Ubiquitin modifications of the nuclear pore complex (NPC) control the architectural plasticity of the nuclear basket, contributing to its tethering to the core NPC, with consequences on the cellular response to DNA damage and telomere recombination.
Abstract
The nuclear pore complex (NPC) serves as both the unique gate between the nucleus and the cytoplasm and a major platform that coordinates nucleocytoplasmic exchanges, gene expression, and genome integrity. To understand how the NPC integrates these functional constraints, we dissected here the posttranslational modifications of the nuclear basket protein Nup60 and analyzed how they intervene to control the plasticity of the NPC. Combined approaches highlight the role of monoubiquitylation in regulating the association dynamics of Nup60 and its partner, Nup2, with the NPC through an interaction with Nup84, a component of the Y complex. Although major nuclear transport routes are not regulated by Nup60 modifications, monoubiquitylation of Nup60 is stimulated upon genotoxic stress and regulates the DNA-damage response and telomere repair. Together, these data reveal an original mechanism contributing to the plasticity of the NPC at a molecular-organization and functional level.
Introduction
In eukaryotic cells, the selective and regulated transport of macromolecules between the cytoplasm and the nucleoplasm occurs across specialized substructures of the nuclear envelope called nuclear pore complexes (NPCs). The NPC is composed of ∼30 different proteins or nucleoporins (Nups) that are highly conserved from yeast to humans and expressed in multiple copies per NPC owing to its eightfold rotational symmetry (Rout et al., 2000; Cronshaw et al., 2002). A combination of structural and biochemical approaches has enabled in silico computational modeling, generating insights into NPC molecular architecture (Alber et al., 2007). In parallel, electron microscopy and cryoelectron tomography led to substantial progress in elucidating the overall architecture of the NPC (Beck et al., 2007). The NPC consists in a central structure or scaffold ring complex localized in the plane of the nuclear envelope and decorated by peripheral extensions: the cytoplasmic filaments and the nuclear basket that extend the NPC domains to the cytoplasm and the nucleus, respectively.
Beyond the core function of NPCs in nucleocytoplasmic transport, the NPC has recently emerged as a key “hub” coordinating diverse nuclear functions. In particular, the nuclear basket substructure has been implicated in genome architecture, gene expression, mRNA surveillance, and DNA-damage response (DDR) and repair (Dieppois and Stutz, 2010; Bermejo et al., 2012; Bukata et al., 2013; Ptak et al., 2014; Guet et al., 2015). The nuclear basket consists in the assembly of five proteins in the yeast Saccharomyces cerevisiae (Nup60, Nup1, Nup2, and the two myosin-like proteins Mlp1 and Mlp2) and three proteins in vertebrates (Nup153 [which may recapitulate both Nup1 and Nup60], Nup50 [Nup2 homolog], and Tpr [Mlps homolog]; Strambio-de-Castillia et al., 1999; Dilworth et al., 2001). Nup60, Nup1, and Nup2 belong to the FG (phenylalanine-glycine) Nups subfamily, characterized by phenylalanine-glycine repeats and acting as docking sites of transport complexes onto the NPC. Nup2/Nup50 actively participates in protein import, cargo release, and karyopherin recycling (Guan et al., 2000; Matsuura and Stewart, 2005), and Nup60/Nup153 mediates Nup2/Nup50 localization at the NPC (Denning et al., 2001; Dilworth et al., 2001). Nup60/Nup153 also plays a major role in the recruitment of Mlps/Tpr at the periphery of the NPC (Hase and Cordes, 2003; Lewis et al., 2007; Niepel et al., 2013). Besides these core components of the yeast nuclear basket, this NPC subcomplex interacts with Ulp1, one of the two yeast SUMO proteases (Zhao et al., 2004). However, the precise organization of the nuclear basket and the mechanisms responsible to tethering to the NPC scaffold are not fully elucidated. Interestingly, it has been shown recently that both Nup60 and Nup1 display an N-terminal amphipathic helix and an adjacent α-helical region, which are both required for an efficient interaction with the core NPC, with a specific role of amphipathic helices in promoting membrane curvature via insertion into the lipid bilayer (Mészáros et al., 2015).
The limited number of NPCs per cell (∼100–200 NPCs per yeast cell) likely imposes a certain coordination of the different functions attributed to the NPC. To dissect at a molecular level how the NPC integrates these functional constraints in time and space represents the next challenge in the biological understanding of this fascinating cellular machine. Dynamic Nup associations, posttranslational or conformational changes, or temporal changes in expression might represent nonexclusive layers of complexity in NPC structure and function. In this respect, we recently reported the systematic ubiquitylation analysis of the budding yeast NPC and found that more than 50% of the Nups are ubiquitylated, mostly by monoubiquitylation, indicating a nondegradative role of this posttranslational modification (PTM; Hayakawa et al., 2012). The NPC cannot be considered as a single entity toward the ubiquitin/proteasome system but is rather the target of multiple ubiquitin-modifying enzymes (Niño et al., 2012).
Here we precisely dissected PTMs of the yeast nuclear basket protein Nup60, monoubiquitylation and SUMOylation, and determined how they control the dynamic organization of the nuclear basket with consequences on DDR and telomere repair. Our results support the hypothesis that dynamic PTM, and in particular ubiquitin modifications, participate to the role of the NPC as a platform orchestrating nuclear functions by regulating its architectural plasticity.
Results
Nup60 is both monoubiquitylated and SUMOylated on distinct lysine targets
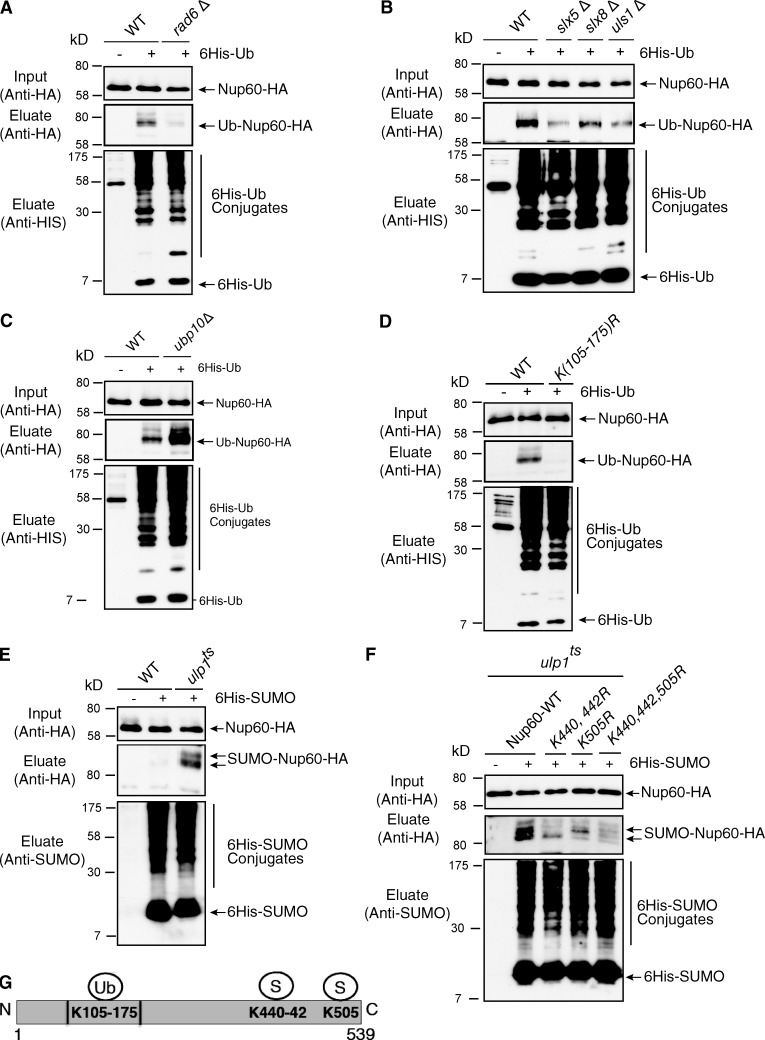
To explore whether PTM, and in particular ubiquitin and ubiquitin-like proteins, might contribute to the role of the NPC in organizing nuclear functions, we focused on Nup60, a component of the nuclear basket modified by monoubiquitylation (Hayakawa et al., 2012), and investigated the molecular mechanisms responsible for this PTM. Ubiquitylation of Nup60 was analyzed in cells expressing genomically functional HA-tagged NUP60 as well as a copper-induced 6His-tagged version of ubiquitin. Modified proteins from denatured cell extracts were purified on a nickel column and analyzed using anti-HA antibodies (Hayakawa et al., 2012). This approach allowed the identification of a ubiquitin-conjugated species of Nup60 corresponding to the monoubiquitylated protein (Fig. 1 A). A systematic screening of mutants for each of the 11 yeast E2 conjugating enzymes indicated that Nup60 ubiquitylation was almost completely inhibited in rad6Δ cells (Fig. 1 A), but not significantly impaired in any of the others E2 mutants (Fig. S1 A). Although Rad6 functions with the Bre1, Rad18, Ubr1, or Ubr2 E3 ubiquitin ligases in S. cerevisiae, Nup60 ubiquitylation was not affected in any of these E3 ligase mutants (Fig. S1 B). Based on the nuclear localization of Nup60, we restricted our search on E3s present in this compartment and in particular at the nuclear periphery. We indeed observed that the level of Nup60 ubiquitylation was decreased in cells deleted for either SLX5 or SLX8, both subunits of the SUMO-targeted ubiquitin ligase (STUbL) reported to localize at the nuclear periphery, at least transiently (Nagai et al., 2008; van de Pasch et al., 2013; Fig. 1 B). A similar inhibition of Nup60 ubiquitylation was also observed upon deletion of ULS1, the second yeast STUbL, suggesting that both STUbL activities are required for the Nup60 ubiquitylation (Uzunova et al., 2007; Fig. 1 B). In addition to ubiquitylation enzymes, we also searched for specific deubiquitylating activity. We screened 12 of the 16 UBPs and identified Ubp10 as the unique ubiquitin protease able to cleave off ubiquitin from Nup60 (Fig. 1 C and not depicted).
Figure 1.
