factor for RA development and severity11. AHR also binds to endogenous ligands, including metabolites of arachidonic acid and tryptophan12. AHR-ligand complex translocates to the nucleus, where it forms a heterodimeric complex with the aryl hydrocarbon nuclear translocator (ARNT, also termed hypoxia-inducible factor-1β), which enables receptor binding to cognate DNA consensus sequences12 including xenobiotic metabolising enzymes of the cytochrome P450 superfamily members CYP1A1, CYP1A2 and CYP1B1 (ref. 10). A non-conventional, ARNT-independent pathway has also been reported13.
Evidence links AHR activation to RA pathogenesis. AHR agonists upregulate IL-1β in human synoviocytes14. AHR also contributes to the differentiation of T helper (Th)17 cells15, and is necessary for their IL-22 production16,17. AHR deficiency in T cells, but not in macrophages, inhibits development of experimental arthritis in mice18. In apparent contradiction, AHR enhances differentiation of regulatory T (Treg) cells16,19 and suppresses experimental autoimmune encephalomyelitis20. AHR stimulation also inhibits TNF-α, IL-6 and IL-1β expression in lipopolysaccharide (LPS)-stimulated macrophages21,22,23. These disparate results hint at cell type-specific circuits that regulate the AHR/ARNT pathway in specific contexts. In this study, we analysed the role of miR-223 as a part of these regulatory circuits in RA-derived macrophages.
Results
Macrophages from RA vs OA patients express distinct sets of miR
Given the effectiveness of current therapies, few RA patients now require replacement surgery, and availability of fresh RA synovial tissue is thus limited. In active RA, numerous neutrophils, lymphocytes and macrophages migrate from synovial tissue and accumulate in the synovial fluid (SF)24. CD14+ macrophage-like cells constitute the majority of inflammatory CD14+ cells isolated from SF25, and these cells display markers compatible with M1-skewed polarisation26. We analysed the miR expression profile in CD14+ cells isolated from SF of three RA patients with active disease. The CD14-based positive selection method rendered >95% CD14+CD3− cell purity that showed no detectable IL-23 expression (not shown). As a reference, we used CD14+ cells isolated from the synovial membrane of three OA patients. OA is a low intensity inflammatory disease, and synovial fluid (SF) from OA patients has very low cellularity, which impedes the isolation of sufficient CD14+ cells to perform the analyses.
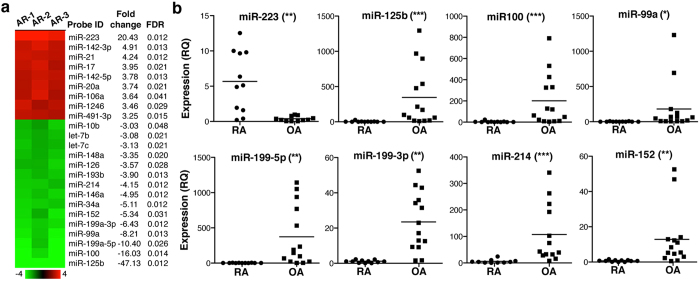
Our analysis identified 24 differentially regulated miR (≥3-fold), with a false discovery rate (FDR) <0.05 (Fig. 1a). Some up- and downregulated miR belong to the same family and are transcribed in a single mRNA; this was the case of clusters miR-17–92 (comprising miR-17, miR-20a and miR-106a), miR-125 (miR-125b, miR-99a, miR-100 and let-7), miR-199 (miR-199a, miR-199b, miR-214) and miR-152/148 (miR-152 and miR-148a).
Figure 1. RA and OA patient macrophages express a specific miR profile.
(a) Heat map showing the differentially expressed miR in CD14+ cells from OA and RA as determined by hybridisation in two-color microarrays. The x-fold change and FDR value are also indicated (n = 3 samples/condition). (b) miR validated by RT-qPCR in an independent set of 11 RA and 14 OA samples. Values are indicated as relative quantification (RQ), using as reference the sample with the lowest 2−ΔΔCt value (RQ = 1). *p < 0.05, **p < 0.01, ***p < 0.001, Mann-Whitney U-test.
Sixteen miR (7 up- and 9 downregulated) were selected for further validation by real-time, quantitative PCR (RT-qPCR) in an independent set of CD14+ cells isolated from SF (RA patients) or synovial tissue (OA patients). miR-223 was the only miR validated as upregulated in RA- compared to OA-derived cells (Fig. 1b). The miR-17–92 cluster and miR-142-5p tended to be upregulated in RA macrophages, but values were not significant (Supplementary Fig. S1). Seven downregulated miR of the miR-125, miR-199 and miR-152 clusters were validated; miR-34a and miR-146a were not confirmed (Supplementary Fig. S1). We focussed further analysis on miR-223, as it was the only miR upregulated in RA-derived cells, given the importance of the targets identified by bioinformatics (see below) and due to its implication in RA pathogenesis27,28,29,30,31.
The Notch pathway regulates miR-223 expression
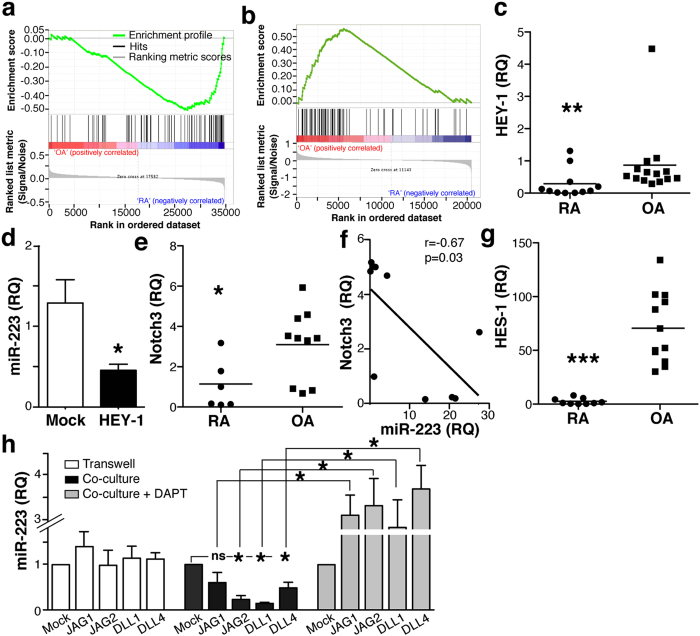
To study miR-223 regulation in RA macrophages, we searched the Ensembl database (v.77) for putative miR-223 promoter-binding transcription factors (TF). Scanning of the 10 Kb upstream of the miR-223 sequence identified two main regulatory regions in which TF binding sites were identified by the ENCODE Project (Supplementary Table S1). Of the TF predicted to bind the miR-223 promoter, we focused on the repressor HEY-1 (hairy/enhancer of split-related -HESR-with YRPW motif protein 1) for several reasons. First, at difference from Srf (serum response factor), Egr1 (early growth response) or TAF1 (TATA box binding protein-associated factor 1), all involved in multiple regulatory programs, HEY-1 is a very specific target of the Notch pathway. In addition, whereas PU.1 is a well-recognised myeloid transcription factor that could upregulate miR-223, Notch pathway and HEY-1 involvement in miR-223 regulation has not been studied in myeloid cells. Finally, study of gene expression profiles for human SF CD14+ cells from RA and tissue macrophages from OA patients (GSE49604) (ref. 32) using gene set enrichment analysis (GSEA) showed diminished Notch signalling in RA compared to OA cells (p < 0.05; Fig. 2a). GSEA also showed a tendency (FDR = 0.28; Fig. 2b) to enriched Notch signalling in human bone marrow-derived cells (BMDC) from OA compared to RA patients33 (GSE27390). Consistent with these analyses, HEY-1 expression was lower in RA than in OA CD14+ cells (Fig. 2c). In HEK-293T cells, HEY-1 overexpression led to significant reduction in miR-223 expression (Fig. 2d), which validated HEY-1 as a miR-223 transcriptional regulator.