Mechanisms of Nup60 ubiquitylation and SUMOylation. Ni-purified 6His-ubiquitin– (Ub) or 6His-SUMO–conjugated forms of Nup60-HA were extracted from cells transformed (+) or not transformed (−) with a plasmid encoding 6His-ubiquitin or 6His-SUMO, respectively, under control of the CUP1 promoter. Cell lysates (top) and Ni-purified material (middle) were examined by Western blotting with an anti-HA antibody. Ubiquitin and SUMO expression and efficiency of purification were controlled using an anti-6His or anti-SUMO antibody, respectively (bottom). (A–D) Analysis of ubiquitin-conjugated forms of genomically HA-tagged Nup60 was performed in WT and rad6Δ cells (A); WT, slx5Δ, slx8Δ, uls1Δ, and strains (B); WT and ubp10Δ cells (C); or WT and nup60 K105-175R mutants (nup60-Ub-KR; D). (E) Analysis of SUMO conjugated forms of genomically HA-tagged Nup60 was performed in WT and ulp1ts cells grown overnight at permissive temperature (25°C) and then shifted to restrictive temperature (37°C) for 3 h. (F) Nup60 SUMOylation was analyzed in ulp1 ts cells expressing HA-tagged WT and indicated nup60 KR mutants as in D. (G) Position of Nup60 lysines conjugated to ubiquitin or SUMO.
A systematic lysine-to-arginine (KR) mutagenesis approach was then used to determine the lysine residues targeted by these enzymes. For this purpose, we developed a library of plasmid-based HA-tagged Nup60 mutants where multiple lysines covering the full-length sequence were mutated to arginines. These plasmids were expressed in nup60Δ cells, and the resulting mutant proteins were tested for ubiquitylation in vivo. Progressive selection of ubiquitin-deficient mutants allowed the identification of a region of eight lysines (between Lys105 and Lys175) responsible for Nup60 ubiquitylation (Fig. 1, D and G). Single point mutations of each of these eight lysines were not sufficient to abolish ubiquitin conjugation (unpublished data), indicating that Nup60 is likely modified on an adjacent lysine when the specific target residue is mutated, a promiscuous behavior already observed in other studies (Iglesias et al., 2010). The K105-175R mutations were genomically integrated to generate a ubiquitin-deficient mutant of Nup60 (nup60-UbKR).
Together, our systematic analysis revealed that Nup60 ubiquitylation on K105-175 lysine residues is achieved by Rad6 as an E2 ubiquitin conjugation enzyme together with two alternative SUMO-dependent E3 ligases, Slx5/Slx8 and Uls1, and that this modification can be reversed by Ubp10.
Besides the core components of the yeast nuclear basket, this NPC subcomplex interacts with Ulp1, one of the two yeast SUMO proteases (Zhao et al., 2004). Together with our findings that Nup60 is a target for STUbL, it was tempting to determine whether Nup60 was not only ubiquitylated but also SUMOylated. For this purpose, SUMOylated proteins were purified from Nup60-HA wild-type (WT) or ulp1 mutant cells expressing a copper-inducible 6His-SUMO protein. Although no SUMO-conjugated form of Nup60-HA could be observed in WT cells, two species of SUMOylated Nup60-HA were detected in both ulp1 thermosensitive mutant (ts) cells at the restrictive temperature and a ΔN338-ulp1 mutant lacking the Ulp1 NPC targeting domain (Li and Hochstrasser, 2003; Figs. 1 E and S1 C). The redundant SUMO ligases, Siz1 and Siz2, were identified as SUMO ligases for Nup60 in the ulp1 ts cellular context (Fig. S1 D). Two SUMOylation sites were characterized in the C terminus of Nup60 on Lys440,442 and Lys505 (Fig. 1, F and G). Both K440,442R and K505R mutations were genomically integrated to generate a SUMO-deficient mutant of Nup60 (nup60-SUMO KR). Surprisingly, nup60-SUMO KR was ubiquitylated to the same extent as the WT protein (Fig. S1 E).
Our results thus indicate that Nup60 is SUMOylated by Siz1 and Siz2 SUMO ligases on two distinct sites and constantly de-SUMOylated by Ulp1. Importantly, Nup60 SUMOylation either is not sufficient for or is independent of Nup60 ubiquitylation by the yeast STUbL enzymes.
Nup60 ubiquitylation controls the plasticity of the nuclear basket organization
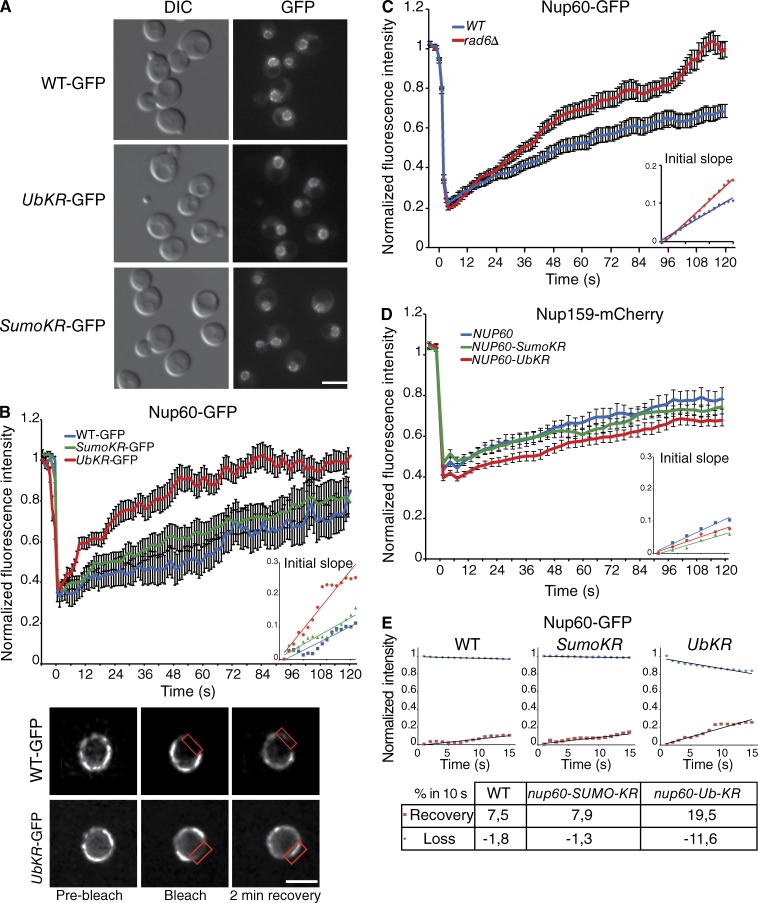
To assess the consequences of ubiquitylation and SUMOylation of Nup60, their consequences on Nup60 localization was investigated using genomically GFP-tagged WT Nup60, nup60-UbKR, and nup60-SUMO-KR. Both WT and mutant proteins displayed a rim-like staining, characteristic of NPC localization (Fig. 2 A), indicating that Nup60 ubiquitylation and SUMOylation are not required for the steady-state localization of Nup60 at the NPC. To further determine whether these PTMs could affect the dynamics of Nup60 at the NPC, fluorescence recovery of Nup60-GFP was analyzed after photobleaching (FRAP). The recovery of Nup60-GFP was relatively slow, as no plateau was reached within 2 min after the bleach (Fig. 2 B). The dynamic recovery was measured by fitting the measurement during the first 15 s postbleach, with a linear equation to determine the initial slope of the curve (Fig. 2, insets). The retrieval of WT Nup60 and nup60-SUMO-KR has been estimated as 7.5% and 7.9% within 10 s, respectively (Table 1). In contrast, the dynamics of nup60-UbKR strain were highly increased, with a plateau reached ∼80 s after bleaching and 19.5% recovery within the first 10 s after photobleaching along a linear fitting (Fig. 2 B and Table 1). The half-times of retrieval were also estimated through an exponential recovery fitting curve and were found to be 68 s in WT (on a 5-min movie [not depicted]), 73 s for nup60-SUMO-KR, and 23 s for nup60-UbKR. Importantly, preventing Nup60 ubiquitylation by deletion of RAD6 also led to instability of Nup60 at the nuclear periphery, supporting the notion that the observed effect for nup60-UbKR is likely related to the absence of ubiquitylation rather than to a conformational effect caused by the nup60-UbKR mutation (Fig. 2 C).
Figure 2.
Nup60 ubiquitylation controls the association dynamics of Nup60 with the NPC. (A) Steady-state localization of GFP-tagged Nup60 in WT, nup60-UbKR, and nup60-SUMO-KR strains. Bar, 5 µm. (B) Mean fluorescence recovery curves after photobleaching for WT (blue, n = 28), nup60-Ub-KR cells (red, n = 31), and nup60-SUMO-KR (green, n = 29). The bottom panel presents a typical example of images acquired during FRAP experiments in cells expressing either Nup60-GFP or nup60-UbKR-GFP. The red rectangle highlights the bleached zone after a 50-ms bleach pulse. Bar, 1.5 µm. (C) Mean fluorescence recovery curves for Nup60-GFP in WT (blue; n = 35) and rad6Δ (red; n = 27) cells. (D) Mean fluorescence recovery curves for Nup159mCh in NUP60 (blue; n = 27), nup60-Ub-KR (red; n = 51), and nup60-SUMO-KR (green; n = 34) cells. (E) Dynamics of fluorescence recovery (red square) compared to the dynamics of the loss of fluorescence in the nonbleached area (blue dot) in WT, nup60-SUMO-KR, and nup60-Ub-KR. The initial slopes presented in the table have been calculated with a linear fit (black line). The error bars in B–D correspond to SEM.
Table 1. Dynamics of recovery of different NPC proteins.
| Cell | Nup60 | Nup2 | Nup159 | Mlp1 | Ulp1 |
|---|---|---|---|---|---|
| WT | 7.5 | 7 | 5 | 5.1 | 2.2 |
| SumoKR | 7.9 | 8.8 | 4.3 | 5.3 | 2.4 |
| UbKR | 19.5 | 17.7 | 3.8 | 4.7 | 2.5 |
Dynamics of recovery of the different NPC proteins measured by FRAP analysis was estimated on the first 15 s of the average curve. The values are expressed as a percentage of recovery within 10 s.
To determine whether Nup60 ubiquitylation regulated the lateral mobility of the entire NPC or rather specifically controlled the association/dissociation of Nup60 with the NPC, the dynamics of the nuclear pore have also been investigated using mCherry-tagged Nup159, a nucleoporin exclusively localized on the cytoplasmic side of the NPC. Nup159 dynamics were affected neither in the Nup60 ubiquitin nor in the SUMO deficient mutants (Fig. 2 D and Table 1). These results thus indicated that ubiquitylation of Nup60 restricts its own mobility and likely participates in the association of Nup60 with the NPC without affecting the overall dynamics of the NPC. In addition, the fluorescence rate recovery that was increased in nup60-UbKR corresponded to a faster decrease of fluorescence of the nonbleached area (Fig. 2 E), indicating that preventing ubiquitylation increases the dynamics of association and dissociation of Nup60 with the NPC rather than mobilizing a soluble pool of Nup60.