Figure 2. Notch activation downregulates miR-223 expression.
(a) Enrichment plot (green) of GSEA using expression profiles of human macrophages isolated from SF of RA (n = 3) and synovial tissue of OA patients (n = 3). (b) GSEA as in a using expression profiles of human BMDC from RA (n = 9) and OA (n = 10) patients. The GSI_Notch gene set was used for GSEA (a,b). The bottom section shows the ranking metric value, which indicates gene correlation with a phenotype. (c) Relative HEY-1 mRNA levels in CD14+ cells from RA and OA patients. (d) miR-223 levels in HEK-293T cells overexpressing HEY-1 or an irrelevant cDNA (GFP; mock). (e) Determination of Notch3 mRNA levels in RA and OA isolated cells. (f) Linear regression analysis of Notch3 and miR-223 levels in RA and OA samples. Only samples with paired determinations of Notch3 and miR-223 were included in the analysis (n = 11). (g) Relative mRNA levels of the Notch effector HES-1 in the indicated samples. (h) Monocytes healthy donors were co-cultured with mock or Notch ligand-expressing OP-9 cells in conditions without (transwell) or with cell-cell contact (co-culture), alone or with the Notch inhibitor DAPT. After cell sorting, monocyte miR-223 levels were determined by RT-qPCR. For (c,e,f,g) each point represents one patient. For (d,h), data are mean ± SEM from triplicate RQ determinations in at least 3 independent experiments. *p < 0.05, **p < 0.01, Mann-Whitney U-test.
Analysis of Notch isoforms showed similar Notch1 and Notch2 levels in OA and RA CD14+ cells (Supplementary Fig. S2). Notch3 mRNA was nonetheless lower in RA than in OA cells (Fig. 2e), and Notch3 mRNA and miR-223 levels showed negative correlation (p = 0.03, r = 0.68; Fig. 2f). Levels of HES-1, another Notch target, were lower in RA cells (Fig. 2g); negative correlation was also found between HES1 and miR-223 levels in our cohort (not shown), which supports GSEA data indicating that the Notch pathway is less active in RA than in OA macrophages.
To determine whether Notch signalling regulates miR-223 expression in myeloid cells, we co-cultured monocytes from healthy donors with OP-9 cells, a bone marrow-derived mesenchymal stem cell line that does not express Notch ligands, or with human Notch ligand-transduced OP-9 subclones; miR-223 was determined in monocytes isolated by cell sorting. OP-9 cells do not express macrophage colony-stimulating factor34, which precluded monocyte expansion and/or differentiation during co-culture.
Monocyte miR-223 expression was unaltered when cells were co-cultured with Notch ligand-expressing OP-9 cells in transwells (Fig. 2h). When cell-cell contact was allowed, OP-9 cells that expressed Jagged2 (JAG2), delta-like1 (DLL1) or delta-like4 (DLL4) induced significant monocyte miR-223 downregulation (Fig. 2h). JAG1-expressing OP-9 cells did not cause miR-223 repression in monocytes (p = 0.14, Mann-Whitney U-test).
Monocytes and OP-9 cells were also co-cultured with DAPT (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester), a gamma-secretase inhibitor that blocks Notch activation35. DAPT reversed the inhibitory effect of the Notch ligands on monocyte miR-223 levels (Fig. 2h). miR-223 expression was slightly although not significantly higher in monocytes co-cultured with DAPT-treated ligand-expressing OP-9 cells compared to OP-9 mock cells (Fig. 2h), suggesting basal Notch-mediated repression of miR-223 in monocytes. These results indicate that Notch signalling represses miR-223 in myeloid cells.
In silico search for miR-223 targets
Although >20 genes have been identified as miR-223 targets in distinct cell types, only four are validated in monocytes/macrophages; these include the IKKα (inhibitor of NF-κB kinase α), the NLRP3 (NOD-like receptor family, pyrin domain containing 3) inflammasome, PKNOX1, and STAT3 (signal transducer and activator of transcription)36,37. miR-223-mediated silencing of NLRP-3, Pknox1, and STAT3 is counterintuitive based on the pro-inflammatory phenotype of RA macrophages. IKKα targeting by miR-223 relieves inhibition of the canonical NF-κB pathway, which enhances production of pro-inflammatory cytokines and chemokines38. We found no differences in ikkα mRNA (Supplementary Fig. S3a) and IKKα staining by immunohistochemistry (IHC) between RA and OA synovial tissues (Supplementary Fig. S3b).
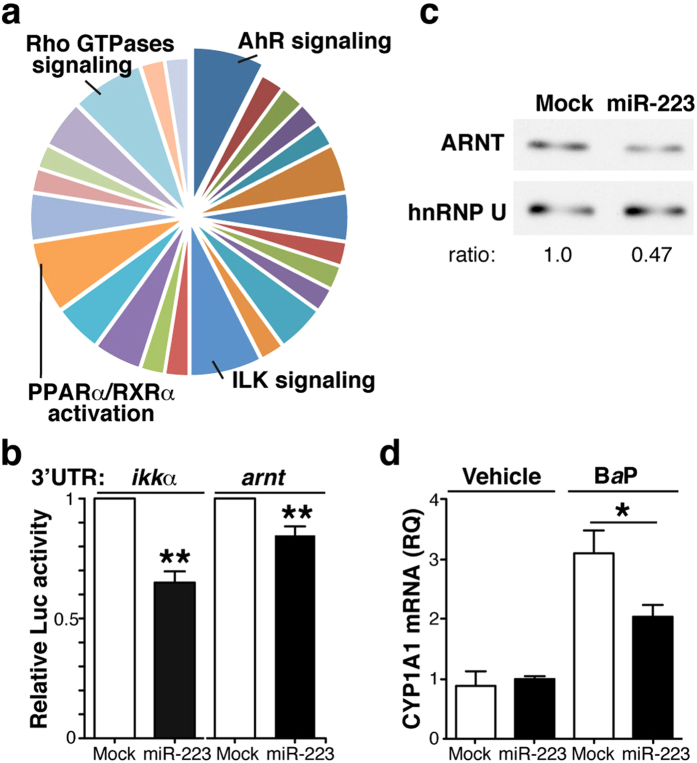
We searched for unknown miR-223 targets using in silico enrichment analysis. Ingenuity pathway analysis (IPA) linked these predicted miR-223 targets to 25 signalling pathways (Supplementary Fig. S4), the most prominent of which were those of the AHR, integrin-linked kinase (ILK), PPAR-α/retinoid X receptor (RXR)-α, and small Rho GTPases (Fig. 3a). We focussed on the AHR pathway because of its relevance in RA pathology. miR-223 was predicted to interact with ARNT, a necessary partner for AHR-driven transcription.
Figure 3. miR-223 target ARNT expression and signalling.
(a) Ingenuity analysis of putative miR-223 targets as determined in silico. Signalling pathways with the highest scores are indicated. (b) Luciferase reporter assays of HEK-293 cells transduced with pre-miR-223 or a control (mock), and transfected with the wild-type ikkα- or the arnt 3′UTR reporter constructs. Luciferase activity was normalised to that of mock-transfected cells. Data shown as mean ± SEM of triplicates in one representative experiment (n = 3). (c) Immunoblot analysis of extracts of HEK-293 cells expressing control or pre-miR-223, probed with anti-ARNT and -hnRNP U (nuclear loading control); the hnRNP U/ARNT ratio in mock-transfected cells is equal to 1. For (b,c), data are representative of at least three experiments. (d) CYP1A1 mRNA levels in mock and pre-miR-223-expressing THP-1 cells treated with BaP or vehicle. Data shown as mean ± SEM of RQ from triplicates in three independent experiments. *p < 0.05, **p < 0.01, Mann-Whitney U-test.
We found 12 transcripts in Ensembl annotated for the human arnt gene, of which only six registered a 3′UTR region. FindTar predicted, with high reliability, four binding sites for miR-223-3p and seven for miR-223-5p in the arnt 3′UTR (Supplementary Table S2); most of these were located in regions of exact sequence identity among the isoforms, which could indicate redundant gene regulation by miR-223.
Identification of ARNT as a miR-223 target
To determine whether the arnt 3′UTR is a miR-223 target, we transduced HEK-293T cells with pre-miR-223 or a mismatched miR (mock), and a cytomegalovirus promoter-driven luciferase construct fused to the arnt 3′UTR region. As a positive control, we used a similar luciferase reporter system under ikkα 3′UTR control, which miR-223 should target. Expression of the miR-223 mimic significantly reduced both ikkα and arnt 3′UTR-driven luciferase expression (Fig. 3b). The large number of miR-223 binding sites on the arnt 3’UTR precluded mutagenesis approaches to confirm silencing specificity. ARNT protein levels were reduced in pre-miR-223-expressing HEK-293T cells (Fig. 3c), which supports miR-223 targeting of arnt mRNA. In addition, pre-miR-223 expression in THP-1 myeloid cells attenuated BaP-induced upregulation of CYP1A1 (Fig. 3d), an AHR effector10. Since pre-miR-223 overexpression did not affect AHR protein levels in these cells (Supplementary Fig. S5), our results suggest that high miR-223 levels functionally impair the AHR/ARNT pathway in myeloid cells by reducing ARNT protein levels.
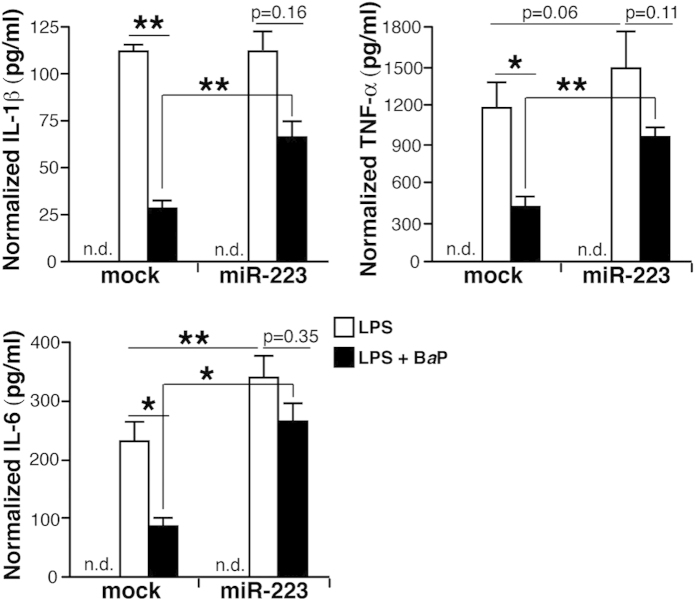
miR-223 counteracts AHR/ARNT-mediated inhibition of pro-inflammatory cytokines
AHR agonists suppress LPS-induced expression of TNF-α, IL-6 and IL-1β21,22,23, which are important cytokines in RA aetiopathogenesis. We quantified the levels of these cytokines in control or pre-miR-223-expressing THP-1 cells exposed to LPS alone or with BaP. Pre-miR-223 expression tended to increase cytokine levels after LPS stimulation, which was significant only for IL-6 (Fig. 4). BaP significantly decreased LPS-induced cytokine expression in mock-transfected cells, but pre-miR-223 partially reversed BaP-induced inhibition (Fig. 4). Moreover, cytokine production was significantly higher in pre-miR-223-expressing THP-1 cells stimulated with LPS+BaP than in controls (Fig. 4). High miR-223 levels thus override AHR-mediated repression of pro-inflammatory cytokines in myeloid cells.
Figure 4. miR-223 reverses AHR/ARNT-induced inhibition of pro-inflammatory cytokines.
ELISA analyses for IL-1β, TNF-α and IL-6 levels in the supernatant of mock- and pre-miR-223-transfected THP-1 cells stimulated with vehicle, LPS or LPS + BaP. Values are expressed as the concentration of cytokine (pg/ml) per mg cell extract. Data shown as mean ± SEM of values in three independent experiments. *p < 0.05, **p < 0.01 Student’s two-tailed t-test; n.d., not detected in vehicle-stimulated.