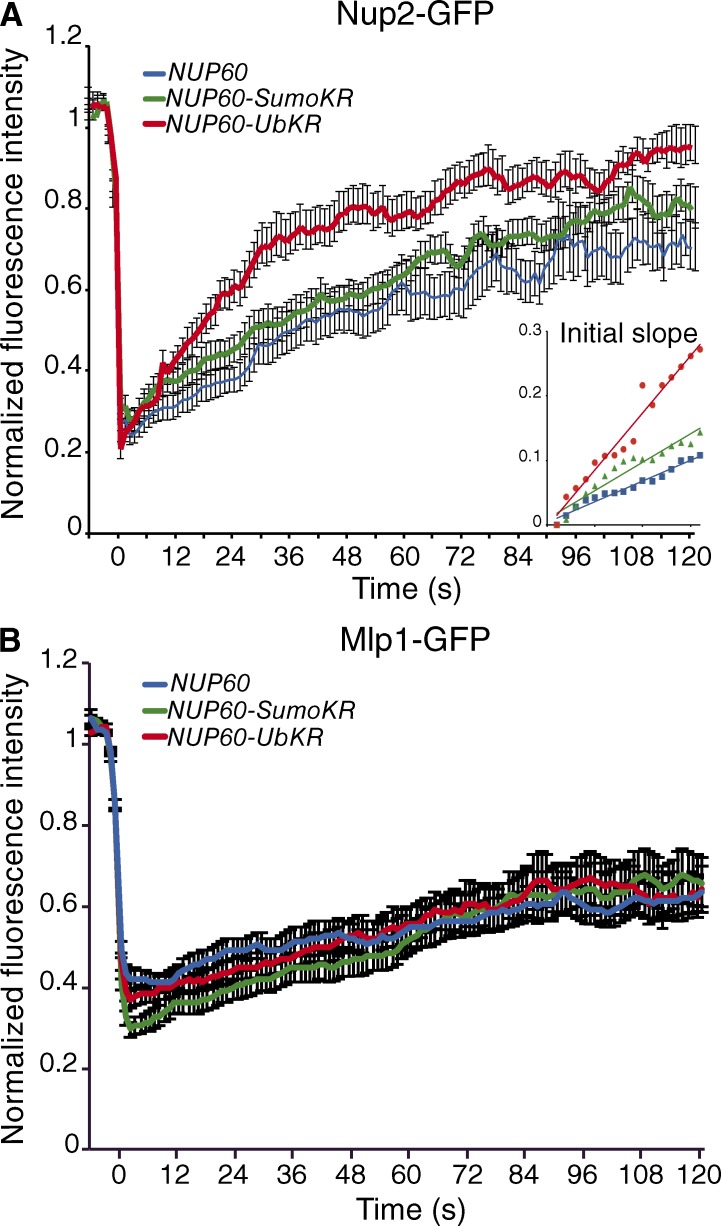
To analyze whether the dynamics of Nup60 more generally control the plasticity of the nuclear basket, steady-state localization and FRAP analysis of GFP-tagged Mlps and Nup2 were investigated in WT, nup60-UbKR, and nup60-SUMO-KR strains. PTMs of Nup60 did not affect the steady-state localization of Mlp1, Mlp2, Nup2, and the nuclear basket–associated Ulp1 at the NPC (Fig. S2, A–C). Whereas the dynamics of Mlp1 (Fig. 3 and Table 1) and also Ulp1 (Table 1) remained unchanged in these strains, Nup2-GFP behaved similarly to Nup60, with faster exchange dynamics in the nup60-UbKR (17.7% per 10 s) than in the WT (7% per 10 s; Fig. 3 and Table 1).
Figure 3.
Nup60 ubiquitylation controls the dynamics of the nuclear basket. Mean fluorescence recovery curve for Nup2-GFP (A) and Mlp1-GFP (B) in NUP60 (blue, n = 31 and n = 31 respectively), nup60-Ub-KR (red; n = 33 and n = 31, respectively), and nup60-SUMO-KR (green; n = 33 and n = 31, respectively) cells. Error bars represent SEM.
Together, these results highlight the role of the ubiquitylation of Nup60 in controlling the dynamics of both Nup60 and its interaction partner, Nup2, at the nuclear basket and reveal an original mechanism allowing for the plasticity of the nuclear basket at an architectural level.
Interaction between ubiquitylated Nup60 and the Y complex, most likely with Nup84, contributes to tether Nup60 at the NPC
The region of Nup60 modified by ubiquitin overlaps with the α-helical region that synergizes the N-terminal amphipathic helix for targeting Nup60 at the NPC (Mészáros et al., 2015). Based on these data, we propose that the N-terminal amphipathic helix would participate in membrane anchoring whereas the α-helical region would mediate interaction with the NPC via the recognition of its ubiquitylation by ubiquitin-binding domains (UBDs).
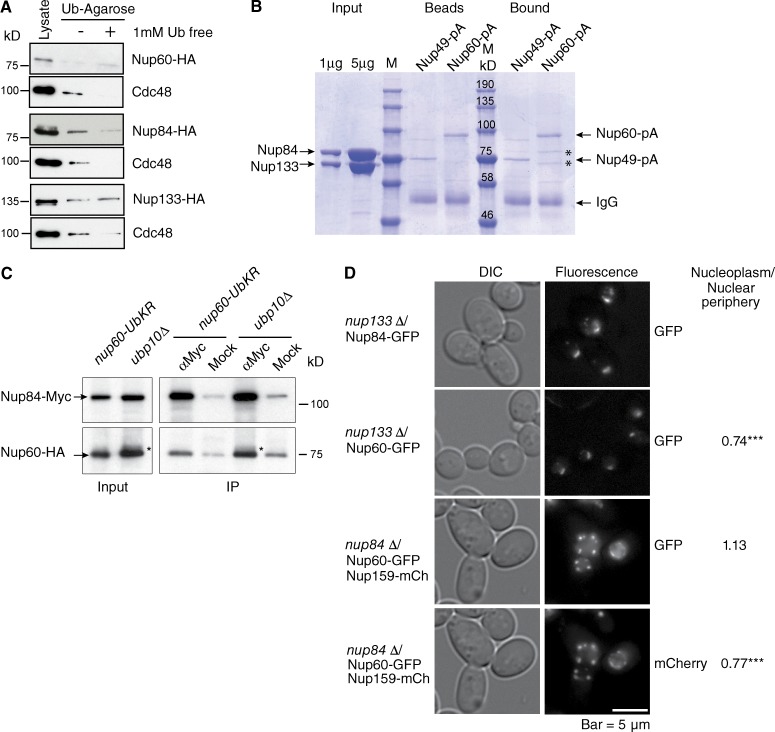
The presence of UBDs allows the independent ubiquitination of E3 ligases through a direct cooperation with ubiquitin-charged E2 enzymes (Hoeller et al., 2007). The candidate UBD-containing Nups are thus supposed to be ubiquitylated themselves. To analyze whether an intramolecular UBD-monoubiquitin could regulate the conformation and dynamic of Nup60 at the NPC, the ability of Nup60 to interact with ubiquitin was thus analyzed. For this purpose, lysates from cells expressing Nup60-HA were subjected to a monoubiquitin affinity column, and the endogenous ubiquitin-dependent AAA ATPase Cdc48 served as an internal control of ubiquitin-binding protein. In addition, an excess of free ubiquitin was used as a competitor to test the specificity of the interaction. As shown in Fig. 4 A, no interaction could be detected between Nup60 and ubiquitin, thus excluding a ubiquitin- or UBD-mediated conformational change of Nup60. According to the molecular architecture of the NPC, the outer ring substructure composed of the Y–Nup84 complex likely represents the best candidate for anchoring the nuclear basket to the core NPC. In addition, mutants of the Nup84 complex share some phenotypes with nuclear basket mutants, particularly in the DDR (Palancade et al., 2007). Screening HA-tagged Nups of the Nup84 complex (Sec13, Seh1, Nup145C, Nup84, Nup85, Nup120, and Nup133) for binding to monoubiquitin affinity column revealed that Nup84 is the unique component of this complex to interact specifically with monoubiquitin (Figs. 4 A and S3), a result consistent with the previously reported ubiquitylation of this protein (Hayakawa et al., 2012).
Figure 4.
Nup84 interacts with monoubiquitin and is required for tethering Nup60 at the NPC. (A) Lysates from cells expressing HA-tagged Nup60, Nup84, or Nup133 were purified on monoubiquitin-coupled agarose beads in the absence (−) or presence (+) of 1 mM ubiquitin. Bound proteins were analyzed by Western blotting using anti-HA or anti-Cdc48 antibodies. (B) Recombinant purified Nup84 complexed with aa 481–1,157 of Nup133 was purified on Nup49-ProtA (Nup49-pA)- or Nup60-ProtA (Nup60-pA)-coupled IgG beads. Input, Nup-coupled beads and bound material were analyzed by SDS-PAGE and Coomassie blue staining. *, bound Nup84/Nup133. (C) Lysates (Input) from nup60-Ub-KR or ubp10Δ cells expressing Nup60-HA and Nup84-Myc were immunoprecipitated (IP) using anti-Myc or mock antibodies and analyzed by Western blotting with anti-Myc and anti-HA antibodies. *, ubiquitylated form of Nup60 accumulated in ubp10Δ cells. (D) Steady-state localization of GFP-tagged Nup60 and Nup84 in nup133Δ (top), and GFP tagged Nup60 and mCherry tagged Nup159 in nup84Δ cells (bottom). Intensity of Nup60-GFP and Nup159-mCherry was determined both in the nucleoplasm and at the nuclear periphery (total nucleus-nucleoplasm) in nup133Δ (n = 69) and nup84Δ (n = 52) cells using ImageJ, and the nucleoplasm/nuclear periphery ratio was calculated in each cell. Results were compared using Student’s t test (***, P < 0.0001).
We then tested whether Nup84 would directly interact with Nup60. For this purpose, Nup60 and Nup49 were genomically tagged with protein A, and corresponding fusion proteins were purified on IgG-coupled magnetic beads before incubation with the recombinant complex between full-length Nup84 and the 481- to 1,157-aa fragment of Nup133. Both components of the Y complex were coproduced to stabilize recombinant Nup84. As shown in Fig. 4 B, a specific interaction of Nup84–Nup133 could be observed with Nup60-ProtA, but not with Nup49-ProtA. Whether this association was affected by Nup60 ubiquitylation was assessed by coimmunoprecipitation of Nup60 with Nup84 in nup60-UbKR and ubp10Δ cells, in which ubiquitylation of Nup60 is either prevented or stabilized, respectively. To both protect the ubiquitin modification and better reflect the in vivo conditions, cells were cross-linked before lysis as previously reported (Vitaliano-Prunier et al., 2012). This approach revealed that in vivo, Nup84 was able to bind nonmodified Nup60, as in vitro, but preferentially interacted with Nup60 when its ubiquitylation is stabilized (Fig. 4 C, note the ubiquitylated Nup60, indicated by the asterisk).