The AHR/ARNT pathway is impaired in RA synovial tissue
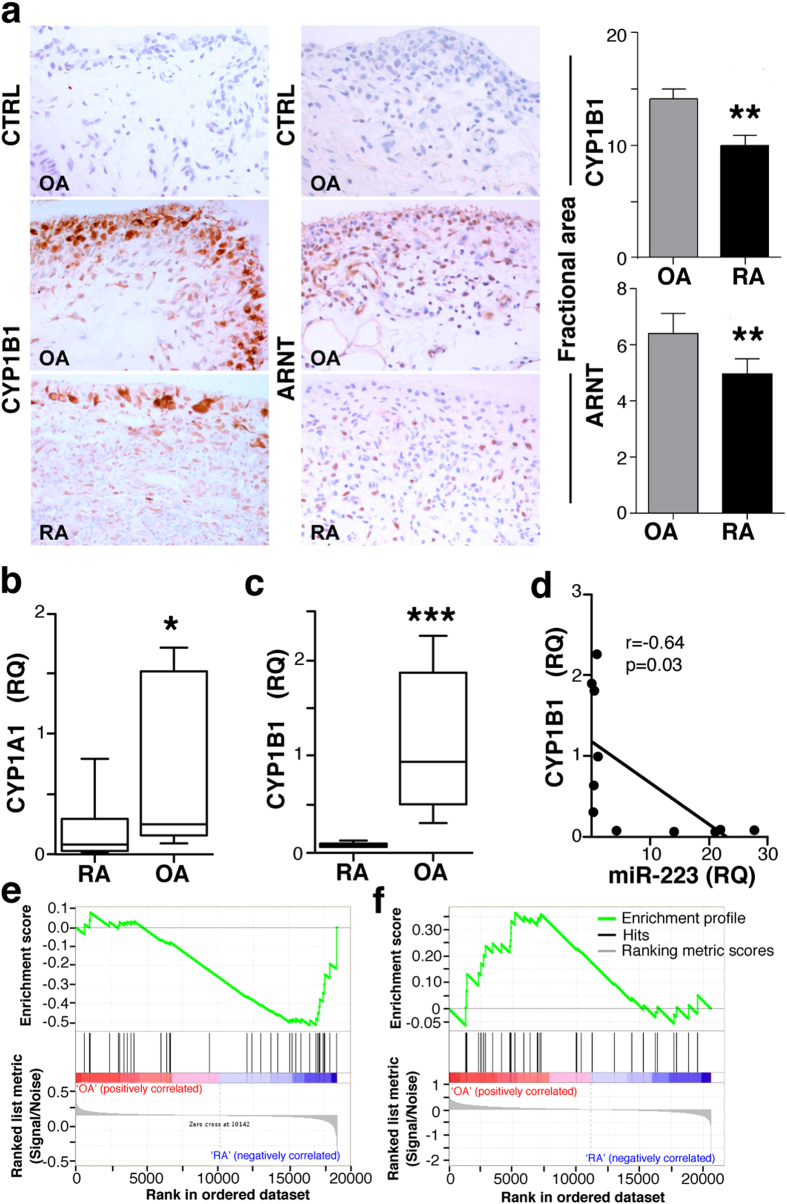
We tested whether miR-223 levels are associated to impaired activation of the AHR/ARNT pathway in RA patients. In synovial tissues from OA and RA patients, we used IHC to analyse the CYP1B1 staining as a functional surrogate marker of AHR/ARNT pathway differences. Double immunofluorescent labelling showed variable CYP1B1 expression by CD68+ lining macrophages in OA and RA synovia, although the frequency of CD68+ cells with very low or undetectable CYP1B1 staining appeared to be higher in RA than in OA samples (Supplementary Fig. S6); CYP1B1 was also detected in other CD68− cell types. IHC indicated profuse cytoplasmic CYP1B1 expression in fibroblastic and large mononuclear cells of lining and sublining areas in RA and OA synovial tissues (Fig. 5a); evaluation showed that the fractional area immunostained by CYP1B1 was larger in OA than in RA tissues (p = 0.002). This increased CYP1B1-stained area correlated with more ARNT+ cell accumulation in lining areas of OA than of RA tissues (p = 0.01; Fig. 5a). The data suggest attenuation of the AHR/ARNT pathway in RA compared to OA patients.
Figure 5. The AHR/ARNT pathway is downmodulated in RA compared to OA.
(a) Immunohistochemical analysis of CYP1B1 (left) and ARNT (right) staining in RA (n = 13) and OA (n = 8) tissues. Staining with an irrelevant antibody is also shown (CTRL). Fractional immunostained area was quantified in digitalised images using ImageJ software. (b,c) CYP1A1 and CYP1B1 levels in cells isolated from RA (n = 6) and OA (n = 8) patients. RQ values were calculated from triplicates for each sample. (d) Linear regression analysis of CYP1B1 mRNA and miR-223 levels in RA and OA samples (n = 11). (e,f) Enrichment plot (green) of GSEA using expression profiles of human SF-derived macrophages (e) from RA (n = 6) and controls (n = 2), or BMDC (f) from RA (n = 9) and OA (n = 10) patients. The AHR gene set was obtained from PubMed (BIOGRID http://www.ncbi.nlm.nih.gov/gene/196). The bottom shows the ranking metric value, which indicates gene correlation with a phenotype.
Analysis of AHR targets also indicated lower CYP1A1 (Fig. 5b) and CYP1B1 mRNA levels (Fig. 5c) in CD14+ cells isolated from RA than from OA patients; CYP1A2 was not expressed in CD14+ cells (not shown). We also found negative correlation in macrophage expression of miR-223 and CYP1A1 (p = 0.05; r = 0.60; Supplementary Fig S7a) and CYP1B1 (p = 0.03, r = 0.64; Fig. 5d), which suggests that miR-223 upregulation in RA CD14+ cells impairs AHR/ARNT-mediated transcription. We found no differences in AHR mRNA levels between OA and RA patients in our cohort (Supplementary Fig. S7b), which suggests that the functional impairment of the AHR/ARNT pathway is a result of reduced ARNT protein levels.
GSEA indicated a tendency to reduced AHR pathway signalling in RA macrophages (FDR = 0.1; GSE49604; Fig. 5e); the AHR gene set enrichment in OA cells was statistically significant when gene expression profiles for BMDC from RA and OA patients were analysed (FDR ≤0.25; Fig. 5f). These data reinforce the idea that the AHR/ARNT pathway is less active in RA than in OA patients.