To further analyze whether Nup84 would be involved in tethering Nup60 at the NPC, we aimed to investigate localization of Nup60-GFP in cells disrupted for NUP84. However, it has been well established that deletion of components of the Y complex, including NUP84 and NUP133, leads to NPC clustering (Palancade et al., 2007). We therefore compared the localization of Nup60GFP in nup84Δ cells with the one observed in nup133Δ. Deletion of NUP133 did not affect the localization of Nup84- or Nup60-GFP at the NPC. In contrast, deleting NUP84 led to a partial delocalization of Nup60-GFP in the nucleoplasm, as evidenced by a 1.5-fold increase in the nucleoplasmic/nuclear periphery intensity ratio (Fig. 4 D). Thus, despite the fact that nup84Δ and nup133Δ both lead to clustering of NPC, only the inactivation of NUP84 is shown to delocalize Nup60 from the NPC to the nucleoplasm.
Altogether, these data indicate that Nup84 participates to the proper interaction of Nup60 with the NPC, most likely via its ability to interact with the ubiquitin moiety of Nup60.
Ubiquitylation of Nup60 does not control the major nucleocytoplasmic transport routes
Whether an alteration of the nuclear basket dynamics induced by Nup60 ubiquitylation could regulate nucleocytoplasmic transport was assessed by the analysis of both the importin-α/β–mediated nuclear import and the exportin1-mediated nuclear export pathways in Nup60 WT and nup60-UbKR mutant cells. For this purpose, the subcellular localization of the reporter proteins consisting in GFP fused to the SV40 T antigen nuclear localization signal (GFP-NLS) and PKI nuclear export sequence (GFP-NLS-NES) were examined. Preventing ubiquitylation of Nup60 did not significantly affect the nuclear localization of GFP-NLS, as observed for nup60Δ (Denning et al., 2001), or the cytoplasmic localization of GFP-NLS-NES (Fig. S4 A). No obvious retention of poly(A)+ RNA investigated by FISH using an oligo-dT probe was observed in WT nuclei, whereas 19% of nup60Δ cells showed a nuclear accumulation of poly(A)+ RNA as previously reported (Skruzný et al., 2009). WT and nup60-UbKR strains displayed a negligible number of cells with RNAs in the nucleus (Fig. S4 B). No further alteration of nucleocytoplasmic transport was observed upon inhibition of SUMOylation (unpublished data).
Together, these results indicated that conjugation of Nup60 to ubiquitin does not regulate the major nuclear transport pathways.
Nup60 ubiquitylation contributes to the cellular response to DNA damage
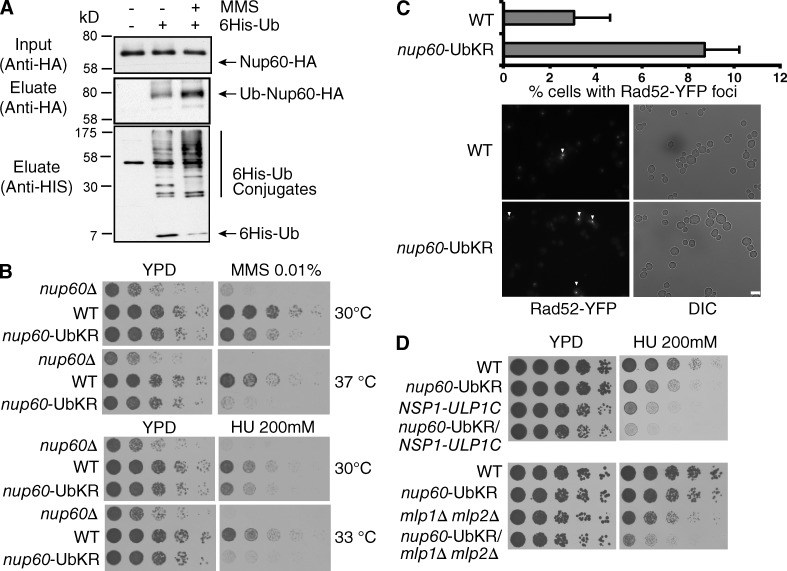
Depletion of distinct members of the Nup84 core complex in yeast leads to synthetic lethality when combined with genes required for double-strand break (DSB) repair through homologous recombination such as in RAD52 (Loeillet et al., 2005). Moreover, mutants of the nuclear basket and the Nup84 complex are highly sensitive to DNA-damaging treatments and accumulate unrepaired DSBs (Loeillet et al., 2005; Palancade et al., 2007). As shown in Figs. 2 and 4, Nup60 ubiquitylation strengthens its binding to Nup84 and thus controls the dynamic of Nup60 at the NPC. To determine whether Nup60 ubiquitylation contributes to the DDR, we first analyzed Nup60 modifications in cells treated the DNA-damaging agent methyl methane sulfonate (MMS). We observed an overall increase of cellular ubiquitylated proteins, including Nup60 (Fig. 5 A). These data indicate that DNA damage would promote an increased association of Nup60 at the NPC via its ubiquitylation. In addition, growth of nup60-UbKR cells was indeed compromised in the presence of both hydroxyurea (HU) and MMS compared with WT cells (Fig. 5 B). Finally, increased sensitivity of nup60-UbKR cells to these genotoxic agents correlated with an increase of unrepaired lesions, followed by the percentage of cells presenting Rad52 foci (Fig. 5 C).
Figure 5.
Nup60 ubiquitylation in the DDR. (A) Ni-purified 6His-Ubiquitin-conjugated proteins were extracted from indicated cells treated with (+) or without (−) MMS (0.2%) and analyzed as in Fig. 1. (B and D) Serial dilutions of WT and mutant cells were spotted on YPD without or with HU or MMS at the indicated concentrations and grown at the indicated temperatures. (C) Microscope analysis of Rad52 foci formation in WT and nup60-UbKR cells expressing Rad52-YFP. Quantified results correspond to mean and SD from four experiments corresponding to four independent transformants, and ∼300 cells were analyzed in each condition for each experiment. ***, P < 0.0005. Bar, 5 µm.
Alteration of DDR previously observed in nup60Δ, mlp1Δ/mlp2Δ cells or mutants of the Nup84 complex was shown to result, at least in part, from NPC mislocalization and destabilization of the SUMO protease Ulp1 (Palancade et al., 2007). However, preventing ubiquitylation or SUMOylation of Nup60 did not affect the steady-state localization (Fig. S2 D) and expression level (not depicted) of Ulp1 at the NPC. Endogenous ULP1 gene was then deleted and complemented by a fusion protein expressing the catalytic domain of Ulp1 fused to the nucleoporin Nsp1 (Nsp1–Ulp1C), in order to force Ulp1 localization at the NPC independently of Nup60/Mlp1-2 (Panse et al., 2003). Introducing nup60-UbKR mutation in NSP1–ULP1C cells still conferred an increased sensitivity to HU or MMS compared with NSP1-ULP1C cells (Fig. 5 C and not depicted). A similar result was obtained when the nup60-UbKR mutation was combined to the double mutant mlp1Δmlp2Δ, in which Ulp1 is not anchored at the NPC (Zhao et al., 2004; Fig. 5 D). Nup60 ubiquitylation thus accumulates upon genotoxic stress and participates to the DNA damage response independently of Ulp1 and Mlp1/2 regulation.
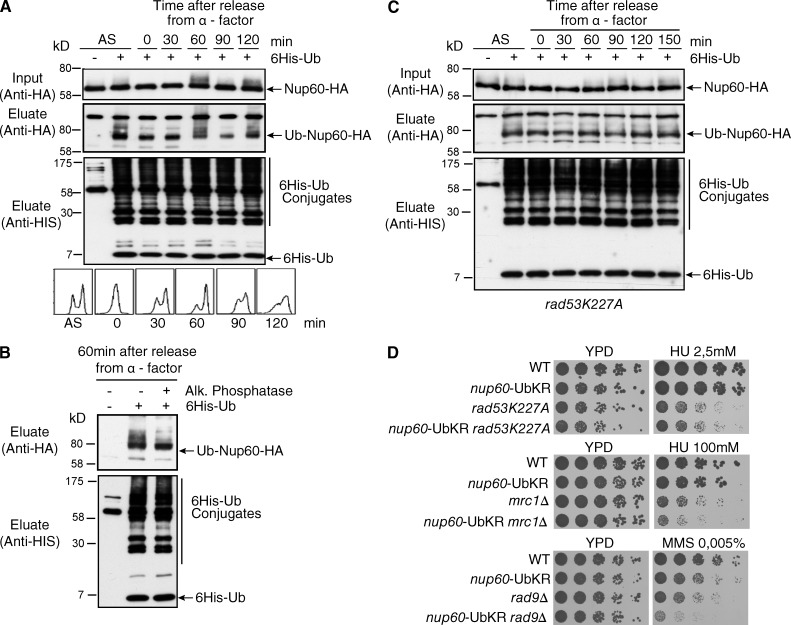
To further characterize the role of Nup60 ubiquitylation, we analyzed whether this modification was regulated during the cell cycle. For this purpose, cells were synchronized in G1 with α-factor and further analyzed for Nup60 ubiquitylation at different time points after α-factor release. Nup60 was monoubiquitylated all along the cell cycle, with a complex pattern observed in lysates from late S/G2 cells (60 min; Fig. 6 A). This ubiquitylation pattern remained unchanged when ubiquitylated Nup60 was purified from G2 cells expressing K48R or K63R 6His-tagged ubiquitin mutants, indicating that Nup60 was not polyubiquitylated (unpublished data). In contrast, treatment with AP eliminated the additional species of ubiquitylated Nup60 and led to the detection of a single monoubiquitylated Nup60 consistent with ubiquitylated Nup60 being phosphorylated in G2 (Fig. 6 B).
Figure 6.
Nup60 ubiquitylation participates in the Rad53-mediated pathway. (A) 6His-Ub-conjugated forms of Nup60-HA were extracted from asynchronous cells (AS) or from cells treated with α-factor for 3 h before release for indicated periods of time and analyzed as in Fig. 1. The bottom panel shows DNA content analysis by flow cytometry. (B) His-Ub protein purified from cells collected 60 min after release from α-factor arrest were treated (+) or not (−) with AP, and Nup60-HA ubiquitylation was analyzed by Western blotting. (C) rad53K227A cells expressing 6His-Ub were synchronized with α-factor for 3 h before release for indicated periods of time and analyzed as in Fig. 1. (D) Serial dilutions of WT and mutant cells were spotted on YPD without or with HU or MMS at the indicated concentrations and grown at 30°C.