miR-223 prevent AHR/ARNT-mediated Notch3 upregulation
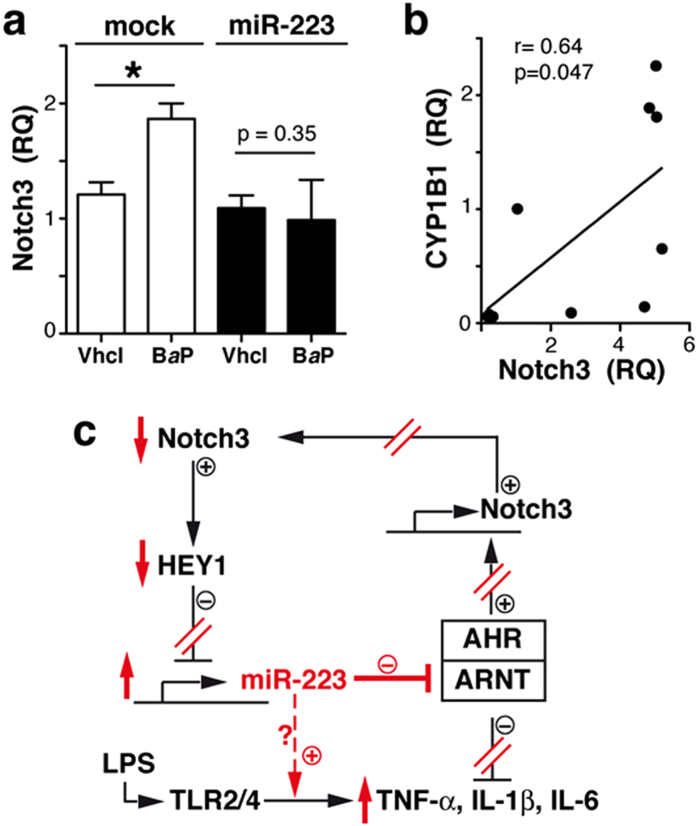
AHR activation upregulates Notch expression in hepatic and intestinal innate immune cells39. We hypothesised that high miR-223 levels in RA cells could reverse its own inhibition by reducing Notch3. To analyse whether miR-223 influences Notch3 expression downstream of AHR in myeloid cells, control miR- or pre-miR-223-transfected THP-1 cells were stimulated with BaP. AHR activation increased Notch3 mRNA levels in THP-1 control cells, but not in those expressing pre-miR-223 (Fig. 6a). Our data suggest a regulatory network that involves Notch3, miR-223 and AHR. This is supported by the positive correlation between Notch3 and CYP1A1 (p = 0.04; r = 0.66; Supplementary Fig S7c) and CYP1B1 mRNA levels (p = 0.047, r = 0.64; Fig. 6b) in our cohort. We therefore propose a model (Fig. 6c) in which high miR-223 levels in RA macrophages impair the AHR/ARNT pathway, which reduces Notch3 expression and prevents the AHR/ARNT-induced inhibition of pro-inflammatory cytokines relevant in RA pathogenesis.
Figure 6. miR-223 prevents AHR/ARNT-induced Notch3 upregulation.
(a) Notch3 mRNA levels in THP-1 cells expressing control (mock) or pre-miR-223 stimulated with BaP or vehicle. Data shown as mean ± SEM of RQ values (triplicates) from three independent experiments. (b) Linear regression analysis of CYP1B1 and Notch3 RQ values in RA and OA samples (n = 10). (c) Proposed model of miR-223 regulation in macrophages; miR-223 activity in RA macrophages indicated in red. *p < 0.05, **p < 0.01, ***p < 0.001, Mann-Whitney U-test.
Discussion
Here we combined gene expression and advanced bioinformatics analyses to demonstrate that (i) miR-223 is selectively upregulated in CD14+ cells from SF of RA patients with active disease; (ii) Notch activation represses miR-223 expression in myeloid cells; (iii) miR-223 targets ARNT, a necessary co-receptor for AHR-dependent transcriptional activity; (iv) miR-223 prevents AHR-mediated inhibition of pro-inflammatory cytokines; and (v) AHR target genes and ARNT protein itself are downmodulated in RA compared to OA synovial tissues. GSEA using gene expression profiles from two independent study32,33 using lesional SF macrophages and BMDC isolated from RA and OA patients, respectively, also detected reduced Notch and AHR signalling in RA-derived cells. Functional impairment of the AHR/ARNT pathway in RA patients might thus be a systemic phenomenon, not limited to lesional macrophages in our patient cohort.
In our analyses, we compared CD14+ cells isolated from synovial fluid (RA) and synovial tissue (OA). Despite their distinct sources, CD14+ cells isolated from SF and tissue should be considered comparable. There is no direct traffic from the bloodstream to the SF, and SF CD14+ cells thus derive from cells detached from the synovial lining layer. Morphological criteria indicate that SF CD14+ cells are not monocytes but macrophage-like cells25 with a genetic program resembling that of M1-skewed macrophages26. miR-223 levels decrease during monocyte-to-macrophage differentiation38, and this reduction leads to higher ikkα mRNA levels in macrophages than in monocytes. In our study, we found no difference in ikkα mRNA levels between RA-CD14+ cells from SF and OA-CD14+ cells from synovial tissue (Supplementary Fig. S3a), which reinforces the idea that CD14+ cells isolated from SF are not monocyte-enriched.
Studies of T cell acute lymphoblastic leukaemia showed that the Notch pathway can repress40 or induce41 miR-223 expression. Our results pinpoint Notch activation, and specifically HEY-1 as a direct repressor of miR-223 in myeloid cells. Reduced Notch signalling in RA macrophages, as evidenced by low HEY-1 and HES-1 levels, might affect miR-223 transcription directly (release of a brake in its promoter) or modify the activity of other miR-223 positive regulators such PU.1 (ref. 6). Indeed, Notch signalling restricts the extent of PU.1 activity in pro-T cells42. The mechanism responsible for impaired Notch signalling in our RA cells is nonetheless unclear. We detected lower Notch3 mRNA levels in RA SF macrophages, but Notch1 and Notch2 were unaltered relative to OA cells. Evidence indicates a non-redundant function of Notch ligands and receptors in cell function43,44. miR-223 repression might thus be a Notch3-mediated event induced by specific ligand interaction. In co-culture experiments, DLL1 and JAG2 inhibited miR-223 to a greater degree than DLL4; JAG1 had no effect. JAG2 induces the strongest Notch3-mediated signals in T cells of all the Notch ligands44, and provides anti-inflammatory signals in lung cancer cells43. Decreased Notch3 levels could also reduce Notch-mediated signalling strength in RA macrophages, leading to a specific differentiation pattern. Signalling strength is a major regulatory factor of Notch-mediated differentiation in human α/β or γ/δ T cells44, endothelial cell fate determination45 and of patterning in inner ear development46, in which Notch signalling strength is determined by competition between DLL1 and JAG1 ligands in adjacent cells. These examples show that combinatorial arrays of receptor-ligand cis- and trans-interactions could affect Notch signals, prompting distinct responses depending on cell or tissue context.
Here we link miR-223 to the AHR pathway, which is implicated in RA pathogenesis47. Although IPA suggested four putative miR-223 targets in the AHR pathway, we focussed on ARNT as a direct AHR regulator. Bioinformatics predicted that miR-223-3p and miR-223-5p both bind with imprecise base-pairing to multiple sites at the ARNT-3′UTR. This is consistent with the translational repression of ARNT, rather than mRNA degradation, observed after pre-miR-223 overexpression. The multiple miR-223 sites at the ARNT-3′UTR resemble other miR, including let-4 (ref. 48), and are usually associated with enhanced specificity and translational suppression. miR-223 overexpression prevented AHR-mediated inhibition of IL-6, TNF-α and IL-1β production in LPS-stimulated myeloid cells, which functionally validates AHR pathway targeting by miR-223.
The reduction of ARNT protein levels and AHR target genes in RA compared to OA synovial tissues suggests a miR-223/ARNT connection for human disease. We propose a model in which elevated miR-223 levels in RA macrophages reduce ARNT translation and thus prevent AHR-driven immunosuppression in these cells (Fig. 6c). Moreover, miR-223 negatively regulates AHR/ARNT-mediated Notch3 expression and could contribute to release of Notch-mediated miR-223 repression, augmenting its levels. miR-223 might also interact via an AHR-independent, TLR-dependent mechanism to boost induction of pro-inflammatory cytokines, particularly IL-6, in myeloid cells. miR-223 upregulation could thus increase RA macrophage sensitivity to pro-inflammatory and reduce their responsiveness to anti-inflammatory signals.
Our results imply that, in RA macrophages, miR-223 is a central regulator of the AHR/ARNT pathway, which might be activated by endogenous (e.g., kynurenic acid) and/or exogenous (e.g., tobacco smoke) ligands that can influence RA aetiology. The AHR pathway leads to pro- and anti-RA activities; AHR activation could promote RA development by inducing Th17 differentiation15 and could simultaneously provide RA-inhibitory signals such as Treg cell differentiation16,19 or impaired macrophage activation21,22,23. Elevated miR-223 levels in RA macrophages would render these cells insensitive to AHR/ARNT-induced inhibition of the production of TNF-α, IL-6 and IL-1β, critical cytokines in RA pathology. Whether miR-223 targets the AHR pathway in other RA synovial inflammatory cells will require further research. For instance, miR-223 is expressed strongly in naïve T cells but is barely detectable in RA Th17 cells27, which might facilitate AHR-driven Th17 cell development. High miR-223 levels correlate with reduced Treg cell numbers in maternal and cord blood of pregnant women exposed to tobacco smoke49, suggesting that miR-223 expression impairs AHR-induced Treg cell differentiation.
In summary, we demonstrate that high miR-223 expression in myeloid cells prevents AHR-induced repression of TNF-α, IL-6 and IL-1β. This miR-223/AHR pathway connection provides conceptual support for miR-223 antagonist-based therapies in RA28, and identifies a means to fine-tune AHR activity in myeloid cells.
Methods
Patients and synovial tissues
For microarray and validation studies, macrophages were obtained by centrifugation on Ficoll-Paque (GE-Healthcare) of SF from RA patients with active knee arthritis, or by collagenase I digestion of synovial tissue from OA patients who had undergone knee joint replacement surgery; both procedures were followed by magnetic separation of CD14+ cells (Miltenyi Biotec). The mean age of RA patients was 63 ± 5 years, and 73% were female; the mean age of OA patients was 73 ± 6 years, and 57% were female. Three patients received only non-steroid anti-inflammatory drug therapy, and four received disease-modifying anti-rheumatic drugs (3 methotrexate and one hydroxychloroquine) plus low dose steroids. The clinical characteristics of RA patients (n = 11) used for miR validation are shown in Supplementary Table S3. None of the RA or OA patients in the microarray and validation cohorts were active smokers at the time of sample collection.
Synovial tissues for IHC studies were derived from arthroscopic knee biopsy of RA patients with active arthritis (n = 13) or by surgical synovectomy of OA patients (n = 8) who underwent knee joint replacement surgery. Tissues were formalin-fixed and paraffin-embedded. Patients gave informed consent and the study was approved by the ethics committees of the Hospital 12 de Octubre (Madrid, Spain) and the Hospital Clinic (Barcelona, Spain). The methods were carried out in accordance with the approved guidelines.
RNA isolation and microarray analyses
RNA was extracted using Trizol reagent (Invitrogen), and integrity assessed in a Bionalyzer 2100 (Agilent). Total RNA (1 μg) from three RA and OA samples was labelled with miRCURY LNA microRNA Hy3/Hy5 Power labelling kit and hybridised onto slides in the human miRCURY LNA microRNA Array Kit v.11.0 (Exiqon). Slides were scanned at 5 μm resolution in an Axon 4000B scanner (Molecular Devices), and spots quantified with GenePix 5.0 software. Raw data (deposited in GEO; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ipilmwaerdujfwr&acc=GSE63745) were normalised and differential expression analysed using the Bioconductor Limma software. FDR was used to correct p-values.
Cell culture
HEK 293T cells (ATCC) were cultured in Dulbecco’s Modified Eagle Medium (DMEM; BioWest, Nuaillé, France) supplemented with 10% foetal calf serum (FCS), 1 mM sodium pyruvate and 2 mM L-glutamine. THP-1 cells (ATCC) were cultured in RPMI 1640 (BioWest) supplemented with 10% FCS, 2 mM L-glutamine, 1% non-essential amino acids and 0.05 mM 2-mercaptoethanol. OP-9 cells and subclones (see below) were cultured in minimum essential medium (MEM)-α (Life Technologies) supplemented with 20% FCS, 1 mM sodium pyruvate, 2 mM L-glutamine and 3 mM sodium bicarbonate. All media also contained penicillin/streptomycin (100 U/mL). All additives were from Sigma-Aldrich.
Transfection of HEK-293T cells and luciferase assay
HEK-293T cells were transfected with human HEY-1 (provided by J.L. de la Pompa; Centro Nacional de Investigaciones Cardiovasculares, Madrid, Spain) using jetPEI (Polyplus), or with pLV-hsa-mir-223 and pLV-mir-control vectors (Biosettia) using the CaCl2 method. Cells were selected with puromycin (3 μg/ml) and transfected with miTarget miRNA Target Sequence (3′UTR) Expression Clones for arnt and ikkα (Labomics). Reporter gene expression was analysed with the Dual-Luciferase Reporter Assay System (Promega). ARNT protein levels were analysed in nuclear extracts using anti-ARNT (clone D28F3; Cell Signaling) and anti-hnRNPU antibody (loading control; Abcam). AHR protein levels were analysed in total cell extracts using anti-AHR (clone RPT1; Thermo Fisher) and anti-β-actin (clone AC-15, Sigma-Aldrich).
Transduction of OP-9 cells with Notch ligands
pLZRS-IRES-eGFP retroviral constructs, empty or encoding human Delta-like1 (DLL1) or Jagged1 (JAG1), were provided by L. Parreira (Instituto de Histologia e Embriologia, Lisbon, Portugal). Human Jagged2 (JAG2) cDNA, PCR-amplified from total RNA isolated from HeLa cells, and human Delta-like4 (DLL4) full-length cDNA (provided by G. Tosato, National Institutes of Health, Bethesda, MD, USA) were subcloned into pLZRS-IRES-eGFP. Individual retroviral constructs were transfected into the packaging 293T Phoenix-Amphotropic cell line (provided by G. Nolan, Stanford University, Stanford, CA, USA), and supernatants obtained from puromycin-selected transfected cells were used to transduce OP-9 cells (ATCC) by centrifugation in the presence of polybrene. Transduced OP-9 cells with equivalent GFP expression levels were isolated by cell sorting (FACSVAntage) 48 h post-transduction.
Co-culture of monocytes with OP-9 cells
Purified CD14+ cells (>95%) from healthy donors were added (1 h, 37 °C) to mock- or Notch ligand-expressing OP-9 cells, in the presence of DMSO (vehicle) or DAPT (Merck Millipore); monocytes were isolated by cell sorting (MoFlo XDP; Beckman Coulter) based on lack of GFP expression. CD14+ cells were also plated in transwells (0.4 μm pore size; Costar) to prevent physical interaction with OP-9 cells during co-culture.