Rad53 (human CHK2), one of the key effector kinases of DDR in S. cerevisiae, phosphorylates Nup60 in vitro (Smolka et al., 2007). Interestingly, ubiquitylated Nup60 was not phosphorylated in cells expressing a rad53K227A kinase-dead mutant (Fig. 6 C). Rad53 activation by Mec1 (human ATR) is mediated by Mrc1 (human CLASPIN) in response to replication stress and by Rad9 (human 53BP1) in response to DNA damage (Branzei and Foiani, 2009). Combining nup60-UbKR with the kinase-defective rad53K227A allele did not affect the growth or HU sensitivity of the rad53K227A mutant (Fig. 6 D). In contrast, introducing the nup60-UbKR mutation in the replication checkpoint mrc1Δ or the DNA-damage checkpoint rad9Δ mutants increased their sensitivity to HU and MMS, respectively (Fig. 6 D).
Altogether, these results argue for a role of Nup60 ubiquitylation in the Rad53-mediated response to DNA damage, independently of Ulp1 and Mlp1-2. The fact that the nup60-UbKR is epistatic to rad53K227A but has additive effects with either rad9Δ or mrc1Δ suggests that the nup60-UbKR acts downstream Mec1/Rad53 and weakens their response.
Nup60 ubiquitylation regulates telomere recombination
Previous work reported that eroded telomeres move from their membrane anchor sites to the NPCs (Khadaroo et al., 2009). In addition, a unique irreparable HO-induced DSB or replication fork–associated breaks relocalize to the NPC (Nagai et al., 2008). These results prompted us to next analyze whether Nup60 ubiquitylation affects telomere recombination.
Telomeres from yeast cells displaying no telomerase activity progressively shorten with each cell cycle until cell population growth rate begins to decline. Most of the cells die or remain arrested, but few cells, called survivors, can escape this arrest by using telomere maintenance pathways that rely on DNA homologous recombination. At the level of cell population, the so-called crisis is defined at the time point when most of the cells, if not all, are arrested at the G2/M transition or dead (IJpma and Greider, 2003). This arrest is thought to result either from a Mec1-dependent DNA-damage checkpoint that is activated by short telomeres being recognized as DSBs or from stalled replication forks (Lisby and Géli, 2009). Two pathways, both requiring Rad52 and the nonessential subunit of DNA Polδ Pol32, operate to produce very rare survivors that escape growth arrest or death. These survivors are classified in two types based on their telomere arrangement and growth characteristics (Teng and Zakian, 1999; Chen et al., 2001). Type I survivors rely on Rad51, whereas type II recombination does not require Rad51 but depends on Rad59 and is favored by the Mec1/Rad53 checkpoint pathway (Chen et al., 2001; Grandin and Charbonneau, 2007).
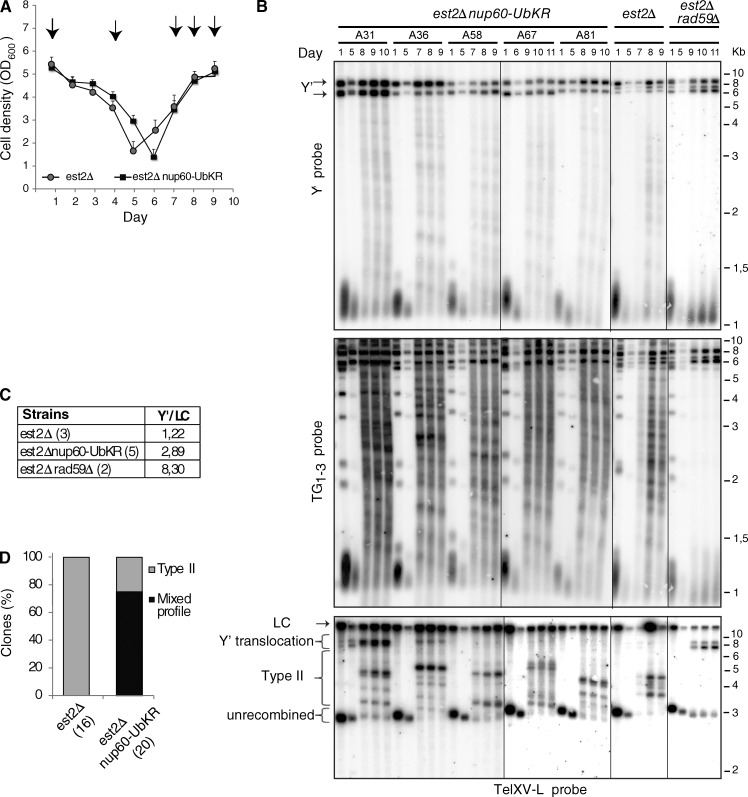
We performed standard senescence assays in liquid cell cultures with telomerase-negative cells (est2Δ) expressing either WT Nup60 or nup60-UbKR and analyzed corresponding survivor types. Preventing ubiquitylation of Nup60 in est2Δ cells did not significantly affect senescence profiles (Fig. 7 A). Type I and type II telomere recombination was monitored by Southern blot assays at different time points of the senescence kinetics using two different probes. Type I survivors have tandem arrays of subtelomeric Y′ elements separated by short tracts of TG1–3 repeats at most chromosome ends and also short terminal TG1–3 repeats (Lundblad and Blackburn, 1993). They can be identified in Southern blot by intense bands corresponding to the amplification of Y′ elements of two size classes, 6.7 (Y′-L) and 5.2 (Y′-S) kb long, found at many chromosome ends. Type II survivors exhibit abnormally elongated terminal TG1–3 repeats, heterogeneous in length, that result from stochastic lengthening events (Teng and Zakian, 1999; Fig. S5 A). Typically, est2Δ cells give rise to type II survivors owing to the slow growth of type I survivors in liquid cultures (Teng and Zakian, 1999). Type II survivors are revealed by a large number of discrete bands, each arising from a distinct recombination event in the population of cells (Figs. 7 B, middle). The nup60-UbKR clones, exemplified by the five indicated clones shown in Fig. 6 (and Fig. S5 B), also produced type II survivors (Fig. 7 B, middle). However, amplification of Y′ element also occurred in a number of nup60-UbKR est2Δ mutants and was maintained in subsequent generations (Figs. 7 B, top). On average, the signal of the Y′ band in the est2Δ nup60-UbKR clones was 2.3 times higher than in the est2Δ corresponding clones (Fig. 7 C). The signal of the Y′ band in the est2Δ rad59Δ mutant, which exhibits a severe type II recombination defect, was increased by sixfold with respect to est2Δ (Churikov et al., 2014). To further confirm that type I telomere recombination was facilitated in the nup60-UbKR mutant, we analyzed the Y′ translocation at a Y′-less telomere with a probe specific for TelXV-L (Churikov et al., 2014). In two nup60-UbKR est2Δ clones, TelXV-L was efficiently converted in a Y′ telomere and maintained in subsequent generations (Fig. 7 B, bottom). Further analysis of a large number of clones indicated that two thirds of the nup60-UbKR est2Δ clones were converted into both type I and type II survivors (mixed profile; Fig. 7 D), as does a nup60Δ mutant (Fig. S5 C). Surprisingly, nup60-UbKR est2Δ type I survivors were not outcompeted by type II survivors.
Figure 7.
Preventing Nup60 ubiquitylation favors the appearance of mixed survivors upon telomerase disruption. (A) Replicative senescence assays were performed in liquid culture by propagating the cells via serial dilutions to 105 cells/ml every 24 h. Each point represents the mean OD600 value for 17 and 21 independent spores of NUP60 and nup60-UbKR strains disrupted for EST2, respectively. SEMs are indicated for each point. (B) Survivor type for the indicated mutants was determined after DNA digestion with XhoI and Southern blot analysis with subtelomeric Y′ (top), telomeric TG1–3 (middle), and TelXV-L (bottom) probes. Representative clones are shown whose senescence profiles are shown in Fig. S5 B. In the bottom panel, fragments that correspond to Y′ translocations of TelXV-L (indicated by arrows) are larger than the fragments corresponding to Y′ amplifications (tandem Y′s revealed by Y′ probe) because the most proximal Y′ fragment includes an extra subtelomeric sequence (not present in tandem Y's). (C) The intensity of the Y′ elements was normalized to a loading control (LC) corresponding to a nonspecific band obtained with the TelXV-L probe (LC) in several est2Δ, nup60-UbKR est2Δ, and est2Δrad59Δ clones. The mean ratio is indicated for each mutant type. (D) The type of survivor profile is represented for all the NUP60 and nup60-UbKR telomerase minus clones. Mixed refers to the presence of type I and type II survivors in the same culture.
These data thus indicate that preventing Nup60 ubiquitylation favors type I recombination and inhibits the counterselection of type I survivors, likely by attenuation of the canonical Mec1-dependent DNA damage checkpoint.
Discussion
During the last decade, growing evidence supported a role of NPCs in diverse cellular functions besides its canonical role as a gateway for nucleocytoplasmic trafficking. In particular, the nuclear basket subcomplex emerged as a key actor participating in and coordinating several nuclear processes, including regulation of gene expression and DNA repair. How NPCs faithfully participate in so many different nuclear functions remains elusive. Evidence presented here reveals the role of PTMs in NPC organization and dynamics and elucidates how preventing PTMs of the NPC selectively impacts the appropriate execution of nuclear processes.
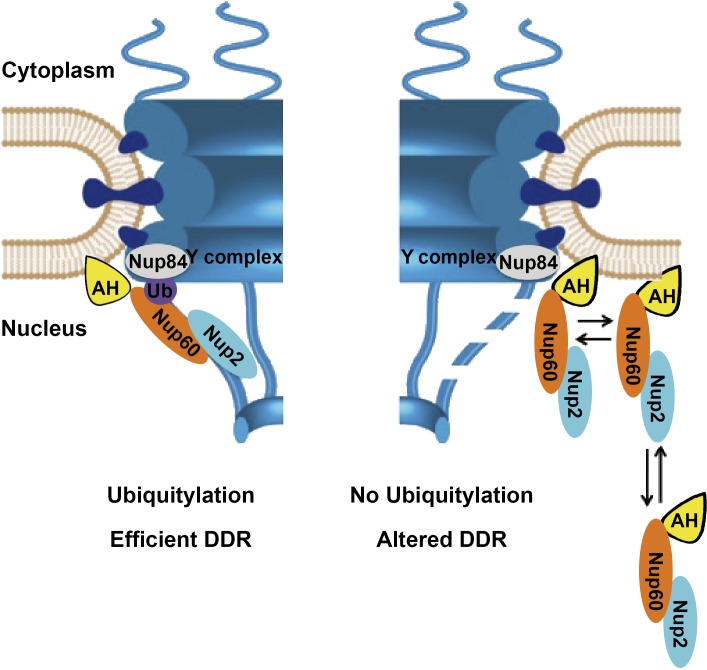
Nup60 PTMs and in particular ubiquitylation alter the dynamics of Nup60 and Nup2 at the NPC, thereby providing a molecular mechanism revealing the functional plasticity of the nuclear basket. Indeed, conjugation of ubiquitin or SUMO to Nup60 did not affect the steady-state localization of Nup60 and other components of the nuclear basket. However, FRAP analysis clearly revealed that preventing ubiquitylation, but not SUMOylation, of Nup60 led to a drastic change in the NPC association and dissociation rate of Nup60 and its partner, Nup2, without affecting the overall dynamics of the NPC. In addition, Nup60 ubiquitylation did not affect the dynamics of the Mlps at the NPC. In contrast to Nup60 or Nup2, which are uniformly distributed in NPCs all over the nucleus, Mlps and the associated Ulp1 enzyme are excluded from NPCs located in the region juxtaposed to the nucleolus (Galy et al., 2004; Zhao et al., 2004). The nuclear basket substructure of the NPC thus does not seem to behave as a single entity. Ubiquitylation of Nup60 is not the unique NPC tethering mode for Nup60, as it has been recently proposed that the N-terminal amphipatic helix directly inserts into the membrane bilayer and induces curvature (Mészáros et al., 2015). Ubiquitin conjugation occurs in an α-helicoidal region described in the same study to function together with the amphipatic helix and to modulate membrane curvature induction by an undescribed mechanism. Results presented here indicate that regulation of NPC tethering promoted by this region results from the ability of ubiquitylated Nup60 to interact with the Y complex, the building block of the outer ring, and most likely with its Nup84 component. Correct positioning of Nup60 at the nuclear basket would thus result from an association with both nuclear membrane and NPC (Fig. 8). Interestingly, Nup60 ubiquitylation is regulated, at least by genotoxic stress, thus opening a new avenue on signaling pathways between cell environment and NPC architectural dynamics.
Figure 8.
Model for a role of Nup60 ubiquitylation in the plasticity of the nuclear basket and its functional consequences. Nup60 is anchored to the nuclear membrane via its N-terminal amphipathic helix (Mészáros et al., 2015) and to the core NPC by the interaction of ubiquitylated Nup60 to Nup84. The combination of both anchoring pathways promotes stable association of Nup60 and Nup2 at the NPC required for an efficient DDR. In contrast, preventing ubiquitylation of Nup60 induces a more dynamic association and dissociation of both Nup60 and Nup2 with the NPC, likely resulting in a partially unstable nuclear basket (dashed fibrils) and altered DDR. The scaffold of this scheme has been adapted from Mészáros et al. (2015).
Nup60 is modified by ubiquitin and SUMO, but both modifications lead to distinct consequences. Nup60 ubiquitylation is mediated by the Rad6 ubiquitin conjugation enzyme and requires the ubiquitin ligases Slx5/Slx8 and Uls1. Slx5/Slx8 is a heterodimeric complex with DNA-binding activity that localizes in the nucleoplasm as well as at the nuclear periphery, with physical interaction between Slx8 and nucleoporin Nup84 being reported (Yang et al., 2006; Nagai et al., 2008). Uls1 belongs to the Swi2-Snf2 family of DNA-dependent ATPases and localizes in the nucleoplasm and the nucleolus (Shirai and Mizuta, 2008). Both Slx5/Slx8 and Uls1 have been classified as STUbLs, enzymes identified in yeast, fly, and human, and characterized by several SUMO-interacting motifs and a RING-finger motif with ubiquitin-ligase activity (Geoffroy and Hay, 2009). The E2 enzyme Ubc4 has been proposed to function together with Slx5/Slx8 and Uls1 to mediate the ubiquitin-proteasome degradation of SUMOylated proteins. Proteasome inhibition or deletion of Slx5, Slx8, Uls1, or Ubc4/Ubc5 accordingly promote accumulation of high molecular weight SUMO conjugates (Uzunova et al., 2007; Xie et al., 2007). However, the identification of in vivo substrates stresses a more complex diversity of Slx5/Slx8-dependent modification pathways and indicates that SUMO conjugation is not always required for Slx5/Slx8-dependent ubiquitylation (Xie et al., 2007; Westerbeck et al., 2014). Here we show that Nup60 ubiquitylation by Slx5/Slx8 and Uls1 was independent of Nup60 or Nup2 SUMOylation and required Rad6 as the specific E2 enzyme with no ubiquitylation defect in ubc4Δ/ubc5Δ cells (Figs. 1 A and S1 A). In addition, Nup60 stability was not affected by ubiquitylation (unpublished data). Together, these data suggest that Slx5/Slx8 and Uls1 can mediate SUMO-independent ubiquitylation that does not always compromise substrate stability.
Preventing Nup60 PTMs did not significantly alter the major nucleocytoplasmic transport routes. Our results rather show that cells disrupted in Nup60 ubiquitylation display an altered DNA damage response. Interestingly, both Slx5/Slx8 and Uls1 E3 ligases are important to maintain genome integrity (Sriramachandran and Dohmen, 2014). Uls1 has been implicated in the replication stress response, as uls1 cells progress slower through the S phase in the presence of MMS and ULS1 deletion confers synthetic lethality to rad52Δ mutant cells (Cal-Bakowska et al., 2011). On the other hand, both SLX5- and SLX8-disrupted cells present gross chromosomal rearrangements and accumulation of DNA damage (Zhang et al., 2006). Interestingly, Slx5/Slx8 has been involved in the relocation of persistently unrepaired DSBs at the nuclear periphery, a process that also requires the Nup84 complex, as well as the Mec1 (ATR)/Tel1 (ATM) kinases (Nagai et al., 2008; Kalocsay et al., 2009; Oza et al., 2009; Horigome et al., 2014). Very recently, Nup84 and Slx5/8 were also shown to suppress CAG repeat instability arising through aberrant homologous recombination (Su et al., 2015). Together, these studies performed in yeast suggest that NPC anchorage sites are associated with DDR pathways.
Here we describe Nup60 as the first target of Slx5/Slx8 and Uls1 at the NPC and could thus participate in the function of these ubiquitin ligases in the maintenance of genome integrity. Increased ubiquitylation of Nup60 upon genotoxic stress would stabilize its interaction with the NPC scaffold via Nup84 and thus likely represents a new link between DDR function and the NPC. Consistent with this idea, our results indicate that Nup60 ubiquitylation functions downstream of Rad53 to reinforce the DDR upon replication stress. This process may belong to a general response to replication stress, including the phosphorylation of Mlp1 that has been proposed to release transcribed genes from the nuclear envelope, thereby reducing topological constraints (Bermejo et al., 2011). Finally, we show that preventing Nup60 ubiquitylation promotes type I recombination of eroded telomeres or alternatively allows their maintenance in survivors (mixed survivors). The weakened checkpoint in the nup60-UbKR could explain the persistence of type I telomeres in survivors.
Together, these results lead us to propose that, via the regulation of Nup60 marks, the nuclear basket of the NPC can organize and control microenvironments at the nuclear internal periphery to favor specific nuclear processes as a function of cell requirements (Fig. 8). Nup60 ubiquitylation is indeed a highly reversible process controlled by Rad6-Slx5/8-Uls1 for ubiquitylation and the ubiquitin protease Ubp10 for deubiquitylation. Different cellular contexts and signaling pathways, such as the genotoxic stress shown here, might affect the Rad6/Ubp10 enzymatic balance to modify Nup60/Nup2 dynamics at the NPC, thus providing clues about the structure–function adaptation of microenvironments at the nuclear periphery.
Materials and methods
Yeast strains, plasmids, and culture
The S. cerevisiae strains and plasmids used in this study are listed in Tables S1 and S2. Yeast cultures were grown at 30°C either in YPD media containing 2% glucose or in synthetic media (SD) with the appropriate supplements. Strains containing the ulp1ts allele were grown at a permissive temperature (25°C) and subsequently shifted to 37°C for 3 h before harvesting for biochemical analysis. For cell growth and drug sensitivity analysis, fivefold serial dilutions of the different strains were spotted on YPD plates without or with HU or MMS at the indicated concentrations and growth at the indicated temperatures.
Chromosomally tagged strains and mutants were constructed using a PCR-based strategy (Longtine et al., 1998). For the systematic KR mutagenesis approach, a Nup60-HA complete KR mutant was chemically synthetized (Genevust) and cloned in SpeI–XhoI sites of the p415 plasmid to generate the p415-nup60-HA-Krall plasmid. A set of yeast plasmids expressing Nup60-HA with different regions of the protein carrying KR mutation was generated using the p415-nup60-HA-KRall as an initial template. The p415-Nup60-HA WT plasmid was obtained by PCR amplification of the genomic DNA fragment encoding Nup60-HA and cloning in SpeI–XhoI sites into p415. Plasmids encoding different Nup60-KR-HA point mutants were constructed from the p415-Nup60-HA using a site-directed mutagenesis kit (QuikChange; Agilent Technologies).
To genomically integrate the Nup60 KR mutations, a PCR fragment was generated containing full-length Nup60-HA (WT, UbKR, or SUMO-KR) followed by the CYC1 terminator, the LEU2 cassette, and 50 nt complementary to the NUP60 locus just downstream of the stop codon. PCR fragments were obtained using plasmid p415-Nup60-HA, p415-Nup60-UbKR-HA, or p415-Nup60-SUMO-KR-HA as a DNA template and the oligonucleotides Int-N60-LEU-F, 5′-ATCAAATAAGCACCGCAAGATATCCTAAAATCGACATCCAATGCATCGTAAATCATTG-3′, and Int-N60-LEU210-R, 5′-GTATTGAGTTGGGCTATACGGTAATTATGTCACGGCTAAAATTTTCATTATTCCTTATCACGTTGAGC-3′. Yeast cells were transformed and selected on restrictive medium plates (DO-LEU). Finally, clones were analyzed by DNA sequencing of full-length NUP60 gene.