Transfection and stimulation of THP-1 cells
THP-1 cells were transfected with the pre-miR-223 (10 μM; Life Technologies), or with a fluorescently-labeled negative control (5 μM) using Amaxa I and the Monocyte Nucleofector Kit (Lonza). Transfected THP-1 cells (~95% based on fluorescence labelling) were stimulated (1–3 h, 37 °C) with BaP (5 μM) or vehicle, followed by LPS (1–8 h, 37 °C). Cytokine levels were determined in supernatants using specific ELISA kits (BioLegend). Concentration was divided by total protein level in cell extracts to correct for cell number in each condition.
qRT-PCR analyses
Total RNA was extracted from CD14+ cells isolated from RA or OA patients, or from cultured cells using TRI Reagent (Sigma-Aldrich). For miR qRT-PCR, retrotranscribed RNA (25 ng) was mixed with specific primers (TaqMan MicroRNA Reverse Transcription Kit; Life Technologies) in an ABI PRISM 7900 HT (Applied Biosystems) using a TaqMan MicroRNA-based system. For quantitative mRNA analyses, total RNA (1 μg) was retrotranscribed with the High Capacity Reverse transcription Kit (Life Technologies) using random primers, and then amplified with the primers listed (Supplementary Table S4) using a Hot FIREPol EvaGreen Plus-based system. The relative quantity (2−ΔΔCt) of each miR or mRNA was calculated using as reference the sample with the highest Ct value after normalisation to U6 Taqman probe (for miR analyses) or β-actin (for mRNA analyses).
Immunolabelling of synovial tissue
For IHC staining, we used a standard indirect avidin-biotin peroxidase method (ABC standard; Vector Laboratories) and developed with diaminobenzidine. Samples were stained with anti-IKKα (ab47453), -CYP1B1 or -ARNT antibodies. Paraffin sections were pretreated by microwave heating in citrate (pH 6.0) or Tris-EDTA buffer (pH 9.0) for CYP1B1 and ARNT detection, respectively. Negative controls were incubated with non-immune IgG. CYP1B1/CD68 double-labelled samples were analysed by simultaneous CYP1B1 immunoperoxidase and CD68 immunofluorescent detection and an Alexa594 goat anti-mouse secondary antibody (Molecular Probes).
Synovial tissue sections were photographed and digitalised using an AxioCam ERc 5S camera and ZEN lite 2012 software on a Zeiss Scope. A1 microscope (Zeiss). Fractional immunostained area was quantified in digitalised images using ImageJ software (http://rsb.info.nih.gov/ij). Data were analysed using Prism software v6.0 (GraphPad). Quantitative variables were compared by the Mann Whitney U-test; a p value <0.05 was considered significant.
GSEA analyses
Raw microarray data corresponding to BMDC from 10 OA and 9 RA patients were obtained from a public database33. Data were normalised using the RMA (quantile) approach and subjected to GSEA (http://www.broadinstitute.org/gsea/downloads.jsp), permutating 1000 times by phenotype. Illumina expression data from the GSE49604 study32 were re-analysed using quantiles normalisation and subjected to GSEA by permutating 1000 times by geneset. Enrichment in both cases was determined using gene sets of Notch (PMID: 19907488) and AHR.
Bioinformatic analyses for identification of miR-223 targets
The sequences of the human 3′-UTR arnt gene, of mature microRNA hsa-miR-223-3p and hsa-miR-223-5p were retrieved from the Ensembl database (release 72, June 2013) and the miRBase (release 19, August 2012). Of the 12 transcripts annotated for the human arnt gene, only 6 for which the 3′-UTR is registered were used in the calculations. Predictions were made using the algorithms miRanda, PITA, FindTar v3.11.12, RNAHybrid, and TargetScan v6 using parameter default values. The sequences of six, seven and eight nucleotides of each miRNA 5′ end were considered as seed, starting from the first, second or third nucleotide. Mismatches were not allowed in the seed. Multiple sequence alignments were performed using ClustalW v2.1.
Additional Information
How to cite this article: Ogando, J. et al. Notch-regulated miR-223 targets the aryl hydrocarbon receptor pathway and increases cytokine production in macrophages from rheumatoid arthritis patients. Sci. Rep. 6, 20223; doi: 10.1038/srep20223 (2016).
Supplementary Material
Acknowledgments
We thank Drs J.L. de la Pompa, L. Parreira, G. Tosato, and G. Nolan for kindly providing reagents, J.M. Franco, G. García-Casado (CNB Gemomics Unit) and J.C. Oliveros for help with microarray data, and C. Mark for editorial assistance. This work was supported by the Spanish Ministry Economy and Competitiveness (SAF2011-24453 and SAF2014-54475-R to SM; SAF2010-15106 and SAF2013-44857-R to MLT), the Comunidad de Madrid (INMUNOTHERCAN; S2010/BMD-2326 to SM and RAPHYME S2010/BMD-2350 to JLP), the Instituto de Salud Carlos III (ISCIII) (Inflammation and Rheumatic Diseases Network, co-financed by FEDER, RD12/0009; and FIS PI12/00439 to JLP). J.O. and D.J.M.H. are recipients of a MINECO and Obra Social “La Caixa” pre-doctoral fellowships, respectively.
Footnotes
Author Contributions S.M. and J.L.P. designed and conceived the study. J.O., M.T., A.D.-A., A.U., L.F., V.M.-R., D.J.M.-H., M.J.G.-L., M.C.C., S.E., J.D.C. and S.M. were involved in data acquisition and analysis. J.O., M.T., D.J.M.-H., M.L.T., I.C., A.P.-M., J.L.P. and S.M. were involved in data interpretation. S.M. wrote the manuscript. J.O., M.T., J.D.C., A.P.-M., M.L.T. and J.L.P. revised critically the manuscript, and all authors approved the final version.