Purification of ubiquitylated and SUMOylated proteins
Cells transformed with a plasmid encoding 6His-Ubiquitin or 6His-SUMO under the CUP1 promoter were grown on selective medium and stimulated overnight with 0.1 mM CuSO4. 100 OD600 of cells were collected and lysed with glass beads in a 20% TCA solution, and the final TCA concentration in the cell lysate was adjusted to 12%. Cell lysates were incubated at 4°C during 45 min, and precipitated proteins were collected by centrifugation. Proteins were resuspended in a solution containing 6 M guanidinium-HCl, 100 mM KH2PO4, 20 mM Tris-HCl, pH 8,0, 100 mM NaCl, 0,1% Triton X-100, and 10 mM imidazole. Purification was performed on Ni-NTA agarose beads (Qiagen) as previously described (Hayakawa et al., 2012), and proteins were analyzed by Western blot using anti-HA (Covance), anti-HIS tag (Millipore), and polyclonal rabbit anti-Smt3 (a gift from B. Palancade, Institut Jacques Monod, Centre National de la Recherche Scientifique, Paris Diderot University, Paris, France) antibodies.
Affinity purifications
For ubiquitin affinity purifications, 100 OD600 cells were collected and lysed with glass beads in 50 mM Hepes, pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100, and 1 mM EDTA. Cell lysates were incubated at 4°C for 3 h with preequilibrated monoubiquitin-agarose beads (Boston Biochem) with or without an excess of ubiquitin (1 mM; Boston Biochem). Bound proteins were analyzed by Western blot using anti-HA (Covance) or anti-Cdc48 (a gift from T. Sommer, Max Delbruck Center, Berlin, Germany).
Coimmunoprecipitation experiments using cross-linked extracts were performed as previously described (Vitaliano-Prunier et al., 2012).
In vitro assay for Nup60–Nup84 interaction
Yeast cells were grown to D600 = 1.5. Cells were collected and rapidly frozen in liquid nitrogen before cryolysis (Alber et al., 2007; Ossareh-Nazari et al., 2010). 1 g grindate was rapidly thawed in ice-cold immunoprecipitation buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM dithiothreitol, 0.5% Triton X-100, 0.1% Tween, and protease inhibitor cocktail). Nup60-ProtA and Nup49-ProtA purification was performed as previously described (Hérissant et al., 2014) except that beads were washed with 20 mM Hepes, pH 7.5, 500 mM NaCl, 1 mM dithiothreitol, 0.5% Triton X-100, 0.1% Tween, and protease inhibitor cocktail. Resulting bead-immobilized Nup60-ProtA and Nup49-ProtA were equilibrated with immunoprecipitation buffer and incubated with 10 µg recombinant yeast Nup84 bound to the 481- to 1,157-aa fragment of Nup133 (a gift from T. Schwartz, Massachusetts Institute of Technology, Cambridge, MA) for 90 min at RT in immunoprecipitation buffer before elution in sample buffer for 5 min at 95°C, SDS-PAGE analysis, and staining with Brillant Blue R-250 (Sigma-Aldrich).
FRAP
Spinning-disk confocal images were taken on a fully motorized inverted microscope (Eclipse Ti-E; Nikon) controlled with MetaMorph software 7.7.8, equipped with the Perfect Focus System (Nikon), a 100×, 1.45-NA Plan Apochromat oil-immersion objective, a piezo stage (Mad City Labs), a spinning-disk unit (CSUX1; Yokogawa), a charge-coupled device camera (CoolSNAP HQ2; Photometrics), and a laser bench (Roper Scientific) with 491- and 561-nm diode lasers (100 mW each; Cobolt). Exposure times for GFP and mCherry were 0.5 s. Laser power was set at 30%, binning at 1, and electronic gain at 3. All images were scaled similarly to their respective controls. The FRAP device iLas 1 was controlled by dedicated software (iLas software; Roper Scientific) integrated into MetaMorph. A 25-pixel-diameter circular region was bleached for 50 ms at 100% laser power, after a prebleach period of five images at 1-s intervals. Postbleach images were acquired every 1 s (or 3 s for Nup159-mCherry and 5 s for Nup60 WT) over a 2-min period (or 5 min for Nup60 WT) and exposed over 0.5 s at 30% laser power, binning at 1, and electronic gain at 3. For fluorescence recovery analysis, images were first registered with the ImageJ plugin TurboReg (Thévenaz et al., 1998), and analysis was performed with ImageJ. In brief, bleaching correction was performed by measuring the GFP intensity decay along the acquisition on the nonbleached area of the cell and on the background. Bleaching correction was individually applied to each bleached region. Bleaching recovery curves were then normalized with the first point before bleaching corresponding to 100% and averaged. Averaged curves were used to study the dynamics of recovery after photobleaching. Fitting with exponential recovery equations was performed only on curves that reached a plateau, and t1/2 was extracted from the fitted curves. On the other curves, the dynamics were estimated by measuring the slope of the initial curves (expressed in percentage of recovery/10 s) on the first 15 s of the recovery, therefore avoiding artifacts on the later time points. Both measurements were performed using the Curve-Fitting tool in ImageJ.
Fluorescence microscopy
Wide-field fluorescence images were acquired using a microscope (DMR; Leica) with a 100× Plan Apochromat HCX oil-immersion objective and a high-sensitivity cooled interlined charge-coupled device camera (CoolSNAP HQ2; Photometrics). Rapid and precise z positioning was accomplished by a piezoelectric motor (LVDT; Physik Instrumente) mounted underneath the objective lens. Maximum-intensity projections were performed using ImageJ software. Identical processing parameters were used in the different conditions. For quantification, eight medial frames over 11 frames (step size, 0.3 µm) of the z stacks were processed as mean intensity projections of raw data. Using the ROI plugin in ImageJ, two oval regions of interest were specified, which allow measurements restricted to the nucleoplasm or of the whole nuclei area respectively, in each cell of the field. For each region of interest (ROI), mean intensities and sizes were measured. Finally, five measurements of the fluorescence background (with a fixed ROI of 950 pixels) were randomly performed in each field. The nucleoplasm signal corresponds to (nucleoplasm mean intensity − fluorescent background intensity) × nucleoplasm ROI size, and the whole nuclear signal is calculated the same way; the peripheral signal corresponds to nuclear-nucleoplasm values.
Telomere analysis
Analysis of senescence and survivor types was performed after deletion of EST2 in each strain. Survivor types for each clone were determined after DNA digestion with XhoI and Southern blot analysis with subtelomeric Y′ TG1–3, or single-telomere (TelVIR) probes (McEachern and Haber, 2006; Churikov et al., 2014).
Online supplemental material
Fig. S1 shows the analysis of Nup60 ubiquitylation and sumoylation in diverse strains mutated for the conjugation machineries. Fig. S2 indicates the steady-state localization of Nup2, Mlps, and Ulp1 in WT, nup60-UbKR, and nup60-SUMO-KR strains. Fig. S3 shows the interaction of components of the Y complex with ubiquitin-agarose. Fig. S4 shows the effect of Nup60 modification on the major nuclear transport pathways. Fig. S5 shows the senescence curves for the individual clones analyzed in Fig. 7 B as well as analysis survivor type of est2Δ nup60Δ compared with est2Δ cells. Table S1 describes the yeast strains used in this study. Table S2 describes the plasmids used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201506130/DC1.
Supplementary Material
Acknowledgments
We thank A. Domingues, J. Drogat, and Suman Maji for their help. We are grateful to T. Schwartz for his generosity in providing recombinant purified Nup84; to members of Dargemont, Salamero, and Géli laboratories, in particular A. Babour, F. Waharte, and Marie-Noelle Simon, for helpful discussions; and to F. Stutz for carefully reading the manuscript.
This work was supported by the Who am I? laboratory of excellence (grant ANR-11-LABX-0071) and the DYNANUP project, both funded by the Investments for the Future program operated by the French National Research Agency (grant ANR-11-IDEX-0005-01), the National Institute of Cancer, and the Cancéropole Ile de France. C. Niño was supported by the Fondation pour la Recherche Medicale. The PICT-IBiSA Imaging core facility is a member of the France-BioImaging national research infrastructure (grant ANR-10-INSB-04). Part of this work was performed on the Nikon Imaging Center at Institut Curie, Centre National de la Recherche Scientifique.
The authors declare no competing financial interests.
Author contributions: C. Niño, D. Guet, S. Brutus, A. Gay, S. Mendiratta, and F. Jourquin performed experiments. C. Niño, D. Guet, J. Salamero, V. Géli, and C. Dargemont designed the experiments and analyzed the data. C. Niño, J. Salamero, V. Géli, and C. Dargemont wrote the manuscript.