References
- Hamilton J. A. & Tak P. P. The dynamics of macrophage lineage populations in inflammatory and autoimmune diseases. Arthritis Rheum 60, 1210–1221 (2009). [DOI] [PubMed] [Google Scholar]
- Tak P. P. et al. Analysis of the synovial cell infiltrate in early rheumatoid synovial tissue in relation to local disease activity. Arthritis Rheum 40, 217–225 (1997). [DOI] [PubMed] [Google Scholar]
- Okamoto H., Hoshi D., Kiire A., Yamanaka H. & Kamatani N. Molecular targets of rheumatoid arthritis. Inflamm Allergy Drug Targets 7, 53–66 (2008). [DOI] [PubMed] [Google Scholar]
- Wijbrandts C. A. et al. Absence of changes in the number of synovial sublining macrophages after ineffective treatment for rheumatoid arthritis: Implications for use of synovial sublining macrophages as a biomarker. Arthritis Rheum 56, 3869–3871 (2007). [DOI] [PubMed] [Google Scholar]
- Valinezhad Orang A., Safaralizadeh R. & Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics 2014, 970607 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansel K. M. RNA regulation of the immune system. Immunol Rev 253, 5–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter D., McInnes I. B. & Kurowska-Stolarska M. Novel regulatory mechanisms in inflammatory arthritis: a role for microRNA. Immunol Cell Biol 90, 288–292 (2012). [DOI] [PubMed] [Google Scholar]
- Pers Y. M. & Jorgensen C. MicroRNA in 2012: Biotherapeutic potential of microRNAs in rheumatic diseases. Nat Rev Rheumatol 9, 76–78 (2013). [DOI] [PubMed] [Google Scholar]
- Miyaki S. & Asahara H. Macro view of microRNA function in osteoarthritis. Nat Rev Rheumatol 8, 543–552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol 35, 307–340 (1995). [DOI] [PubMed] [Google Scholar]
- Kallberg H. et al. Smoking is a major preventable risk factor for rheumatoid arthritis: estimations of risks after various exposures to cigarette smoke. Ann Rheum Dis 70, 508–511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger B., Hirota K., Duarte J. & Veldhoen M. External influences on the immune system via activation of the aryl hydrocarbon receptor. Semin Immunol 23, 99–105 (2011). [DOI] [PubMed] [Google Scholar]
- Huang G. & Elferink C. J. A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol Pharmacol 81, 338–347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki A. et al. Polycyclic aromatic hydrocarbon increases mRNA level for interleukin 1 beta in human fibroblast-like synoviocyte line via aryl hydrocarbon receptor. Biol Pharm Bull 27, 407–410 (2004). [DOI] [PubMed] [Google Scholar]
- Kimura A., Naka T., Nohara K., Fujii-Kuriyama Y. & Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Nat Acad Sci USA 105, 9721–9726 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana F. J. et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453, 65–71 (2008). [DOI] [PubMed] [Google Scholar]
- Veldhoen M. et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453, 106–109 (2008). [DOI] [PubMed] [Google Scholar]
- Nakahama T. et al. Aryl hydrocarbon receptor deficiency in T cells suppresses the development of collagen-induced arthritis. Proc Nat Acad Sci USA 108, 14222–14227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetoh L. et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol 11, 854–861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana F. J. et al. An endogenous aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress experimental autoimmune encephalomyelitis. Proc Nat Acad Sci USA 107, 20768–20773 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A. et al. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J Exp Med 206, 2027–2035 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine H. et al. Hypersensitivity of aryl hydrocarbon receptor-deficient mice to lipopolysaccharide-induced septic shock. Mol Cell Biol 29, 6391–6400 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huai W. et al. Aryl hydrocarbon receptor negatively regulates NLRP3 inflammasome activity by inhibiting NLRP3 transcription. Nat Commun 5, 4738 (2014). [DOI] [PubMed] [Google Scholar]
- Freemont A. J. & Denton J. Atlas of synovial fluid cytopathology (Springer-Science, 1991). [Google Scholar]
- Segura E. et al. Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity 38, 336–348 (2013). [DOI] [PubMed] [Google Scholar]
- Soler Palacios B. et al. Macrophages from the synovium of active rheumatoid arthritis exhibit an activin A-dependent pro-inflammatory profile. J Pathol 235, 515–526 (2015). [DOI] [PubMed] [Google Scholar]
- Fulci V. et al. miR-223 is overexpressed in T-lymphocytes of patients affected by rheumatoid arthritis. Hum Immunol 71, 206–211 (2010). [DOI] [PubMed] [Google Scholar]
- Li Y. T. et al. Brief report: amelioration of collagen-induced arthritis in mice by lentivirus-mediated silencing of microRNA-223. Arthritis Rheum 64, 3240–3245 (2012). [DOI] [PubMed] [Google Scholar]
- Shibuya H. et al. Overexpression of microRNA-223 in rheumatoid arthritis synovium controls osteoclast differentiation. Mod Rheumatol 23, 674–685 (2013). [DOI] [PubMed] [Google Scholar]
- Filkova M. et al. Association of circulating miR-223 and miR-16 with disease activity in patients with early rheumatoid arthritis. Ann Rheum Dis 73, 1898–1904 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M. C. et al. Increased miR-223 expression in T cells from patients with rheumatoid arthritis leads to decreased insulin-like growth factor-1-mediated interleukin-10 production. Clin Exp Immunol 177, 641–651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S. et al. Identification of key regulators for the migration and invasion of rheumatoid synoviocytes through a systems approach. Proc Nat Acad Sci USA 111, 550–555 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. M. et al. Abnormal networks of immune response-related molecules in bone marrow cells from patients with rheumatoid arthritis as revealed by DNA microarray analysis. Arthritis Res Ther 13, R89 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H. et al. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature 345, 442–444 (1990). [DOI] [PubMed] [Google Scholar]
- Geling A., Steiner H., Willem M., Bally-Cuif L. & Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep 3, 688–694 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind F. et al. NLRP3 inflammasome activity is negatively controlled by miR-223. J Immunol 189, 4175–4181 (2012). [DOI] [PubMed] [Google Scholar]
- Chen Q. et al. Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1beta production in macrophages by targeting STAT3. PloS One 7, e42971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T. et al. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol 11, 799–805 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S. et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol 13, 144–151 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusscott S., Kuchenbauer F., Humphries R. K. & Weng A. P. Notch-mediated repression of miR-223 contributes to IGF1R regulation in T-ALL. Leuk Res 36, 905–911 (2012). [DOI] [PubMed] [Google Scholar]
- Kumar V. et al. Notch and NF-kB signaling pathways regulate miR-223/FBXW7 axis in T-cell acute lymphoblastic leukemia. Leukemia 28, 2324–2335 (2014). [DOI] [PubMed] [Google Scholar]
- Franco C. B. et al. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci USA 103, 11993–11998 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. et al. Distinct biological roles for the notch ligands Jagged-1 and Jagged-2. The J Biol Chem 284, 17766–17774 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Walle I. et al. Specific Notch receptor-ligand interactions control human TCR-alphabeta/gammadelta development by inducing differential Notch signal strength. J Exp Med 210, 683–697 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Norton L. et al. Notch signal strength controls cell fate in the haemogenic endothelium. Nat Commun 6, 8510 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic J. et al. Ligand-dependent Notch signaling strength orchestrates lateral induction and lateral inhibition in the developing inner ear. Development 141, 2313–2324 (2014). [DOI] [PubMed] [Google Scholar]
- Nguyen N. T. et al. Aryl hydrocarbon receptor and kynurenine: recent advances in autoimmune disease research. Front Immunol 5, 551 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I. & Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 (1993). [DOI] [PubMed] [Google Scholar]
- Herberth G. et al. Maternal and cord blood miR-223 expression associates with prenatal tobacco smoke exposure and low regulatory T-cell numbers. J Allergy Clin Immunol 133, 543–550 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.