Footnotes
Abbreviations used in this paper:
- DDR
- DNA-damage response
- DSB
- double-strand break
- HU
- hydroxyurea
- KR
- lysine to arginine
- MMS
- methyl methane sulfonate
- NPC
- nuclear pore complex
- Nup
- nucleoporin
- PTM
- posttranslational modification
- ROI
- region of interest
- STUbL
- SUMO-targeted ubiquitin ligase
- ts
- thermosensitive mutant
- UBD
- ubiquitin-binding domain
- WT
- wild type
References
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007. Determining the architectures of macromolecular assemblies. Nature. 450:683–694. 10.1038/nature06404 [DOI] [PubMed] [Google Scholar]
- Beck M., Lucić V., Förster F., Baumeister W., and Medalia O.. 2007. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 449:611–615. 10.1038/nature06170 [DOI] [PubMed] [Google Scholar]
- Bermejo R., Capra T., Jossen R., Colosio A., Frattini C., Carotenuto W., Cocito A., Doksani Y., Klein H., Gómez-González B., et al. 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 146:233–246. 10.1016/j.cell.2011.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R., Lai M.S., and Foiani M.. 2012. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol. Cell. 45:710–718. 10.1016/j.molcel.2012.03.001 [DOI] [PubMed] [Google Scholar]
- Branzei D., and Foiani M.. 2009. The checkpoint response to replication stress. DNA Repair (Amst.). 8:1038–1046. 10.1016/j.dnarep.2009.04.014 [DOI] [PubMed] [Google Scholar]
- Bukata L., Parker S.L., and D’Angelo M.A.. 2013. Nuclear pore complexes in the maintenance of genome integrity. Curr. Opin. Cell Biol. 25:378–386. 10.1016/j.ceb.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Cal-Bakowska M., Litwin I., Bocer T., Wysocki R., and Dziadkowiec D.. 2011. The Swi2-Snf2-like protein Uls1 is involved in replication stress response. Nucleic Acids Res. 39:8765–8777. 10.1093/nar/gkr587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Ijpma A., and Greider C.W.. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819–1827. 10.1128/MCB.21.5.1819-1827.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churikov D., Charifi F., Simon M.N., and Géli V.. 2014. Rad59-facilitated acquisition of Y′ elements by short telomeres delays the onset of senescence. PLoS Genet. 10:e1004736 10.1371/journal.pgen.1004736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw J.M., Krutchinsky A.N., Zhang W., Chait B.T., and Matunis M.J.. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158:915–927. 10.1083/jcb.200206106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D., Mykytka B., Allen N.P., Huang L., Al Burlingame, and Rexach M.. 2001. The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol. 154:937–950. 10.1083/jcb.200101007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G., and Stutz F.. 2010. Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression. J. Cell Sci. 123:1989–1999. 10.1242/jcs.053694 [DOI] [PubMed] [Google Scholar]
- Dilworth D.J., Suprapto A., Padovan J.C., Chait B.T., Wozniak R.W., Rout M.P., and Aitchison J.D.. 2001. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 153:1465–1478. 10.1083/jcb.153.7.1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V., Gadal O., Fromont-Racine M., Romano A., Jacquier A., and Nehrbass U.. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 116:63–73. 10.1016/S0092-8674(03)01026-2 [DOI] [PubMed] [Google Scholar]
- Geoffroy M.C., and Hay R.T.. 2009. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 10:564–568. 10.1038/nrm2707 [DOI] [PubMed] [Google Scholar]
- Grandin N., and Charbonneau M.. 2007. Control of the yeast telomeric senescence survival pathways of recombination by the Mec1 and Mec3 DNA damage sensors and RPA. Nucleic Acids Res. 35:822–838. 10.1093/nar/gkl1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T., Kehlenbach R.H., Schirmer E.C., Kehlenbach A., Fan F., Clurman B.E., Arnheim N., and Gerace L.. 2000. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell. Biol. 20:5619–5630. 10.1128/MCB.20.15.5619-5630.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guet D., Burns L.T., Maji S., Boulanger J., Hersen P., Wente S.R., Salamero J., and Dargemont C.. 2015. Combining Spinach-tagged RNA and gene localization to image gene expression in live yeast. Nat. Commun. 6:8882 10.1038/ncomms9882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase M.E., and Cordes V.C.. 2003. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell. 14:1923–1940. 10.1091/mbc.E02-09-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa A., Babour A., Sengmanivong L., and Dargemont C.. 2012. Ubiquitylation of the nuclear pore complex controls nuclear migration during mitosis in S. cerevisiae. J. Cell Biol. 196:19–27. 10.1083/jcb.201108124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérissant L., Moehle E.A., Bertaccini D., Van Dorsselaer A., Schaeffer-Reiss C., Guthrie C., and Dargemont C.. 2014. H2B ubiquitylation modulates spliceosome assembly and function in budding yeast. Biol. Cell. 106:126–138. 10.1111/boc.201400003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller D., Hecker C.M., Wagner S., Rogov V., Dötsch V., and Dikic I.. 2007. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol. Cell. 26:891–898. 10.1016/j.molcel.2007.05.014 [DOI] [PubMed] [Google Scholar]
- Horigome C., Oma Y., Konishi T., Schmid R., Marcomini I., Hauer M.H., Dion V., Harata M., and Gasser S.M.. 2014. SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol. Cell. 55:626–639. 10.1016/j.molcel.2014.06.027 [DOI] [PubMed] [Google Scholar]
- Iglesias N., Tutucci E., Gwizdek C., Vinciguerra P., Von Dach E., Corbett A.H., Dargemont C., and Stutz F.. 2010. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 24:1927–1938. 10.1101/gad.583310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IJpma A.S., and Greider C.W.. 2003. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell. 14:987–1001. 10.1091/mbc.02-04-0057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M., Hiller N.J., and Jentsch S.. 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell. 33:335–343. 10.1016/j.molcel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Khadaroo B., Teixeira M.T., Luciano P., Eckert-Boulet N., Germann S.M., Simon M.N., Gallina I., Abdallah P., Gilson E., Géli V., and Lisby M.. 2009. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat. Cell Biol. 11:980–987. 10.1038/ncb1910 [DOI] [PubMed] [Google Scholar]
- Lewis A., Felberbaum R., and Hochstrasser M.. 2007. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J. Cell Biol. 178:813–827. 10.1083/jcb.200702154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.J., and Hochstrasser M.. 2003. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 160:1069–1081. 10.1083/jcb.200212052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., and Géli V.. 2009. DNA damage response to eroded telomeres. Cell Cycle. 8:3617–3618. 10.4161/cc.8.22.9945 [DOI] [PubMed] [Google Scholar]
- Loeillet S., Palancade B., Cartron M., Thierry A., Richard G.F., Dujon B., Doye V., and Nicolas A.. 2005. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair (Amst.). 4:459–468. 10.1016/j.dnarep.2004.11.010 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Lundblad V., and Blackburn E.H.. 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 73:347–360. 10.1016/0092-8674(93)90234-H [DOI] [PubMed] [Google Scholar]
- Matsuura Y., and Stewart M.. 2005. Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 24:3681–3689. 10.1038/sj.emboj.7600843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M.J., and Haber J.E.. 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75:111–135. 10.1146/annurev.biochem.74.082803.133234 [DOI] [PubMed] [Google Scholar]
- Mészáros N., Cibulka J., Mendiburo M.J., Romanauska A., Schneider M., and Köhler A.. 2015. Nuclear pore basket proteins are tethered to the nuclear envelope and can regulate membrane curvature. Dev. Cell. 33:285–298. 10.1016/j.devcel.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M.B., Roberts T.M., Brown G.W., Varela E., Hediger F., Gasser S.M., and Krogan N.J.. 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 322:597–602. 10.1126/science.1162790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M., Molloy K.R., Williams R., Farr J.C., Meinema A.C., Vecchietti N., Cristea I.M., Chait B.T., Rout M.P., and Strambio-De-Castillia C.. 2013. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol. Biol. Cell. 24:3920–3938. 10.1091/mbc.E13-07-0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niño C.A., Hayakawa A., Dargemont C., and Babour A.. 2012. Mapping ubiquitin modifications reveals new functions for the yeast nuclear pore complex. Cell. Logist. 2:43–45. 10.4161/cl.19720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B., Bonizec M., Cohen M., Dokudovskaya S., Delalande F., Schaeffer C., Van Dorsselaer A., and Dargemont C.. 2010. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep. 11:548–554. 10.1038/embor.2010.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P., Jaspersen S.L., Miele A., Dekker J., and Peterson C.L.. 2009. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 23:912–927. 10.1101/gad.1782209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B., Liu X., Garcia-Rubio M., Aguilera A., Zhao X., and Doye V.. 2007. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell. 18:2912–2923. 10.1091/mbc.E07-02-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse V.G., Küster B., Gerstberger T., and Hurt E.. 2003. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 5:21–27. 10.1038/ncb893 [DOI] [PubMed] [Google Scholar]
- Ptak C., Aitchison J.D., and Wozniak R.W.. 2014. The multifunctional nuclear pore complex: a platform for controlling gene expression. Curr. Opin. Cell Biol. 28:46–53. 10.1016/j.ceb.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., and Chait B.T.. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651. 10.1083/jcb.148.4.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai C., and Mizuta K.. 2008. SUMO mediates interaction of Ebp2p, the yeast homolog of Epstein-Barr virus nuclear antigen 1-binding protein 2, with a RING finger protein Ris1p. Biosci. Biotechnol. Biochem. 72:1881–1886. 10.1271/bbb.80131 [DOI] [PubMed] [Google Scholar]
- Skruzný M., Schneider C., Rácz A., Weng J., Tollervey D., and Hurt E.. 2009. An endoribonuclease functionally linked to perinuclear mRNP quality control associates with the nuclear pore complexes. PLoS Biol. 7:e8 10.1371/journal.pbio.1000008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka M.B., Albuquerque C.P., Chen S.H., and Zhou H.. 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. USA. 104:10364–10369. 10.1073/pnas.0701622104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriramachandran A.M., and Dohmen R.J.. 2014. SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta. 1843:75–85. 10.1016/j.bbamcr.2013.08.022 [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel G., and Rout M.P.. 1999. Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol. 144:839–855. 10.1083/jcb.144.5.839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X.A., Dion V., Gasser S.M., and Freudenreich C.H.. 2015. Regulation of recombination at yeast nuclear pores controls repair and triplet repeat stability. Genes Dev. 29:1006–1017. 10.1101/gad.256404.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S.C., and Zakian V.A.. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083–8093. 10.1128/MCB.19.12.8083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenaz P., Ruttimann U.E., and Unser M.. 1998. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7:27–41. 10.1109/83.650848 [DOI] [PubMed] [Google Scholar]
- Uzunova K., Göttsche K., Miteva M., Weisshaar S.R., Glanemann C., Schnellhardt M., Niessen M., Scheel H., Hofmann K., Johnson E.S., et al. 2007. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282:34167–34175. 10.1074/jbc.M706505200 [DOI] [PubMed] [Google Scholar]
- van de Pasch L.A., Miles A.J., Nijenhuis W., Brabers N.A., van Leenen D., Lijnzaad P., Brown M.K., Ouellet J., Barral Y., Kops G.J., and Holstege F.C.. 2013. Centromere binding and a conserved role in chromosome stability for SUMO-dependent ubiquitin ligases. PLoS One. 8:e65628 10.1371/journal.pone.0065628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaliano-Prunier A., Babour A., Hérissant L., Apponi L., Margaritis T., Holstege F.C., Corbett A.H., Gwizdek C., and Dargemont C.. 2012. H2B ubiquitylation controls the formation of export-competent mRNP. Mol. Cell. 45:132–139. 10.1016/j.molcel.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerbeck J.W., Pasupala N., Guillotte M., Szymanski E., Matson B.C., Esteban C., and Kerscher O.. 2014. A SUMO-targeted ubiquitin ligase is involved in the degradation of the nuclear pool of the SUMO E3 ligase Siz1. Mol. Biol. Cell. 25:1–16. 10.1091/mbc.E13-05-0291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Kerscher O., Kroetz M.B., McConchie H.F., Sung P., and Hochstrasser M.. 2007. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 282:34176–34184. 10.1074/jbc.M706025200 [DOI] [PubMed] [Google Scholar]
- Yang L., Mullen J.R., and Brill S.J.. 2006. Purification of the yeast Slx5-Slx8 protein complex and characterization of its DNA-binding activity. Nucleic Acids Res. 34:5541–5551. 10.1093/nar/gkl685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Roberts T.M., Yang J., Desai R., and Brown G.W.. 2006. Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair (Amst.). 5:336–346. 10.1016/j.dnarep.2005.10.010 [DOI] [PubMed] [Google Scholar]
- Zhao X., Wu C.Y., and Blobel G.. 2004. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167:605–611. 10.1083/jcb.200405168 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.